Abstract
Cyclotides are a unique class of ribosomally synthesized cysteine-rich miniproteins characterized by a head-to-tail cyclized backbone and three conserved disulfide-bonds in a knotted arrangement. Originally they were discovered in the coffee-family plant Oldenlandia affinis (Rubiaceae) and have since been identified in several species of the violet, cucurbit, pea, potato, and grass families. However, the identification of novel cyclotide-containing plant species still is a major challenge due to the lack of a rapid and accurate analytical workflow in particular for large sampling numbers. As a consequence, their phylogeny in the plant kingdom remains unclear. To gain further insight into the distribution and evolution of plant cyclotides, we analyzed ~300 species of >40 different families, with special emphasis on plants from the order Gentianales. For this purpose, we have developed a refined screening methodology combining chemical analysis of plant extracts and bioinformatic analysis of transcript databases. Using mass spectrometry and transcriptome-mining, we identified nine novel cyclotide-containing species and their related cyclotide precursor genes in the tribe Palicoureeae. The characterization of novel peptide sequences underlines the high variability and plasticity of the cyclotide framework, and a comparison of novel precursor proteins from Carapichea ipecacuanha illustrated their typical cyclotide gene architectures. Phylogenetic analysis of their distribution within the Psychotria alliance revealed cyclotides to be restricted to Palicourea, Margaritopsis, Notopleura, Carapichea, Chassalia, and Geophila. In line with previous reports, our findings confirm cyclotides to be one of the largest peptide families within the plant kingdom and suggest that their total number may exceed tens of thousands.
Keywords: cyclotides, peptidomics, transcriptomics, ipecac, Rubiaceae, Carapichea, Notopleura, Psychotrieae, Palicoureeae
INTRODUCTION
Bioactive peptides occur in all organisms from bacteria to plants to mammals. These diverse natural peptide libraries often exhibit activities against a range of molecular targets offering starting points for drug development.1 In particular, the discovery of circular peptides is of great interest to peptide engineering as cyclization confers them with increased stability.2,3
One particular class of circular peptides are plant cyclotides. They are disulfide-rich peptides of about 30 amino acids in size that contain a head-to-tail cyclized backbone and six conserved cysteine residues forming three disulfide bonds in a knotted arrangement. This unique structural feature, known as the cyclic cystine-knot motif (CCK),4 confers them a tightly packed three-dimensional fold and makes them notably stable against thermal, chemical, and enzymatic degradation.5 Their physiological function appears to be as part of the plant defense system, based on observations that they are antifouling,6 insecticidal,7,8 anthelmintic,9 and molluscicidal agents.10 Besides these, cyclotides exhibit several other bioactivities including uterotonic,11-15 anti-neurotensin,16 antibacterial,17 anti-HIV,18 anticancer,19 and immunosuppressive properties.20 Their structural framework, sequence diversity, and range of bioactivities make them interesting scaffolds for agricultural and pharmaceutical applications.21
In the 1970s, Gran11 reported for the first time the occurrence of this class of polypeptides in the Rubiaceae plant Oldenlandia affinis. An aqueous decoction of this plant is used in traditional African medicine, applied orally or intravaginally for its oxytocic activity.13,15 Since then, a number of cyclotides have been discovered, but knowledge of their occurrence within flowering plants still remains limited.22 To date, every species of the violet family (Violaceae)23 investigated, and several species of the coffee family (Rubiaceae),22 as well as species of the cucurbit family (Cucurbitaceae)24 are known to contain cyclotides. Recently, Clitoria ternatea (Fabaceae) and Petunia × hybrida (Solanaceae) were reported to produce cyclotides.25,26 Already in 2006, the presence of cyclotide-like genes within species of the Poaceae family has been reported,27 and this has been recently confirmed by the identification of several acyclic peptides from Panicum laxum.28 As previously suggested,22,29 cyclotides seem to be widely distributed and diverse as their predicted number in Rubiaceae alone is greater than 50,000.24 On the other hand, cyclotides are one of the biggest classes of ribosomally synthesized peptides in plants, and their distribution and evolution within the plant kingdom still remains unclear. The number of cyclotides varies within species and as recently shown one single species can express more than 70 unique cyclotides.20,22 Altogether well over 200 cyclotide-sequences have been characterized so far (www.cybase.org.au).30
On the basis of their unique chemical and biophysical properties, the following peptidomic identification criteria have been proposed earlier: (i) molecular weight in the mass range between 2500 and 4000 Da, (ii) late elution properties on reversed-phase high performance liquid chromatography (RP-HPLC) as well as (iii) six conserved cysteines.22 In addition, the typical fragmentation pattern of tandem mass spectrometry (MS/MS) spectra of native and enzymatically digested peptides were monitored as additional criteria to distinguish cyclotides from linear peptides.25 Furthermore, it is possible to de novo sequence cyclotides within complex mixtures such as crude plant extracts.31 However, screening of plant species for the occurrence of cyclotides is still laborious and often can lead to ambiguous results, due to other plant peptides (e.g., cysteine-rich defense peptides) that interfere with the screening and identification criteria or due to limited availability of sample.22,32
Therefore, we investigated 296 species of 43 different plant families (Supporting Information Table S1) to gain novel insights into cyclotide presence and evolution in flowering plants, with focus on the Rubiaceae family. According to their unique properties and previously defined identification criteria, we refined the screening protocol for the identification of novel cyclotide-containing plants by analyzing crude and chemically modified plant extracts using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). Additionally, we used a transcriptome-mining approach to identify cyclotide precursor proteins and mature cyclotide sequences to help shed light on the evolution and phylogenetic distribution of this class of circular miniproteins.
MATERIALS AND METHODS
Plant Material
The following plant material has been collected for this study: 133 plant samples were obtained from the National Botanical Garden of Belgium, 67 plant samples were collected in Nigeria, 62 plant samples were collected in different regions of Costa Rica, including the tropical research station La Gamba, four samples were collected in the Botanical Garden of the University of Vienna, nine samples were collected in Panama, and 18 samples in Ethiopia. Voucher specimens were deposited in the Herbaria of the National Botanical Garden of Belgium (BR), University of Vienna (WU, HG, AB, and BS) and the Forestry Herbarium Ibadan (FHI). Carapichea ipecacuanha (Brot.) L. Andersson herbarium material was obtained from the Museum of Natural History Vienna (voucher specimen W 1922-0008476) and a sample of Radix Ipecacuanhae pulvis was kindly supplied from the drug collection of the Department of Pharmacognosy, University of Vienna (WU 0067845). All plant species, author information and their collection/deposition IDs have been reported in Supporting Information Table S1.
Extraction and C18-Flash Chromatography
Dried plant material (0.5 to 2 g) was ground before solvent extraction with 5 mL of a mixture of methanol: dichloromethane (1:1; v/v) for 18–24 h under continuous agitation at 25°C. After adding 2.5 mL of water, samples were centrifuged for 3 min and the supernatant diluted to less than 10% (v/v) methanol. These extracts were further processed with solid-phase extraction C18 columns (Phenomenex, Aschaffenburg, Germany) and eluted with 5 mL of 20% and 80% solvent B (90% (v/v) acetonitrile, 0.045% (v/v) trifluoroacetic acid in double distilled water), respectively, to separate the potential cyclotide-containing fraction from hydrophilic compounds. After freeze-drying, the 80% B fractions, yielding between 1 and 5 mg, samples were stored at −20°C. For cyclotide identification experiments, small aliquots were redissolved in 0.1M NH4HCO3 (Sigma-Aldrich, St. Louis, MO) or solvent A (0.05% (v/v) trifluoroacetic acid in double distilled water), what is referred to as crude extract and samples were immediately used for further experiments to avoid side-chain modifications such as deamidation of asparagine.
RP-HPLC Fractionation and Peptide Purification
In case of poor MS/MS fragmentation during cyclotide sequencing of crude extracts, selected samples were fractionated on a Dionex Ultimate 3000 HPLC unit (Dionex, Amsterdam, The Netherlands). Depending on the available amount of extracts, we used either preparative (Jupiter; 250 × 21.2 mm, 10 μm, 300 Å, Phenomenex), semipreparative (250 × 10 mm), or analytical (250 × 4.6 mm) Kromasil C18 columns (5 μm; 100 Å) with linear gradients of 0.1%–1% min−1 solvent B at flow rates of 8, 3, and 1 mL min−1, respectively. Collected fractions were freeze-dried and stored at 4°C for subsequent analysis and −20°C for long-term storage.
Reduction, Alkylation and Digestion of Extracts
Disulfide bond reduction of peptides in the crude extracts was performed by adding 2 μL of a freshly prepared 0.2M solution of dithiothreitol (Sigma-Aldrich, St. Louis, MO) to a 20 μL aliquot of peptide sample dissolved in 0.1M NH4HCO3 followed by incubation for 30 min at 60°C in the dark. Cysteine residues were alkylated by adding 4 μL of freshly prepared 0.5M solution of iodoacetamide (Sigma-Aldrich, St. Louis, MO) to the reduced peptide samples and incubated for 10 min at 25°C. Reduced and alkylated peptide samples were enzymatically digested by adding 2 μL of either 0.5 μg μL−1 endoproteinase GluC (Sigma-Aldrich, St. Louis, MO) or 0.1 μg μL−1 trypsin (Sigma-Aldrich, St. Louis, MO) and were incubated for 3 h at 37°C. The reaction was quenched by adding 1 μL of trifluoroacetic acid to the samples. Before MS analysis, samples were desalted using C18 Zip-Tips™ (Millipore, Billerica, MA) and stored at 4°C for subsequent analysis and −20°C for long-term storage.
MALDI-TOF/TOF Analysis and Peptide Sequencing
Analysis of crude, reduced-alkylated, and digested samples were performed on a MALDI-TOF/TOF 4800 Analyzer (AB Sciex, Framingham, MA) operated in reflector positive mode acquiring 2000–10,000 total shots per spectrum with a laser intensity of 4000. MS and MS/MS experiments were performed using α-cyano-hydroxy cinnamic acid 5 mg mL−1 in 50% (v/v) acetonitrile as matrix. A total of 0.5 −L of each sample was mixed with 3 μL of matrix and 0.5 μL of the mixture was spotted onto the plate. Spectra were acquired and processed using 4800 Analyzer Software. Cyclotides were identified by manual sequencing. The MS/MS spectra were examined and peptides were sequenced based on N-terminal b-ion and C-terminal y-ion fragmentation. Database searching of MALDI-TOF/TOF MS/MS data was performed using the ProteinPilot™ software and the Paragon algorithm 4.0 together with a custom-made ERA database for the identification of cyclotides.33 The disulfide connectivity of CysI-IV, CysII-V, and CysIII-VI and the isobaric amino acid Leu and Ile were assigned based on homology with known sequences.34
Transcriptome-Mining for Cyclotide Precursors
Transcriptome data of Carapichea ipecacuanha (recently transferred from Psychotria ipecacuanha (Brot.) Stokes) were accessed via the 1KP-project (www.onekp.com). Growth conditions and preparation of in vitro plantlet and root culture of C. ipecacuanha were as published previously.35 RNA was isolated using a Trizol/RNAqueous-Midi Kit (Ambion, Paisley, UK) method.36 Samples were sequenced as an indexed RNAseq library on an Illumina GA II machine (Illumina, San Diego, CA). Sequencing was paired-end (73 bp + 75 bp) and produced a total of over 2.3 Gbp. The sequences were assembled into scaffolds using SOAPdenovo-Trans (Beijing Genomics Institute, China). The BLAST searches were against these scaffolds. The C. ipecacuanha data for in vitro plantlet (ID: BQEQ) and root culture (ID: JOPH) were accessed via tBLASTn using the Oldenlandia affinis precursor Oak1 (UniProt KB entry: P56254) sequence as query. Resulting hits and contig sequences were translated and annotated based on homology to known cyclotide precursors from Chassalia chartacea Craib37 using a similar methodology as described by Gruber and Muttenthaler.38 All transcriptome contig sequence IDs were listed in Supporting Information Table S2. Sequence alignments were prepared using ClustalW (http://www.genome.jp/tools/clustalw/) and boxshade (www.ch.embnet.org/software/BOX_form.html). Nucleotide sequence data of precursors caripe A2 and B2 are available in the GenBank database under the accession numbers KC807202 and KC811328, respectively.
Cyclotide Modeling and Sequence Alignment
Modeling of the cyclotides caripe 1 and caripe 2 was performed using the CycloMod tool on CyBase (www.cybase.org.au).30 Structural images were prepared using PyMol by aligning the two novel cyclotide models using the “super” command. Coloring of surface representations was done based on the hydrophobicity scale as described by Eisenberg et al.39
RESULTS
Since the first discovery of cyclotides in the Rubiaceae plant Oldenlandia affinis, few plant families have been identified that contain species which produce cyclotides. To expand the current knowledge about the occurrence and sequence diversity of cyclotides with special focus on their distribution and phylogenetic relationship within Rubiaceae plants, a total number of 296 plant species has been investigated using crude and chemically modified plant extracts as well as bioinformatics tools that resulted in the discovery of novel cyclotide-containing species and sequences.
Chemical Screening Protocol Revisited
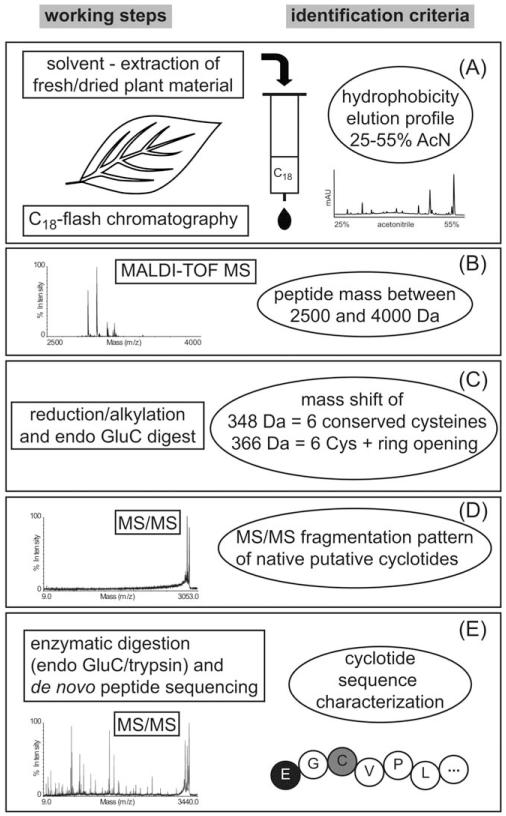
Based on previously established identification criteria,22,25 such as hydrophobicity, mass range, cysteine content, and stability of native peptides during collision-induced dissociation MS/MS, we refined the screening methodology with the aim to combine the two most crucial features, i.e., rapid analysis and accurate cyclotide discovery (see Figure 1). After solvent extraction of homogenized plant material (e.g., a small piece of plant tissue), a single prepurification step was performed by solid-phase extraction over prepacked C18 columns. The resulting samples, which are referred to as crude extracts were initially analyzed by MALDI-TOF MS. All extracts that yielded mass peaks within the expected range of 2500 to 4000 Da were subjected to chemical modification to verify the presence of three disulfide bonds and six cysteines, respectively. Therefore aliquots of the crude extracts were reduced and alkylated using iodoacetamide, which in the case of six cysteines led to a distinct mass shift of 348 Da. In parallel, crude extracts were analyzed by analytical RP-HPLC to confirm the presence of late-eluting, hydrophobic peptide peaks.
FIGURE 1. Cyclotide screening methodology.
This scheme illustrates the refined experimental workflow and identification criteria that led to the discovery of novel cyclotide-containing species and peptide sequences. All plant extracts were analyzed stepwise to assess their (A) hydrophobicity, (B) mass range, (C) disulfide bonds/cysteine content and circular backbone, and (D) MS/MS fragmentation pattern of native peptides. Only plants for which we obtained partial sequence tags or full cyclotide sequences (E) have been assigned unambiguously as novel cyclotide-containing species.
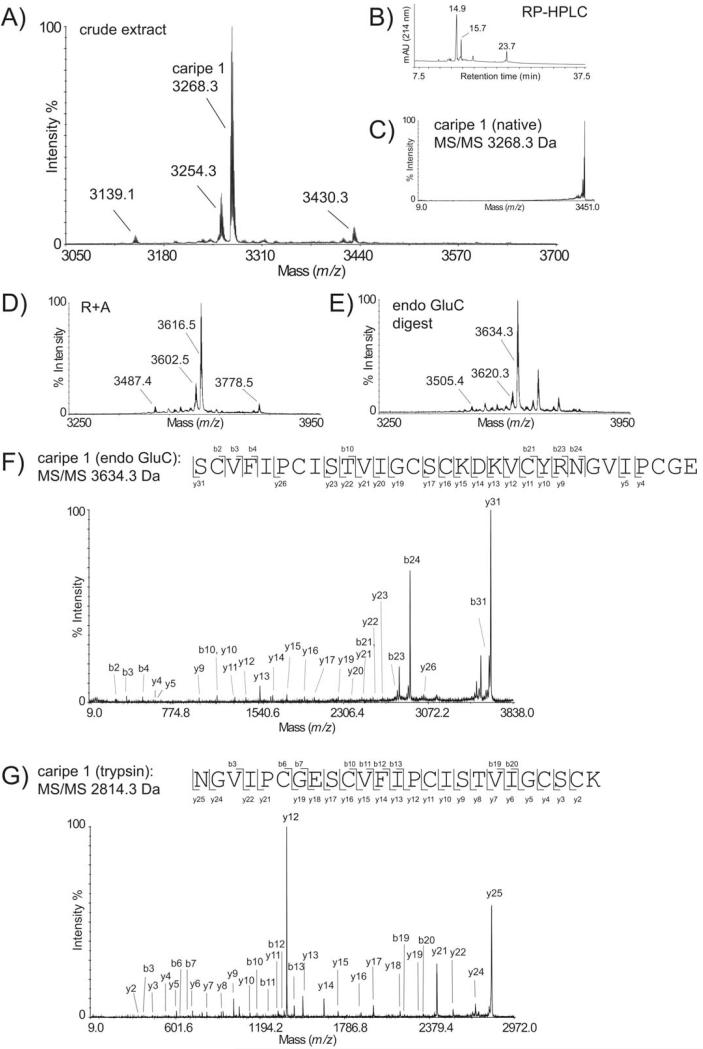
As plants contain other disulfide-rich peptides besides cyclotides, such as knottins or defensins that may fulfill the above described criteria of mass range, hydrophobicity, and cysteine content, it was necessary to consider additional and more stringent analysis criteria that reflect distinct cyclotide features such as the circular backbone. Therefore, the MS/MS fragmentation patterns of putative cyclotides were analyzed, because spectra of head-to-tail circular peptides lack fragmentation, whereas spectra of linear peptides display multiple fragmentions.25 Furthermore, most cyclotides contain a single conserved glutamic acid in loop 1 and therefore enzymatic digestion with endoproteinase GluC yielded single-site cleaved peptides, which in the case of backbone cyclized peptides can be identified by the addition of water and thus in a resultant mass shift of + 18 Da. Plants containing acyclic cyclotide homologs as well as samples containing peptides without glutamic acid or more than one glutamic acid residue may be identified as “false-negatives.” As a final decision criterion for the accurate discovery of cyclotide plant species and to exclude any potential cyclic peptides containing a noncystine-knotted arrangement of three disulfide bonds, it is essential to characterize at least partial amino acid sequences that match current cyclotide features. Partial peptide sequences that consist of at least two adjacent inter-cysteine loops are necessary for a reliable identification of cyclotides. Hence, if one or more peptides in the crude plant extracts satisfied all the above set criteria, i.e., mass, hydrophobicity, and cysteine content, the respective masses have been selected as precursor ions for tandem MS experiments. Cyclotide de novo sequencing was performed on endoproteinase GluC and trypsin digests from crude extracts or if necessary fractions obtained from RP-HPLC that contained complex cyclotide mixtures without the need for laborious purification or isolation of single peptides.31 The applied methodology, i.e., each step of the chemical and analytical screening procedure, is exemplarily shown for the root powder sample of Carapichea ipecacuanha (Figure 2). This species has passed all screening criteria and thus allowed sequencing of the novel cyclotide caripe 1. To accelerate de novo peptide sequencing, the automated ERA tool33 was implemented into the screening protocol, but this approach did not yield any cyclotide sequence apart from the known cyclotides vibi B and cycloviolacin O22, which have been identified in Palicourea tetragona.
FIGURE 2. Characterization of the novel cyclotide caripe 1 from Carapichea ipecacuanha.
The MALDI-MS (A) and RP-HPLC trace (B) of Carapichea ipecacuanha is shown. The putative cyclotide mass of 3268.3 Da was selected as precursor for MS/MS experiments (C). The reduced and alkylated (R1A) (D), and endoproteinase GluC digested (E) extract is shown. MALDI-TOF/TOF sequencing of the precursor masses 3634.9 Da (F) and 2814.9 Da (G) from an endoproteinase GluC and a trypsin digest, respectively, allowed the characterization of the novel cyclotide caripe 1 based on observed b- and y-ions. All shown masses are monoisotopic. MS evidence for the existence of cyclotides in all other positive cyclotide plants can be found in the Supporting Information section (Figures S1-S9).
Novel Cyclotide-Producing Plants and Their Sequences
Based on the known distribution of cyclotide-containing species among flowering plants and using a refined screening workflow as described above, 296 plants belonging to 43 different families were analyzed. It was possible to positively identify nine novel cyclotide-containing species that all belong to the tribe Palicoureeae within the Rubiaceae family. These species are Carapichea ipecacuanha, Chassalia curviflora, Notopleura capacifolia, Palicourea tetragona, Psychotria brachiata, P. deflexa, P. poeppigiana, P. solitudinum, and P. suerensis (Figure 2, Supporting Information Figures S1-S9). Herein, for the first time, species of the genera Notopleura and Carapichea were identified to contain cyclotides. Using de novo peptide sequencing, we characterized four full-length sequences and seven sequence tags whereof four peptides belong to the bracelet subfamily, five to the Möbius subfamily, and four cannot be classified due to the lack of a full loop 5 sequence. Apart from novel sequence motifs and variations within loops 2, 3, and 5, we also identified two previously known cyclotides, i.e., cycloviolacin O22 and vibi B. An alignment of obtained sequences is shown in Table I.
Table I. Novel Cyclotide-Containing Rubiaceae Species and Corresponding Sequences.
| Sequencea |
MW(Da)b |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | ID | Peptide | Loop | 1 | 2 | 3 | 4 | 5 | 6 | Exp. | Calc. | Δ(ppm) | Subfamily | |||||||
| Carapichea ipecacuanha | WU0067845 | caripe 1 | cyclo- | GVIP | C | GES | C | VFIP | C | IST-VIG | C | S | C | K-DKV | C | YRN | 3267.3 | 3267.5 | 61.2 | Bracelet |
| (Brot.) L. Andersson | caripe 2 | G-IP | C | GES | C | VFIR | C | TITALLG | C | S | C | S-NNV | C | YKN | —c | 3243.4 | — | Bracelet | ||
| caripe 3 | X-IP | C | GES | C | VFIP | C | ISAVVGS | C | S | C | --NKV | C | YNN | — | n.a.d | — | Bracelet | |||
| caripe 4 | --LI | C | SST | C | LRIP | C | LSPR--- | C | T | V | R-HHI | C | YLN | — | 3080.4 | — | Bracelet | |||
| caripe 5 | X | C | GES | C | VFIP | C- | FTSV--G | C | S | C | K-DKV | C | YRN | — | n.a. | — | Bracelet | |||
| caripe 6 | GAI- | C | TGT | C | FRNP | C | LSRR--- | C | T | C | R-HYI | C | YLN | — | 3199.4 | — | Bracelet | |||
| Chassalia curviflora (Wall.) Thwaites | HG, 1074 | chacur 1 | GLPV | C | GET | C | VGGT | C | ---NTPG | C | T | C | S-WPI | C | TRN | 2903.9 | 2904.1 | 89.5 | Möbius | |
|
Notopleura capacifolia (Dwyer) C.M. Taylor |
HG, 2907083 | notcap 1 | X | C | X-S | C | VW-X | C | ITSPSAG | C | K | C | X | C | X | 3368.9 | n.a. | — | — | |
| Psychotria brachiata Sw. | BS PYB1 | psybra 1 | GLPI | C | GET | C | TLGT | C | ---NTPG | C | T | C | S-WPI | C | TKN | 2947.9 | 2948.2 | 105.2 | Möbius | |
| Palicourea tetragona Ruiz&Pav. | HG, 24070811 | cO22e | GLPI | C | GET | C | VGGT | C | ---NTPG | C | T | C | S-WPV | C | TRN | 2903.9 | 2904.2 | 99.9 | Möbius | |
| vibi B | GLPV | C | GET | C | FGGT | C | ---NTPG | C | T | C | S-YPI | C | TRN | 2928.9 | 2929.1 | 92.2 | Möbius | |||
| paltet 1 | GLPI | C | GET | C | FTGT | C | ---NTPG | C | T | C | S-YPV | C | TRN | 2972.9 | 2973.2 | 100.9 | Möbius | |||
| Psychotria deflexa DC. | HG, 22070810 | psydef1 | X | C | X | C | X | C | ---NTSG | C | T | C | KW-X | C | TRX | 3123.9 | n.a. | — | — | |
| psydef 2 | X | C | XES | C | WTSN | C | ---FTSP | C | X | C | X-HP | C | TRX | 3303.9 | n.a. | — | — | |||
| Psychotria poeppigiana Müll. Arg. | HG, 3007081 | psypoe 1 | GSVI | C | GET | C | FTTV | C | ---NTPG | C | Y | C | GAY-X | C | TRN | 3133.9 | n.a. | — | — | |
| Psychotria solitudinum Standl. | HG, 2607083 | psysol 1 | X | C | X | C | X | C | ---YTPG | C | T | C | GSYFV | C | N-X | 3089.9 | n.a. | — | Bracelet | |
| Psychotria suerensis Donn. Sm. | AB 16021005 | psysue 1 | X | C | X | C | X | C | X---IAG | C | S | C | SSALL | C | V-X | 3190.4 | n.a. | — | Bracelet | |
| psysue 2 | X | C | X | C | X | C | X---IAG | C | S | C | SSALL | C | V-X | 3229.3 | n.a. | — | Bracelet | |||
| CYS | I | II | III | IV | V | VI | ||||||||||||||
Sequences are aligned based on conserved Cys residues (bold), isobaric amino acids Leu/Ile were assigned based on homology to published sequences, X represents one or multiple unidentified amino acid residues according to respective inter-cysteine loop sequences, i.e. loop 1: n=3-6, 2: 4-8, 3: 3-10, 4: 1, 5: 4-8, 6: 5-13 amino acid residues, according to Griindemann et al.21.
Listed molecular weights display monoisotopic masses, exp. experimental, calc. calculated, Δ mass difference exp. and calc.
Not determined.
n.a. not applicable, partial sequence.
Cycloviolacin O22.
Besides successful identification of novel cyclotide-containing plants, 22 samples fulfilled all of the aforementioned screening criteria except the presence of cyclotides could not be confirmed by peptide sequencing, and these species were therefore treated as cyclotide absent plant species to avoid “false-positive” results (Supporting Information Table S1). Limited sample amount and high complexity of crude extracts often does not allow unambiguous confirmation of cyclotide expression by peptide sequencing. One way to overcome this problem is peptide sequence discovery at a genetic level.
Identification of Cyclotides and Their Precursors in Transcriptome Datasets
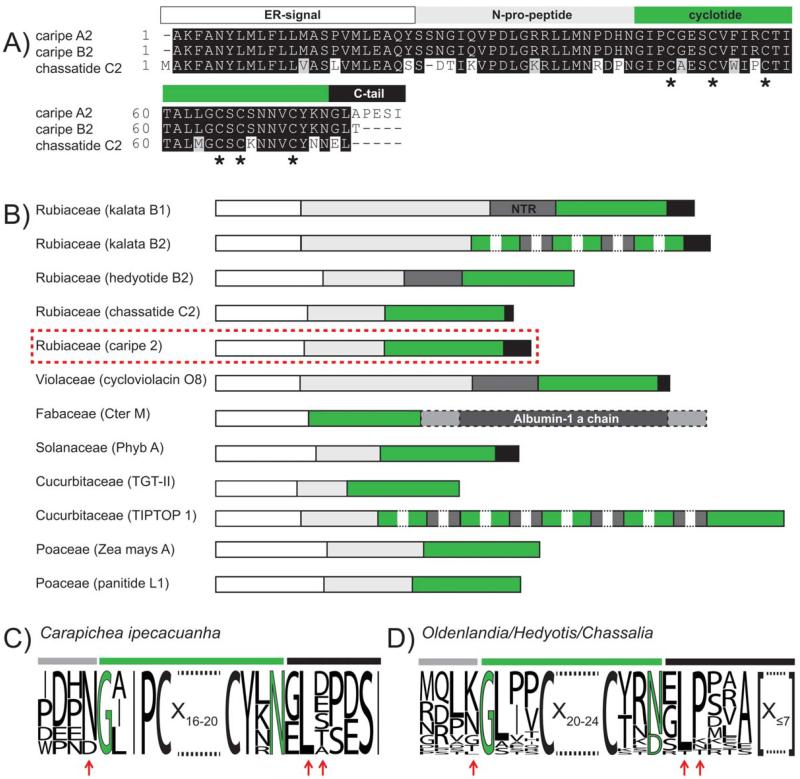
In the era of genomics and transcriptomics, analyzing publicly available transcriptome datasets can be a useful tool for the discovery of peptide sequences.38 One such rich source of available transcriptome datasets has been provided by the 1KP consortium, which analyzed over 1000 different plant species. Based on the use of prereleased datasets, we performed tBLASTn analysis of known cyclotide precursors against available datasets of Rubiaceae species. Within Carapichea ipecacuanha we discovered nine precursor proteins, and these novel precursors show high similarity to previously identified precursors of Chassalia chartacea37 (Figure 3A and Supporting Information Figure S10). The analysis of Carapichea precursors yielded an additional five putative cyclotide sequences of this species named caripe 2 to caripe 6 (Table I). Interestingly two identified sequences (caripe 4 and 6) lack the conserved glutamic acid residue within loop 1 of the cyclotide domain. Sequence comparison of Carapichea precursors to Oak1 from Oldenlandia affinis and chassatide C2 from Chassalia chartacea indicated their extensive homology to cyclotides, whereas no homology could be found to the gene structure of linear knottins such as TGT-II from the towel gourd Luffa cylindrica (Supporting Information Figure S10). Comparison of the Carapichea precursors (Figure 3C) to those of other Rubiaceae species (Figure 3D) confirmed high homology to the recently discovered precursors of Chassalia chartacea.37 This is true in particular for the two residues following their proto-C-terminus, i.e., a glycine or glutamic acid followed by a highly conserved leucine residue and the amino acid immediately preceding the proto-N-terminus of the cyclotide domain (Figures 3C and 3D).
FIGURE 3. Comparison of novel Carapichea and known cyclotide precursors.
(A) A boxshade alignment of the precursors caripe A2 (plantlet, GenBank ID: KC807202), caripe B2 (root, Gen-Bank ID: KC811328), and their closest homolog precursor sequence (according to BLASTp analysis), chassatide C2 (UniProtKB ID: I0B6F2) is shown. Based on this comparison, both Carapichea sequences lack only the N-terminal methionine residue. Conserved cysteine residues are indicated with asterisks at the bottom of the alignment. (B) An overview of the architecture of cyclotide precursor proteins among different plant families is shown. Typical cyclotide precursor domains are highlighted with colors, i.e., ER signal (white), N-terminal pro peptide (light-grey), N-terminal repeats (NTR, dark-grey), mature cyclotide or knottin sequence (TGT-II and TIPTOP 1) (green), C-terminal tail sequence (black), albumin-1 a-chain (dark-grey, dashed box), and linking peptides (light-grey, dashed box). For comparison of processing residues, a sequence alignment in the form of a sequence logo for nine Carapichea ipecacuanha precursors (C) and for 25 Rubiaceae precursors (D), including those from Oldenlandia, Hedyotis, and Chassalia spp. is shown. Residues near the N- and C-terminal processing sites that show intra- or inter-species homology have been indicated by red arrows. Numbers indicate the residue length of amino acids of mature cyclotides or C-tail residues, respectively, that are not displayed for illustration purpose.
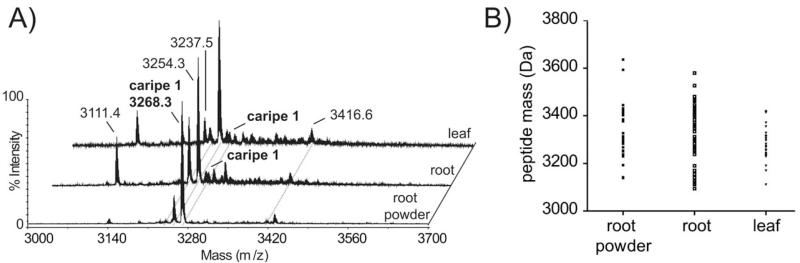
The predicted presence of cyclotides, obtained by transcriptome analysis of Carapichea plantlet and root culture was compared with our findings from MS analysis. In addition to the root powder sample, we investigated two different tissues from a herbarium sample (i.e., root and leaf). A comparison of these three samples is shown in Figure 4. The different samples, i.e., unprocessed and powdered root, showed a similar cyclotide expression pattern, as judged by the presence of similar peptide masses. The cyclotide caripe 1 is present in all three samples, and the most abundant one in the root powder sample.
FIGURE 4. MALDI-MS comparison of different Carapichea ipecacuanha tissues and samples.
(A) Offset-aligned MALDI-MS traces of two types of plant tissues from the same Herbarium sample (W-1922-0008476), i.e., leaf (back) and root (middle) as well as root-powder (front, WU-0067845) are shown. Most abundant peptide masses are labeled, and their presence in all three samples has been indicated by dashed lines. (B) A comparison of the distribution of observed peptide masses in all three samples is shown as a dot-chart. Displayed monoisotopic masses were obtained from DataExplorer™ software with signal-to-noise limit set to 5. Caripe 1 has a mass of 3268.3 Da and is highlighted in bold text.
To compare representative cyclotides identified by MS (caripe 1) and transcriptome-mining (caripe 2), we constructed three-dimensional models and analyzed the structural properties of these two cyclotides (Figure 5). Both peptides share fundamental cyclotide characteristics including the β-hairpin in loop 5, a distorted triple-stranded β-sheet and several conserved amino acids including a glutamic acid (E) in loop 1 involved with hydrogen bond interactions40 and an asparagine (N) or aspartic acid (D) in loop 6 presumed vital for cyclization. The 310 helix typical for bracelet cyclotides can be found in the model of caripe 2. From the hydrophobicity analysis, it is obvious that residues within loops 2 and 3 contribute to a hydrophobic surface, whereas loops 5 and 6 contribute to a hydrophilic surface thus conferring the peptides an overall amphiphilic nature.
FIGURE 5. Structural models of the cyclotides caripe 1 and caripe 2.
(A) The sequence alignment of two novel cyclotides caripe 1 and 2 from Carapichea ipecacuanha is presented. The typical six cysteine residues are highlighted in yellow and are labeled with roman numerals and the six inter-cysteine loops are labeled on top of the alignment. Three-dimensional models of caripe 1 (B) and caripe 2 (C) show cyclotide-typical secondary structure elements, i.e., two anti-parallel β-sheets (in loops 4 + 5) and a helix (in loop 3). Surface representations of caripe 1 and caripe 2 are colored in hydrophobicity scale according to Eisenberg et al.,39 to illustrate the amphiphilic nature of these peptides. Typical hydrophobic parts of cyclotides are loops 2 and 3, whereas the hydrophilic parts of the molecules can be found on the opposite site in loops 5 and 6. Representative amino acids, i.e., hydrophobic residues I3, F11 and V27 (caripe 1) and I2, F10 and V27 (caripe 2), as well hydrophilic residues P13 (caripe 1) and R12 (caripe 2) are labeled. The cyclotide models have been prepared using the CycloMod tool on CyBase and illustrations have been prepared with PyMol (see Materials and Methods section for details).
DISCUSSION
The identification of novel cyclotide-containing plant species still is a major challenge lacking a rapid and accurate analysis methodology in particular for large sampling numbers. To overcome these major challenges, current screening methods have been improved. The presented approach included the combination of chemical analysis with transcriptome-mining, which triggered the discovery of several novel cyclotide plants within Gentianales. Based on the phylogeny of cyclotide-bearing species in Rubiaceae, the distribution and evolution of cyclotides has been refined.
Cyclotides are a large class of bioactive plant compounds displaying enormous sequence diversity. Hence, it is of great interest to shed light on their distribution and natural sequence variation. Their unique biophysical and molecular properties, i.e., six conserved cysteines that form three disulfide bonds and a cyclized backbone, have been used to identify cyclotides in plants. Standard procedures include RP-HPLC and MS analysis to investigate hydrophobicity, mass range, and cysteine content of unknown peptides. To determine the cyclic nature of peptides in the sampled plants, enzymatic digests with endoproteinase GluC were performed. As most cyclotides usually contain a conserved glutamic acid in loop 1,4 digests typically yield single-site cleaved peptides that show a distinct mass shift of + 18 Da. This mass shift has been used as an additional cyclotide identification criterion. Cyclotides which (i) contain multiple glutamic acid residues, (ii) lack this residue, or (iii) have a linear backbone41 may be tested “false-negative” based on the proposed criteria. The stability of the CCK may be used to gain further confidence in the discovery of cyclotides. Comparison of MS/MS fragmentation pattern of crude and digested precursors can be used to distinguish between linear and head-to-tail cyclized peptides.25 The typical knotted and cyclic back-bone structure of cyclotides prohibits direct MS/MS analysis due to the lack of free N- and C-termini that are necessary for retaining charges. This circumstance has been implemented in our screening workflow and hence a comparison of the MS/MS fragmentation pattern of crude and digested extracts of all samples has been performed. After chemical modification of disulfide bonds by reduction and alkylation and the “creation” of free N- and C-termini by enzymatic cleavage, various fragment ions of the linearized peptides may be observed. By comparison, other disulfide-rich naturally occurring linear peptides, such as defensins, are amenable to amino acid fragmentation by direct MS/MS of the nonmodified crude samples prior digestion. Thus, the fragmentation pattern criterion adds another level of confidence in the decision of whether a sample contains cyclotides or not. As described above, some plant samples that have satisfied the screening criteria A–D (Figure 1) have not been confirmed to contain cyclotide sequences, due to limited sample amount or complexity of the crude peptide extracts (Supporting Information Table S1). To minimize the discovery of “false-positive” hits, only plant species for which full or partial sequences were obtained were classified as novel cyclotide-containing species (Table I) and the identification of sequence tags “typical” for cyclotides is essential to our methodology. As manual de novo peptide sequencing is time-consuming, we tried to implement the previously reported, automatic ERA database tool33 to aid rapid identification of putative cyclotide sequences in the plant samples. However, besides two peptides matching known cyclotide sequences in one species, no other hits were identified (Table I). Following customization and updating of the ERA database with manually obtained cyclotides sequences, the automatic approach confirmed some of the de novo sequenced peptides (data not shown). Typical proteomics database algorithms such as Paragon can only assign MS/MS sequence tags that show high homology to those in the database (i.e. one or two residue substitutions) and consequently peptide sequences with lesser similarity to known cyclotides may be missed by the database search. As judged from our results, the automated ERA approach is only useful to identify already known peptides; if sequences differ significantly from known ones that have been deposited in the database or in case of weak MS/MS fragmentation automatic database search using the ERA-tool does not help to accelerate the discovery of novel cyclotides.
With more sequences being published, including those from bioinformatics approaches an updated ERA database may become a useful alternative to de novo peptide characterization. In particular, transcriptome-mining has great potential for the discovery of novel peptides.38 As an advantage over MS-based peptide discovery, transcriptome sequencing provides the correct primary sequence and hence additional identification of isobaric residues becomes obsolete. Furthermore, not only the mature peptides will be characterized, but this approach may yield partial or even full length precursor sequences, which allows the analysis of processing sites involved in cyclotide biosynthesis. On the other hand, transcriptome sequencing has disadvantages in providing accurate information about the cyclic or linear nature of the identified peptides. Therefore, a combination of chemical and analytical identification procedures with high-throughput transcriptome sequence analysis provides the best results for rapid and accurate cyclotide discovery.
Using this combined approach, we identified 16 cyclotide sequences or partial sequence tags, including two previously known sequences (Table 1). The novel full-length cyclotides contain typical Möbius or bracelet characteristics and have been classified accordingly. All novel cyclotide sequences display a certain degree of sequence variation, when compared with frequently occurring residues in known cyclotides, in particular within loops 2, 3, and 5. This is as expected because these loops are known to have the highest amino acid variability, and this underlines the high plasticity of the cyclotide framework.42 A comparison of the three-dimensional structural models from two Carapichea ipecacuanha cyclotides, caripe 1 (identified by MS sequencing) and caripe 2 (identified by transcriptome-mining), confirms the highly amphiphilic nature of these two peptides, typical for cyclotides. Further-more, both novel cyclotides display other structural characteristics of bracelet cyclotides such as the β-hairpin in loop 5, a distorted triple-stranded β-sheet, a short 310 helix, and a glutamic acid residue in loop 1 involved in hydrogen bond interactions. The absence of the helix motif in the model of caripe 1 is most likely a modeling-artifact as the comparison of the structures of cycloviolacin O2 and kalata B5 show high similarity in amino acid sequence of loop 3.40,43 Interestingly, two putative cyclotides identified from the transcriptome dataset, i.e., caripe 4 and caripe 6, lack the conserved glutamic acid in loop 1. These cyclotides contain a serine and glycine, respectively, at the corresponding position. The nucleotide coding triplets of these amino acids have been confirmed unambiguously by transcript read depth analysis (Supporting Information Table S2). In addition, transcriptome data of all mature caripe cyclotide sequences were independently obtained from two different tissue samples, i.e., in vitro plantlet and root culture. Besides the Momordica cochinchinensis squash trypsin inhibitors and kalata B12, these peptides represent to our knowledge the only cyclotides lacking this residue, which has a key role for the stability of cyclotides.40 In the future, it will be interesting to determine whether the replacement of the glutamic acid residue has any structural effects or consequences for the overall stability of these novel cyclotides. As a prerequisite, we compared the precursor proteins of Carapichea with known cyclotide and knottin precursors. Selected precursor proteins of other Rubiaceae species (Figures 3C and 3D) as well as the squash trypsin inhibitors TGT-II and MCoTI-II (Supporting Information Figure S10B) indicate higher similarity to the sequences of cyclotide precursors than to the knottin precursor architecture. In particular, they display high amino acid homology within the C-terminal tail, known to be important for the in planta cyclization.44
Cyclotides are typically produced as precursor proteins and are post-translationally processed including the excision of the cyclotide domain, oxidative folding,45 and head-to-tail cyclization.46,47 The detailed analysis of the different cyclotides precursors is of great interest to fully comprehend their in planta synthesis. From all plant families that are known to contain cyclotides, respective precursor proteins have been reported. An overview of the currently known cyclotide precursor architecture is given in Figure 3B. During the screening procedure, we found precursor sequences obtained from in vitro plantlet and root culture that predicted the presence of cyclotides in Carapichea ipecacuanha. The roots of this species are used to make “syrup of ipecac,” an important traditional medication known for its powerful emetic activity.48 When analyzing Herbarium samples by amino acid sequencing, we indeed identified cyclotides in this species. However, it appears (see Figure 4) that genetic heterogeneity,49 different growth conditions of the samples, and different types of tissues, i.e., native and unprocessed collections in the wild, traditionally processed plant material as well as in vitro tissue cultures from the laboratory may explain the differences in the cyclotide content.43,50-52 The combined MS analysis of diverse plant samples together with transcriptome-mining not only helped to identify novel peptides but also provided information about the gene architecture to aid the characterization of processing mechanisms involved in cyclotide biosynthesis.
To further evaluate the importance of specific amino acid residues at the processing sites of Rubiaceae cyclotide precursor proteins, we performed an alignment of 25 precursors, including those from Oldenlandia affinis, Chassalia chartacea, Hedyotis biflora, and those newly identified in Carapichea ipecacuanha (Figures 3C and 3D). As recently shown for Oldenlandia affinis a C-terminal asparagine within the cyclotide domain as well as an adjacent small amino acid residue followed by leucine/isoleucine is crucial for correct bioprocessing, in particular cyclization.44 Analysis of the residues preceding the proto-N-terminus of the cyclotide domain revealed a conserved asparagine/aspartic acid in the precursor sequences of Carapichea ipecacuanha. The asparagine residue is also highly conserved in previously identified precursors from Chassalia chartacea. Only three peptides have a different residue, i.e., a leucine (chassatide C8) or a glycine (chassatide C7 and C17) at the N-terminal site of the cyclotide domain. However, Oldenlandia affinis and Hedyotis biflora contain a highly conserved lysine residue at this position and only precursors for Oak 7, 8, and 9 contain different residues, namely a threonine (Oak 8) or a glycine (Oak 7 and 9) (Figure 3D). Overall this variation supports the hypothesis that the residue immediate preceding the conserved N-terminal residues, e.g., glycine and leucine/isoleucine is not crucial for cyclization as has been suggested previously.44 On the other hand, the first residues of the C-tail are presumably vital for cyclization and are usually conserved across species.44 The residue at the second position adjacent to the cyclotide domain is a leucine in all precursors of C. ipecacuanha and in most Rubiaceae cyclotide precursor. Indeed, this residue (leucine or isoleucine) is highly conserved throughout all known cyclotide precursor except those from Fabaceae species. The first residue of the C-tail in Carapichea ipecacuanha precursors is either glutamic acid or glycine. Glutamic acid in this position has been previously reported in chassatide C2 (Rubiaceae) and in Phyb I (Solanaceae), but in other precursors is often a small residue (e.g., alanine, serine, or glycine). These findings allow speculations about species or family-selective mechanisms or processing enzymes involved in cyclotide biosynthesis, which needs to be addressed in future studies. The third residue in the C-terminal tail region is only conserved in Oldenlandia affinis precursor proteins and is always a proline with one exception, namely the precursor of Oak 9, which lacks the C-tail at all. Precursors of Chassalia chartacea, Hedyotis biflora, and Carapichea ipecacuanha display various residues at this position including asparagine, aspartic acid, lysine, alanine, threonine, glutamic acid, and serine (Figures 3C and 3D). According to previous studies, only the first two C-terminal residues are presumed crucial for a correct in planta cyclization37,44 and further residues beyond position two do not affect peptide processing. Similar residues, e.g., an asparagine at the N- and a glycine at the C-terminus of the peptide domain have recently been found to be present within Momordica cochinchinensis precursors encoding for the cyclotide MCoTI-II, pointing out similar mechanisms underlying cyclization of peptides within different plant families.53 In summary, cyclotide precursors including those encoding the novel C. ipecacuanha proteins show high similarities within one species and appear to have family specific variations (Figure 3). Whether this implies that there are also family specific variations in the biosynthesis pathways and in particular the cyclization mechanism has yet to be determined.
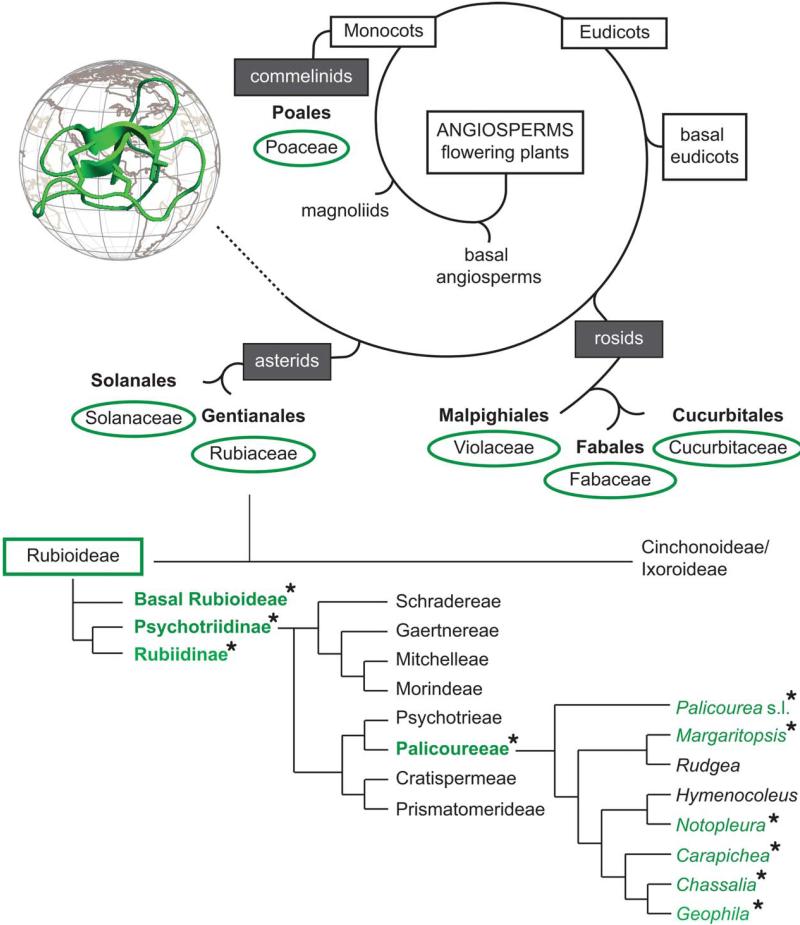
Besides comparing different cyclotide precursors by molecular sequence analysis, it was important to determine the distribution and phylogenetic relationship of cyclotide expressing species. Hence, we analyzed the distribution of cyclotide-containing plants with regards to their botanical relationship. Since the first discovery of cyclotides in Oldenlandia affinis (Rubiaceae), few families have been identified to contain cyclotides (Figure 6). However, all species of the Violaceae analyzed so far contain cyclotides, the distribution within other families is not clear. With special emphasis on the analysis of Rubiaceae, we identified nine novel plants from this family to express cyclotides. Rubiaceous species have previously been established as a prominent source of cyclotides. As to now, they have been found in species of several tribes within the subfamily Rubioideae (Figure 6), namely they have been reported for the tribes Hedyotideae, Lasiantheae, Palicoureeae, and Psychotrieae.22,56 So far it has been suggested that Psychotria cyclotides are not restricted to a specific clade. Especially in Asia, Australasia, and the Pacific region, Psychotria has been treated in a broader sense, including taxa belonging to both Psychotrieae and Palicoureeae.57 To assess the systematic value of cyclotide distribution, the correct placement of a given species is crucial. To render Psychotria a monophyletic group, species of the subgenus Heteropsychotria need to be excluded from the genus Psychotria, and thus should be transferred to Palicourea within the Palicoureeae.54,55,58,59 Consequently, the assignment of all cyclotide-positive Psychotria species from this study (Table I) should be checked by means of morphological and phylogenetic studies and revised as needed (Figure 6).
FIGURE 6. Phylogenetic analyses of cyclotide-containing plants and distribution in Rubiaceae.
An overview of the distribution of cyclotides among angiosperms is schematically shown. Cyclotide-containing plant families, known for both monocotyledon and dicotyledon plants (grey boxes) are highlighted with green circles. A detailed analysis of the distribution and phylogenetic relation of cyclotide-containing species within Rubiaceae is shown in the lower part. The presence and absence of cyclotide-containing species within the listed genera (right column, italic font) are indicated with asterisk and green colored font. Cyclotides have been discovered within the tribes Lasiantheae (basal Rubioideae), Spermacoceae (Rubiidinae), and Palicoureeae. Rubiaceae phylogeny is according to the literature.54,55
With the exception of Psychotria punctata22 that belongs to the tribe Psychotrieae, the presented results suggest that within the Psychotria alliance, i.e., a taxonomic group comprising inter alia the tribes Psychotrieae and Palicoureeae, cyclotides are restricted to species of the tribe Palicoureeae. However, no sequences have been reported for P. punctata22 (Supporting Information Table S1), yet, and hence it remains doubtful whether this species actually expresses cyclotides. Thus, the occurrence of cyclotides within the genus Psychotria sensu stricto and the Psychotrieae has to be questioned. Additional evidence for the restriction of cyclotide plants to the Palicoureeae is the observation that all 24 analyzed species from the Psychotrieae in this study do not contain cyclotides. Within the Palicoureeae, cyclotides have previously been found in the genera Chassalia, Geophila, and Palicourea sensu lato (including species of the subgenus Heteropsychotria). This study adds two further cyclotide producing genera, namely Notopleura and Carapichea to the list of cyclotide-containing plants. Furthermore, we propose that cyclotides also occur in the genus Margaritopsis, based on reassignment of the Micronesian plant Psychotria leptothyrsa, which was discovered by Gerlach et al.56 and was recently shown to belong to the Margaritopsis clade on the basis of DNA sequence data.60 Two genera, i.e., Rudgea and Hymenocoleus remain the last blank spots on the distribution map of cyclotides within Palicoureeae (Figure 6), but based on current knowledge about the occurrence of cyclotides and genetic relationships we expect species of those genera to contain cyclotides. Overall our findings confirm previous suggestions about the abundance of cyclotides within flowering plants and they may indeed form the largest protein class within the plant kingdom.22,29 Yet, apart from their wide distribution among angiosperms, cyclotides within Rubiaceae plants seem to be restricted to specific tribes.
In conclusion, the use of a refined chemical screening methodology together with bioinformatics approaches such as transcriptome-mining and automated peptide database search efficiently accelerates the discovery of novel cyclotides. Further information about their respective genes allows deeper insights into the distribution, evolution, and diversity of this unique class of circular miniproteins. Furthermore, the expansion of information about the range of natural sequences of cyclotides will facilitate their ongoing applications as pharmaceutical61 and agricultural62 bioactive agents, as well as pharmaceutical grafting frameworks,63 as described elsewhere in this special issue of Biopolymers Peptide Science.
Supplementary Material
Acknowledgments
Contract grant sponsor: Austrian Science Fund
Contract grant numbers: FWF-P22889 and P24743
Contract grant sponsor: iPlant (NSF, National Science Foundation)
Contract grant number: DBI-0735191
Contract grant sponsors: Ernst-Mach Scholarship from the Austrian Agency for International Mobility and Cooperation in Education, Science and Research (OeAD), Alberta Ministry of Enterprise and Advanced Education, Alberta Innovates Technology Futures (AITF), Innovates Centre of Research Excellence (iCORE), Musea Ventures, and BGI-Shenzhen
The authors thank all individuals who supported the collection and taxonomic identification of plant species. In particular, they are grateful to Nuria García Hernández for help during plant extraction.
Footnotes
Additional Supporting Information may be found in the online version of this article
REFERENCES
- 1.Gruber CW, Muttenthaler M, Freissmuth M. Curr Pharm Des. 2010;16:3071–3088. doi: 10.2174/138161210793292474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Craik DJ. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- 3.Trabi M, Craik DJ. Trends Biochem Sci. 2002;27:132–138. doi: 10.1016/s0968-0004(02)02057-1. [DOI] [PubMed] [Google Scholar]
- 4.Craik DJ, Daly NL, Bond T, Waine C. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 5.Colgrave ML, Craik DJ. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 6.Göransson U, Sjogren M, Svangard E, Claeson P, Bohlin L. J Nat Prod. 2004;67:1287–1290. doi: 10.1021/np0499719. [DOI] [PubMed] [Google Scholar]
- 7.Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA. Proc Natl Acad Sci USA. 2008;105:1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber CW, Cemazar M, Anderson MA, Craik DJ. Toxicon. 2007;49:561–575. doi: 10.1016/j.toxicon.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Colgrave ML, Kotze AC, Huang YH, O’Grady J, Simonsen SM, Craik DJ. Biochemistry. 2008;47:5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- 10.Plan MRR, Saska I, Cagauan AG, Craik DJ. J Agric Food Chem. 2008;56:5237–5241. doi: 10.1021/jf800302f. [DOI] [PubMed] [Google Scholar]
- 11.Gran L. Medd Nor Farm Selsk. 1970;12:173–180. [Google Scholar]
- 12.Gran L. Acta Pharmacol Toxicol (Copenh) 1973;33:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 13.Gran L. Lloydia. 1973;36:174–178. [PubMed] [Google Scholar]
- 14.Gran L, Sandberg F, Sletten K. J Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- 15.Gran L, Sletten K, Skjeldal L. Chem Biodiversity. 2008;5:2014–2022. doi: 10.1002/cbdv.200890184. [DOI] [PubMed] [Google Scholar]
- 16.Witherup KM, Bogusky MJ, Anderson PS, Ramjit H, Ransom RW, Wood T, Sardana M. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 17.Pranting M, Loov C, Burman R, Göransson U, Andersson DI. J Antimicrob Chemother. 2010;65:1964–1971. doi: 10.1093/jac/dkq220. [DOI] [PubMed] [Google Scholar]
- 18.Ireland DC, Wang CK, Wilson JA, Gustafson KR, Craik DJ. Biopolymers. 2008;90:51–60. doi: 10.1002/bip.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burman R, Svedlund E, Felth J, Hassan S, Herrmann A, Clark RJ, Craik DJ, Bohlin L, Claeson P, Göransson U, Gullbo J. Biopolymers. 2010;94:626–634. doi: 10.1002/bip.21408. [DOI] [PubMed] [Google Scholar]
- 20.Gründemann C, Koehbach J, Huber R, Gruber CW. J Nat Prod. 2012;75:164–175. doi: 10.1021/np200722w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henriques ST, Craik DJ. Drug Discov Today. 2010;15:57–64. doi: 10.1016/j.drudis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Göransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E, Craik DJ. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Göransson U, Burman R, Gunasekera S, Stromstedt AA, Rosengren KJ. J Biol Chem. 2012;287:27001–27006. doi: 10.1074/jbc.R111.300129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 25.Poth AG, Colgrave ML, Philip R, Kerenga B, Daly NL, Anderson MA, Craik DJ. ACS Chem Biol. 2010;6:345–355. doi: 10.1021/cb100388j. [DOI] [PubMed] [Google Scholar]
- 26.Poth AG, Mylne JS, Grassl J, Lyons RE, Millar AH, Colgrave ML, Craik DJ. J Biol Chem. 2012;287:27033–27046. doi: 10.1074/jbc.M112.370841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulvenna JP, Mylne JS, Bharathi R, Burton RA, Shirley NJ, Fincher GB, Anderson MA, Craik DJ. Plant Cell. 2006;18:2134–2144. doi: 10.1105/tpc.106.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen GK, Lian Y, Pang EW, Nguyen PQ, Tran TD, Tam JP. J Biol Chem. 2013;288:3370–3380. doi: 10.1074/jbc.M112.415356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsen SM, Sando L, Ireland DC, Colgrave ML, Bharathi R, Göransson U, Craik DJ. Plant Cell. 2005;17:3176–3189. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaas Q, Craik DJ. Biopolymers. 2010;94:584–591. doi: 10.1002/bip.21424. [DOI] [PubMed] [Google Scholar]
- 31.Hashempour H, Koehbach J, Daly NL, Ghassempour A, Gruber CW. Amino Acids. 2013;44:581–595. doi: 10.1007/s00726-012-1376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber CW. Biopolymers. 2010;94:565–572. doi: 10.1002/bip.21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colgrave ML, Poth A, Kaas Q, Craik DJ. Biopolymers. 2010;94:592–601. doi: 10.1002/bip.21400. [DOI] [PubMed] [Google Scholar]
- 34.Ireland DC, Clark RJ, Daly NL, Craik DJ. J Nat Prod. 2010;73:1610–1622. doi: 10.1021/np1000413. [DOI] [PubMed] [Google Scholar]
- 35.Nomura T, Quesada AL, Kutchan TM. J Biol Chem. 2008;283:34650–34659. doi: 10.1074/jbc.M806953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson MT, Carpenter EJ, Tian Z, Bruskiewich R, Burris JN, Carrigan CT, Chase MW, Clarke ND, Covshoff S, Depamphilis CW, Edger PP, Goh F, Graham S, Greiner S, Hibberd JM, Jordon-Thaden I, Kutchan TM, Leebens-Mack J, Melkonian M, Miles N, Myburg H, Patterson J, Pires JC, Ralph P, Rolf M, Sage RF, Soltis D, Soltis P, Stevenson D, Stewart CN, Jr., Surek B, Thomsen CJ, Villarreal JC, Wu X, Zhang Y, Deyholos MK, Wong GK. PLoS One. 2012;7:e50226. doi: 10.1371/journal.pone.0050226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen GK, Lim WH, Nguyen PQ, Tam JP. J Biol Chem. 2012;287:17598–17607. doi: 10.1074/jbc.M111.338970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruber CW, Muttenthaler M. PLoS One. 2012;7:e32559. doi: 10.1371/journal.pone.0032559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eisenberg D, Schwarz E, Komaromy M, Wall R. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 40.Göransson U, Herrmann A, Burman R, Haugaard-Jönsson LM, Rosengren KJ. ChemBioChem. 2009;10:2354–2360. doi: 10.1002/cbic.200900342. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen GKT, Zhang S, Wang W, Wong CTT, Nguyen NTK, Tam JP. J Biol Chem. 2011;286:44833–44844. doi: 10.1074/jbc.M111.290296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark RJ, Daly NL, Craik DJ. Biochem J. 2006;394:85–93. doi: 10.1042/BJ20051691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plan MR, Rosengren KJ, Sando L, Daly NL, Craik DJ. Biopolymers. 2010;94:647–658. doi: 10.1002/bip.21409. [DOI] [PubMed] [Google Scholar]
- 44.Conlan BF, Colgrave ML, Gillon AD, Guarino R, Craik DJ, Anderson MA. J Biol Chem. 2012;287:28037–28046. doi: 10.1074/jbc.M112.347823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruber CW, Cemazar M, Clark RJ, Horibe T, Renda RF, Anderson MA, Craik DJ. J Biol Chem. 2007;282:20435–20446. doi: 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- 46.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 47.Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, Craik DJ. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 48.World Health Organisation [accessed 17.12.2012];WHO Monographs on Selected Medicinal Plants. 2007 Available at http://apps.who.int/medici-nedocs/documents/s14213e/s14213e.pdf#page212.
- 49.de Sousa Queiroz C, de Carvalho Batista FR, de Oliveira LO. Mol Phylogenet Evol. 2011;59:293–302. doi: 10.1016/j.ympev.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 50.Seydel P, Gruber CW, Craik DJ, Dornenburg H. Appl Microbiol Biotechnol. 2007;77:275–284. doi: 10.1007/s00253-007-1159-6. [DOI] [PubMed] [Google Scholar]
- 51.Trabi M, Craik DJ. Plant Cell. 2004;16:2204–2216. doi: 10.1105/tpc.104.021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trabi M, Svangard E, Herrmann A, Göransson U, Claeson P, Craik DJ, Bohlin L. J Nat Prod. 2004;67:806–810. doi: 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- 53.Mylne JS, Chan LY, Chanson AH, Daly NL, Schaefer H, Bailey TL, Nguyencong P, Cascales L, Craik DJ. Plant Cell. 2012;24:2765–2778. doi: 10.1105/tpc.112.099085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nepokroeff M, Bremer B, Sytsma KJ. Syst Bot. 1999;24:5–27. [Google Scholar]
- 55.Robbrecht E, Manen JF. Syst Geogr Pl. 2006;76:85–146. [Google Scholar]
- 56.Gerlach SL, Burman R, Bohlin L, Mondal D, Göransson U. J Nat Prod. 2010;73:1207–1213. doi: 10.1021/np9007365. [DOI] [PubMed] [Google Scholar]
- 57.Andersson L. Syst Geogr Plants. 2001;71:73–85. [Google Scholar]
- 58.Borhidi A. Acta Bot Hung. 2011;53:241–250. [Google Scholar]
- 59.Taylor CM, Lorence DH, Gereau RE. Novon. 2010;20:481–492. [Google Scholar]
- 60.Barrabé L, Buerki S, Mouly A, Davis AP, Munzinger J, Maggia L. Taxon. 2012;61:1251–1268. [Google Scholar]
- 61.Gerlach SL, Yeshak M, Göransson U, Roy U, Izadpanah R, Mondal D. Biopolymers (Pept Sci) 2013;100:471–479. doi: 10.1002/bip.22325. [DOI] [PubMed] [Google Scholar]
- 62.Malagón D, Botterill B, Gray DJ, Lovas E, Duke M, Gray C, Kopp SR, Knott LM, McManus DP, Daly NL, Mulvenna J, Craik DJ, Jones MK. Biopolymers (Pept Sci) 2013;100:461–470. doi: 10.1002/bip.22229. [DOI] [PubMed] [Google Scholar]
- 63.Poth AG, Chan LY, Craik DJ. Biopolymers (Pept Sci) 2013;100:480–491. doi: 10.1002/bip.22284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.