Abstract
Objective
Early HIV infection is characterized by a dramatic depletion of CD4 T cells in the gastrointestinal tract and translocation of bacterial products from the gut into the blood. In this study, we evaluated if gut bacterial profiles were associated with immune status before and after starting antiretroviral therapy (ART).
Design
We evaluated the gut microbiota of men recently infected with HIV (n = 13) who were participating in a randomized, double-blind controlled trial of combination ART and maraviroc versus placebo and who were followed for 48 weeks.
Methods
To evaluate the gut microbiota of participants, we pyrosequenced the bacterial populations from anal swabs collected before and longitudinally after the initiation of ART. Associations of the gut flora with clinical variables (lymphocyte profiles and viral loads), activation and proliferation markers in peripheral blood mononuclear cells and gut biopsies (measured by flow cytometry) and markers of microbial translocation (lipopolysaccharide and soluble CD14) were performed by regression analyses using R statistical software.
Results
Using pyrosequencing, we identified that higher proportions of Lactobacillales in the distal gut of recently HIV-infected individuals were associated with lower markers of microbial translocation, higher CD4% and lower viral loads before ART was started. Similarly, during ART, higher proportions of gut Lactobacillales were associated with higher CD4%, less microbial translocation, less systemic immune activation, less gut T lymphocyte proliferation, and higher CD4% in the gut.
Conclusion
Shaping the gut microbiome, especially proportions of Lactobacillales, could help to preserve immune function during HIV infection.
Keywords: gut-associated lymphoid tissue, gut microbiome, immune activation, microbial translocation, pyrosequencing
Introduction
The gut-associated lymphoid tissue (GALT) is a crucial part of the immunological network that maintains the integrity of the gastrointestinal (GI) tract against gut microbes [1]. Early HIV infection results in substantial depletion of CD4 T cells, preferentially in the GALT [2–4]. Consequences of this depletion include mucosal immune dysfunction, increased permeability of the gut and ultimately translocation of bacterial products [5], which contribute to chronic immune activation [6–8]. Immune activation is one of the strongest predictors of HIV disease progression [9–11]. Antiretroviral therapy (ART) seems to at least partially restore gut integrity [12–14] with marginal reduction in microbial translocation, but not to the levels seen in HIV-uninfected persons [6,7].
The GI tract is colonized by numerous commensal microorganisms, which can be identified by next-generation sequencing (NGS). NGS of the gut microbiome has revealed that certain disease states, such as inflammatory bowel disease, HIV, and others may be associated with modified gut flora [15–17]. Certain microbes may interact with the GALT to preserve gut integrity [18], thereby decreasing the likelihood of translocation of microbial products [19]. For example, consumption of probiotics and specifically Lactobacillales may modulate inflammatory responses, eradicate potential pathogens, and reduce gut permeability [19–24]. Manipulation of the gut flora may therefore benefit immune recovery during HIV infection. In this study, we conducted a metagenomics analysis to longitudinally characterize the changes of the gut microbiome during acute and early HIV infection and examined the effects of ART on this microbiome by associating clinical and immunological factors before starting and during ART.
Material and methods
Study cohort
Eligible participants were men who had sex with men (MSM) co-enrolled in the San Diego Primary Infection Cohort (n = 13) and a randomized, double-blind controlled trial of combination ART and maraviroc versus placebo. The Institutional Review Board of our center approved this study and all participants provided written informed consent. All patients initiated ART within 2 weeks of study enrollment with a combination of tenofovir, emtricitabine and ritonavir-boosted atazanavir, with or without maraviroc. The double-blind clinical trial is ongoing with all patients remaining blinded to maraviroc use. Anal swabs, blood, semen, peripheral lymphocyte profiles, and HIV levels (Amplicor, Roche) were collected at baseline (within a week before the initiation of ART) and approximately every 4 weeks thereafter for 48 weeks. A proportion of participants consented to repeat colonoscopies to obtain mucosal biopsies of the rectosigmoid colon and terminal ileum. Epidemiological, behavioral risk and HIV-related data were also collected from the participants. We determined estimated duration of infection (EDI) using results of serologic and virologic tests as described previously [25].
DNA extraction and viral quantification from peripheral blood mononuclear cells, stool, and semen
Genomic DNA was extracted from 5 million peripheral blood mononuclear cells (PBMCs) for each timepoint using QIAamp DNA Mini Kit (Qiagen) per manufacturer’s protocol. Extracted DNA was eluted in 100 μl elution buffer and total proviral HIV-1 DNA was quantified by real-time PCR in an ABI 7900HT thermocycler (Applied Biosystems) with virus-specific PCR primers and two DNA-locked nucleic acids (LNA) detection probes as previously published [26]. Cellular input was normalized with beta-actin PCR as previously described [27] and results were expressed in HIV DNA copies per 1 million actin cells equivalents.
Stool DNA from anal swabs was extracted using the QIAamp Stool DNA kit (Qiagen) per manufacturer’s protocol except that the elution was performed in 200 μl incubated for 5 min before the final spin. DNA extraction and quantification of cytomegalovirus (CMV) in seminal plasma and stool DNA was done as previously published [28].
Amplification of bacterial DNA and pyrosequencing
Amplification of the V6 hypervariable region of the 16 s rDNA gene was carried out in a 50 μl reaction using the highly purified Amplitaq Gold Low DNA polymerase (Applied Biosystem) to reduce bacterial contamination as manufacturer’s protocol with primers previously described [15]. The cycling conditions followed were: initial activation at 93°C for 15 min; 30 cycles of 95°C for 30 s, 57°C for 30 s and 72°C for 1 min; followed by a final extension at 72°C for 10 min. Samples were run in duplicates and a 1% agarose gel electrophoresis was used to confirm the ~110 bp size of product. Duplicate samples were combined and purified immediately after reaction (Qiagen PCR Purification Kit). We used the Agilent 2100 BioAnalyzer to quantify and assess purity of DNA. Amplicon pyrosequencing (Roche 454 FLX Titanium) was performed using standard protocols [29].
Classification of bacteria
We considered bacterial sequences with at least 90 continuous base pairs, which contained a quality score of at least 20 [30–32] for metagenomics analyses. We classified sequences to the order level using the Ribosomal Database Project [33]. We used the tool ESPRIT [34] to assign operational taxonomic units (OTU) based on genetic distance to unclassified sequences. With a cut-off of 10%, we built a consensus sequence for each OTU and classified it using small subunit rRNA taxonomy and alignment pipeline (STAP) [35]. We evaluated orders of bacteria in common across all samples and sequences that were unique to an individual were categorized as ‘Other’.
Microbial translocation markers
Limulus Amebocyte Lysate QCL-1000 and Quantikine ELISA Human sCD14 Immunoassay were used to measure plasma lipopolysaccharide (LPS) and soluble CD14 (sCD14), respectively, following manufacturer’s protocols.
Flow cytometry
Fresh whole blood was collected at weeks 0,12, 24, and 48 and processed using density gradient centrifugation to obtain PBMCs. Cells were incubated with antibodies for surface marker staining, before fixation and permeabilization for intracellular assays (eBioscience). We used the following antibody combinations to evaluate immune activation and proliferation in T cells: HLA-DR–FITC, CD45RO–PE, CD38–PE-Cy7, CD27–APC, CD3–APC-Cy7, CD4–PerCP-Cy5.5 and CD8–Pacific Blue, and Ki67–FITC, CD45RO–PE, CD27–APC, CD3–APC-Cy7, CD4–PerCP-Cy5.5 and CD8–Pacific Blue, (BD Biosciences). These measures were used to assess naïve (CD45RO−CD27+), central memory (CD45RO+CD27+) and effector memory (CD45RO+CD27−) in CD4 and CD8 T-cell subsets.
Rectosigmoid junction and the terminal ileum gut biopsies were available for a subgroup of nine patients at weeks 0 and 48 and interval biopsies at weeks 12 or 24 for a proportion of the subgroup. Tissue samples were incubated with collagenase and DNAse before passage through a cellular strainer (PGC Scientifics) and viably stored at −140°C. We assayed proliferation of collected tissue on viably thawed mucosal mononuclear cells using the following combination of antibodies: Ki67–FITC, CD45RO–PE, CD27–APC, CD3–APC–Cy7, CD4–PerCP-Cy5.5, CD8–Pacific Blue (BD Biosciences) and Aqua-L-D (Invitrogen) All samples were run on the BD FACSCanto (BD Biosciences) and data were analyzed with FlowJo software (Tree Star Inc.).
Statistical analyses
Methods of unsupervised clustering, statistical tests and regression analyses were implemented utilizing R statistical software. A two-tailed Mann–Whitney test was used to assess statistical difference between groups and comparison of bacterial proportions between groups was performed using Fisher exact test.
Normality of each order of bacteria and clinical variables was tested using a Shapiro test with a significance of P < 0.05. We evaluated cross-sectional associations between gut bacterial profiles (GBP) and clinical and immunological variables using fixed effects linear models. We utilized mixed effects linear models for analysis of longitudinal data to adjust for repeated measurements (packages lme4 and languageR).
We modeled each participant’s GBP as a vector , where xi corresponds to every order of bacteria classified, excluding the category ‘Other’. We calculated the sample variance of a GBP generalizing the sample variance formula for vector calculations. We measured intra-patient variability calculating the sample variance of the GBP at all timepoints for that particular patient. We computed inter-patient variability taking all the samples at each timepoint available.
Results
Study cohort
Study participants were 10 Caucasians and three Asians with an average age of 33 years and EDI of 6.5 weeks. All participants started ARTwithin 1 week of enrollment and were followed for 48 weeks. A summary of the clinical variables measured at baseline is provided in Table 1. We excluded two participants (L and M) from the baseline analysis because of antibiotic use shortly before the collection of their stool samples.
Table 1.
Demographics and clinical variables at baseline.
| PID | Racea | Ethnicityb | Age | EDI (weeks) |
Viral load (log10 copies/ml) |
CD4 absolute (cells/μl) |
CD4% | CD8 absolute (cells/μl) |
CD8% | CD4/CD8 ratio |
|---|---|---|---|---|---|---|---|---|---|---|
| A | A | NH | 23 | 2 | 5.78 | 276 | 10 | 1890 | 70 | 0.15 |
| B | A | NH | 28 | 2 | 3.10 | 493 | 24 | 856 | 45 | 0.58 |
| C | C | NH | 35 | 3 | 6.78 | 525 | 9 | 5216 | 85 | 0.10 |
| D | C | H | 22 | 3 | 6.99 | 91 | 9 | 728 | 70 | 0.13 |
| E | A/C | NH | 39 | 3 | 4.15 | 692 | 29 | 1387 | 55 | 0.50 |
| F | C | NH | 52 | 3 | 5.36 | 374 | 28 | 732 | 53 | 0.51 |
| G | C | NH | 21 | 3 | 5.93 | 916 | 37 | 1082 | 44 | 0.85 |
| H | C | NH | 28 | 4 | 7.00 | 520 | 11 | 3334 | 71 | 0.16 |
| I | C | H | 28 | 10 | 3.89 | 501 | 33 | 775 | 51 | 0.65 |
| J | C | NH | 26 | 11 | 5.25 | 614 | 33 | 776 | 42 | 0.79 |
| K | C | H | 33 | 12 | 4.40 | 971 | 34 | 1086 | 38 | 0.89 |
| L | C | NH | 40 | 14 | 4.15 | 383 | 30 | 558 | 42 | 0.69 |
| M | C | NH | 55 | 14 | 3.77 | 913 | 40 | 714 | 31 | 1.28 |
| Average | 33.1 | 6.46 | 5.12 | 559.15 | 25.15 | 1471.85 | 53.62 | 0.56 |
A, Asian; C, Caucasian.
H, Hispanic; NH, non-Hispanic. Participants are ordered according to estimated duration of infection (EDI).
Classifications of gut bacterial profiles
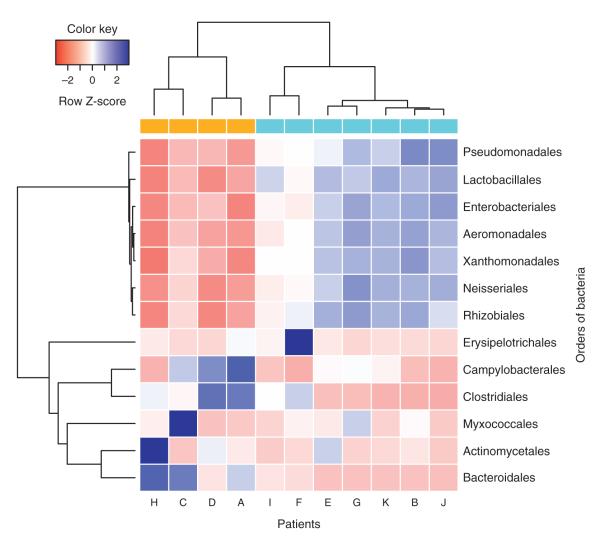
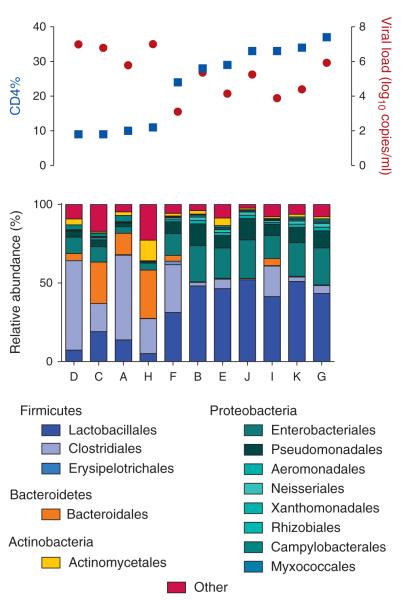
The study classified proportions of distal gut bacterial flora at the order level (13 orders) using the V6 region of the bacterial 16s rDNA. The overall median intra-patient variability in GBP was significantly lower than inter-patient variability (P = 0.006), as reported previously [36]. An unsupervised clustering analysis of baseline orders of bacteria revealed two distinct GBP (Fig. 1). The first GBP cluster (Group 1) showed significantly lower proportions of Lactobacillales, Enterobacteriales, Pseudomonadales, Xanthomonadales, Aeromonadales, Rhizobiales, and Neisseriales (P = 0.006 for all) and higher proportions of Bacteroidales (P = 0.01) and Clostridiales (P = 0.04) compared with the second GBP cluster (Group 2) (Fig. 1 and Supplementary Fig. 1, http://links.lww.com/QAD/A335). Interestingly, participants in these two groups also differed significantly by the percentage of CD4 (CD4%) and viral load at baseline, but there were no differences in markers of microbial translocation, LPS and sCD14 (Supplementary Table 1, http://links.lww.com/QAD/A336). Although Group 1 had low CD4% (median = 9.5%) and high viral load (median = 6.90 HIV RNA log10 copies/ml), Group 2 had high CD4% (median = 33%) and low viral load (median = 4.39 HIV RNA log10 copies/ml, P = 0.01, for both CD4% and viral load) (Fig. 2 and Supplementary Table 1, http://links.lww.com/QAD/A336). We observed the most profound differences between groups in the proportion of Lactobacillales. Group 1 with low CD4% and high viral load had low Lactobacillales, while Group 2 with high CD4% and low viral load had high Lactobacillales (median 10.6 vs. 46.5%) (Supplementary Fig. 1, http://links.lww.com/QAD/A335). As the proportion of gut Lactobacillales has been associated with mode of delivery at birth [37], we also examined reported differences between vaginal versus cesarean delivery of participants and found no differences between groups (P = 0.48).
Fig. 1. Unsupervised clustering before antiretroviral therapy (ART).
Gut bacterial profiles separated our participants in two main groups colored as gold and blue representing participants with low and high CD4% (Group 1 vs. Group 2), respectively. Orders of bacteria are separated into two main groups clustered by correlation. Overall, bacteria in the same cluster are positively correlated, whereas bacteria in different clusters are negatively correlated. Blue and red correspond to enrichment or depletion on proportions of bacteria, respectively.
Fig. 2. Overview of participants’ CD4%, viral load (VL) and gut bacterial profiles (GBP) at baseline.
Participants’ CD4% and VL are colored blue and red, respectively, and their corresponding GBP is in the bottom. Participants with lower CD4% and higher VL exhibit lower proportions of Lactobacillales.
Immune and clinical correlates of Lactobacillales before antiretroviral therapy
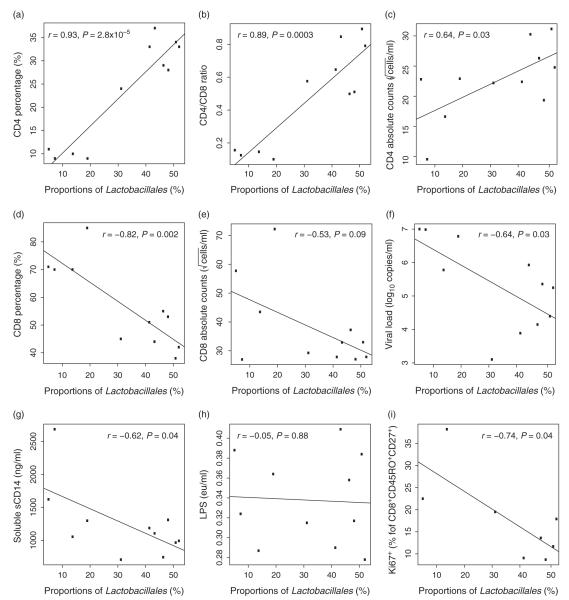
Before initiating ART, proportions of gut Lactobacillales were significantly correlated with CD4% (P = 2.8 × 10−5), CD4/CD8 T-cell ratio (P = 0.0003) and CD4 cell count (P = 0.03), and negatively associated with CD8% (P = 0.002), but only a negative trend with CD8 cell count (P = 0.09, Fig. 3a–e). Proportions of Lactobacillales were also negatively correlated with viral load (P = 0.03) and sCD14 (P = 0.04, Fig. 3f–g), but there was no association with LPS (P = 0.88, Fig. 3h). Despite an association with a marker of microbial translocation (sCD14), there was no observed relationship between Lactobacillales and either CD4 and CD8 lymphocyte activation (measured as HLA-DR+ or CD38+ T cells) or gut CD4 lymphocyte proliferation (percentage of Ki67+ of CD4 T cells, data not shown). There was, however, a negative association between higher proportion of gut Lactobacillales and lower gut CD8 lymphocyte proliferation in the central memory subset (percentage of Ki67+ of CD8+CD45RO+CD27+, P = 0.04, Fig. 3i). Although other orders of bacteria also showed associations with clinical and immunological variables, most consistent associations were found with Lactobacillales (Supplementary Table 2, http://links.lww.com/QAD/A336).
Fig. 3. Associations of Lactobacillales with clinical and immunological variables at baseline and before antiretroviral therapy (ART).
There is a positive association with (a) CD4%, (b) CD4/CD8 ratio, (c) CD4 cell count and a negative correlation (or trend) with (d) CD8%, (e) CD8 cell count, (f) viral load, (g) soluble CD14, and (i) Gut CD4+ T-cell proliferation. (h) There was no correlation between Lactobacillales and lipopolysaccharide (LPS). All these correlations suggest that higher proportions of Lactobacillales are beneficial for the host in the absence of ART.
Since duration of HIV infection is associated with immune activation [38] and participants had variable EDI at baseline, we evaluated if EDI influenced the associations between baseline proportions of Lactobacillales and lymphocyte activation in the blood. As might be expected, participants with a more recent EDI (≤4 weeks or acute phase of the infection, n = 8) at baseline showed a higher T-cell activation and proliferation than participants with longer EDI at baseline (>4 weeks, n = 3, Supplementary Fig. 2, http://links.lww.com/QAD/A335); however, baseline proportions of Lactobacillales were not associated with EDI (data not shown).
Immune and clinical correlates of Lactobacillales during antiretroviral therapy
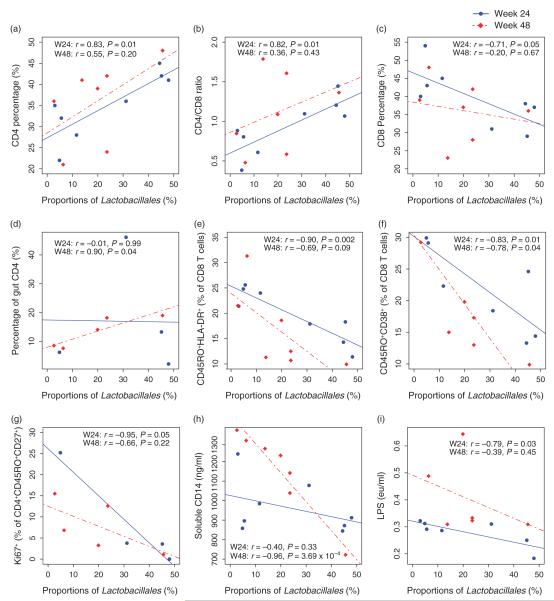
To determine the impact of ART on the associations between proportions of Lactobacillales, clinical variables (viral load, CD4%, CD4 cell count, CD8% and CD8 cell count), markers of translocation (sCD14 and LPS), and lymphocyte activation (HLA-DR, CD38) and proliferation (Ki67) markers, we performed regression analyses for all variables at each study timepoint, longitudinally including all time-points and cross-sectionally at weeks 24 and 48. As expected, ART was very effective at suppressing viral load, and only one participant (Patient H) had detectable viral load at weeks 24 and 48 (2.70 and 1.91 HIV RNA log10 copies/ml, respectively). Similarly, all participants increased their CD4% during ART (median +9%, range 4–25%, P = 0.001) (Supplementary Fig. 3A, http://links.lww.com/QAD/A335), and there was a trend for proportions of Lactobacillales to be positively associated with CD4% gains (P = 0.07) (Supplementary Fig. 3A, http://links.lww.com/QAD/A335). Although all participants demonstrated an increase in CD4% and a decrease in viral load during ART, those who started with low CD4% (Group 1), maintained lower CD4% compared with Group 2 participants and could still be identified by their GBP (Supplementary Fig. 4, http://links.lww.com/QAD/A335). Further, the relationships between Lactobacillales and CD4%, CD4/CD8 ratio and CD8% remained consistent with baseline results at week 24 but not at week 48 (P = 0.01, P = 0.01, P = 0.05, respectively, Fig. 4a–c); however, there were no associations between proportions of Lactobacillales and CD4 or CD8 T-cell count at either week 24 or 48 (data not shown). After 48 weeks of ART, increased proportions of Lactobacillales were positively correlated with increased CD4% in the gut (P = 0.04, Fig. 4d). As detectable viral load may have confounded the analysis, we also excluded Patient H from cross-sectional analysis, and Lactobacillales became positively associated with CD4 cell count at weeks 24 and 48 (r = 0.78, P = 0.04 and r = 0.82, P = 0.04, respectively). All other correlations maintained significant levels except with CD4% at week 48 of the gut, likely due to a power issue (data not shown).
Fig. 4. Associations of Lactobacillales with clinical and immunological variables after ART.
Week 24 is represented by solid dots and lines, and week 48 by empty dots and dotted lines. At week 24, there is a positive association with (a) CD4%, (b) CD4/CD8 ratio and a negative correlation with (c) CD8%, as observed at baseline. Additionally, higher gut Lactobacillales are associated with higher CD4% in the gut (d), less CD8+ T-cell activation (e, f), less CD4+ T-cell proliferation (g), and less microbial translocation (h, i). Associations were independent of ART and suggest that higher proportions of Lactobacillales are associated with better immune health.
Evaluation of lymphocyte activation demonstrated a strong negative correlation between the proportions of Lactobacillales and CD8 T-cell activation in the blood after 24 weeks of ART (CD45RO+HLA-DR+, P = 0.002, CD45RO+CD38+, P = 0.01, Fig. 4e–f). After 48 weeks of ART, the proportions of Lactobacillales were still negatively associated with CD38+ lymphocyte activation (P = 0.04), but only a trend remained for HLA-DR+ (P = 0.09) in CD8 T cells (Fig. 4e–f). The proliferation of central memory CD4 T cells in the gut was negatively associated with Lactobacillales at week 24 of ART (percentage of Ki67+ of CD4+CD45RO+CD27+, P = 0.05), but not after 48 weeks (P = 0.22, Fig. 4g). Regarding microbial translocation, the negative association between sCD14 and Lactobacillales observed at baseline was lost at week 24 but regained after 48 weeks of ART (P = 3.7 × 10−4, Fig. 4h). Throughout the study, sCD14 was inversely correlated with Lactobacillales (P = 0.02) and time on ART (P = 0.04) (Supplementary Fig. 3B, http://links.lww.com/QAD/A335). In contrast, LPS showed an isolated negative association at week 24 (P = 0.03, Fig. 4i). Other orders of bacteria also showed significant associations with these factors at week 24 but most were not significant by week 48 (Supplementary Table 3, http://links.lww.com/QAD/A336). Similar to above, exclusion of Patient H only decreased the significance of correlations with activation markers (HLA-DR and CD38) at week 48, possibly a power limitation (data not shown).
HIV latent reservoir and cytomegalovirus shedding
As changes in immune activation may impact the HIV latent reservoir [39], we also evaluated HIV DNA levels in PBMCs, but we did not observe any associations between HIV proviral DNA and Lactobacillales after 24 or 48 weeks of ART. Proviral DNA did not correlate with CD8 T-cell activation (Supplementary Fig. 5A-B, http://links.lww.com/QAD/A335). There was a strong negative correlation, however between proviral DNA levels and activation of CD4 lymphocytes (CD45RO+HLA-CD38+, P = 0.007) at week 24 of ART, but not at week 48 (Supplementary Fig. 5C-D, http://links.lww.com/QAD/A335). We also investigated the relationship between CMV shedding in the gut and the genital tract and Lactobacillales, as the presence and magnitude of CMV shedding may influence immune activation [28] and all participants were CMV antibody positive. Only two patients (D and H) had detectable levels of CMV in rectal swabs. Patient D had 2.41 and 2.14 log10 copies/swab at weeks 16 and 24 and patient H had 1.46 log10 copies/swab at week 4. Due to limited sample availability, only nine semen samples were screened for CMV shedding and Patient D exhibited high levels of CMV (4.49 log10 copies/swab). The eight remaining samples showed no evidence of CMV, though no samples were available for Patient H. Interestingly, both patients (D and H) exhibited the lowest overall proportion of Lactobacillales (<10%) throughout infection and Patient D in particular had the lowest CD4% after 48 weeks of ART.
Discussion
This is the first study to identify associations between specific GBP and higher CD4 cell count and CD4%, lower viral load, less CD4 T-cell proliferation in the gut and less evidence of microbial translocation in untreated HIV infection. All of these factors have been previously associated with better HIV disease outcomes [40,41]. The associations between GBP, CD4%, immune activation, and markers of bacterial translocation continued, albeit weakly, during ART that suppressed viral load. The important caveat of this study is that the associations between changes in the GBP and the HIV disease markers cannot determine causality.
The human gut prevents the translocation of commensal bacteria through physical barriers (e.g. epithelial tight junctions), biochemical agents (e.g. antibacterial peptides and mucus), and immune mechanisms (e.g., secretory IgA and Toll-like receptor mediated sensing, oxidative bursts) [1,42–45]. During early HIV infection, gut populations of Bifidobacteria and Lactobacillus species are reduced [46], the GALT is depleted [7], the gut barrier is compromised and translocation of bacterial products can occur. The translocation of these products is associated with HIV disease progression, most likely through persistent immune activation [6,7]. This study aimed to further evaluate these connections by observing multiple factors during acute and early HIV infection and by treating HIV infection at the earliest stages possible, and identifying correlates associated with optimal immune recovery and preservation. The current study extends previous observations [47] by linking the constitution of the gut microbiome itself to immunologic and virologic dynamics during recent HIV infection and subsequent ART, specifically identifying that higher proportions of gut Lactobacillales are associated with markers that are predicative of better HIV outcomes including higher CD4 percentage, lower viral load, and less evidence of microbial translocation. Moreover, the higher proliferation of central memory cells in GALT, as a surrogate marker of antigen stimulation, was less likely to occur in participants with higher proportions of Lactobacillales suggesting a favorable gut immune health. As Lactobacillales can modulate anti-inflammatory responses and immune cells (e.g., T-regulatory cells), improve gut integrity and reduce gut permeability in other conditions [20–24,48], it is interesting that this bacterial order would be identified as the main component of GBP associated with markers of improved HIV outcome. However, it still needs to be investigated whether these GBP are metabolically associated or represent only a biomarker of a favorable state.
There are a number of limitations that should be considered. First, we conducted this investigation in the setting of a randomized double-blind controlled trial of maraviroc versus placebo combined with standard ART, but the overall study remains blinded to maraviroc use because participant enrollment continues. Maraviroc use is not thought to have contributed significantly to the study observations as associations between GBP and clinical and immunological variables were identified at baseline, before the start of ART for all participants. In general, these associations persisted during follow-up, and unblinding or modification of the parent study is thought to be unnecessary. However, as maraviroc inhibits CCR5, it could theoretically alter the composition of T cells in the GALT, influencing our observations after the start of ART, and this will need to be assessed after the unblinding of the study. Second, although NGS allowed us to conduct large-scale metagenomic analyses of the distal gut microbiome, the analysis is limited by the classification of sequences. To focus our study on the main drivers of gut bacteria across all participants, we only considered bacterial populations shared across individuals. Bacterial sequences that were not identified in all participants were classified as ‘Other’, and these ‘Other’ populations may be very informative and warrant further investigation. Along these lines, the study only considered classification at the order level and a more granular classification may provide additional insight. To this end, all bacterial sequences have been publically deposited at http://mepac.ucsd.edu. Third, the observation that sCD14 is more highly correlated with measures of microbial translocation than LPS has been previously reported [7,49], and could be related to the EDI. Microbial translocation occurs in early phases of infection, thus levels of LPS do not increase until later HIV stages, while levels of sCD14 increase earlier during the course of infection [7]. Alternatively, plasma inhibitors [50] were observed that might have interfered with LPS assay measurements. Fourth, in limited number of samples, we investigated if CMV shedding in the distal GI tract or semen demonstrated similar associations to classified GBP. Although, the associations between rectal CMV shedding and lower CD4 T-cell count and percentage were provocative in two of the participants, the numbers are too small to draw conclusions for a role of CMV to impact gut microbiota, but should provide testable hypotheses for future investigations.
It is increasingly evident that the human immune system is linked to the GI system [48], but guidance regarding how this information should influence clinical care is lacking, especially for HIV infection. Although the gut microbiome is influenced by methods of birth delivery, host genetics, household contacts, age, geography, and so on [51], it is unknown if the microbiome can be shaped with agents like prebiotics, probiotics, or targeted antibiotics for beneficial outcomes. This study determined that increasing Lactobacillales in the gut could be important for recovering and preserving immune system function during HIV infection and a promising clinical target may be HIV-infected individuals who are able to suppress their viral load with ART, but are unable to sufficiently recover their CD4 T-cell count.
Supplementary Material
Acknowledgements
We are grateful to all the participants in the San Diego Primary Infection Cohort, the CFAR Flow Cytometry, Genomics, Translational Virology Cores and Joanne Santangelo who personally collected clinical data from participants.
J.P.S. performed the amplification and sequencing of bacterial DNA experiments, participated in the data analyses for this study, performed the statistical analyses, and wrote the primary version of the article. S.G. performed the CMV and HIV proviral DNA experiments and participated in the data analyses. M.M. analyzed the flow cytometry, and other clinical and immunological data. C.A.S. and M.Y.K. designed the antibody panel for flow cytometry and participated in the data analyses. S.R.V. participated in the data analyses. D.P. obtained the gut biopsies. J.A.Y. obtained the demographic, epidemiological and HIV-related data of participants. P.S.J. performed microbial translocation markers experiments. S.J.L. and D.M.S. enrolled participants. D.D.R., S.J.L., and D.M.S. designed the present study, participated in data analysis. All authors read and approved the final article.
This work was supported by the Department of Veterans Affairs and by grants from the National Institutes of Health: AI69432, AI043638, MH62512, MH083552, AI077304, AI36214, AI047745, AI74621, AI080353, DA034978, U19AI090970; the California HIV Research Program grant RN07-SD-702 and the James B. Pendleton Charitable Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
Footnotes
Conflicts of interest J.P.S., S.G., M.M., C.A.S., M.Y.K., S.R.V., D.P., P.S.J., J.A.Y. and S.J.L. do not have any commercial or other associations that might pose a conflict of interest. D.D.R. has served as a consultant for Bristol-Myers Squibb, Gilead Sciences, Merck & Co, Monogram Biosciences, Biota, Chimerix, Tobira, and Gen-Probe. D.M.S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe.
In limited form some of these data were presented at the Fred Hutchinson Cancer Research Center retreat from June 14–15, 2012 in Seattle, Washington.
References
- 1.Shulzhenko N, Morgun A, Hsiao W, Battle M, Yao M, Gavrilova O, et al. Crosstalk between B lymphocytes, microbiota and the intestinal epithelium governs immunity versus metabolism in the gut. Nat Med. 2011;17:1585–1593. doi: 10.1038/nm.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenchley JM, Knox KS, Asher AI, Price DA, Kohli LM, Gostick E, et al. High frequencies of polyfunctional HIV-specific T cells are associated with preservation of mucosal CD4 T cells in bronchoalveolar lavage. Mucosal Immunol. 2008;1:49–58. doi: 10.1038/mi.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paiardini M, Frank I, Pandrea I, Apetrei C, Silvestri G. Mucosal immune dysfunction in AIDS pathogenesis. AIDS Rev. 2008;10:36–46. [PubMed] [Google Scholar]
- 6.Jiang W, Lederman MM, Hunt P, Sieg SF, Haley K, Rodriguez B, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 8.Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, Wurcel A, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS One. 2008;3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchetti G, Cozzi-Lepri A, Merlini E, Bellistri GM, Castagna A, Galli M, et al. Microbial translocation predicts disease progression of HIV-infected antiretroviral-naive patients with high CD4+ cell count. AIDS. 2011;25:1385–1394. doi: 10.1097/QAD.0b013e3283471d10. [DOI] [PubMed] [Google Scholar]
- 10.Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narvaez AB, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 12.Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, Shacklett BL, et al. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J Virol. 2006;80:8236–8247. doi: 10.1128/JVI.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macal M, Sankaran S, Chun TW, Reay E, Flamm J, Prindiville TJ, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 2008;1:475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- 14.Mehandru S, Poles MA, Tenner-Racz K, Jean-Pierre P, Manuelli V, Lopez P, et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2012;109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acheson DW, Luccioli S. Microbial-gut interactions in health and disease. Mucosal immune responses. Best Pract Res Clin Gastroenterol. 2004;18:387–404. doi: 10.1016/j.bpg.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Gori A, Rizzardini G, Van’t Land B, Amor KB, van Schaik J, Torti C, et al. Specific prebiotics modulate gut microbiota and immune activation in HAART-naive HIV-infected adults: results of the ‘COPA’ pilot randomized trial. Mucosal Immunol. 2011;4:554–563. doi: 10.1038/mi.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bengmark S. Use of some pre, pro- and synbiotics in critically ill patients. Best Pract Res Clin Gastroenterol. 2003;17:833–848. doi: 10.1016/s1521-6918(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 21.Sanders ME. Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol. 2011;45(Suppl):S115–S119. doi: 10.1097/MCG.0b013e318227414a. [DOI] [PubMed] [Google Scholar]
- 22.Servin AL. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev. 2004;28:405–440. doi: 10.1016/j.femsre.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Nissen L, Chingwaru W, Sgorbati B, Biavati B, Cencic A. Gut health promoting activity of new putative probiotic/protective Lactobacillus spp. strains: a functional study in the small intestinal cell model. Int J Food Microbiol. 2009;135:288–294. doi: 10.1016/j.ijfoodmicro.2009.08.027. [DOI] [PubMed] [Google Scholar]
- 24.Girardin M, Seidman EG. Indications for the use of probiotics in gastrointestinal diseases. Dig Dis. 2011;29:574–587. doi: 10.1159/000332980. [DOI] [PubMed] [Google Scholar]
- 25.Little SJ, Frost SD, Wong JK, Smith DM, Pond SL, Ignacio CC, et al. Persistence of transmitted drug resistance among subjects with primary human immunodeficiency virus infection. J Virol. 2008;82:5510–5518. doi: 10.1128/JVI.02579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Althaus CF, Gianella S, Rieder P, von Wyl V, Kouyos RD, Niederost B, et al. Rational design of HIV-1 fluorescent hydrolysis probes considering phylogenetic variation and probe performance. J Virol Methods. 2010;165:151–160. doi: 10.1016/j.jviromet.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Gianella S, Anderson CM, Vargas MV, Richman DD, Little SJ, Morris SR, et al. Cytomegalovirus DNA in semen and blood is associated with higher levels of proviral HIV DNA. J Infect Dis. 2013;207:898–902. doi: 10.1093/infdis/jis777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gianella S, Strain MC, Rought SE, Vargas MV, Little SJ, Richman DD, et al. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol. 2012;86:1307–1315. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, et al. Ausubel F, et al., editors. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19:Unit 19 10 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, et al. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W, et al. ESPRIT: estimating species richness using large collections of 16S rRNA pyrosequences. Nucleic Acids Res. 2009;37:e76. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu D, Hartman A, Ward N, Eisen JA. An automated phylogenetic tree-based small subunit rRNA taxonomy and alignment pipeline (STAP) PLoS One. 2008;3:e2566. doi: 10.1371/journal.pone.0002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 39.Yukl SA, Gianella S, Sinclair E, Epling L, Li Q, Duan L, et al. Differences in HIV burden and immune activation within the gut of HIV-positive patients receiving suppressive antiretroviral therapy. J Infect Dis. 2010;202:1553–1561. doi: 10.1086/656722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham NM. The role of immunologic and viral markers in predicting clinical outcome in HIV infection. AIDS. 1996;10(Suppl 5):S21–S25. doi: 10.1097/00002030-199612005-00004. [DOI] [PubMed] [Google Scholar]
- 41.Fahey JL, Taylor JM, Detels R, Hofmann B, Melmed R, Nishanian P, et al. The prognostic value of cellular and serologic markers in infection with human immunodeficiency virus type 1. N Engl J Med. 1990;322:166–172. doi: 10.1056/NEJM199001183220305. [DOI] [PubMed] [Google Scholar]
- 42.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–667. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MA, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science. 2009;325:617–620. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cunningham-Rundles S, Ahrne S, Johann-Liang R, Abuav R, Dunn-Navarra AM, Grassey C, et al. Effect of probiotic bacteria on microbial host defense, growth, and immune function in human immunodeficiency virus type-1 infection. Nutrients. 2011;3:1042–1070. doi: 10.3390/nu3121042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellis CL, Ma ZM, Mann SK, Li CS, Wu J, Knight TH, et al. Molecular characterization of stool microbiota in HIV-infected subjects by panbacterial and order-level 16S ribosomal DNA (rDNA) quantification and correlations with immune activation. J Acquir Immune Defic Syndr. 2011;57:363–370. doi: 10.1097/QAI.0b013e31821a603c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kosiewicz MM, Zirnheld AL, Alard P. Gut microbiota, immunity, and disease: a complex relationship. Front Microbiol. 2011;2:180. doi: 10.3389/fmicb.2011.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kramski M, Gaeguta AJ, Lichtfuss GF, Rajasuriar R, Crowe SM, French MA, et al. Novel sensitive real-time PCR for quantification of bacterial 16S rRNA genes in plasma of HIV-infected patients as a marker for microbial translocation. J Clin Microbiol. 2011;49:3691–3693. doi: 10.1128/JCM.01018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.