Stem cells are highly specialized cells endowed with unlimited replicative self-renewing potential. These cells are capable of either limited multipotent (adult stem cells, ASC) or unlimited pluripotent (embryonic stem cells, ESC) differentiation to somatic cell lineages. Control of this differentiation process holds great promise in areas such as tissue regeneration and the treatment of chronic degenerative diseases. The medical exploitation of this phenomenon is carried out using stem cells derived from different sources (e.g., ASC/ESC), developmental stages (e.g., mesenchymal cells), cellular reprogramming (e.g., induced pluripotent stem cells), or even nonstem cells in a process known as transdifferentiation (Fig. 1). Despite this wide diversity, all of the differentiation steps that drive these cells to the desired somatic cell lineage rely on the directed manipulation of endogenous pathways. This process is achieved primarily by direct ligand signaling to cell-surface receptors, or use of distinct combinations of transcription factors derived from these signals. Now in PNAS, Melidoni et al. (1), building upon earlier reports (2, 3), raise the exciting possibility of using combinatorial library-derived antibodies intimately linked to stem-cell phenotype as a powerful tool in controlling these differentiation events.
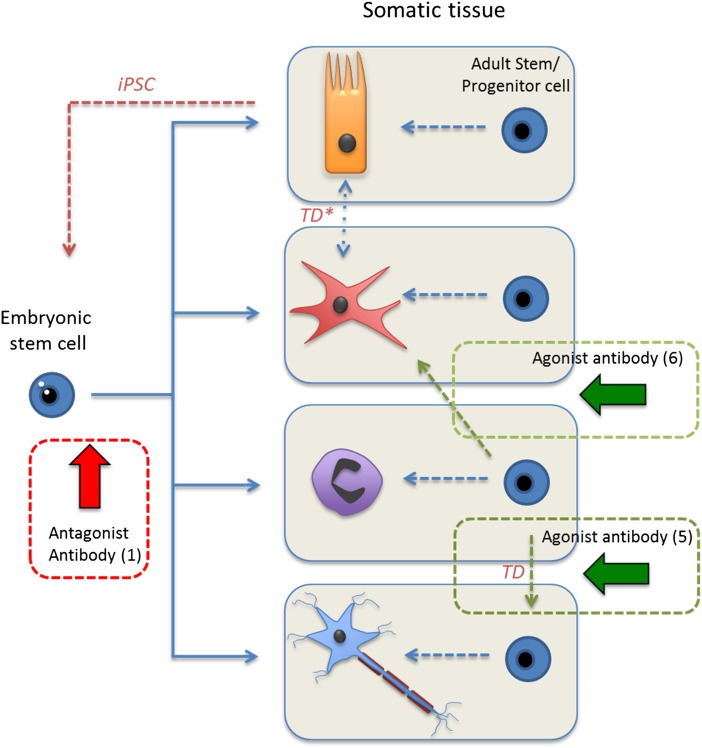
Fig. 1.
Stem-cell differentiation pathways. Stem cells (embryonic and adult) are shown in blue. Somatic cells shown are from top to bottom: epithelial (orange), dendrite (red), granulocyte (purple), and neuron (blue). The approximate points at which the different studies discussed are indicated alongside their appropriate reference (numbers in parenthesis) with antagonist signaling in red and agonist in green. iPSC, induced pluripotent stem cell; TD, transdifferentiation. *, TD can occur either between nonstem cells or differentiated (progenitor) stem cells to outwith their established differentiation pathway. Although grouped together in this figure, progenitor cells are generally considered to be stem cells that have undergone significant commitment to a specific somatic lineage with concomitant diminishment of both multipotency and replicative potential.
The use of antibodies provides technical advantages over traditional techniques thanks to their intrinsic stability and precise specificity. However, an even greater benefit lies within the antibody selection method itself. These antibodies are derived from combinatorial libraries that contain a vast repertoire of possible antibody combinations. This diversity means that as a whole, such libraries contain members that can influence every possible cell-receptor–signaling event of a target cell, including potentially unforeseen synergistic phenomena. Coupling antibody selection to a desirable cell phenotypic trait enables significant opportunities in stem-cell manipulation.
The conception and realization of combinatorial antibody libraries is arguably the most important advance in immunochemistry since monoclonal antibodies were discovered (4). Combinatorial antibody libraries are one of the most widely used of all biochemical libraries. Indeed, they were the first biochemically generated libraries and were the source of the term “combinatorial libraries” (5, 6). Antibodies have now taken center stage in therapeutics and, in this age of proteomics, access to an antibody diversity system where the number of antibodies matches the number of targets is crucial.
The creation of combinatorial antibody libraries involved the imaginative leap that naturally expressed heavy- and light-chain fragments that make up the intact antibody repertoire could be cloned and reassociated in a combinatorial fashion to generate large numbers of useful antibodies. Furthermore, the introduction of diversity at the crucial antigen-binding hypermutation loops, by synthetic mutation, led to the creation of libraries whose diversity far exceeds that of its starting immune repertoire. Thus, the synthesis of combinatorial antibody libraries unrestricted by the limitations of tolerance can result in antibody combinations normally unobtainable in vivo. Importantly, linking these libraries to replicative bacteriophage provided a powerful and easy to use methodological platform by linking phenotype (i.e., antibody binding) to genotype (i.e., antibody cDNA), facilitating simultaneous antibody selection and recovery. The first combinatorial antibody libraries were actually expressed in phage using the Lambda system to link recognition and replication (5). However, as the field anticipated the need for even larger numbers, other phage systems such as M13 and yeast were engineered to express antibody libraries (7, 8) each with its own contextual advantage. Almost everything that has happened in combinatorial libraries is derived, directly or indirectly, from the pioneering work of these early reports (5, 6).
The application of combinatorial library technology provides a wealth of novel antibodies against a wide range of antigens, including previously inaccessible or challenging targets. Combinatorial libraries are profoundly changing many fields and perhaps one of the most exciting discoveries is in its ability to shape cellular development, such as stem cells. To achieve this result with some degree of efficiency, the antibody selection procedure from the combinatorial library, with allowance for potential counterintuitive antibody sequence combinations (2), is coupled to the acquisition of the desired cellular phenotype itself.
Richard Lerner’s group reported the first effective application of phage-derived anti
Coupling antibody selection to a desirable cell phenotypic trait enables significant opportunities in stem-cell manipulation.
bodies to stem-cell reprogramming (2, 3).These researchers used a combination of a cell-receptor–preselected phage library coupled to a lentiviral in vivo recombination system that allowed direct cell-surface expression of these focused libraries within receptor-positive progenitor cells. This process enabled the successful isolation of unique monoclonal antibody fragments capable of agonistically transdifferentiating these progenitor cells (3). Turning the focus of this concept upon ESCs, work reported in PNAS by Melidoni et al. (1) elegantly demonstrates that direct conditionally controlled stem-cell expression of a preselected ES receptor phage library coupled to a fluorescent reporter can be used to isolate monoclonal antibodies that antagonistically inhibit ES differentiation. Finally, it is worth noting that both of these studies chose to begin antibody selection by focusing on a specific receptor known to be important in stem cell differentiation. A recent paper has now described the use of unbiased libraries, with the potential to influence any receptor present in the target cells, during the successful isolation of an antibody that efficiently differentiates bone marrow progenitors into dendritic cells (9).
These proof-of-principle studies open the way to further developments in stem-cell reprogramming technology. The potential of this new technology seems only limited by parameters, such as the choice of preselection targets, the means of antibody library combination/delivery and, importantly, choice of phenotypic monitoring (1, 10). As detailed in these studies, the power of these selection systems derives from the fact that they are autocrine-based. These emerging and complementary reprogramming technologies should catalyze the development of antibody-directed stem-cell differentiation.
Acknowledgments
M.D. and M.C. are funded by The UK Medical Research Council (CiC3), The Wellcome Trust (094041/Z/10/Z), The International AIDS Vaccine Initiative Neutralizing Antibody Center, and The Center for HIV/AIDS Vaccine Immunology. R.A.D. is the co-chairman of the Britain Israel Research and Academic Exchange Partnership Regenerative Medicine Initiative.
Footnotes
The authors declare no conflict of interest.
See companion article on page 17802.
References
- 1.Melidoni AN, Dyson MR, Wormald S, McCafferty J. Selecting antagonistic antibodies that control differentiation through inducible expression in embryonic stem cells. Proc Natl Acad Sci USA. 2013;110:17802–17807. doi: 10.1073/pnas.1312062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H, Wilson IA, Lerner RA. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc Natl Acad Sci USA. 2012;109(39):15728–15733. doi: 10.1073/pnas.1214275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J, Zhang H, Yea K, Lerner RA. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc Natl Acad Sci USA. 2013;110(20):8099–8104. doi: 10.1073/pnas.1306263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köhler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 5.Huse WD, et al. Generation of a large combinatorial library of the immunoglobulin repertoire in phage lambda. Science. 1989;246(4935):1275–1281. doi: 10.1126/science.2531466. [DOI] [PubMed] [Google Scholar]
- 6.McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature. 1990;348(6301):552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 7.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 8.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology. 1997;15(6):553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 9.Yea K, et al. Converting stem cells to dendritic cells by agonist antibodies from unbiased morphogenic selections. Proc Natl Acad Sci USA. 2013;110(37):14966–14971. doi: 10.1073/pnas.1313671110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, et al. Selecting agonists from single cells infected with combinatorial antibody libraries. Chem Biol. 2013;20(5):734–741. doi: 10.1016/j.chembiol.2013.04.012. [DOI] [PubMed] [Google Scholar]