Abstract
A major challenge in the field of high-throughput proteomics is the conversion of the large volume of experimental data that is generated into biological knowledge. Typically, proteomics experiments involve the combination and comparison of multiple data sets and the analysis and annotation of these combined results. Although there are some commercial applications that provide some of these functions, there is a need for a free, open source, multifunction tool for advanced proteomics data analysis. We have developed the Visualize program that provides users with the abilities to visualize, analyze, and annotate proteomics data; combine data from multiple runs, and quantitate differences between individual runs and combined data sets. Visualize is licensed under GNU GPL and can be downloaded from http://proteomics.mcw.edu/visualize. It is available as compiled client-based executable files for both Windows and Mac OS X platforms as well as PERL source code.
Keywords: Bioinformatics, Protein-automated identification, Quantitative analysis, Software
Modern mass spectrometers generate large volumes of data that make confident protein identifications notoriously difficult. Beyond that, a proteomics experiment generally requires that investigators combine and compare the results from multiple runs or groups of runs. These results then need to be further annotated and analyzed to determine quantitative differences and changes in biological pathways. There is currently no open source, multifunction stand alone tool for advanced proteomics data analysis that can provide the necessary functionality required by most proteomics laboratories.
In order to fill this void, we have developed a comprehensive proteomics data analysis tool, Visualize, which addresses these challenges. To make Visualize easy to use, we have constructed it as a workstation application with an intuitive graphical user interface. We have also made it compatible with standard proteomics data formats as well as data generated from different instrumental platforms, database search algorithms, and the Trans-Proteomic Pipeline (TPP) [1]. To allow for a wide range of experimental approaches, we have designed Visualize to be able to carry out quantitative proteomics using spectral counting, SILAC Peptide Count Ratio Analysis (SPeCtRA) [2], and MS/MS isobaric mass tag (iTRAQ [3]/TMT [4]) methods. The program is freely distributed to users as complied executable files for Windows and Mac OS X systems as well as the complete PERL source code.
The Visualize program was developed using ActiveState Perl v5.8.8 and compiled using ActiveState PDK version 8.0.1 PerlApp compiler. The graphical user interface is provided by the use of Perl Tk modules and runs natively on the Windows platform and under X11 for the Mac OS X platform.
The Visualize program allows users to interact with their data in two different modes: direct mode and batch mode. In direct mode, the user can open an individual results file and perform analysis and visualization of the data contained within a single file. In batch mode, data from multiple results files or lists of files, comprising whole experiments, can be combined, quantitated, or annotated automatically. We have developed the .ez2 file format that is described in more detail in the Supporting Information material. The.ez2 format is based on the Zip compressed file and contains XML and YAML files containing peptide, spectra, and chromatogram information as well as the.out and.dta files corresponding to the peptide identifications reported. Ez2 files can be created using the included Epitomize program or by carrying out searches with the ViPDAC system or from files produced by the TPP [5].
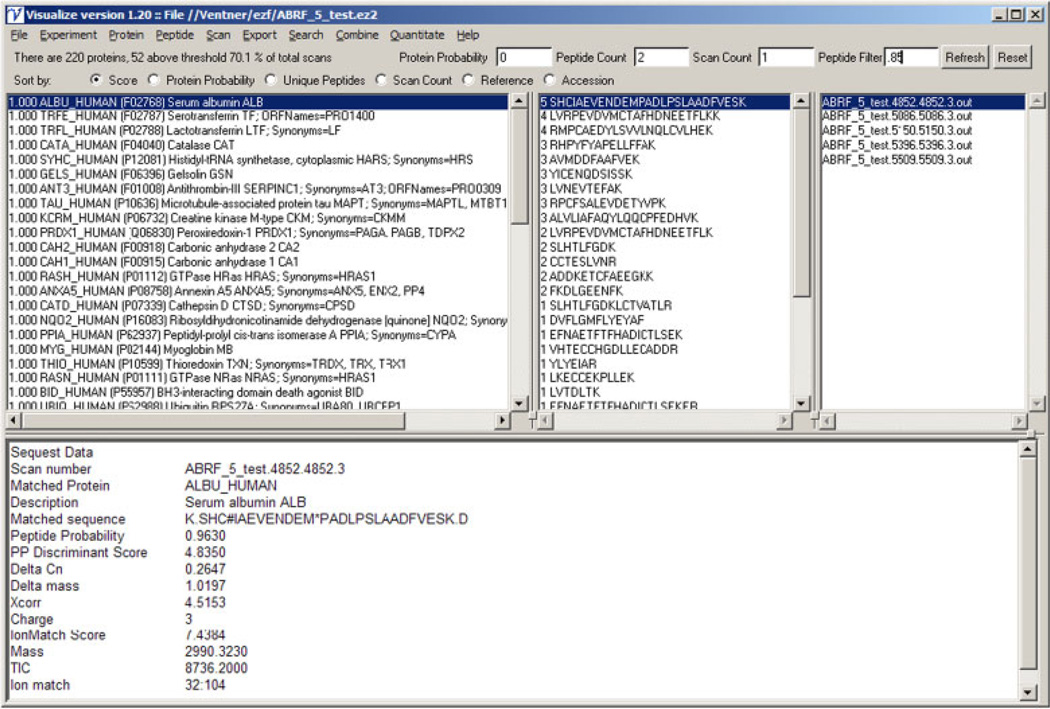
Figure 1 shows the main Visualize window in direct mode. This window is divided into four frames, each representing a view of a different level of the data. The protein frame (top left) shows the list of proteins identified with the protein probability score shown on the left and the description shown on the right. The peptide frame (top middle) shows the complete list of individual peptides identified for the protein selected in the protein frame. The number of spectra associated with each peptide is shown on the left side of this frame. The scan frame (top right) provides a link to the spectra associated with the selected peptide. The data frame (bottom) shows key results from the search that identified the spectra. Above these frames are a series of filter and display options. The user can apply filters at the level of protein probability, peptide count, scan count, and peptide probability as well as determine the order in which the proteins are shown. The analysis and display options in single file mode are divided into menus representing the different levels of the experimental hierarchy: Experiment, Protein, Peptide, and Scan.
Figure 1.
Direct mode visualization.
Under the Experiment menu, the options are divided into the functional groups Overview, Analyze, Display, Maintenance, and Experimental Notes. The selections in the Overview submenu include options to create reports that encompass data from the entire run. The Experimental Dashboard provides a single page graphical synopsis of the run. The Experimental Summary provides a more detailed view of all of the proteins identified in the run and the Spectral Summary provides a detailed view of each of the spectra in the file. The Protein Groups allows the user to collapse the peptide assignments to minimalize the protein list. Options under the Analysis submenu allow the user to carry out analysis of the group of proteins identified in the run. If the sample has been searched against a UniProt database [6], the Species/Gene Summary will summarize the species and genes observed based on protein IDs. The Show Serum Proteins option identifies the most common serum proteins in the sample based on an internal list of the most common serum proteins based on the study of Anderson [7].
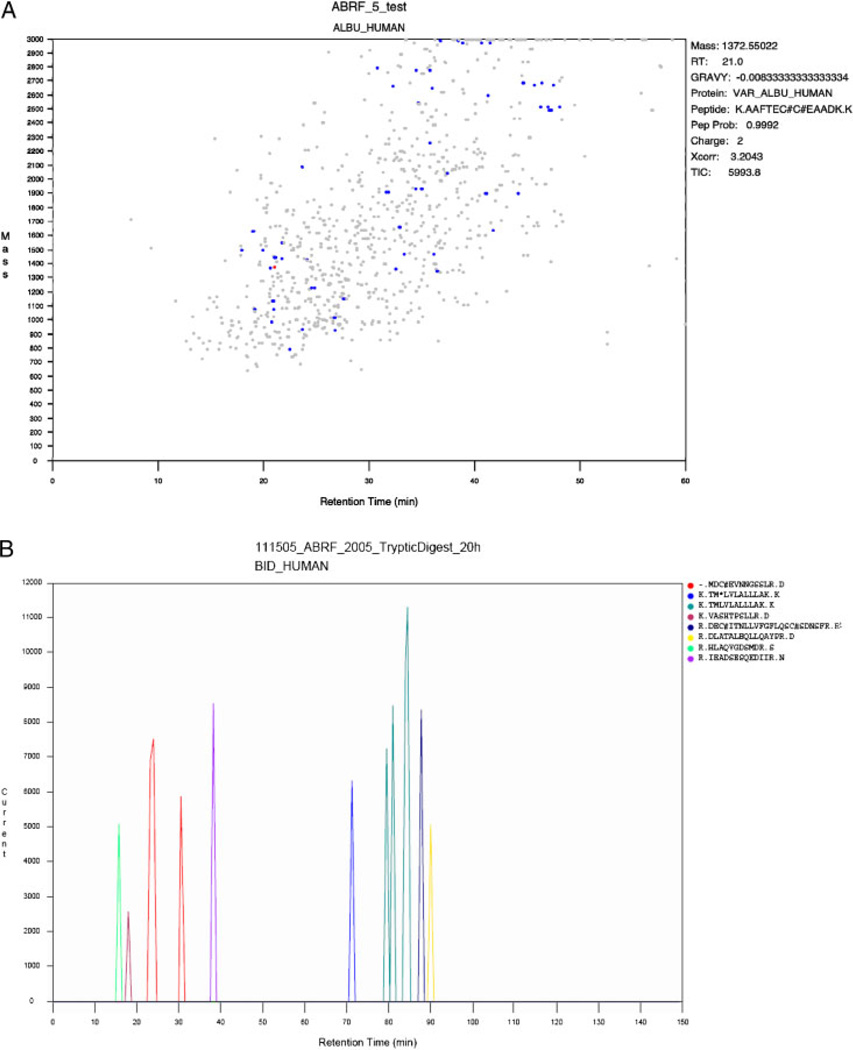
This ability to identify proteins within the sample that belongs to protein groups is extended with the Show Selected Proteins option. The Cleavage Site option shows the count and percent for the cleavage site of each of the peptides identified. This can be very useful in identifying nontryptic cleavages in samples in which endogenous proteolysis may be important. The Amino Acid Analysis option allows the user to view the physiochemical properties of the group of proteins observed in the run. It calculates the molecular weight, pI, and GRAVY (GRand AVerage hYdropathicity [8]) score as well as the amino acid composition for each protein. The Modification Analysis shows the modified peptides found in the search, sorted by modification type. Using local databases, the proteins found in the run can be mapped to either the KEGG pathways [9] or the GO Ontologies [10]. GO annotation is available using either the standard qualitative method based on the lists of proteins observed or an alternative quantitative approach based on spectral counting. Alternatively, the list of identified proteins can also be directly submitted to either the DAVID [11] or the Reactome [12] annotation resources. The Display submenu provides a number of options to generate graphic views of the data. The Peptide Probability model option displays the graph of the data used by the Bayesian classifier to assign peptide probability scores (manuscript in preparation). The Discriminant Score Histogram option displays the distribution of discriminant scores for the current run. In addition to the total, it also shows the plot of the distribution of the scores calculated for correct and random assignments. The FDR versus Prob graph displays the global False Discovery Rate (FDR) versus the local False Discovery Rate (peptide probability score). From these data, users can infer what peptide probability score corresponds to a 5% FDR and use this value for filtering the data. The Chromatogram shows the total ion current of the MS scans versus the elution time (Fig. 2B). As for most of the graphs produced by Visualize, the user can change axis and dynamically zoom in and out of any part of the graph and export the graph to an .eps file for inclusion in publications and presentations.
Figure 2.
Scan map peptide profile outputs.
The Scanmap option allows the user to obtain a detailed view of the elution profile at the level of individual proteins and peptides (Fig. 2A and B). Figure 2A shows the experiment level view. Each scan resulting in a peptide identification is mapped based on its parent ion mass and elution time and the user can examine the data for the identification and plot the elution of each of the peptides of a selected protein (Fig. 2B). In this view, the user can directly observe how peptide modifications affect elution times. The Calculated versus Observed retention time (RT) graph option is similar to the Scanmap option. The 1-D gel option creates a simulation of a physical 1-D gel from the list of identified proteins. The protein masses are used to calculate protein mobilities and the number of spectral counts for each protein is used to calculate the protein intensities. The 2-D gel option is analogous to the 1-D gel option, plotting proteins based on their calculated mass and pI.
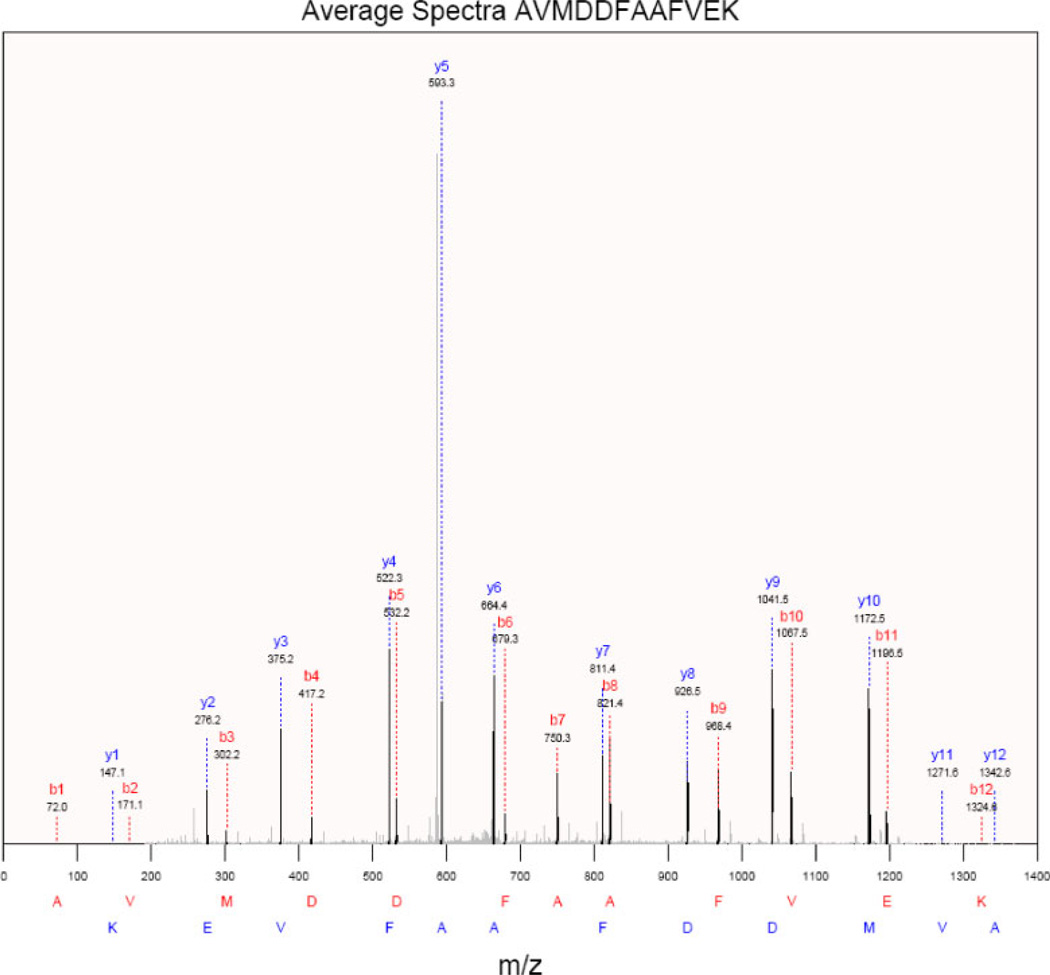
Once a protein has been selected in the protein frame of the main window, the Protein menu becomes active. The Protein Coverage option calculates and displays the protein coverage for each amino acid residue in the currently selected protein. The option to automatically link to a number of protein resources such as Apropos, IPI, iHOP, UniProt, KEGG and String is also available. The Peptide menu is activated when a peptide has been selected in the peptide frame and contains options specific to analysis at the peptide level. To find related sequences in NCBI databases, the Blast Peptide Sequence option can be used to automatically carry out and retrieve the results of a Blast search [13] of the peptide sequences using the NCBI servers. Since many peptides are often identified in multiple spectra, it is useful to see the average data for the spectra that identified a given peptide. This graph can be saved as a .pdf file (Fig. 3) or exported as a composite.dta file.
Figure 3.
Average spectra display.
The Scan menu provides the user with the ability to visualize more detailed information about the currently selected scan. The original .dta and .out file from SEQUEST searches can be viewed. The data contained in individual files can be exported in a number of different formats including lists of protein names or accession numbers for import into other tools, Excel files, and pdfs of selected spectra to meet publication requirements. The data can be searched for protein ID’s or accession numbers, description or the text of the underlying out files. The files can also be filtered to remove proteins below a score threshold, proteins that belong to a specific group, such as serum proteins, or proteins that are in common with another file, for example to remove nonspecific proteins from an affinity selection experiment.
Many of the functions of Visualize work on more than one file at a time or on a file of file names. The Combine menu contains options for combining data from multiple .ez2 files to create a new file. This is useful if the files represent technical replicates, fractions of a sample, or searches done with different algorithms or parameters. The Search option is used to combine data from different searches of the same data. The algorithm selects the best result for each spectrum to create the combined .ez2 file. This can be used to integrate results from different search engines or parameter sets. The Quantitate menu lists various methods for quantitating and comparing multiple files. The submenus are divided based on the quantitative method used. The Spectral Counting submenu allows the user to choose between simple quantitation and more sophisticated analyses based on “label-free” or spectral counting methods [14, 15]. For pairs of samples, an Excel file containing ratios with p-values can be exported. Quantitation can also be performed using either the SPeCtRA [2] method based on spectral counting of SILAC-labeled peptides or by using isobaric mass tags such as the iTRAQ [3] system.
Although there are a number of commercial programs that provide some of the analytical functions described above, Visualize combines them into a single open source package running as a stand-alone workstation application. Since the .ez2 file format allows for the inclusion of all of the data required for analysis and annotation, they are self-contained. This avoids the need to run a web server to visualize and analyze data and simplifies data security in that the .ez2 files can be distributed with the Visualize program.
MassSieve [16] is a free and open source program that allows users to import search results from MASCOT, OMSSA, and X!Tandem. The user can examine the proteins and peptides observed and examine the relationships between the protein and the peptide sequences. Results from multiple runs can be combined but there is no option to quantitatively compare the runs. Unlike Visualize, Mass-Sieve does not have options to examine the MS data underlying the results or to use other data to annotate the results. Programs such as xtandemparser [17], OMSSA Parser [18], and MascotDatfile [19] allow the user to parse and examine the results from X!Tandem, OMSSA, or MASCOT. Unlike MassSieve, these programs are very focused on the MS data involved in the peptide identification, but do not allow the user to view the data as a list of proteins or peptides identified. In contrast to Visualize, there are no quantitation or annotation options present in these programs and they are limited to output only from a single algorithm. Peptizer [20] is another multiplatform Java-based proteomics analysis program. Like these programs, Peptizer is focused on the MS details of peptide identification and not on the proteins or biology of the experiment. It allows the user to use and create sophisticated methods to validate protein identifications but does not allow for quantitation or annotation of the protein results.
Visualize is focused on the experiment rather than the details of the peptide assignments or identifications. Although this and other information is also available from Visualize to meet the needs of the mass spectrometrist, the overall goal of Visualize is to allow the biologist end user to use the search results to answer biologically relevant questions such as which proteins are present in a sample, what groups or pathways do they belong to and how do the proteins and groups change between samples. Visualize brings together the tools to examine complex proteomics data sets at both the low level of MS spectra and the identification details as well as the high levels of quantitation, analysis, and annotation to produce the output and figures required to communicate the results in presentations, publications, and applications.
Supplementary Material
Acknowledgments
The authors thank Bassam Wakim and the members of the Biotechnology and Bioengineering Center for program testing, bug reports and feature requests. This work was supported by NHLBI proteomics contract N01-HV-28182 to A. S. G.
Abbreviations
- SPeCtRA
SILAC Peptide Count Ratio Analysis
- TPP
Trans-Proteomic Pipeline
Footnotes
The authors have declared no conflict of interest.
References
- 1.Deutsch EW, Mendoza L, Shteynberg D, Farrah T, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker SJ, Halligan BD, Greene AS. Quantitative analysis of SILAC data sets using spectral counting. Proteomics. 2010;10:1408–1415. doi: 10.1002/pmic.200900684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 4.Thompson A, Schafer J, Kuhn K, Kienle S, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 5.Deutsch EW, Shteynberg D, Lam H, Sun Z, et al. Trans-Proteomic Pipeline supports and improves analysis of electron transfer dissociation data sets. Proteomics. 2010;10:1190–1195. doi: 10.1002/pmic.200900567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A, Apweiler R, Wu CH, Barker WC, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson NL. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 9.Ogata H, Goto S, Sato K, Fujibuchi W, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Vastrik I, D’Eustachio P, Schmidt E, Joshi-Tope G, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 14.Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst. 2006;131:1335–1341. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller LN, Brusniak M-Y, Mani DR, Aebersold RH. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J. Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- 16.Slotta DJ, McFarland MA, Markey SP. MassSieve: panning MS/MS peptide data for proteins. Proteomics. 2010;10:3035–3039. doi: 10.1002/pmic.200900370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muth T, Vaudel M, Barsnes H, Martens L, Sickmann A. XTandem Parser: an open-source library to parse and analyse X!Tandem MS/MS search results. Proteomics. 2010;10:1522–1524. doi: 10.1002/pmic.200900759. [DOI] [PubMed] [Google Scholar]
- 18.Barsnes H, Huber S, Sickmann A, Eidhammer I, Martens L. OMSSA Parser: an open-source library to parse and extract data from OMSSA MS/MS search results. Proteomics. 2009;9:3772–3774. doi: 10.1002/pmic.200900037. [DOI] [PubMed] [Google Scholar]
- 19.Helsens K, Martens L, Vandekerckhove J, Gevaert K. MascotDatfile: an open-source library to fully parse and analyse MASCOT MS/MS search results. Proteomics. 2007;7:364–366. doi: 10.1002/pmic.200600682. [DOI] [PubMed] [Google Scholar]
- 20.Helsens K, Timmerman E, Vandekerckhove J, Gevaert K, Martens L. Peptizer, a tool for assessing false positive peptide identifications and manually validating selected results. Mol. Cell. Proteomics. 2008;7:2364–2372. doi: 10.1074/mcp.M800082-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.