Abstract
Anion-exchange chromatography resolves human plasma low-density lipoprotein (LDL) into 5 subfractions, with increasing negative surface charge in the direction of L1 to L5. Unlike the harmless L1 to L4, the exclusively atherogenic L5 is rejected by the normal LDL receptor (LDLR) but endocytosed into vascular endothelial cells through the lectin-like oxidized LDL receptor-1 (LOX-1). Analysis with SDS-PAGE and 2-dimensional electrophoresis showed that the protein framework of L1 was composed mainly of apolipoprotein (apo) B100, with an isoelectric point (pI) of 6.620. There was a progressively increased association of additional proteins, including apoE (pI 5.5), apoAI (pI 5.4), apoCIII (pI 5.1), and apo(a) (pI 5.5), from L1 to L5. LC/MSE was used to quantify protein distribution in all subfractions. On the basis of weight percentages, L1 contained 99% apoB-100 and trace amounts of other proteins. In contrast, L5 contained 60% apoB100 and substantially increased amounts of apo(a), apoE, apoAI, and apoCIII. The compositional characteristics contribute to L5’s electronegativity, rendering it unrecognizable by LDLR. LOX-1, which has a high affinity for negatively charged ligands, is known to mediate the signaling of proinflammatory cytokines. Thus, the chemical composition–oriented receptor selectivity hinders normal metabolism of L5, enhancing its atherogenicity through abnormal receptors, such as LOX-1.
Keywords: low-density lipoprotein, apolipoproteins, electronegative, LDL receptor, LOX-1, atherosclerosis
INTRODUCTION
Low-density lipoprotein (LDL) is normally metabolized by liver and vascular cells via the LDL receptor (LDLR); homozygous or heterozygous genetic defects in LDLR expression lead to plasma LDL accumulation and premature coronary artery disease (CAD) as well as other atherosclerotic vascular abnormalities [1, 2]. But elevation of plasma LDL cholesterol (LDL-C) alone may not be sufficient to induce vascular changes and it has been thought that LDL modification, such as oxidation, plays an important role in generating LDL’s atherogenicity [3, 4]. However, oxidized LDL equivalent to what has been produced experimentally in vitro has not been isolated from human plasma for mechanistic scrutiny. Evidence is now accumulating that LDL that carries a greater negative charge than the majority of LDL particles may be responsible for atherogenicity in dyslipidemia [5–8]. For simplicity, these highly negatively charged LDL particles have been called “electronegative” LDL, although in reality it is a relative term and should not be used to imply that the other LDL particles are “electropositive.”
By use of anion-exchange chromatography, we have been able to divide LDL obtained from the plasma of patients with increased cardiac risks (hypercholesterolemia, type 2 diabetes mellitus, smoking) into 5 subfractions, L1 to L5, with increasing electronegativity [8–11]. L5, the most negatively charged, is the only subfraction that can induce endothelial dysfunction in cultivated arteries and atherogenic responses in cultured vascular cells. It also impairs normal differentiation of endothelial progenitor cells [7, 8, 10–14]. Of importance, L5 is not recognized by LDLR and blocking LDLR does not reduce L5’s proapoptotic effects on vascular endothelial cells (ECs) [7]. In fact, L5 signals through and is internalized by lectin-like oxidized LDL receptor-1 (LOX-1) in both ECs and endothelial progenitor cells (EPCs) [7, 8]. This suggests that, beyond absolute LDL-C surplus in the plasma, the proportion of abnormal LDL, such as L5, that signals differently from normal LDL, such as L1, is an important factor for atherosclerosis. Thus, it is important to define the chemical basis for L5’s receptor affinity and hence its biological activities.
L5 AND ELECTRONEGATIVE LDL
Since the initial characterization of human atheroma-derived LDL by Hoff, Gotto, and associates in the USA, the term “electronegative LDL” has been used to describe LDL with fast relative electrophoretic mobility on agarose gel [15–17]. By use of fast protein liquid chromatography (FPLC) through ion-exchange columns, Avogaro and colleagues in Italy divided human plasma LDL dichotomously into electropositive LDL(+) and electronegative LDL(−) [18]. Since then, several groups, in particular, the team led by Sánchez-Quesada in Spain, have described the chemical composition and functional characteristics of LDL(−), isolated by a similar protocol [6, 19–33]. Using a different protocol, we chromatographically divided human plasma LDL into L1 to L5, with L5 representing a pure and highly negatively charged LDL entity [9, 10].
SAMPLE PREPARATION AND L5 PURIFICATION
Whole blood removed from 6 hyperlipidemic (LDL-C > 160 mg/dL) adult subjects with the approval of the internal review board at Baylor College of Medicine, Houston, Texas, USA, was protected by 1% penicillin/streptomycin and citrate phosphate dextrose adenine-1 from bacterial contamination and coagulation. The plasma obtained was treated with Complete Protease Inhibitor Cocktail (Roche; Cat. No. 05056489001; 1 tablet/100 mL) to prevent protein degradation. LDL (d=1.019–1.063 g/mL) was then isolated by sequential potassium bromide density centrifugation and treated with 5 mM EDTA and nitrogen to avoid ex vivo oxidation [34]. LDL was further divided into L1 to L5 against graded salt gradient through anion-exchange columns by FPLC as previously described in detail [9, 10].
2-DIMENSIONAL ELECTROPHORESIS
Protein contents of LDL subfractions were analyzed by 2-dimensional electrophoresis. Twice delipidated (1:1 EtAc+EtOH, 0.3 mL/30 μg LDL protein) L1 and L5 particles were centrifuged for 30 min (14000 rpm, 4°C). After removal of the solution, the lipoprotein pellet was resuspended in 30 μL H2O. Samples were incubated in ZOOM IPGRunner Cassette with Strip, 1X ZOOM 2D Protein Solubilizer 1, 1X Protease Inhibitor Cocktail, 20 mM DTT, and 3.5 mM Tris base at pH 7.4 for 2 hours. Two-dimensional-PAGE (isoelectrofocusing, equilibrating, performing) was performed by ZOOM IPGRunner, ZOOM Equilibration Tray and XCell SureLock Mini-Cel according to the user manual. The 4–20% 2-dimensional gels were then stained with SYPRO Ruby Protein Gel Stain (Ex/Em: 280, 450/610 nm).
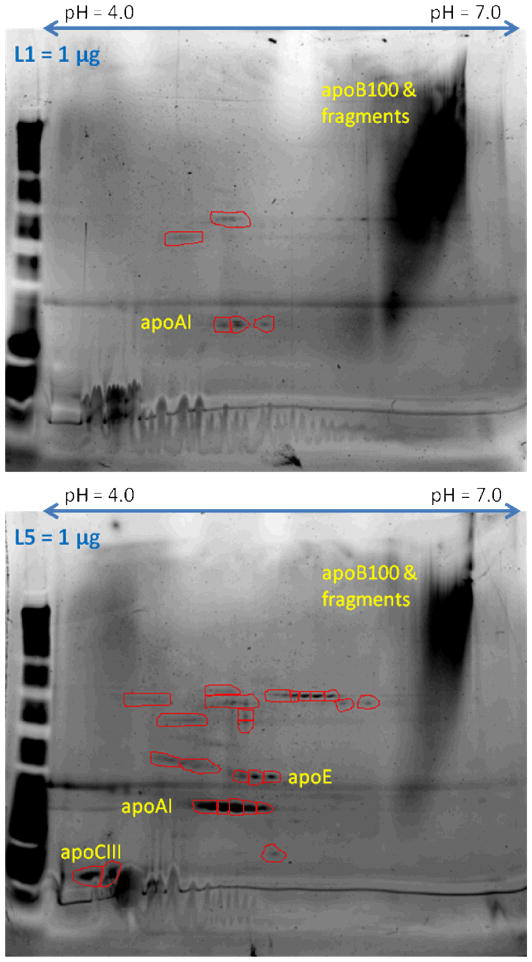
As shown in Fig. 1, the protein framework of L1 was composed mainly of apolipoprotein (apo) B100, which has an isoelectric point (pI) of 6.620. In contrast, L5 is associated with apoE (pI 5.5), apoAI (pI 5.4), apoCIII (pI 5.1), and apo(a) (pI 5.5), in addition to apoB100. Among the low-PI proteins, apoCIII is known to be atherogenic by itself [35]. Apo(a) is a main constituent of Lp(a), which is infamous for its negative charge, although its atherogenicity remains controversial [36].
Fig. 1.
SYPRO Ruby Protein Gel Stain–treated 2-dimensional electrophoresis of L1 and L5. L1 contains mainly apoB100, whose PI value is close to 7.0, with scant other proteins. In contrast, for each unit of total protein, L5 contains a smaller proportion of apoB100 but greater quantities of other proteins with lower PI values.
LC/MSE ANALYSIS AND UNIQUE PROTEIN COMPOSITION OF L5
Because the proportional increases of these low-pI proteins may contribute to the overall negative charge of L5, we quantified the protein contents in L1 to L5 subfractions by use of quantitative proteomics techniques utilizing serially coupled liquid chromatography data-independent parallel-fragmentation mass spectrometry (LC/MSE) [37] Such analysis has been shown to be highly quantitative with respect to both relative and/or absolute (when incorporating spiked internal peptide standards in the data collection/analysis procedures) protein abundance in complex protein mixtures [37–39]. Quantitative analysis was performed essentially as previously described [39], except on a Waters Synapt HDMS mass spectrometer (Waters Corporation, MA, USA). In brief, total proteins isolated from each LDL subfraction were first digested with trypsin, and the resulting tryptic peptides were chromatographically separated on a Nano-Acquity separations module (Waters Corporation, MA, USA) incorporating a 50 fmol-on-column tryptic digest of yeast alcohol dehydrogenase as the internally spiked protein quantification standard. Peptide elution was executed through a 75 μm × 25 cm BEH C-18 column under gradient conditions at a flow rate of 300 nL/min over 30 min at 35°C. The mobile phase was composed of acetonitrile as the organic modifier and formic acid (0.1% v/v) for molecule protonation. Mass spectrometry was performed on a Synapt HDMS instrument equipped with a nano-electrospray ionization interface and operated in the data-independent collection mode (MSE). Parallel ion fragmentation was programmed to switch between low (4 eV) and high (15–45 eV) energies in the collision cell, and data were collected from 50 to 2000 m/z utilizing glu-fibrinopeptide B as the separate data channel lock mass calibrant. Data were processed with ProteinLynx GlobalServer v2.4 (Waters) [40]. Deisotoped results were searched for protein association from the Uniprot (www.uniprot.org) human protein database (v15.12; containing 34,786 entries).
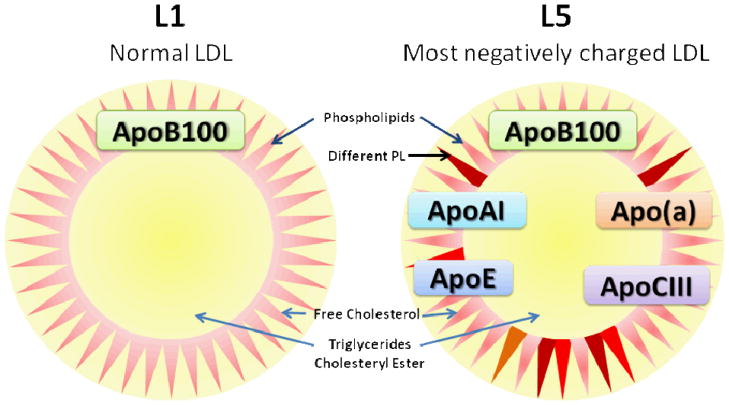
As shown in Table 1, L1 contained 99.71±0.37% apoB100 (n = 6) on the basis of weight percentage. Minute amounts of albumin, apoE, and apoAI were detected. In contrast, apoB100 represented only 61.26±21.44% of protein weight in L5. Approximately 30% of L5 protein weight was occupied by apo(a). ApoE and apoAI rose to about 3% and 2%, respectively. Sizable amounts of albumin, apoCIII, apoJ, and paraoxonase 1 (PON1) were also measured. All these non-apoB100 proteins, in particular apo(a), have <6.0 pI values. L5 was found to be associated with platelet-activating acetylhydrolase (PAF-AH), also known as lipoprotein-associated phospholipase A2 (Lp-PLA2) [41]. Although this enzyme has a pI value of 7.3, it represented only 0.03% of L5 proteins and should not factor significantly in L5’s pI value and hence electrical charge. In consideration of value variations among the samples, median values for each protein detected are also provided for clarity (Tables 1 and 2). On the basis of abundance relative to 1 mol of apoB100, minimal amounts of albumin, apoE, and apoAI were detected in L1. In contrast, L5 contained about 0.8 mol of apo(a) (median 0.326) and up to 0.5 mol each of apoE, apoAI, and apoCIII, as well as detectable apoJ, PAF-AH, and PON1 (Table 2). On the particle surface—although the total content of phospholipids is similar between L5 and L1—their components differ, as analyzed by liquid chromatography electrospray tandem mass spectrometry (LC-ESI/MS/MS; unpublished data). For clarity, L5 and L1 compositions are schematically summarized as shown in Fig. 2. Small amounts of amyloid A4 and complement C3 were also detected in L5 but not L1 (data not shown).
Table 1.
Protein distribution in L1 and L5 according to weight percentage
| Description (μg/μg LDL100%) | L1 (Mean±SD) | L5 (Mean±SD) | P value* | L5 (Median) | pI (pH) |
|---|---|---|---|---|---|
| ApoB100 | 99.71±0.37% | 61.26±21.44% | 0.007 | 68.72% | 6.6 |
| Apo(a) | ND | 30.88±23.34% | 21.79% | 5.5 | |
| ApoE | 0.14±0.31% | 2.56±1.93% | 0.075 | 2.17% | 5.5 |
| ApoAI | 0.03±0.03% | 1.57±0.92% | 0.009 | 1.88% | 5.4 |
| ApoCIII | ND | 0.46±0.57% | 0.25% | 5.1 | |
| ApoJ | ND | 0.27±0.33% | 0.15% | 5.8 | |
| PAF-AH | ND | 0.03±0.06% | 0.00% | 7.3 | |
| PON1 | ND | 0.21±0.32% | 0.11% | 4.9 | |
| Albumin | 0.05±0.03% | 0.64±0.51% | 0.491 | 0.85% | 5.9 |
PAF-AH: platelet-activating factor acetylhydrolase; PON1: paraoxonase 1;
ND: not detected.
n = 6.
Table 2.
Protein distribution in L1 and L5 measured by mol concentration relative to 1 mol apoB100
| Description (mol/mol apoB100) | L1 (Mean±SD) | L5 (Mean±SD) | P value* | L5 (Median) | pI (pH) |
|---|---|---|---|---|---|
| ApoB100 | 6.6 | ||||
| Apo(a) | ND | 0.805±1.016 | 0.326 | 5.5 | |
| ApoE | 0.020±0.023% | 0.525±0.317 | 0.011 | 0.428 | 5.5 |
| ApoAI | 0.052±0.091% | 0.402±0.167 | 0.005 | 0.421 | 5.4 |
| ApoCIII | ND | 0.348±0.353 | 0.298 | 5.1 | |
| ApoJ | ND | 0.041±0.047 | 0.033 | 5.8 | |
| PAF-AH | ND | 0.004±0.006 | 0.000 | 7.3 | |
| PON1 | ND | 0.036±0.054 | 0.019 | 4.9 | |
| Albumin | 0.004±0.006% | 0.095±0.096 | 0.157 | 0.090 | 5.9 |
PAF-AH: platelet-activating factor acetylhydrolase; PON1: paraoxonase 1;
ND: not detected.
n = 6.
Fig. 2.
Schematic summary of L1 and L5 compositions. PL: phospholipid.
L5 VS. LDL(−)
Our findings in protein distribution discrepancies between L5 and L1 are largely compatible with those between LDL(−) and LDL(+) recently reported by Bancells et al. from the Sánchez-Quesada group [33], although notable differences do exist. These differences may be a result of the variations in the different chromatographic separation methods used between the two centers. L5 is a product of the progressive increase of salt gradient during anion-exchange chromatography and represents a small but highly negatively charged LDL, whereas LDL(−) is a product of dichromatic division of total LDL [9, 10, 18, 33]. The major differences between the L5/L1 and LDL(−)/LDL(+) pairs lie in their differential contents of apoE and apoAI. Measured by either ELISA or LC-ESI/MS/MS, LDL(−) contains approximately 5 fold of apoE and 4 fold of apoAI in comparison with LDL(+) [33]. In contrast, L5 possessed 24-fold greater apoE and 8-fold greater apoAI compared with L1 in the current study. The significance of markedly greater apoE and apoAI in L5 is being investigated in our laboratory. Another major disparity between the L5/L1 and LDL(−)/LDL(+) pairs is the high presence of apo(a) in L5, whereas apo(a) is not detectable by ELISA and is detected only in a minute amount by LC-ESI/MS/MS in LDL(−) [33]. The density of apo(a)-containing particles ranges between 1.020 and 1.120 g/mL, most of them between 1.050 and 1.120 g/mL [42]. The LDL density range used by Bancells et al. to separate LDL(−) from LDL(+) is 1.019 to 1.050 g/mL, whereas we subfractionated LDL in the range of 1.019 to 1.063 g/mL. This probably explains why a negligible amount of apo(a) is present in LDL(−) and L5 was found to be associated with a large amount of this particle.
APO(a) AND L5
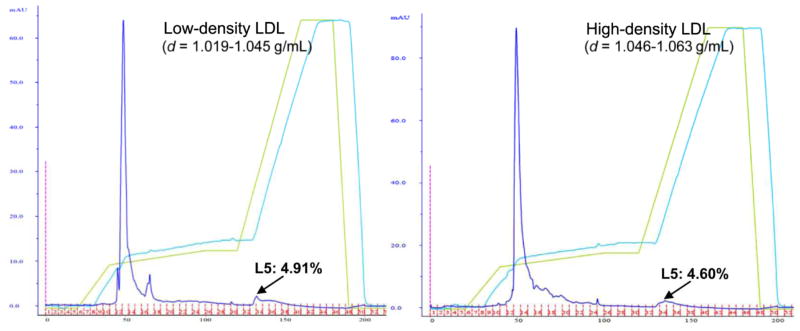
The significance of apo(a) abundance in L5 is yet to be fully determined. However, its presence should not necessarily be interpreted as being equivalent to being in the effective form of Lp(a), which consists of an LDL-like particle and apo(a) covalently bound to the particle’s apoB100. We observed a relatively low sequence coverage of the apo(a) molecule in our LC/MSE data (only 2.29–8.65% amino acid sequence coverage), and most of the apo(a) peptides observed were identified as being located, structurally, in the C-terminal portion of the apo(a) molecule. Thus, the abundance of apo(a) may be overestimated on a weight basis, as the MS evidence may indicate that a shortened alternative length form of apo(a) is associated with L5. Plasma Lp(a) concentrations are highly heritable and mainly controlled by the apo(a) gene, LPA [43, 44]. As noted above, the density ranges are 1.050 to 1.100 g/mL for Lp(a) and 1.019 to 1.063 g/mL for LDL. To determine whether the presence of a high quantity of apo(a) implies that most L5 particles reside in the high density range, we examined the L5 distribution in low-density (1.019 to 1.045 g/mL) and high-density (1.046 to 1.063 g/mL) LDL particles. An equal distribution (4.60% to 4.91 %) of L5 was eluted in both preparations (Fig. 3). This result, compatible with our previous finding using equilibrium density gradient ultracentrifugation [14], suggests that the association with apo(a) does not indicate that the L5 particles possess Lp(a)-like physical characteristics. However, the contribution of apo(a) to the overall electronegativity may play an important role in L5’s receptor selectivity, as discussed below.
Fig. 3.
Equal distribution of L5 in low and high density ranges of LDL shown in anion-exchange chromatographic tracings. Data are representative of 3 independent studies.
SURFACE ELECTRICAL CHARGE AND RECEPTOR SELECTIVITY BETWEEN L1 AND L5
As discovered and well described by Brown and Goldstein, the receptor for normal LDL on human cells is LDLR of the apoB/E receptor family [1, 45]. LDLR contains a domain at the NH2-terminal portion of the receptor that consists of cysteine-rich repeating units. A cluster of negatively charged amino acids occurs near the C-terminus of each repeat. These charged residues are complementary to a cluster of positively charged residues of the receptor-binding domain, first shown to include residues 3345 through 3381 [46] and later identified to include a cluster of basic residues between 3359 and 3367 [47]. The electrostatic attraction between the positively charged apoB100 domain of normal LDL and the negatively charged binding residues of LDLR leads to LDL endocytosis and subsequent digestion in the lysosomes [2, 48].
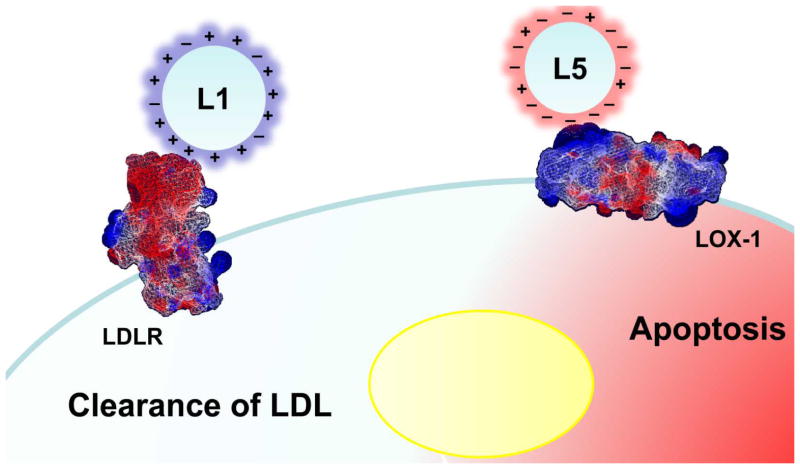
The positively charged ε-amino groups of the lysine residues on apoB100 play an important role in the binding between normal LDL and LDLR [49]. Derivatization of the lysine residues, as occurs in copper oxidation, redirects the binding affinity of the modified LDL from LDLR toward scavenger receptors of macrophages [50]. Recent work using 2-dimensional nuclear magnetic resonance (2D-NMR) spectroscopy revealed conformational differences between LDL(−) and LDL(+) involving changes in the basicity of the lysine residues on apoB100, which may contribute to the reduced affinity of LDL(−) for LDLR [51]. In our settings, L5, which is not recognized by LDLR, competes with copper-oxidized LDL for internalization through LOX-1 in vascular ECs [7], as well as signals through LOX-1 in EPCs [8]. LOX-1 is a type II membrane protein that structurally belongs to the C-type lectin family and does not share homology with the class A or B scavenger receptors for oxidized LDL [52]. The positively charged cysteine-rich lectin-like domain at the C-terminal plays a key role in the receptor’s affinity for modified LDL, which involves interaction of this region and the negatively charged domains on the modified LDL particles. Our preliminary data suggest that the interaction between L5 and LOX-1 involves modified residues on apoB100 and other apolipoproteins. The detailed structural changes are still under investigation, but the highly negatively charged surface of L5 attributable to the assembly of multiple acidic apolipoproteins facilitates the electrostatic attraction between L5 and LOX-1, which operates in the opposite direction of that between L1 and the normal LDLR. Hence, results are totally different in the receiving cells. A highly simplified but conceptually clear comparison of the electrical charge–oriented receptor selection between L1 and L5 is schematized in Fig. 4.
Fig. 4.
Schematic summary of electrical charge–oriented receptor selectivity of L1 and L5. L1 is endocytosed by the normal LDLR through interaction between the basic residues of apoB100 on L1 (normal LDL) and the negatively charged residues of LDLR, resulting in LDL degradation and utilization of cholesterol and phospholipids for biological purposes. In contrast, L5 (modified LDL) is internalized by means of the interaction between the negatively charged surface of L5 and the positively charged lectin-like domain of LOX-1, resulting in cell apoptosis.
CONCLUSIONS
In summary, an LDL particle is spherical and comprises an apolipoprotein framework, neutral lipids (triglycerides, cholesteryl esters) in the core, and other lipids (phospholipids, free cholesterol) on the surface. SDS-PAGE and 2-dimensional electrophoresis showed that the protein framework of L1 is composed mainly of apolipoprotein (apo) B100, with an isoelectric point (pI) of 6.620. The protein composition of the LDL particle changes as the chromatographic subfractions become more electronegative. The more electronegative subfractions have increased levels of additional proteins in the LDL particle, including apoE (pI 5.5), apoAI (pI 5.4), apoCIII (pI 5.1), and apo(a) (pI 5.5), and a concomitant decrease in overall mole abundance of apoB100. Protein analysis using LC/MSE revealed that L1 contains 99% apoB100 and trace amounts of other proteins, while L5 contains 60% apoB100 and substantially increased amounts of apo(a), apoE, apoAI, and apoCIII. Other minor proteins identified in L5 but not L1 include apoJ, PON1, PAF-AH, albumin, serum amyloid A4, and complement C3. L1 represents a pure form of normal LDL and is endocytosed through the well-described LDLR pathway, to deliver its content to the cells. In contrast, L5 is not recognized by the normal LDLR but is internalized by LOX-1, resulting in apoptosis of ECs. Both L1 to LDLR and L5 to LOX-1 interactions involve, although not exclusively, electrostatic attraction, but in the opposite direction to the other pair.
Acknowledgments
This work was supported in part by Research Grant 1-04-RA-13 from the American Diabetes Association, Grant HL-63364 from the National Institutes of Health, Mao-Kuei Lin Research Fund of Chicony Electronics in Taiwan, and Taiwan Department of Health Clinical Trial and Research Center of Excellence DOH99-TD-B-111-004. The authors are grateful to Miss Suzanne Simpson for editorial assistance.
References
- 1.Brown MS, Goldstein JL. Science. 1974;185:61–63. doi: 10.1126/science.185.4145.61. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JL, Brown MS. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witztum JL, Steinberg D. Trends Cardiovasc Med. 2001;11:93–102. doi: 10.1016/s1050-1738(01)00111-6. [DOI] [PubMed] [Google Scholar]
- 4.Libby P. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 5.Bancells C, Benitez S, Ordonez-Llanos J, Oorni K, Kovanen PT, Milne RW, Sanchez-Quesada JL. J Biol Chem. 2010 doi: 10.1074/jbc.M110.175315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bancells C, Villegas S, Blanco FJ, Benitez S, Gallego I, Beloki L, Perez-Cuellar M, Ordonez-Llanos J, Sanchez-Quesada JL. J Biol Chem. 2010;285:32425–32435. doi: 10.1074/jbc.M110.139691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J, Yang JH, Burns AR, Chen HH, Tang D, Walterscheid JP, Suzuki S, Yang CY, Sawamura T, Chen CH. Circ Res. 2009;104:619–627. doi: 10.1161/CIRCRESAHA.108.190116. [DOI] [PubMed] [Google Scholar]
- 8.Tang D, Lu J, Walterscheid JP, Chen HH, Engler DA, Sawamura T, Chang PY, Safi HJ, Yang CY, Chen CH. J Lipid Res. 2008;49:33–47. doi: 10.1194/jlr.M700305-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Chen CH, Jiang T, Yang JH, Jiang W, Lu J, Marathe GK, Pownall HJ, Ballantyne CM, McIntyre TM, Henry PD, Yang CY. Circulation. 2003;107:2102–2108. doi: 10.1161/01.CIR.0000065220.70220.F7. [DOI] [PubMed] [Google Scholar]
- 10.Yang CY, Raya JL, Chen HH, Chen CH, Abe Y, Pownall HJ, Taylor AA, Smith CV. Arterioscler Thromb Vasc Biol. 2003;23:1083–1090. doi: 10.1161/01.ATV.0000071350.78872.C4. [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Jiang W, Yang JH, Chang PY, Walterscheid JP, Chen HH, Marcelli M, Tang D, Lee YT, Liao WS, Yang CY, Chen CH. Diabetes. 2008;57:158–166. doi: 10.2337/db07-1287. [DOI] [PubMed] [Google Scholar]
- 12.Yang CY, Chen HH, Huang MT, Raya JL, Yang JH, Chen CH, Gaubatz JW, Pownall HJ, Taylor AA, Ballantyne CM, Jenniskens FA, Smith CV. Atherosclerosis. 2007;193:283–291. doi: 10.1016/j.atherosclerosis.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 13.Abe Y, Fornage M, Yang CY, Bui-Thanh NA, Wise V, Chen HH, Rangaraj G, Ballantyne CM. Atherosclerosis. 2007;192:56–66. doi: 10.1016/j.atherosclerosis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Chen HH, Hosken BD, Huang M, Gaubatz JW, Myers CL, Macfarlane RD, Pownall HJ, Yang CY. J Lipid Res. 2007;48:177–184. doi: 10.1194/jlr.M500481-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Hoff HF, Karagas M, Heideman CL, Gaubatz JW, Gotto AM., Jr Atherosclerosis. 1979;32:259–268. doi: 10.1016/0021-9150(79)90169-2. [DOI] [PubMed] [Google Scholar]
- 16.Hoff HF, Gaubatz JW. Atherosclerosis. 1982;42:273–297. doi: 10.1016/0021-9150(82)90157-5. [DOI] [PubMed] [Google Scholar]
- 17.Clevidence BA, Morton RE, West G, Dusek DM, Hoff HF. Arteriosclerosis. 1984;4:196–207. doi: 10.1161/01.atv.4.3.196. [DOI] [PubMed] [Google Scholar]
- 18.Avogaro P, Bon GB, Cazzolato G. Arteriosclerosis. 1988;8:79–87. [PubMed] [Google Scholar]
- 19.Chappey B, Myara I, Benoit MO, Maziere C, Maziere JC, Moatti N. Biochim Biophys Acta. 1995;1259:261–270. doi: 10.1016/0005-2760(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 20.Demuth K, Myara I, Chappey B, Vedie B, Pech-Amsellem MA, Haberland ME, Moatti N. Arterioscler Thromb Vasc Biol. 1996;16:773–783. doi: 10.1161/01.atv.16.6.773. [DOI] [PubMed] [Google Scholar]
- 21.Sanchez-Quesada JL, Perez A, Caixas A, Ordonmez-Llanos J, Carreras G, Payes A, Gonzalez-Sastre F, de Leiva A. Diabetologia. 1996;39:1469–1476. doi: 10.1007/s001250050600. [DOI] [PubMed] [Google Scholar]
- 22.Cordoba-Porras A, Sanchez-Quesada JL, Gonzalez-Sastre F, Ordonez-Llanos J, Blanco-Vaca F. J Mol Med. 1996;74:771–776. doi: 10.1007/s001090050079. [DOI] [PubMed] [Google Scholar]
- 23.Vedie B, Jeunemaitre X, Megnien JL, Myara I, Trebeden H, Simon A, Moatti N. Arterioscler Thromb Vasc Biol. 1998;18:1780–1789. doi: 10.1161/01.atv.18.11.1780. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Quesada JL, Otal-Entraigas C, Franco M, Jorba O, Gonzalez-Sastre F, Blanco-Vaca F, Ordonez-Llanos J. Am J Cardiol. 1999;84:655–659. doi: 10.1016/s0002-9149(99)00411-7. [DOI] [PubMed] [Google Scholar]
- 25.Fabjan JS, Abuja PM, Schaur RJ, Sevanian A. FEBS Lett. 2001;499:69–72. doi: 10.1016/s0014-5793(01)02523-6. [DOI] [PubMed] [Google Scholar]
- 26.Benitez S, Sanchez-Quesada JL, Ribas V, Jorba O, Blanco-Vaca F, Gonzalez-Sastre F, Ordonez-Llanos J. Circulation. 2003;108:92–96. doi: 10.1161/01.CIR.0000072791.40232.8F. [DOI] [PubMed] [Google Scholar]
- 27.Ziouzenkova O, Asatryan L, Sahady D, Orasanu G, Perrey S, Cutak B, Hassell T, Akiyama TE, Berger JP, Sevanian A, Plutzky J. J Biol Chem. 2003;278:39874–39881. doi: 10.1074/jbc.M306786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Quesada JL, Benitez S, Ordonez-Llanos J. Curr Opin Lipidol. 2004;15:329–335. doi: 10.1097/00041433-200406000-00014. [DOI] [PubMed] [Google Scholar]
- 29.Barros MR, Bertolami MC, Abdalla DS, Ferreira WP. Atherosclerosis. 2006;184:103–107. doi: 10.1016/j.atherosclerosis.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira JA, Sevanian A, Rodrigues RJ, Apolinario E, Abdalla DS. Clin Biochem. 2006;39:708–714. doi: 10.1016/j.clinbiochem.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Bancells C, Sanchez-Quesada JL, Birkelund R, Ordonez-Llanos J, Benitez S. J Lipid Res. 2010;51:2947–2956. doi: 10.1194/jlr.M005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brunelli R, Balogh G, Costa G, De Spirito M, Greco G, Mei G, Nicolai E, Vigh L, Ursini F, Parasassi T. Biochemistry. 2010;49:7297–7302. doi: 10.1021/bi100715f. [DOI] [PubMed] [Google Scholar]
- 33.Bancells C, Canals F, Benitez S, Colome N, Julve J, Ordonez-Llanos J, Sanchez-Quesada JL. J Lipid Res. 2010 doi: 10.1194/jlr.M009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aviram M. Biochem Med. 1983;30:111–118. doi: 10.1016/0006-2944(83)90013-3. [DOI] [PubMed] [Google Scholar]
- 35.Kawakami A, Osaka M, Tani M, Azuma H, Sacks FM, Shimokado K, Yoshida M. Circulation. 2008;118:731–742. doi: 10.1161/CIRCULATIONAHA.108.784785. [DOI] [PubMed] [Google Scholar]
- 36.Tziomalos K, Athyros VG, Wierzbicki AS, Mikhailidis DP. Curr Opin Cardiol. 2009;24:351–357. doi: 10.1097/HCO.0b013e32832ac21a. [DOI] [PubMed] [Google Scholar]
- 37.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Silva JC, Denny R, Dorschel CA, Gorenstein M, Kass IJ, Li GZ, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Anal Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 39.Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, Langridge JI. Proteomics. 2009;9:1683–1695. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 40.Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 41.Chen CH. Curr Opin Lipidol. 2004;15:337–341. doi: 10.1097/00041433-200406000-00015. [DOI] [PubMed] [Google Scholar]
- 42.Scheffer PG, Bakker SJ, Heine RJ, Teerlink T. Clin Chem. 1997;43:1904–1912. [PubMed] [Google Scholar]
- 43.McLean JW, Tomlinson JE, Kuang WJ, Eaton DL, Chen EY, Fless GM, Scanu AM, Lawn RM. Nature. 1987;330:132–137. doi: 10.1038/330132a0. [DOI] [PubMed] [Google Scholar]
- 44.Utermann G, Menzel HJ, Kraft HG, Duba HC, Kemmler HG, Seitz C. J Clin Invest. 1987;80:458–465. doi: 10.1172/JCI113093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown MS, Goldstein JL. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 46.Yang CY, Chen SH, Gianturco SH, Bradley WA, Sparrow JT, Tanimura M, Li WH, Sparrow DA, DeLoof H, Rosseneu M, et al. Nature. 1986;323:738–742. doi: 10.1038/323738a0. [DOI] [PubMed] [Google Scholar]
- 47.Law A, Scott J. J Lipid Res. 1990;31:1109–1120. [PubMed] [Google Scholar]
- 48.Brown MS, Goldstein JL. Proc Natl Acad Sci U S A. 1979;76:3330–3337. doi: 10.1073/pnas.76.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weisgraber KH, Innerarity TL, Mahley RW. J Biol Chem. 1978;253:9053–9062. [PubMed] [Google Scholar]
- 50.Steinbrecher UP. J Biol Chem. 1987;262:3603–3608. [PubMed] [Google Scholar]
- 51.Blanco FJ, Villegas S, Benitez S, Bancells C, Diercks T, Ordonez-Llanos J, Sanchez-Quesada JL. J Lipid Res. 2010;51:1560–1565. doi: 10.1194/jlr.D002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sawamura T, Kume N, Aoyama T, Moriwaki H, Hoshikawa H, Aiba Y, Tanaka T, Miwa S, Katsura Y, Kita T, Masaki T. Nature. 1997;386:73–77. doi: 10.1038/386073a0. [DOI] [PubMed] [Google Scholar]