Abstract
The SWI/SNF family of ATP-dependent chromatin-remodeling factors plays a central role in eukaryotic transcriptional regulation. In yeast and human cells, two subclasses have been recognized: one comprises yeast SWI/SNF and human BAF, and the other includes yeast RSC and human PBAF. Therefore, it was puzzling that Drosophila appeared to contain only a single SWI/SNF-type remodeler, the Brahma (BRM) complex. Here, we report the identification of two novel BRM complex-associated proteins: Drosophila Polybromo and BAP170, a conserved protein not described previously. Biochemical analysis established that Drosophila contains two distinct BRM complexes: (i) the BAP complex, defined by the presence of OSA and the absence of Polybromo and BAP170, and (ii) the PBAP complex, containing Polybromo and BAP170 but lacking OSA. Determination of the genome-wide distributions of OSA and Polybromo on larval salivary gland polytene chromosomes revealed that BAP and PBAP display overlapping but distinct distribution patterns. Both complexes associate predominantly with regions of open, hyperacetylated chromatin but are largely excluded from Polycomb-bound repressive chromatin. We conclude that, like yeast and human cells, Drosophila cells express two distinct subclasses of the SWI/SNF family. Our results support a close reciprocity of chromatin regulation by ATP-dependent remodelers and histone-modifying enzymes.
ATP-dependent chromatin-remodeling complexes (remodelers) regulate gene expression by destabilizing nucleosome structures to allow transcription factors to gain access to DNA (2, 23, 25, 27, 33, 35, 44). These complexes contain an Swi2p/Snf2p-like ATPase and use the energy of ATP hydrolysis to alter the structures of nucleosomes. Chromatin-remodeling factors have been isolated from a variety of organisms, ranging from yeasts and flies to humans. Typically, they have been found to comprise highly conserved multiprotein complexes implicated in transcriptional regulation. Chromatin-remodeling complexes can be divided into several main groups, characterized by different core ATPase subunits and associated factors. Named after their defining ATPase, they are referred to as the Swi2p/Snf2p, ISWI, Mi-2, and INO80 families of remodelers (2, 35).
The yeast Saccharomyces cerevisiae contains two closely related chromatin-remodeling complexes, ySWI/SNF (switching defective and sucrose nonfermenting) and RSC (remodel the structure of chromatin) (2, 6, 8, 47, 52). RSC and ySWI/SNF are similar in structure, sharing two subunits and containing at least four other homologous components (5, 53). The central ATPase-remodeling motor of RSC is Sth1p, a protein closely related to the Swi2p/Snf2p subunit of the ySWI/SNF complex. Despite these structural similarities and comparable in vitro chromatin-restructuring activities (4), there are a number of important functional differences between ySWI/SNF and RSC (47, 52, 53). First, RSC function is required for yeast viability, while ySWI/SNF function is not. Consistent with potentially broader roles in chromatin dynamics, RSC is very abundant, while ySWI/SNF is not. Moreover, genome-wide gene expression studies revealed that these remodelers regulate different, largely nonoverlapping sets of target genes. Although originally remodelers were thought of as activators of transcription, it has been found that ySWI/SNF and RSC may also be involved in gene repression (33).
The SWI/SNF family of remodelers is evolutionarily highly conserved, and homologous complexes have been identified in Drosophila and mammals. Human cells contain two distinct Swi2p/Snf2p-like ATPase subunits, hBRM and BRG1 (reviewed in references 2, 26, 41, and 53). These subunits are homologous and orthologous to yeast Swi2p/Snf2p and Sth1p, respectively. In contrast to yeast and mammalian cells, Drosophila cells contain only a single protein corresponding to Swi2p/Snf2p and Sth1p; this protein is Brahma (BRM) (46, 48). Most of the other subunits of the SWI/SNF-related complexes are also well conserved from yeasts to humans and constitute related complexes in yeast, Drosophila, and human cells (43, 53). The Drosophila SWI/SNF-related complex is named BAP (for “Brahma-associated proteins”), while human SWI/SNF-related complexes are referred to as hSWI/SNF or BAF (for “BRG1- or hBRM-associated factors”) and PBAF (for “Polybromo-associated BAF”) (53).
Two types of SWI/SNF complexes that differ only in a few subunits can be recognized in human cells: (i) the SWI/SNF-α or BAF complex, containing either hBRM or BRG1 and defined by the presence of BAF250 and the absence of Polybromo, and (ii) the SWI/SNF-β or PBAF complex, containing BRG1 and Polybromo but lacking BAF250/p270 and hBRM. BAF250/p270 is related to yeast Swi1p, a ySWI/SNF subunit without a corresponding protein in RSC (40). Conversely, BAF180/Polybromo is structurally related to the Rsc1, Rsc2, and Rsc4 proteins but lacks a counterpart in ySWI/SNF. Thus, based on the conserved structural motifs in these hallmark subunits, yeast and mammalian SWI/SNF-type remodelers fall into two subclasses: one comprising SWI/SNF and BAF (SWI/SNF-α) and the other represented by RSC and PBAF (SWI/SNF-β) (53, 57). Interestingly, a number of studies on human remodelers have revealed functional differences between BAF and PBAF (29, 53, 57).
The recognition of two major subclasses of SWI/SNF-type remodelers in both yeast and human cells suggested a strict evolutionary conservation. Therefore, it is puzzling that Drosophila appears to contain only a single SWI/SNF-type remodeler, the BRM complex. In contrast to yeast and mammalian cells, Drosophila cells have only a single motor protein, BRM, related to both Swi2p/Snf2p and Sth1p. BRM was originally discovered as a dominant suppressor of Polycomb (PC) and therefore was classified as a trithorax group (trxG) protein (48). The trxG of activators, together with their antagonists, the PC group of repressors, maintains the correct expression of many developmental regulators (31, 46). Two BRM-associated proteins, Moira (MOR) and OSA, are also encoded by trxG genes (10, 11, 24). OSA is the homologue of yeast Swi1 and human BAF250/p270, hallmarks of the SWI/SNF-BAF subclass of remodelers. MOR is homologous to yeast Swi3p and Rsc8p and to human BAF170 and BAF155, common components of BAF and PBAF. All other identified BRM-associated subunits are equally related to their SWI/SNF-BAF and RSC-PBAF counterparts (43, 53). Thus, so far only a tentative relationship has been established between the BRM complex and SWI/SNF-BAF but not RSC-PBAF.
The aim of this study was to determine the relationship between the Drosophila BRM complex and the SWI/SNF-BAF or RSC-PBAF subclass of remodelers. We purified BRM and its associated factors and identified two novel subunits by mass spectrometric (MS) analysis: Drosophila Polybromo and BAP170. Biochemical analysis revealed that, like yeast and human cells, Drosophila cells also contain two distinct subclasses of SWI/SNF-type ATP-dependent remodelers, which we named BAP and PBAP. The BAP complex is defined by the presence of OSA and the absence of Polybromo and BAP170. The PBAP complex lacks OSA but contains Polybromo as a distinguishing subunit. Thus, the distinction between the SWI/SNF-BAF and RSC-PBAF complexes is maintained throughout eukaryotic evolution. Moreover, BAP170 is found exclusively in the PBAP complex and not in the BAP complex. To determine potentially differential targeting of BAP and PBAP, we determined the genome-wide distributions of OSA and Polybromo on larval salivary gland polytene chromosomes. Our analyses revealed that BAP and PBAP display distinct patterns of distribution. The chromatin association of BRM and that of its antagonist, PC, appear to be mutually exclusive. Interestingly, whereas BRM complexes associate predominantly with regions of open, hyperacetylated chromatin, PC binding sites are largely devoid of hyperacetylation. Thus, there is a very good correlation between a global mark of chromatin status and the binding of the epigenetic regulatory complexes.
MATERIALS AND METHODS
Purification and MS identification of BRM-associated proteins.
Nuclear extracts derived from wild-type Drosophila melanogaster embryos (0 to 12 h) were prepared essentially as described by Heberlein and Tjian (18). The nuclear extracts were concentrated by chromatography on a POROS-heparin (PerSeptive Biosystems) column equilibrated with HEMG/100 buffer (25 mM HEPES-KOH [pH 7.6], 0.1 mM EDTA, 12.5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol [DTT], 0.2 mM AEBSF [(α-aminoethyl)benzenesulfonyl fluoride], 1 μM pepstatin, 0.01% Nonidet P-40 [NP-40], 100 mM KCl), followed by a step elution with HEMG/400 buffer (HEMG/100 buffer with 400 mM KCl instead of 100 mM KCl). The heparin- 400 mM KCl fraction (H0.4 fraction) contained the vast majority of BRM, as determined by Western immunoblotting (data not shown). BRM was purified further on the basis of the Western blot analysis. The H0.4 fraction was loaded onto an 800-ml Sephacryl S-300 column (elution volume, 300 ml) (Pharmacia) equilibrated and developed with HEMG/100 buffer. BRM eluted in the void and was purified further on a Bioscale Q10 (Bio-Rad) column developed with a linear salt gradient of 100 to 600 mM KCl. The majority of BRM eluted from the Q10 column at 200 to 280 mM KCl.
Fractions were pooled (240 mM KCl) and incubated with protein A beads(Pharmacia) coated with affinity-purified rabbit anti-BRM antibodies as described previously (24). Proteins retained on the beads after extensive washes with HEMG/800 buffer (HEMG/100 buffer with 800 mM KCl instead of 100 mM KCl and 0.1% NP-40 instead of 0.01% NP-40) were subjected to preparative sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) on an 8% gel. Following Coomassie blue staining, gel slices containing each polypeptide band were excised, and proteins were digested in gel with trypsin (Roche Molecular Biochemicals) in 50 mM ammonium bicarbonate (Sigma). Before matrix-assisted laser desorption ionization (MALDI)-time-of-flight (TOF) analysis, peptides were concentrated by using μC18-ZipTips (Millipore) and eluted directly onto the MALDI target in 1 μl of a saturated solution of α-cyanohydroxycinnamic acid in 50% acetonitrile.
Peptides were analyzed by using a Voyager DE-STR MALDI-TOF mass spectrometer (Applied Biosystems) operated in the Reflectron mode at a 20-kV accelerating voltage. Tandem MS measurements were obtained with an electrospray ionization (ESI) quadrupole TOF (Q-TOF) instrument (Micromass Ltd., Manchester, United Kingdom) operating in the positive-ion mode and equipped with a Z-spray nano-ESI source. Nano-ESI needles were prepared from borosilicate glass capillaries (Kwik-Fil; World Precision Instruments Inc., Sarasota, Fla.) on a P-97 puller (Sutter Instrument Co., Novato, Calif.). The needles were coated with a gold layer by using an Edwards Scancoat model 501 sputter coater (at 40 mV and 1 kV for 200 s). The capillary voltage was 1,500 V, and the cone voltage was 40 V. Collision energy was optimized for individual peptides for optimal fragmentation.
For protein identification, Mascot software (www.matrixscience.com) was used for database searches for both peptide mass fingerprinting and peptide sequence tagging. The obtained sequences and identified peptides are shown in Fig. 1C. Sequence alignments were performed by using BLAST searches; manual editing and sequence motifs were identified by using SMART software (http://smart.embl-heidelberg.de).
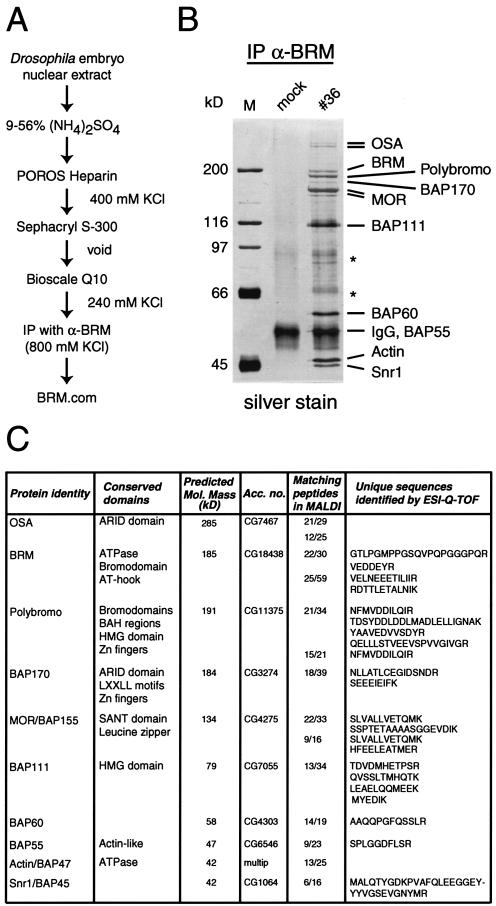
FIG. 1.
Purification and characterization of the BRM complex from Drosophila embryo nuclear extracts. (A) Outline of the chromatographic scheme used to purify Drosophila BRM and associated proteins. IP, immunoprecipitation; α-BRM, antibodies to BRM; BRM.com, Brahma complex. (B) Polypeptide composition of the BRM complex. Peak fractions from the Bioscale Q10 column were combined and incubated with beads coated with affinity-purified anti-BRM antibodies. Mock-treated beads were used as a negative control. Proteins retained on the beads after extensive washes with a buffer containing 800 mM KCl and 0.1% NP-40 were resolved by SDS-PAGE on 8% polyacrylamide gels and stained with silver stain. The nine abundant proteins that consistently copurified with BRM are designated. Degradation products are indicated by asterisks. No polypeptides smaller than 45 kDa were detected on gels with a higher percentage of polyacrylamide (data not shown). Lane M, marker proteins; lane #36, fraction 36. IgG, immunoglobulin G. (C) Summary of BRM-associated proteins identified by MS. Experimental details are described in Materials and Methods. The conserved domains and the predicted molecular (Mol.) masses of the identified proteins are indicated. The numbers of tryptic peptides out of the total identified by MALDI-TOF peptide mass fingerprinting which gave unique matches are shown, along with the GenBank accession numbers (Acc. no.) of the corresponding gene loci; multip, multiprotein. Unique sequence tags identified through tandem MS sequencing by ESI with a Q-TOF instrument (ESI-Q-TOF) also are shown.
Biochemical procedures.
For glycerol gradient sedimentation, the Drosophila embryo nuclear extracts were diluted in 37.5 mM HEPES-KOH (pH 7.9)-90 mM KCl- 6.25 mM MgCl2- 0.05 mM EDTA- 0.75 mM DTT- 0.05 mM phenylmethylsulfonyl fluoride- 0.25 mM sodium metabisulfite- 5% glycerol. Approximately 5 mg (in 600 μl) of nuclear extract was applied to a 10-ml, 10 to 25% gradient of glycerol in HEMG/100 buffer containing 100 mM KCl, 0.1 mM phenylmethylsulfonyl fluoride, and 0.5 mM sodium metabisulfite. Samples were centrifuged in a Beckman SW40Ti rotor at 32,000 rpm for 24 h at 4°C. Twenty-three 500-μl fractions were collected from the top (no. 1) to the bottom (no. 23) of the gradient. Fractions were separated by SDS-PAGE and analyzed by immunoblotting with anti-BRM, anti-Polybromo, anti-MOR, anti-OSA, and anti-BAP170 antibodies. The antibodies directed against BRM (24) and OSA (50) were previously described. The anti-Polybromo antibodies were generated by immunization of rabbits with bacterially expressed glutathione S-transferase fusion polypeptides corresponding to residues 1 to 500 and 877 to 1654 of Polybromo encoded by plasmids PAK224 and PAK225. The anti-MOR antibodies were generated by immunization of rabbits with bacterially expressed glutathione S-transferase fusion polypeptides corresponding to the full-length protein. The anti-BAP170 antibodies were generated by immunization of rabbits with bacterially expressed six-histidine-tagged polypeptides corresponding to residues 600 to 849 of BAP170 encoded by the pRSETB (Invitrogen)-derived plasmid pPVBAP170G. The anti-BRM and anti-Polybromo antibodies were affinity purified prior to use. For immunoprecipitation, affinity-purified antibodies were cross-linked to protein A or G beads by using dimethylpimelimidate essentially as described previously (16). Affinity resins were incubated with the H0.4 fraction for 2 h at 4°C in HEMG/200 buffer (HEMG/100 buffer with 200 mM KCl instead of 100 mM KCl and 0.1% NP-40 instead of 0.01% NP-40). Following extensive washes with excess HEMG/800 buffer and a brief final wash with HEMG/200 buffer, bound proteins were eluted in SDS sample buffer, resolved by SDS-PAGE, and visualized by silver staining or Western blotting.
Polytene chromosome immunostaining.
The analysis of polytene chromosomes was performed essentially as described previously (1, 60) with a number of minor modifications. Briefly, dissection of salivary glands from third-instar Canton S larvae was performed with 0.7% NaCl, followed by fixation for 10 min with 45% acetic acid- 1.85% formaldehyde. After squashing, slides were frozen in liquid N2, the coverslips were snapped off, and then the slides were stored in 100% methanol at −20°C until use (up to 1 week). The slides were washed in phosphate-buffered saline (PBS) for 10 min, washed in PBS-1% Triton X-100 (PBST) for 10 min, and then blocked for 10 min in blocking buffer (PBST with 1% bovine serum albumin). The slides were incubated with primary antibodies (anti-acetylphosphate, 1:100 [Abcam ab76]; purified anti-Polybromo, 1:15; anti-OSA, 1:20; anti-MOR, 1:100; purified anti-BRM, 1:50 [see above for details]; and anti-PC, 1:100) (34) in blocking buffer for 1 h at room temperature. The slides then were rinsed in PBS, washed three times for 10 min each time in PBST, and incubated with the appropriate secondary antibodies (Alexafluor; Molecular Probes) in blocking buffer for 1 h at room temperature. The slides then were rinsed in PBS and washed three times for 10 min each time in PBST. The slides were rinsed in PBS again before being mounted in mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) counterstain (Vectashield with DAPI; H-1200; Vector Laboratories). Images were captured by using a Leica DM-RXA microscope with Texas red, fluorescein isothiocyanate, and DAPI filters (program ColorProc 2003) and processed by using Photoshop.
RESULTS
Identification of two novel BRM-associated proteins.
One of the primary aims of this study was to determine whether there is one single or multiple distinct BRM complexes in Drosophila. We first sought to determine whether there were any remaining uncharacterized subunits of the BRM complex. To isolate proteins associated with BRM, we purified BRM from Drosophila embryo nuclear extracts by a combination of conventional column chromatography and immunopurification (Fig. 1A). The elution profiles of BRM with the various columns were monitored by Western immunoblotting with polyclonal antibodies against the BRM protein. The final step in the purification scheme was carried out with affinity-purified antibodies directed against BRM coupled to protein A beads. The immunopurification was performed under stringent conditions, involving washes with a buffer containing 800 mM NaCl and 0.1% NP-40. Analysis of the final fractions by SDS-PAGE followed by silver staining consistently revealed over 10 protein bands, ranging from 250 to 45 kDa (Fig. 1B). To determine the identities of the proteins associated with BRM, the immunopurified proteins were resolved by SDS-PAGE and stained with Coomassie blue. The protein bands were excised, digested with trypsin, and subjected to MS. MALDI-TOF peptide mass fingerprinting and tandem MS sequencing by ESI with a Q-TOF instrument resulted in the unambiguous identification of 10 proteins (Fig. 1C). In addition to the eight known subunits of the BRM complex, we identified two novel BRM-associated proteins: the Drosophila homologue of Polybromo (CG11375) and a novel protein that we have named BAP170 (CG3274). The latter protein is conserved from flies to mammals but has not been described elsewhere as a component of an SWI/SNF-related complex.
Drosophila Polybromo is closely related to human BAF180 and the yeast Rsc1, Rsc2, and Rsc4 proteins (8, 39, 57). Sequence alignments and searches for structural motifs (http://smart.embl-heidelberg.de) (Fig. 2A) revealed that Polybromo comprises a multitude of potential protein-protein interaction domains, including six bromodomains, two bromo-adjacent homology (BAH) domains, and a high-mobility-group (HMG) box (32). All of these domains are conserved in Drosophila, chicken, Caenorhabditis elegans, and human Polybromo. Furthermore, two putative C2H2 zinc fingers were identified in Drosophila and C. elegans Polybromo, but these appeared to be absent in chicken and human Polybromo.
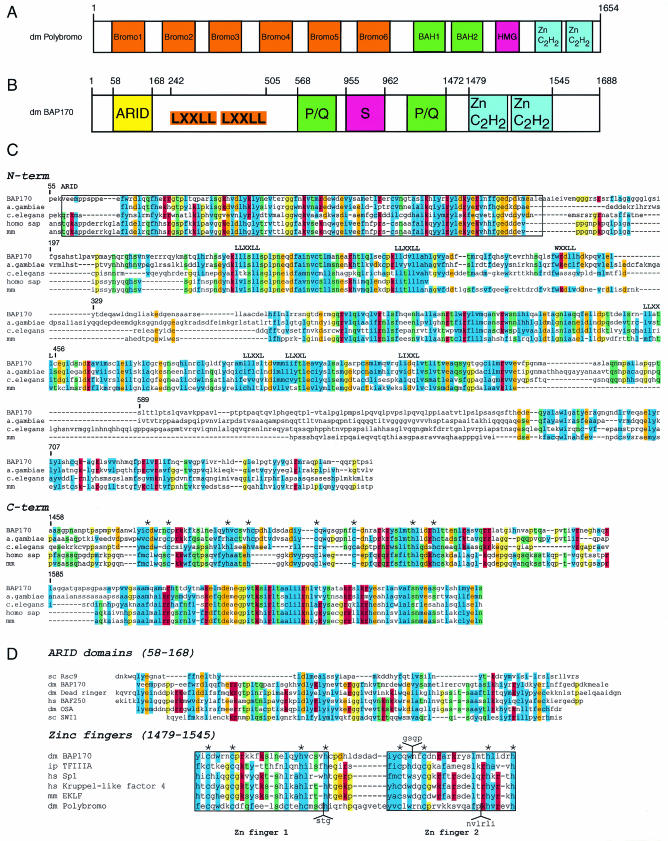
FIG. 2.
Domain structures of Drosophila Polybromo and BAP170. (A) Schematic representation of the functional domains of D. melanogaster (dm) Polybromo (GenBank accession no. NP_651288). Abbreviations: Bromo, bromodomain; Zn C2H2, C2H2 zinc finger motif. (B) Schematic representation of the domain structure of D. melanogaster (dm) BAP170. Indicated are the ARID region, consensus and variant LXXLL sequence motifs (where L is leucine and X is any amino acid) implicated in binding to hormone receptors or coactivators, domains rich in proline and glutamine residues (P/Q), a serine-rich region (S), and two putative C2H2 zinc finger motifs. (C) Alignment of the conserved sequence domains of Drosophila BAP170 (accession no. NM_136372) with homologous predicted amino acid sequences of A. gambiae (accession no. EAA04740), C. elegans (accession no. NP_495679), Homo sapiens (homo sap; accession no. XM_292131 and XP_300559), and Mus musculus (mm; accession no. BAB24929, XM_128029.3, and BAC28898) identified by a BLAST search with the full-length BAP170 sequence. The major conserved blocks in the N- and C-terminal (term) parts of the proteins are shown. Indicated are the conserved ARID region (boxed sequence), several consensus and variant LXXLL sequence motifs, and the potential zinc-coordinating residues (marked with asterisks) of canonical and variant C2H2 zinc finger motifs. Amino acids are given in the single-letter code, and conserved residues are color coded (hydrophobic, blue; acidic, orange; basic, red; polar, green; and glycine or proline, yellow). Please note that a complete human sequence is not available. (D) Sequence alignment of the ARID region and zinc finger domain of BAP170 with cognate structures from other Drosophila (dm), human (hs), S. cerevisiae (sc), Ictalurus punctatus (ip), and mouse (mm) proteins. Residues involved in coordinating zinc are marked with asterisks. Sequence alignments and structural domain predictions were performed with http://smart.embl-heidelberg.de/smart/show_motifs.pl and http://www.ebi.ac.uk/clustalw/.
Like all other BRM-associated proteins, the BAP170 protein is evolutionarily highly conserved and present in many species, such as Anopheles gambiae, C. elegans, mice, and humans (Fig. 2B and C). It is particularly noteworthy that this protein contains several conserved motifs associated with transcriptional regulation. The N terminus contains an AT-rich interaction domain (ARID) (55) which has also been found in yeast Swi1p and Drosophila OSA (Fig. 2D). Unlike OSA, however, BAP170 does not display additional sequence homology with ySwi1, suggesting that OSA is its closest relative in flies. Furthermore, it should be noted that Rsc9 also contains a region with similarity to the ARID region (13) and therefore may be related to BAP170. Intriguingly, BAP170 contains several consensus as well as variant LXXLL (where L is leucine and X is any amino acid) sequence motifs. LXXLL motifs are protein-protein interaction surfaces, first identified in transcriptional coactivators of nuclear hormone receptors (30). Canonical and variant LXXLL motifs have also been implicated to mediate binding to other coactivators, such as HAT. Furthermore, BAP170 contains regions rich in proline (P) and glutamine (Q) residues, and there is a serine-rich region. The conserved C terminus contains one canonical C2H2 zinc finger motif and a second putative zinc finger motif in which the spacing between the two cysteines is larger than those in the classical TFIIIA and Sp1 transcription factors (Fig. 2D).
In summary, our analysis of BRM-associated proteins revealed two novel subunits, BAP170 and Drosophila Polybromo. The identification of Polybromo raises the important issue of whether Drosophila cells, like yeast and human cells, express two distinct ATP-remodeling complexes rather than a single complex, as is currently believed.
Drosophila contains two distinct BRM complexes, BAP and PBAP.
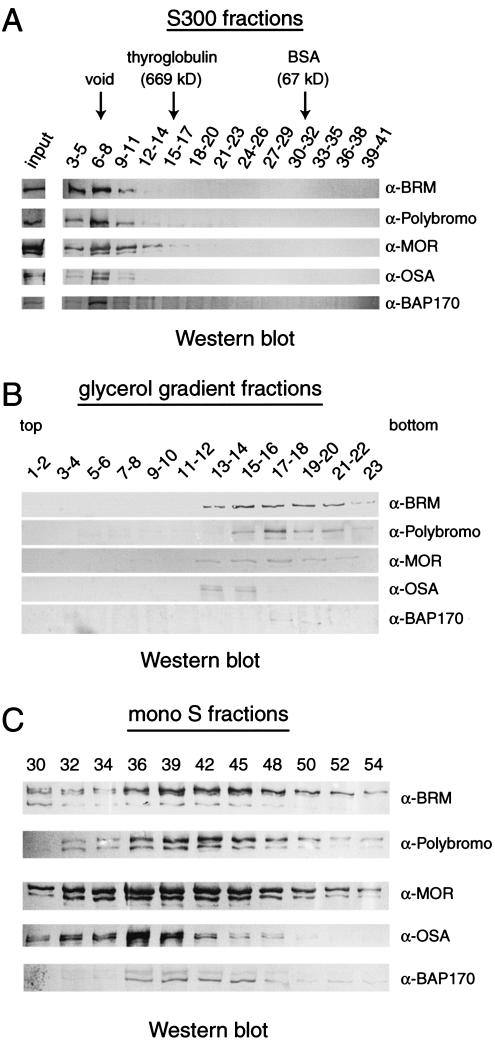
To establish whether Polybromo and BAP170 exist predominantly as part of a large multiprotein complex, we first raised antisera directed against these two novel BRM-associated proteins. The resulting antibodies recognized the predicted endogenous protein bands first identified by MS (data not shown but see below). Next, we analyzed Drosophila embryo nuclear extracts by Sephacryl S-300 size-exclusion chromatography (Fig. 3A). The eluted fractions were resolved by SDS-PAGE and analyzed by immunoblotting with antibodies directed against BRM, Polybromo, MOR, OSA, or BAP170. All of these BRM-associated proteins were present in fractions corresponding to a molecular mass of 2 MDa or higher. None of the proteins could be detected in eluted fractions predicted to contain free subunits, suggesting that they reside predominantly in a very large multiprotein complex. As an alternative method, we fractionated Drosophila embryo nuclear extracts by glycerol gradient sedimentation (Fig. 3B). Again, all BRM-associated proteins sedimented with a velocity indicative of a large protein complex. However, to our surprise, we observed that the pattern of fractionation of OSA was clearly distinct from those of Polybromo and BAP170. The majority of OSA was detected in fractions 13 through 16, while Polybromo and BAP170 were present in fractions 15 through 22. All of these fractions also contained BRM and MOR, which displayed a broad range of migration. The glycerol gradient sedimentation experiment suggested that there might be more than one BRM complex. This notion was supported further by Western blot analysis of the BRM-containing fractions obtained from the final, Mono-S column during purification of endogenous BRM by conventional column chromatography (24). BRM and MOR were both present in the fractions containing Polybromo, BAP170, or OSA. However, Polybromo and BAP170 again displayed elution profiles which were distinct from that of OSA (Fig. 3C). Taken together, the results of the chromatographic analyses of BRM-associated proteins suggested that there are two distinct BRM complexes, containing either OSA or Polybromo and BAP170 as unique subunits.
FIG. 3.
Polybromo and BAP170 are present in a high-molecular-mass complex. (A) Drososphila embryo nuclear extracts were fractionated by Sephacryl S-300 size-exclusion chromatography. The indicated fractions were combined and resolved by SDS-PAGE, followed by immunoblotting with antibodies (α) directed against BRM, Polybromo, MOR, OSA, and BAP170. All of these BRM-associated proteins were present in fractions corresponding to molecular masses of 2 MDa or greater. The elution of the voided volume (void) and the elution of the known markers thyroglobulin (669 kDa) and bovine serum albumin (BSA) (67 kDa) are indicated. (B) Embryo nuclear extracts were centrifuged through a glycerol gradient, and the different fractions collected were examined for the presence of BRM, Polybromo, MOR, OSA, and BAP170 by immunoblotting. (C) The pooled Bioscale Q10 fractions containing BRM were purified further by Mono-S column chromatography. A relevant selection of the fractions analyzed by immunoblotting with antibodies directed against BRM, Polybromo, MOR, OSA, and BAP170 is shown.
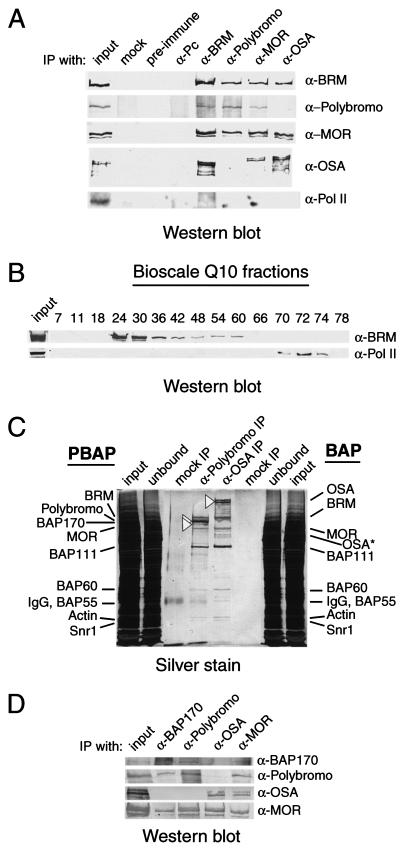
To test this idea directly, we performed a series of coimmunoprecipitation experiments (Fig. 4A). Antibodies directed against Polybromo coimmunoprecipitated BRM and MOR, but not OSA. Conversely, MOR and BRM, but not Polybromo, were found in a complex with OSA. As expected, preimmune antibodies or antibodies directed against PC did not reveal any component of a BRM complex. The possible association of SWI/SNF-type remodelers with an RNA polymerase (Pol) II holoenzyme has been the subject of debate in recent years. Such an association was first reported for yeast cells (56) and later challenged (7). Moreover, conflicting reports have emerged regarding the existence of a mammalian holoenzyme containing Pol II and hBRM or BRG1 (9, 38, 54). To investigate whether such a holoenzyme might exist in Drosophila, we investigated whether Pol II was present in immunoprecipitates of BRM complexes but failed to detect it using antibodies against the second largest Pol II subunit (Fig. 4A, bottom panel). Furthermore, during protein purification by conventional column chromatography, Pol II and BRM were not copurified (Fig. 4B). Thus, at least under the conditions used in our experiments, we failed to obtain direct evidence for a tight association between BRM complexes and Pol II.
FIG. 4.
Polybromo and OSA define two distinct BRM chromatin-remodeling complexes. (A) Drosophila embryo nuclear extracts were incubated with control protein A beads (mock); with protein A beads coated with preimmune, anti-PC, anti-BRM, anti-Polybromo, or anti-MOR antibodies (α); or with protein G beads coated with anti-OSA antibodies. Following coimmunoprecipitation (IP), samples were resolved by SDS-PAGE, followed by Western blotting to determine whether BRM, Polybromo, MOR, OSA, or the second largest subunit of RNA Pol II (IIc; 140 kDa) was present. (B) The presence of BRM or RNA Pol II in the indicated Bioscale Q10 column fractions was determined by immunoblotting. The chromatographic purification scheme is outlined in Fig. 1A. It should be noted that the majority of RNA Pol II already has been separated from BRM on the Sephacryl S-300 column (data not shown). The majority of BRM eluted from the Bioscale Q10 column at about 240 mM KCl, whereas RNA Pol II eluted at about 450 mM KCl. We failed to detect any overlap in theirelution profiles. (C) OSA and Polybromo form part of two distinct BRM chromatin-remodeling complexes. Drosophila embryo nuclear extracts, concentrated by step elution from a POROS-heparin column (H0.4 fraction), were incubated with protein A beads coated with either affinity-purified rabbit polyclonal antibodies directed against Polybromo or monoclonal antibodies directed against OSA. Proteins retained on the beads after extensive washes with a buffer containing 800 mM KCl and 0.1% NP-40 were resolved by SDS-PAGE on 8% polyacrylamide gels and stained with silver stain. Open triangles indicate the presence of OSA in the anti-OSA immunoprecipitate (BAP complex) and the presence of Polybromo and BAP170 in the anti-Polybromo immunoprecipitate (PBAP complex). The other BRM-associated proteins are components of both the BAP and the PBAP complexes. OSA*, OSA breakdown products. The input, the unbound material, and the beads (mock IP) in the immunoprecipitation reactions are shown. IgG, immunoglobulin G. (D) BAP170 is found solely in the Polybromo-containing complex. Drosophila embryo nuclear extracts were incubated with protein A beads coated with antibodies directed against BAP170, Polybromo, OSA, or MOR. The immunoprecipitates were resolved by SDS-PAGE, followed by Western blotting with antibodies directed against BAP170, Polybromo, OSA, or MOR.
Next, we used antibodies directed against either Polybromo or OSA to purify associated proteins. The immunoprecipitates were resolved by SDS-PAGE and stained with silver stain. Although Polybromo and OSA were mutually exclusive subunits of two distinct BRM complexes, most of the other proteins were shared. The only exception was BAP170, which was found solely in the Polybromo-containing complex (Fig. 4C and D). To reflect the presence of two distinct BRM complexes in Drosophila cells and their evolutionary relationship to those in mammalian and yeast cells, we named the OSA-containing complex BAP and the Polybromo-containing complex PBAP. BAP170 is also a distinguishing subunit of the PBAP complex. These results demonstrated that, like yeast and human cells, Drosophila cells do indeed express distinct SWI/SNF-BAF and RSC-PBAF complexes.
BAP and PBAP are differentially targeted in vivo.
To investigate the possibility that the BAP and PBAP complexes might perform distinct functions, we sought to determine their distributions on chromatin in vivo. First, we examined the pattern of binding of Polybromo on larval salivary gland polytene chromosomes (Fig. 5). Polybromo is associated with several hundreds of sites, consistent with a general role in chromatin regulation. Interestingly, Polybromo was found predominantly at the interband regions of open, less condensed chromatin, which were stained weakly with DAPI. In contrast, it appeared to be excluded from heterochromatic regions, such as the chromocenter or inactive chromosome 4 (Fig. 5). In agreement with earlier results obtained for BRM by Armstrong et al. (1), we observed that BRM and OSA were also widely distributed on polytene chromosomes and were associated predominantly with the interband regions (data not shown). Such a widespread distribution along polytene chromosomes is suggestive of global functions in transcription for BRM, Polybromo, and OSA.
FIG. 5.
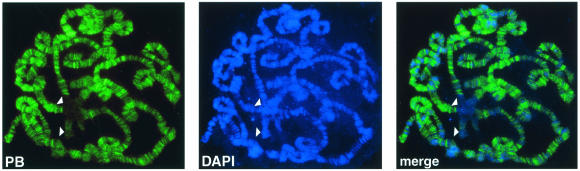
Polybromo is found predominantly in regions of open chromatin on salivary gland polytene chromosomes. The distribution of Polybromo on wild-type polytene chromosomes was determined by indirect immunofluorescence with affinity-purified polyclonal antibodies (green) directed against the protein. DNA was visualized by DAPI staining (blue). The arrowheads indicate the inactive chromocenter and chromosome 4. (Left panel) Indirect immunofluorescence with an anti-Polybromo (PB) antibody (green). (Middle panel) DAPI-stained DNA (blue). (Right panel) Merge image revealing the predominant localization of Polybromo at the interband regions of open chromatin, which are stained weakly with DAPI.
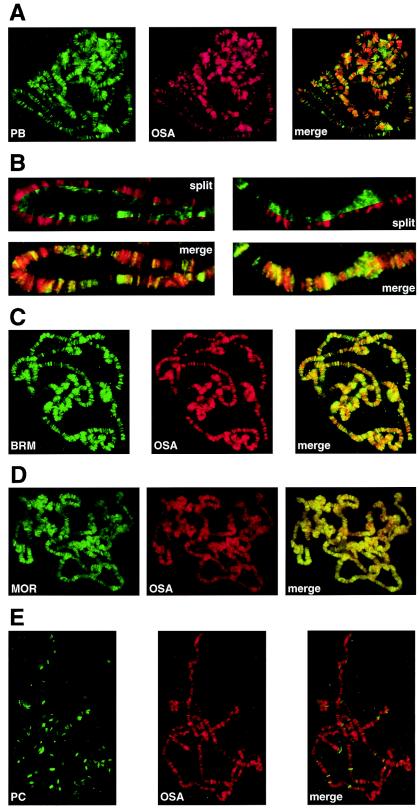
To examine the targeting of the BAP and PBAP complexes, we compared the chromosomal distributions of their respective defining components, OSA and Polybromo. As shown in Fig. 6A and B, Polybromo and OSA displayed clearly distinct, albeit overlapping patterns of distribution on polytene chromosomes. We consistently observed this dissimilar pattern of binding for OSA and Polybromo on distinct polytene chromosome preparations. Although they were colocalized on multiple sites, there were many other sites at which either Polybromo only or OSA only could be detected. Typically, in locations at which both were present, either OSA or Polybromo appeared to be of a clearly higher abundance. Thus, the relative levels of OSA and Polybromo varied from site to site, suggesting differential mechanisms of targeting. In contrast, the universal subunits of the BRM complexes, BRM itself and MOR (Fig. 6C and D), were present at more or less all locations at which OSA was detected. PC is an epigenetic repressor of transcription which acts antagonistically against the trxG proteins of the BRM complex. Consistent with these data and with earlier results obtained for BRM (1), the OSA and PC binding sites on polytene chromosomes were mutually exclusive (Fig. 6D). Likewise, we found that Polybromo was not colocalized with the PRC1 component Posterior Sex Combs (data not shown).
FIG. 6.
Polybromo and OSA display distinct but overlapping patterns of distribution on polytene chromosomes. The distributions of Polybromo, OSA, BRM, MOR, and PC on wild-type polytene chromosomes were determined by indirect immunofluorescence with affinity-purified polyclonal antibodies directed against Polybromo, BRM, MOR, and PC (all green) and monoclonal antibodies directed against OSA (red). (A) Localization of Polybromo (PB) (green) and OSA (red). (B) Higher magnifications of two areas of panel A. The split and merge images illustrate the distinct patterns of distribution of OSA and Polybromo. (C) Localization of BRM (green) and OSA (red). (D) Localization of MOR (green) and OSA (red). (E) Localization of PC (green) and OSA (red).
Taken together, the results of our analysis of the genome-wide distributions of Polybromo and OSA by immunolocalization on salivary gland polytene chromosomes revealed that the BAP and PBAP complexes are differentially targeted in vivo. These findings suggest that these distinct remodelers each control the expression of a unique set of target genes. This situation is reminiscent of that in yeast cells, where the ySWI/SNF and RSC complexes each form parts of distinct regulatory circuits.
BRM, but not PC, associates with hyperacetylated chromatin domains.
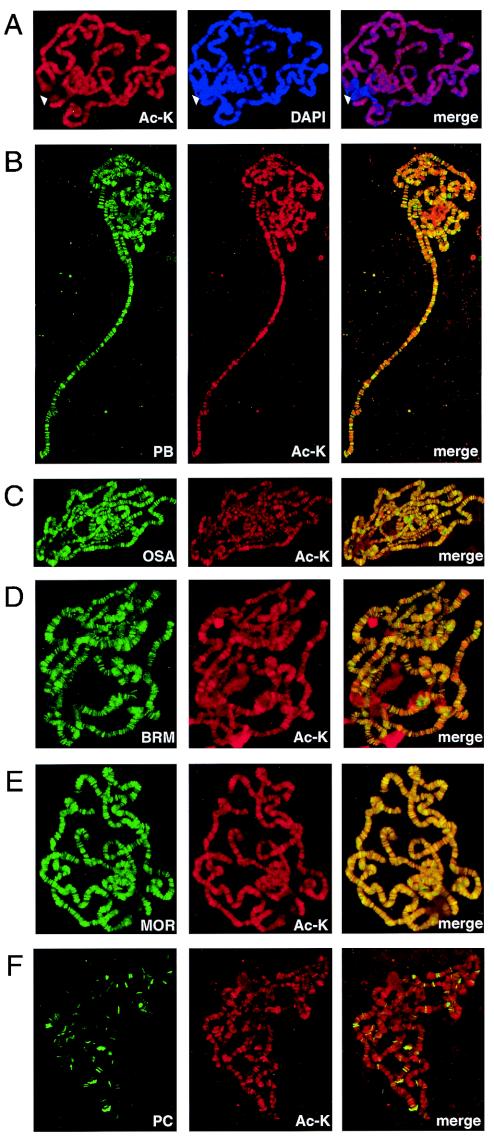
Having established the existence of two distinct subclasses of remodeling complexes which are differentially distributed on chromatin, we sought to establish further whether these complexes are associated with transcriptionally active or silent regions of the genome. Generally speaking, hyperacetylation of the amino-terminal histone tail domains and other chromatin-associated proteins correlates with active regions of the genome. We performed immunolocalization studies using an antibody directed against acetyllysine residues and antibodies directed against various components of the BAP and PBAP complexes. As shown in Fig. 7A, the anti-acetyllysine antibody revealed a multitude of regions of hyperacetylation widely distributed throughout the genome. The sites of hyperacetylation correlated predominantly with the interband regions of open chromatin, which were stained weakly with DAPI (Fig. 7A). Strikingly, both Polybromo and OSA were overwhelmingly associated with regions of hyperacetylated chromatin (Fig. 7B and C). Likewise, the chromosomal distributions of the universal subunits BRM and MOR showed predominantly colocalization with hyperacetylated chromatin (Fig. 7D and E). In contrast, the BRM antagonist PC bound only hypoacetylated chromatin (Fig. 7F).
FIG. 7.
The BRM chromatin-remodeling complexes, but not the PC repressor, associate with regions of hyperacetylated chromatin. (A) Regions of open chromatin are hyperacetylated. The distribution of acetyllysine residues (Ac-K) on polytene chromosomes was determined by indirect immunofluorescence with a sheep polyclonal antibody (red). DNA was visualized by DAPI staining (blue). The arrowhead indicates the inactive chromocenter. The merge image reveals the predominant localization of Ac-K at the interband regions, which are stained weakly with DAPI. (B to E) Colocalization of Polybromo (PB)(B), OSA (C), BRM (D), and MOR (E) (all green) with hyperacetylated chromatin (Ac-K) (red). (F) PC is associated with regions that are not acetylated. PC (green) binds hypoacetylated chromatin, which is stained weakly with antibodies directed against Ac-K (red).
Thus, chromosome painting with a global marker of chromatin modification reveals a clear relationship between chromatin acetylation and the binding of BRM remodelers. Conversely, chromatin hyperacetylation is absent from binding sites for the PC repressor of gene transcription. These results emphasize the close reciprocity of chromatin regulation by ATP-dependent remodelers, acetyltransferases and deacetylases, and PC repression.
DISCUSSION
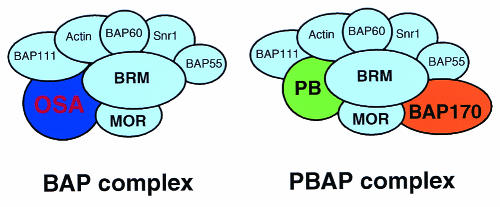
Recent studies emphasized a more extensive structural and functional diversification among chromatin-remodeling complexes than was previously appreciated. Here, we report the identification of two novel BRM-associated proteins, which allowed us to define two distinct Drosophila BRM chromatin-remodeling complexes: BAP and PBAP (Fig. 8). BAP is the Drosophila counterpart of ySWI/SNF and human BAF-SWI/SNF-α, whereas PBAP is related to yeast RSC and human PBAF-SWI/SNF-β. BAP and PBAP share seven identical subunits, which include the central ATPase BRM, actin, and an actin-like protein (BAP55). The distinguishing subunits, unique for each subclass of remodeler, are OSA in the BAP complex and Polybromo and BAP170 in the PBAP complex. Determination of the genome-wide distributions of OSA and Polybromo on larval salivary gland polytene chromosomes revealed differential targeting of BAP and PBAP. Combined with the results of studies of yeast and mammalian cells, these results suggest evolutionarily conserved structural and functional differences between the SWI/SNF-BAF-BAP and RSC-PBAF-PBAP subfamilies of remodelers.
FIG. 8.
Drosophila contains two distinct BRM complexes: BAP and PBAP. To determine the relationship between the BRM complex and the SWI/SNF-BAF or RSC-PBAF subclass of remodelers, we purified BRM and its associated factors and identified two novel subunits: Drosophila Polybromo and BAP170. These findings in turn allowed us to define two distinct Drosophila BRM chromatin-remodeling complexes: BAP and PBAP. The former is the Drosophila counterpart of SWI/SNF-BAF, and the latter corresponds to RSC-PBAF. BAP and PBAP share seven identical subunits (light blue), which include the central ATPase BRM, trxG protein MOR, actin, and an actin-like protein (BAP55). The distinguishing subunits are OSA in the BAP complex and Polybromo (PB) and BAP170 in the PBAP complex. Determination of the genome-wide distributions of OSA and Polybromo on larval salivary gland polytene chromosomes revealed differential targeting of BAP and PBAP. These results suggest an evolutionarily conserved structural and functional differentiation between the SWI/SNF-BAF-BAP and the RSC-PBAF-PBAP subfamilies of remodelers.
Polybromo and BAP170 are both evolutionarily highly conserved proteins. Polybromo appears to be a homologue of the yeast Rsc1, Rsc2, and Rsc4 proteins and of human BAF180 (5, 53, 57). Likewise, BAP170 is highly conserved among flies, C. elegans, and mammals, a fact suggesting essential cellular functions. BAP170 contains a putative ARID region DNA binding motif like those in OSA, yeast Swi1p, and yeast Rsc9. Because OSA, but not BAP170, shows additional sequence homology to Swi1p outside the ARID region (10), it seems likely that OSA represents the Swi1p orthologue in higher eukaryotes. Consequently, it is an attractive possibility that yeast Rsc9 is distantly related to the PBAP component BAP170. However, we could not identify additional regions of sequence homology between these two proteins outside the ARID region. Because of the very tight association with the fly PBAP complex, we suspect that the BAP170 homologues in other species will also be shown to form parts of PBAP-related complexes. Yeast RSC is much more abundant than ySWI/SNF (7), whereas in human cells, BAF appears to be more abundant than PBAF (53). We estimate from our purifications that BAP and PBAP are approximately equally abundant in Drosophila embryos. Moreover, OSA and Polybromo bind comparable numbers of chromosomal sites on polytene chromosomes, suggesting a similar number of target genes for BAP and PBAP. Thus, BAP and PBAP may play equally broad roles in chromatin regulation in Drosophila, suggesting that the SWI/SNF family of complexes has become more widely utilized throughout evolution.
Like its human and yeast counterparts, Polybromo harbors a multitude of putative protein-protein interaction domains: six bromodomains and two BAH domains. Bromodomains are conserved in eukaryotes and frequently are found in chromatin binding proteins and in nearly all nuclear histone acetyltransferases (32). These domains have been shown to mediate the recognition of specific acetylated lysine residues in histone tails (17, 22, 42). Therefore, it is tempting to speculate that the bromodomains in Polybromo play a critical role in targeting of the PBAP complex. BRM is the only other bromodomain-containing protein present in both the BAP and the PBAP complexes. Deletion of its bromodomain, however, affects neither BRM function nor chromatin binding (1, 14). Furthermore, it should be noted that not only PBAP but also BAP (lacking Polybromo) is found associated with open, hyperacetylated chromatin. Therefore, Polybromo by itself cannot be exclusively responsible for the recognition of hyperacetylated chromatin. Nevertheless, in light of the distinct chromosomal distributions of OSA and Polybromo, it seems likely that these proteins somehow direct BAP and PBAP to differentially modified chromatin domains. Future studies will be directed at discovering the mechanism of selective targeting of BAP and PBAP. Another potential interface with chromatin in Polybromo is formed by two conserved BAH domains. These motifs are also present in other chromatin-associated proteins and have been implicated in critical protein-protein interactions (8, 59). Furthermore, Polybromo has two potential DNA binding domains: a highly conserved HMG box (49) and two, albeit poorly conserved, C2H2-type zinc fingers (28). An attractive hypothesis is that Polybromo acts as a specialized anchoring subunit. For example, its bromodomains could mediate histone tail recognition, the BAH domains might contact transcription factors, and the HMG domain and the putative zinc fingers might stabilize DNA binding.
Like Polybromo, BAP170 contains multiple conserved motifs, which may also play a role in PBAP targeting. First, there is an N-terminal ARID region. ARID regions have been implicated in sequence-specific as well as sequence-independent DNA binding (10, 15, 19, 55). Although OSA regulates gene expression in a promoter-selective manner, its ARID region does not mediate sequence-specific DNA binding (10, 51). It is possible that the ARID region acquires specificity through interactions with other cofactors. For instance, the ARID region in the human OSA homologue BAF250/p270 was recently implicated in the transcriptional coactivation of hormone receptors (21). The highly conserved C terminus of BAP170 contains a second putative DNA binding domain: a double zinc finger motif comprising a canonical C2H2 zinc finger and another zinc finger in which the spacing between the two cysteine residues is larger (28). Finally, BAP170 contains several consensus as well as variant LXXLL motifs. These motifs may mediate ligand-dependent binding to hormone receptors or interactions with coactivators such as the histone acetyltransferase CBP (12, 20, 30). A functional characterization of the biochemical properties of Polybromo and BAP170 is expected to provide valuable insights into the mechanism of selective targeting of the PBAP chromatin-remodeling complex.
Immunolocalization on larval salivary gland polytene chromosomes revealed that OSA and Polybromo, the defining subunits of BAP and PBAP, display distinct, albeit overlapping, genome-wide distributions. Interestingly, the relative amounts of Polybromo and OSA on distinct sites are highly dissimilar. While on some locations Polybromo and OSA appear to be almost mutually exclusive, on other sites both are present. When OSA and Polybromo are colocalized, their relative abundances often appear quite distinct. Thus, it seems that on some locations, either Polybromo or OSA largely dictates the recruitment of PBAP or BAP, respectively. On sites where both complexes are present, they may be tethered through any of their subunits. Indeed, Collins et al. found that OSA is not required for BRM localization on chromatin per se (10). A corollary of these localization studies is that some genes may be exclusively regulated by either BAP or PBAP. For other genes, both BRM complexes may be involved in regulation. Taken together, our findings support the notion that, like SWI/SNF-BAF and RSC-PBAF, BAP and PBAP have distinct regulatory functions.
We found that both PBAP and BAP complexes preferentially associate with regions of open, hyperacetylated chromatin. In contrast, the BRM antagonist PC displays an inverse pattern of chromosome binding, localizing predominantly at sites of closed, hypoacetylated chromatin devoid of BRM. These findings reinforce the close interrelationship between chromatin modulation by ATP-dependent remodelers and chromatin modulation by enzymes that catalyze covalent protein modifications (17, 35). Our results correlate very well with the findings of Armstrong and colleagues, who showed that BRM marks nearly all transcriptionally active chromatin on polytene chromosomes (1). These researchers also established that BRM is required for most RNA Pol II transcription in salivary gland nuclei.
An important question raised by these studies is how the BRM complexes are targeted. Because we as well as Armstrong et al. (1) failed to detect a direct association with Pol II, we prefer the idea that BRM remodelers are targeted by a combination of recruitment by sequence-specific DNA binding proteins and recognition of a local chromatin environment (4, 17, 24, 36, 37, 44, 58). An example of the former mechanism is the selective BRM complex recruitment by the Zeste trxG transcriptional activator (24). A number of different studies have provided evidence for stabilization of the association of a remodeler with chromatin by histone acetylation or methylation (3, 17, 35). The bromodomain, BAH, SANT, and other conserved protein-protein interaction motifs in BAP and PBAP are prime candidates for mediating binding to specifically modified histones (32). Finally, the various ARID, zinc finger, SANT/Myb repeat, and HMG box DNA binding motifs present in the BRM complexes may direct association with areas of open chromatin where DNA is more accessible (45). SWI/SNF-type remodelers have been implicated in transcriptional repression (33). However, the virtually exclusive association of BRM with open but not with silent chromatin suggests that a role in repression may be transient, for example, through facilitating repressor binding to target genes. It should be noted, however, that salivary gland cells are terminally differentiated and that dynamic processes may not be evident in polytene immunolocalization studies.
In summary, we have presented evidence that, like yeast and human cells, Drosophila cells contain a SWI/SNF-BAF- and RSC-PBAF-type remodeling complex. Intriguingly, Drosophila contains only a single Swi2p/Snf2p-related ATPase, BRM, which is present in both BAP and PBAP. In fact, OSA, Polybromo, and BAP170 are the only distinguishing subunits between BAP and PBAP. The identification of Polybromo and BAP170 described here hopefully will allow discovery of the mechanism of differential target gene selection by BAP and PBAP. We anticipate that such studies will reveal further regulatory diversification and selectivity among SWI/SNF-type remodelers and provide insight into the cross talk between chromatin modulation by ATP-dependent remodelers and chromatin modulation by enzymes that catalyze covalent histone modifications.
Acknowledgments
We thank Jessica Treisman for the generous gift of antibodies directed against OSA; Gill Chalkley for the Sephacryl S-300 column fractions; and Jesper Svejstrup, David Baker, Colin Logie, Eric Kalkhoven, and Jan van der Knaap for discussions and critical reading of the manuscript.
This work was supported in part by NWO Chemical Sciences grant 700.52.312.
REFERENCES
- 1.Armstrong, J. A., O. Papoulas, G. Daubresse, A. S. Sperling, J. T. Lis, M. P. Scott, and J. W. Tamkun. 2002. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 21:5245-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 3.Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, L. A., C. Logie, E. Bonte, P. B. Becker, P. A. Wade, A. P. Wolffe, C. Wu, A. N. Imbalzano, and C. L. Peterson. 2000. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 275:18864-18870. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, B. R., H. Erdjument-Bromage, P. Tempst, F. Winston, and R. D. Kornberg. 1998. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol. Cell 2:639-651. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R., Y. J. Kim, M. H. Sayre, B. C. Laurent, and R. D. Kornberg. 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91:1950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 9.Cho, H., G. Orphanides, X. Sun, X. J. Yang, V. Ogryzko, E. Lees, Y. Nakatani, and D. Reinberg. 1998. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 18:5355-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, R. T., T. Furukawa, N. Tanese, and J. E. Treisman. 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18:7029-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosby, M. A., C. Miller, T. Alon, K. L. Watson, C. P. Verrijzer, R. Goldman-Levi, and N. B. Zak. 1999. The trithorax group gene moira encodes a brahma-associated putative chromatin-remodeling factor in Drosophila melanogaster. Mol. Cell. Biol. 19:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas, P. B., I. W. Cheney, D. Liao, V. Bowrin, W. Byam, S. Pacchione, R. Kobayashi, P. Yaciuk, and E. Moran. 1998. p300/CREB binding protein-related protein p270 is a component of mammalian SWI/SNF complexes. Mol. Cell. Biol. 18:3596-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 14.Elfring, L. K., C. Daniel, O. Papoulas, R. Deuring, M. Sarte, S. Moseley, S. J. Beek, W. R. Waldrip, G. Daubresse, A. DePace, J. A. Kennison, and J. W. Tamkun. 1998. Genetic analysis of brahma: the Drosophila homolog of the yeast chromatin remodeling factor SWI2/SNF2. Genetics 148:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory, S. L., R. D. Kortschak, B. Kalionis, and R. Saint. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 18.Heberlein, U., and R. Tjian. 1988. Temporal pattern of alcohol dehydrogenase gene transcription reproduced by Drosophila stage-specific embryonic extracts. Nature 331:410-415. [DOI] [PubMed] [Google Scholar]
- 19.Herrscher, R. F., M. H. Kaplan, D. L. Lelsz, C. Das, R. Scheuermann, and P. W. Tucker. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067-3082. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Z., J. Li, L. M. Sachs, P. A. Cole, and J. Wong. 2003. A role for cofactor-cofactor and cofactor-histone interactions in targeting p300, SWI/SNF and Mediator for transcription. EMBO J. 22:2146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue, H., T. Furukawa, S. Giannakopoulos, S. Zhou, D. S. King, and N. Tanese. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 277:41674-41685. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 23.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 24.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 25.Katsani, K. R., T. Mahmoudi, and C. P. Verrijzer. 2003. Selective gene regulation by SWI/SNF-related chromatin remodeling factors. Curr. Top. Microbiol. Immunol. 274:113-141. [DOI] [PubMed] [Google Scholar]
- 26.Klochendler-Yeivin, A., C. Muchardt, and M. Yaniv. 2002. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 12:73-79. [DOI] [PubMed] [Google Scholar]
- 27.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 28.Laity, J. H., B. M. Lee, and P. E. Wright. 2001. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11:39-46. [DOI] [PubMed] [Google Scholar]
- 29.Lemon, B., C. Inouye, D. S. King, and R. Tjian. 2001. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 414:924-928. [DOI] [PubMed] [Google Scholar]
- 30.Leo, C., and J. D. Chen. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1-11. [DOI] [PubMed] [Google Scholar]
- 31.Mahmoudi, T., and C. P. Verrijzer. 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20:3055-3066. [DOI] [PubMed] [Google Scholar]
- 32.Marmorstein, R. 2001. Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2:422-432. [DOI] [PubMed] [Google Scholar]
- 33.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 34.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 36.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 38.Neish, A. S., S. F. Anderson, B. P. Schlegel, W. Wei, and J. D. Parvin. 1998. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 26:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicolas, R. H., and G. H. Goodwin. 1996. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene 175:233-240. [DOI] [PubMed] [Google Scholar]
- 40.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olave, I. A., S. L. Reck-Peterson, and G. R. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodeling. Annu. Rev. Biochem. 71:755-781. [DOI] [PubMed] [Google Scholar]
- 42.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125:3955-3966. [DOI] [PubMed] [Google Scholar]
- 44.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 45.Quinn, J., A. M. Fyrberg, R. W. Ganster, M. C. Schmidt, and C. L. Peterson. 1996. DNA-binding properties of the yeast SWI/SNF complex. Nature 379:844-847. [DOI] [PubMed] [Google Scholar]
- 46.Simon, J. A., and J. W. Tamkun. 2002. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12:210-218. [DOI] [PubMed] [Google Scholar]
- 47.Sudarsanam, P., and F. Winston. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16:345-351. [DOI] [PubMed] [Google Scholar]
- 48.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. Brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68:561-572. [DOI] [PubMed] [Google Scholar]
- 49.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ′architectural' DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 50.Treisman, J. E., A. Luk, G. M. Rubin, and U. Heberlein. 1997. Eyelid antagonizes wingless signaling during Drosophila development and has homology to the Bright family of DNA-binding proteins. Genes Dev. 11:1949-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vazquez, M., L. Moore, and J. A. Kennison. 1999. The trithorax group gene osa encodes an ARID-domain protein that genetically interacts with the brahma chromatin-remodeling factor to regulate transcription. Development 126:733-742. [DOI] [PubMed] [Google Scholar]
- 52.Vignali, M., A. H. Hassan, K. E. Neely, and J. L. Workman. 2000. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 20:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274:143-169. [DOI] [PubMed] [Google Scholar]
- 54.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 55.Wilsker, D., A. Patsialou, P. B. Dallas, and E. Moran. 2002. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 13:95-106. [PubMed] [Google Scholar]
- 56.Wilson, C. J., D. M. Chao, A. N. Imbalzano, G. R. Schnitzler, R. E. Kingston, and R. A. Young. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84:235-244. [DOI] [PubMed] [Google Scholar]
- 57.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Z., M. K. Hayashi, O. Merkel, B. Stillman, and R. M. Xu. 2002. Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing. EMBO J. 21:4600-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zink, B., and R. Paro. 1989. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature 337:468-471. [DOI] [PubMed] [Google Scholar]