Significance
Glucose transporters mediate the exchange of glucose and related hexoses in living cells. In humans, these transporters (known as GLUT) are involved in several diseases, including cancer and diabetes. The glucose transporter from Staphylococcus epidermidis (GlcPSe) has high sequence homology to human GLUT, is specific for glucose, and is inhibited by human GLUT inhibitors. The crystal structure of GlcPSe, along with site-directed mutagenesis and transport-activity studies, provide insight into the mechanism of glucose transport.
Keywords: major facilitator superfamily, membrane protein, GLUT, sugar transporter, solute-carrier 2A
Abstract
Glucose transporters are required to bring glucose into cells, where it is an essential energy source and precursor in protein and lipid synthesis. These transporters are involved in important common diseases such as cancer and diabetes. Here, we report the crystal structure of the Staphylococcus epidermidis glucose/H+ symporter in an inward-facing conformation at 3.2-Å resolution. The Staphylococcus epidermidis glucose/H+ symporter is homologous to human glucose transporters, is very specific and has high avidity for glucose, and is inhibited by the human glucose transport inhibitors cytochalasin B, phloretin, and forskolin. On the basis of the crystal structure in conjunction with mutagenesis and functional studies, we propose a mechanism for glucose/H+ symport and discuss the symport mechanism versus facilitated diffusion.
Glucose and related sugars are vital to most living cells as a source of both energy and carbon. In animal cells, facilitated diffusion of glucose and related monosaccharides occurs through members of the solute-carrier 2A or glucose transport (GLUT) family. In humans, 14 members of the GLUT family have been identified. Despite significant sequence homology (19–65% identity) (Table S1), they differ in tissue distribution, level of expression, substrate specificity, and affinity, presumably to suit the physiological needs of a particular tissue (1, 2). GLUTs have been implicated in GLUT deficiency syndrome (3), Fanconi-Bickel syndrome (4), cancer (5, 6) and diabetes (7). Except for GLUT13, which is a myo-inositol/H+ symporter (8), and GLUT12, which functions as a glucose/H+ symporter (9), all others presumably are uniporters that catalyze equilibration of glucose across the membrane. In addition to human GLUTs, glucose transporters are important for understanding glucose uptake and metabolism during sugar fermentation and alcohol production via a myriad of hexose transporters in yeasts (10). In plants, monosaccharide and sucrose transporters play a fundamental role in stress responses and developmental processes including seed germination and balanced growth (11, 12).
GLUTs belong to the major facilitator superfamily (MFS), one of the largest protein families containing more than 10,000 members (www.tcdb.org). However, 3D structures are available for only nine MFS proteins (13–21), and none is a true glucose transporter. The most accepted model for transport by MFS proteins is alternating access in which the substrate-binding site is alternatively exposed to either side of the membrane (22). The available MFS X-ray crystal structures are consistent with such a model and represent inward-facing (open to the cytoplasmic side) [as in LacY (15), GlpT (16), PepTSo (19), and NarK (20)], outward-facing (open to the periplasmic side) [as in FucP (18) and XylE (13)], and occluded conformations [as in EmrD (17), OxlT (14), and NarU (21)].
Recently the crystal structures of XylE, a glucose transporter homolog from Escherichia coli, in outward-facing, substrate-bound conformation (13), and inward-facing conformation (23) were determined. Here we report the inward-facing, unliganded structure of a Staphylococcus epidermidis glucose/H+ symporter (GlcPSe) by single anomalous dispersion methods. GlcPSe shares high sequence identity (27–34%) and homology (49–58%) with the human GLUTs (Table S1), is highly specific for glucose, and is inhibited by the well-characterized inhibitors of human GLUTs phloretin, cytochalasin B, and forskolin. In contrast to GlcPSe, XylE transports xylose but not glucose, which is an inhibitor, and is impervious to inhibition by cytochalasin B (13, 24). On the basis of the GlcPSe structure and functional studies of wild-type and mutant transporters, we identify residues from helices 1 and 4 that are important for glucose/H+ symport (most bacterial glucose transporters) versus uniport (most mammalian GLUTs) and propose a kinetic mechanism for glucose/H+ symport.
Results
With the goal of determining the 3D structure of a glucose transporter, we screened more than 50 human, archaeal, and bacterial genes and proteins based on BLAST searches of human GLUTs and their homologs for protein overexpression, purification, transport activity, crystallization, and X-ray diffraction. S. epidermidis had a high score in BLAST searches against many human GLUTs (Table S2). Because the gene (ZP-12295482.1) product exhibited significant specific activity for glucose transport, we named it “GlcPSe.” Diffracting crystals (up to 5 Å) were obtained for several different bacterial gene products. Among all the transporters that yielded diffracting crystals, GlcPSe had the most robust glucose transporter activity.
GlcPSe Is a Glucose/H+ Symporter.
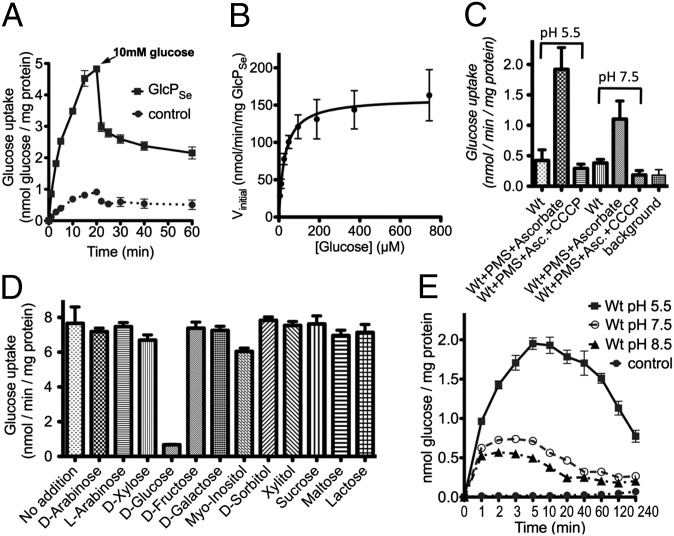
We tested GlcPSe for glucose transport using whole cells, right-side-out (RSO) vesicles, and proteoliposomes (Fig. 1). Glucose transport was linear for at least 5 min (Fig. 1A). The Km (29 ± 4 μM) and Vmax (160 ± 6 nmol⋅min−1⋅mg protein−1) for glucose transport were determined in whole cells and RSO vesicles (Fig. 1B). Other carbohydrates were examined for their ability to compete with glucose for transport, but no inhibitor of glucose transport was identified (Fig. 1D). Furthermore, no transport activity of 14C-labeled fructose, galactose, xylose, myo-inositol, or arabinose was detected. Thus, GlcPSe is a glucose transporter with high kinetic affinity (i.e., low Km) and specificity for substrate.
Fig. 1.
Glucose transport activity of GlcPSe. (A) Glucose uptake was measured with 30 µM of 14C-glucose, in RSO vesicles with GlcPSe (GlcPSe) or without GlcPSe (control). After 15 min glucose uptake reached steady state. To determine the ratio of H+:glucose symport, 10 mM of unlabeled glucose was added after 20 min. (B) Michaelis–Menten plot of GlcPSe glucose transport. Transport activity was measured at a total glucose concentration of 5–750 µM. Error bars indicate SD from nine data points of both RSO vesicles and intact cells. Km (29 ± 4 μM) and Vmax (160 ± 6 nmol⋅min−1⋅mg protein−1) were determined by nonlinear regression enzyme kinetics. The y-axis was normalized based on GlcPSe concentration as determined from Western blotting. (C) Glucose uptake in RSO vesicles for GlcPSe (Wt) in various conditions. The electrochemical H+ gradient, ∆µ̃H+, was generated by adding 0.2 mM phenazine methosulfate (PMS) and 20 mM Ascorbate (Asc). The transport reaction was initiated by the addition of 25 µM 14C-glucose. The effects of pH (5.5 or 7.5) and the protonophore CCCP (10 μM) on glucose uptake were examined. Background is RSO vesicles without GlcPSe. (D) Substrate specificity of GlcPSe. Glucose uptake activity was measured in intact cells, in the presence of 20 mM of a selected sugar, in triplicate. Transport reaction was initiated by the addition of 25 µM 14C-glucose. (E) Entrance counterflow glucose transport assay of GlcPSe proteoliposomes preloaded with 10 mM glucose. The reaction was initiated by the 50-fold dilution of proteoliposomes in 100 μL of 50 mM KPi, at pH 5.5, 7.5, or 8.5, containing 4.3 μM 14C-glucose. Liposomes preloaded with 10 mM glucose served as control.
Generally, bacterial carbohydrate transporters such as GalP (25), FucP (18), LacY (15), and XylE (13) are H+ symporters. However, human GLUTs are preponderantly uniporters that catalyze equilibration but not accumulation. To test whether GlcPSe is dependent on the electrochemical H+ gradient (∆µ̃H+; interior negative and/or alkaline) for the active transport of glucose, we examined the effect of carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a protonophore that abolishes ∆µ̃H+, on the activity of GlcPSe in RSO vesicles where ∆µ̃H+ was generated by using ascorbate and phenazine methosulfate (26). Glucose transport by RSO vesicles containing GlcPSe is markedly inhibited by CCCP (Fig. 1C), and the protonophore is more effective at pH 7.5 than at 5.5 because of its pKa (27). Thus, GlcPSe is a glucose/H+ symporter, and the stoichiometry is likely 1:1 (Fig. 1A; see calculation in Materials and Methods). Proteoliposomes reconstituted with purified GlcPSe exhibit entrance counterflow activity for glucose with an optimum at acidic pH (Fig. 1E).
Effect of Human GLUT Inhibitors on the Glucose Transport of GlcPSe.
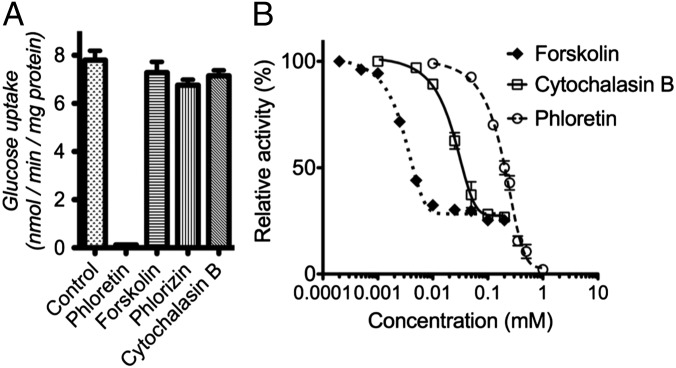
Previous work with human GLUTs identified several inhibitors of glucose transport: cytochalasin B, phloretin, forskolin, and phlorizin (28, 29). Among these, phloretin fully inhibited the transport activity of GlcPSe in whole cells and RSO vesicles (Fig. 2). Cytochalasin B and forskolin partially inhibited GlcPSe (75%) in RSO vesicles. Partial inhibition by cytochalasin B has been reported for the glucose transporter from Trypanosoma brucei (30). The phloretin IC50 was 153 ± 25 μM (Fig. 2B). The relative IC50 for cytochalasin B and forskolin was 18.1 ± 6.3 μM, and 2.35 ± 0.41 μM, respectively (Fig. 2B). Phlorizin did not inhibit the glucose transport significantly at the concentrations used (up to 1 mM).
Fig. 2.
Effect of various inhibitors on glucose transport of GlcPSe. The inhibition was measured (A) in E. coli JM1100 cells at 0 (for control) or 200 µM inhibitor or (B) in RSO vesicles at various inhibitor concentrations. The transport activity was initiated by the addition of 50 µM C14-labeled glucose. Cytochalasin B inhibits by binding to the cytoplasmic side of the protein; thus it showed no inhibition in whole cells, which are impenetrable to cytochalasin B. However, RSO vesicles are permeable to cytochalasin B and forskolin.
Inward-Facing Conformation.
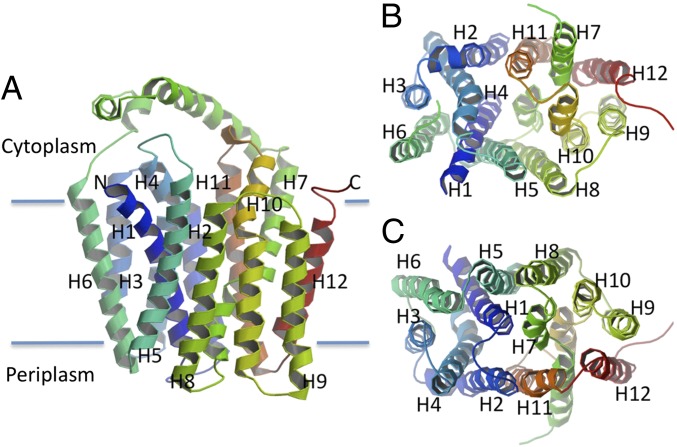
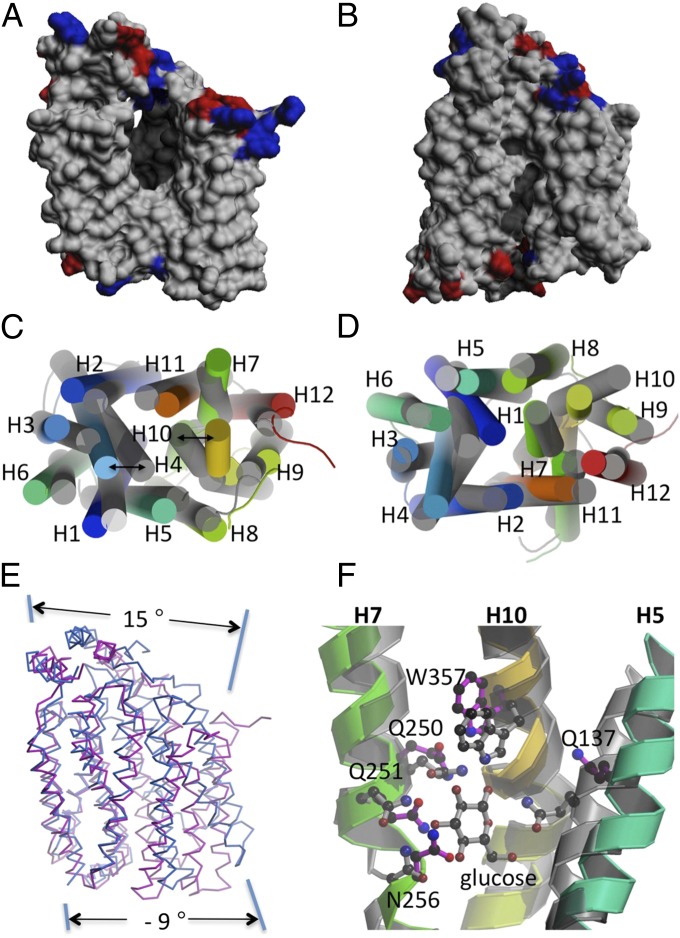
The structure of GlcPSe (Table S3 and Fig. S1) exhibits the trademark topology of MFS proteins: 12 transmembrane helices organized into two six-helix bundles, with the N and C domains related by pseudo two-fold symmetry (Fig. 3). The N and C domains can be superimposed with an rmsd of 2.8 Å. They are connected by a long cytoplasmic loop located between transmembrane (TM) helices 6 and 7, and the N and C termini are on the cytoplasmic face of the membrane. Each corresponding domain from the outward-facing conformation of XylE (13) and the inward-facing conformation of GlcPSe can be superimposed with an rmsd of 1.7 Å for the N domains and 2.3 Å for the C domains. Therefore, the transition from the inward- to the outward-facing conformations appears to involve general rigid body movements of the two halves (Movie S1). The outward-facing conformation of GlcPSe was modeled on the basis of XylE [Protein Data Bank (PDB) ID code 4GC0] (13), and the inward- and outward-facing conformations of GlcPSe were compared. On the cytoplasmic side, the TM helices of GlcPSe are moved further away from the center of the molecule in the inward-facing conformation than in the outward-facing conformation, with helices 4 and 10 moving the most (6 Å, compared with an average of 3 Å for the overall structure; Fig. 4C). On the periplasmic side, the helices of the inward-facing conformation in general are moved toward the center of molecule relative to the outward-facing conformation (Fig. 4D). The transition from the inward-facing to the outward-facing conformation involves a relative rotation of the N and C domains of ∼24° around an axis parallel to the membrane, passing through the middle of the molecule and close to the glucose-binding site (Fig. 4E, Fig. S2, and Movie S1). In the outward-facing conformation of GlcPSe, R71 (helix 3) and R369 (helix 11) form a salt bridge with E122 (helix 4) that is absent in the inward-facing conformation of GlcPSe (Fig. S3). All these residues are conserved in human GLUTs, GlcPSe, and XylE (Fig. S1). Furthermore, mutation of E122 to Ala in GlcPSe inactivated the symporter (Fig. S4). We speculate that this mutant probably is locked in the inward-facing conformation.
Fig. 3.
Overall structure of the inward-facing conformation of GlcPSe. (A) Side-view. Twelve TM helices are arranged as two twofold pseudosymmetrical domains connected by a cytoplasmic loop made up of three helical segments. (B) Cytoplasmic view. The cytoplasmic loop was omitted for clarity. TM helices 4 and 10 are internal, making up the cavity toward the cytoplasm. (C) Periplasmic view. TM helices 1 and 7 are internal; helices 4 and 10 are moved toward the membrane, away from the cavity.
Fig. 4.
Comparison of the inward- and outward-facing conformations of GlcPSe. The outward-facing conformation of GlcPSe was modeled on the basis of the XylE structure (PDB ID code 4GC0; see SI Materials and Methods). (A) Side-view of the cavity in GlcPSe, inward-facing conformation, unliganded. For clarity helices 5 and 8 were omitted. (B) Side-view of the modeled outward-facing conformation. Helices 5 and 8 were omitted for clarity. (C and D) Cytoplasmic (C) and periplasmic (D) views of the superposition between GlcPSe in the inward-facing conformation (rainbow) and the outward-facing conformation (gray). (E) Side-view of the superposition between GlcPSe in the inward-facing conformation (red) and the outward-facing conformation (blue). The superposition is based on the N domain, to show the movement in the C domain better. (F) The glucose-binding site in the inward-facing conformation (rainbow) and outward-facing conformation of GlcPSe (gray). Residues from the outward-facing conformation are shown in gray; residues of the inward-facing conformation are magenta.
The Glucose-Binding Site.
A central amphipathic cavity ∼20 Å deep is formed by helices 1, 4, 7, and 10 (Figs. 3 and 4A); this cavity is larger than that observed for the outward-facing conformation of XylE (Fig. 4B). The glucose-binding site was identified by comparison with the XylE structure (13) and was corroborated by biochemical findings with GLUT1 (31–33) as well as mutagenesis of GlcPSe. GlcPSe mutants Q137A, Q250A, Q251A, N256A, and W357A, which correspond respectively to residues Q161, Q282, Q283, N288, and W388 in human GLUT1, exhibit no transport activity (Fig. S4). Because the corresponding positions in GLUT1 are critical for glucose binding, it is likely that the residues in GlcPSe also are involved in glucose binding. The glucose-binding sites in the inward-facing (Fig. S5A) and in the modeled outward-facing structure of GlcPSe are shown in Fig. 4F. Residues Q250, Q251, and N256 are in approximately the same location in both structures; however, Q137 and W357 are removed completely from the glucose-binding site of GlcPSe inward-facing structure.
The H+-Binding Site.
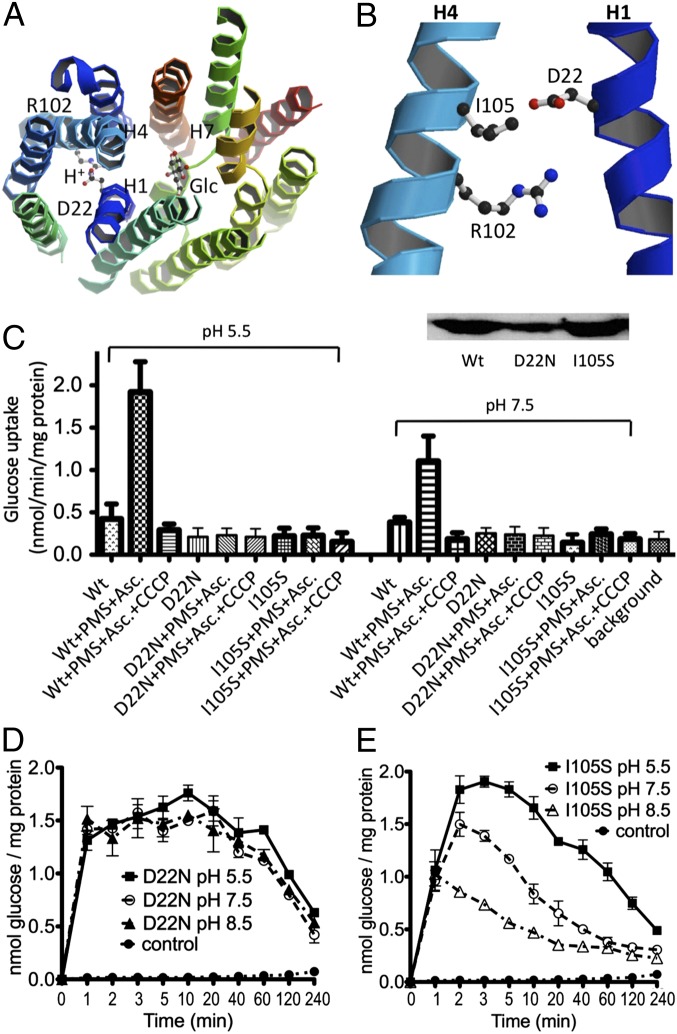
Based on sequence homology between GlcPSe and previously characterized bacterial MFS transporters, residues with potential H+-binding capabilities were identified (18, 25). Mutagenesis coupled with transport assays revealed that D22 is an important residue involved in H+ binding (Fig. 5 A and B). Unlike XylE, GlcPSe has only two charged residues around the postulated H+-binding site (Fig. 5B and Fig. S5B). Given the close proximity of these charged residues in the crystal structure, D22 from helix 1 may form a salt bridge with R102 from helix 4 (Fig. 6B). In human GLUTs the residue corresponding to R102 is fully conserved, and D22 is a neutral residue Asn (in GLUT1, -3, -4, -5, -7, -9, and -11) or an acidic Asp (in GLUT2 and -13) or Glu (in GLUT10 and -12) (Fig. S1). Mutant R102A in GlcPSe exhibited no glucose transport in whole cells, RSO vesicles, or proteoliposomes (Fig. S4), indicating that R102 is an important residue for glucose transport. Mutant D22N in GlcPSe had glucose counterflow activity in proteoliposomes, although active glucose uptake in RSO vesicles was insignificant (Fig. 5 C and D). Therefore, D22N GlcPSe may not be an H+ symporter but may catalyze uniport as does GLUT1. Thus, in agreement with previous findings with other carbohydrate-H+ symporters such as LacY (34), FucP (18), or GalP (25), GlcPSe also contains an acidic residue that plays a key role in H+ symport. Interestingly, GLUT12 and -13, which have Glu and Asp, respectively, in the corresponding position of D22, have been shown to function as H+ symporters, whereas GLUT2 is a uniporter even though it has Asp in the position corresponding to D22 in GlcPSe. However, based on modeling of GLUT2, S161 (helix 4) is in close proximity to D27 (D22 in GlcPSe). S161 of GLUT2 is I105, I140, or V162 in GlcPSe, GLUT12, or GLUT13, respectively. Similar to mutant D22N, mutant I105S in GlcPSe showed no significant active glucose transport in RSO vesicles (Fig. 5C) but had glucose counterflow activity in proteoliposomes (Fig. 5E). Atom Hγ of S105 presumably is positioned near the carboxyl group in D22. Thus, the D22 side chain can H-bond to S105 and is not available to form the salt bridge with R102 (Fig. 6D). Blockade of active glucose transport in RSO vesicles containing the I105S mutant and attenuation of the pH effect in counterflow, compared with wild type, indicate that this residue can alter H+ symport activity.
Fig. 5.
The H+-binding site and glucose transport assay in D22N and I105S GlcPSe. (A) Relative location of the H+- and glucose-binding sites. (B) Close-up of the H+-binding site. (C) Active glucose transport assay for D22N and I105S GlcPSe in RSO vesicles. For comparison, the activity of the wild-type GlcPSe (Wt) is included. The effects of pH (5.5 and 7.5) and the protonophore CCCP (10 μM) on the transport activity are shown. The electrochemical H+ gradient was generated using ascorbate (Asc) and PMS. (Inset) Western blotting of wild-type, D22N, and I105S GlcPSe RSO vesicles. The transport activity was normalized by the amount of protein present in RSO vesicles. (D and E) Entrance counterflow glucose transport assay of D22N (D) and I105S (E) GlcPSe proteoliposomes preloaded with 10 mM glucose, at pH 5.5, 7.5, and 8.5 (50 mM KPi).
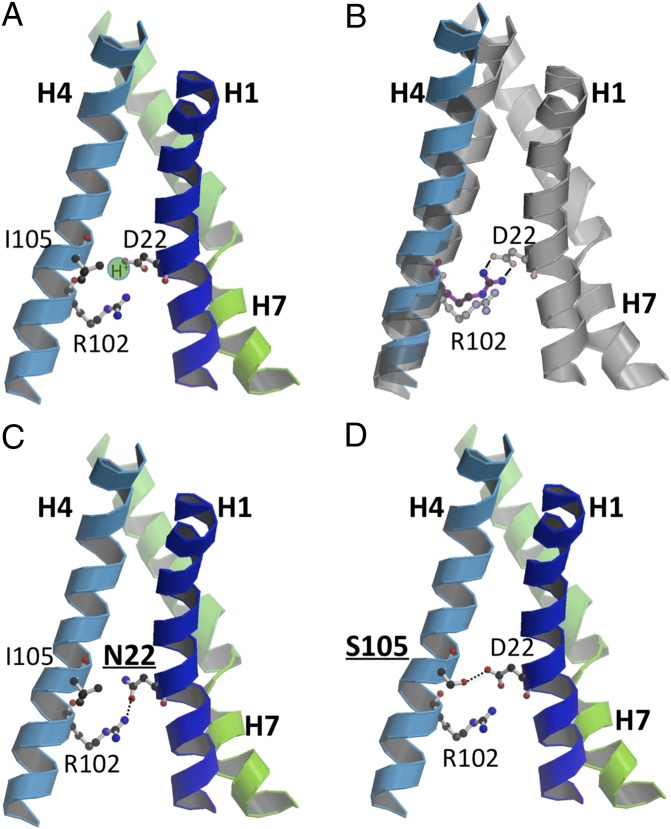
Fig. 6.
Modeled H+-binding site in the inward-facing conformation of GlcPSe. (A) D22 from helix 1 is protonated. (B) In the absence of a proton, D22 and R122 form a salt bridge that moves helix 4, pushing other helices to open wide the active site cavity. The new position of helix 4 upon salt bridge formation is shown in blue; the position of helix 4 in the protonated state is shown in gray. (C) Interactions around the H+-binding site in the D22N mutant of GlcPSe. (D) Interactions around the H+-binding site in the I105S mutant of GlcPSe. S105 H-bonds to D22, with Hγ approximately occupying the H+-binding site.
Discussion
Glucose transporters are ubiquitous and important for essential cellular processes. Involvement of human GLUTs in diabetes and cancer makes it imperative to obtain 3D structures so that the function of these fundamental transporters can be better understood and controlled by potential effectors. In our efforts to determine the 3D structure of human GLUTs or their homologs, we have identified a glucose transporter from S. epidermidis that is unusually specific for glucose and is inhibited by well-characterized inhibitors of human GLUTs: phloretin, forskolin, and cytochalasin B (Figs. 1 and 2) (35). Sequence identity and homology of GlcPSe to human GLUTs are 27–34% and 49–58%, respectively (Table S1). GlcPSe and XylE share 33% identity in amino acid sequences. Therefore, the structure of GlcPSe is a good homology model for human GLUTs and, along with the structures of XylE (16, 25), will allow modeling of various human GLUTs in different conformations.
Given the sequence identity in amino acid sequences between GlcPSe and XylE, the outward-facing (substrate-bound occluded) conformation of GlcPSe can be inferred on the basis of the XylE structure. The transition between the inward-facing and outward-facing conformations involves a 24° relative rotation of the N and C domains around an axis that passes through the substrate-binding site, suggesting that translocation of glucose is coupled with conformational changes associated with the transition between the outward- and inward-facing conformations.
In the absence of an H+, the salt bridge between R102 in helix 4 and D22 in helix 1 opens the cavity housing the glucose-binding site by linking two far-distant residues from helices 1 and 4, which in turn push out surrounding helices (Fig. 6B). At low pH, protonated D22 can “release” R102 and loosen the packing between helices 1 and 4, lowering the energetic barrier of the transition between the inward and outward conformations (Fig. 6A). The involvement of D22 in H+ binding probably is similar in GLUT12 (which has Glu at this position) and GLUT13 (with Asp at this position), which have been shown to be H+ symporters. On the other hand, GLUT1, -3, -4, -5, -7, -9, and -11 have Asn in the corresponding position of D22 of GlcPSe and are uniporters. Mutation D22N in GlcPSe affected H+/glucose symport, as attested by the lack of active glucose transport in RSO vesicles and loss of a pH effect on entrance counterflow in proteoliposomes. Thus, the presence of an acidic residue at the corresponding position of D22 in GlcPSe seems to be indicative of H+ symport. The exception to this rule is GLUT2, which has an Asp in the corresponding position of D22 in GlcPSe but is a uniporter. Upon closer inspection of the GLUT2 structure modeled on the basis of GlcPSe, we found that this Asp can H-bond to an adjacent Ser residue (corresponding to I105 in GlcPSe) (Fig. 6D). With GlcPSe mutant I105S (which mimics the “H+-binding site” in GLUT2), glucose transport becomes insensitive to ∆µ̃H+ (Fig. 5C), and entrance counterflow is significantly less affected by pH (activity is 35% higher at pH 5.5 than at pH 7.5) (Fig. 5E) than in wild type (activity 300% higher at pH 5.5 than at 7.5) (Fig. 1E), suggesting that the I105S mutation affects H+ symport. Therefore, in establishing the type of transport—uniport versus symport—the presence of Asp/Glu in the corresponding position 22 in the GlcPSe sequence is not sufficient; the environment in the H+-binding site (e.g., a Ser residue close to the important carboxylate) matters as well.
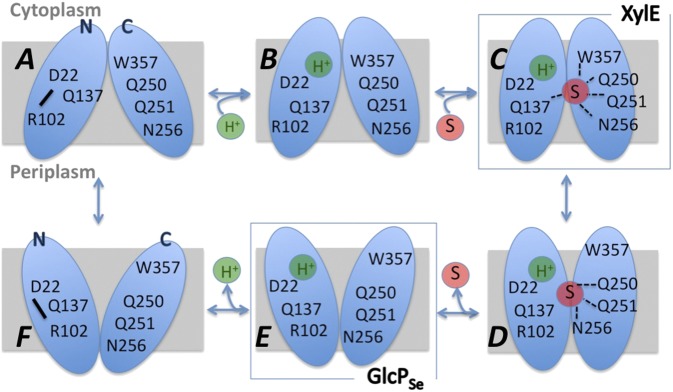
We propose a mechanism of glucose transport for GlcPSe, adapted from that proposed for LacY (15), the most intensively studied MFS protein, with the differences that the location of the H+-binding site is changed and that the protonated and deprotonated protein conformations are distinct (Fig. 7). Thus, in the absence of a H+, D22 and R102 form a salt bridge to juxtapose helices 1 and 4, thereby opening the substrate cavity wide (Fig. 7A). When the H+ binds to D22, the salt bridge is disrupted, and helices 1 and 4 rearrange to decrease the size of the cavity (Fig. 7B), which in turn lowers the energetic barrier of the transporter’s conformations, so that when glucose binds, it is translocated rapidly (Fig. 7 C and D). Glucose binds through residues of helices 5 (Q137), 7 (Q250, Q251, N256), and 10 (W357), probably bringing the N (helix 5) and C (helices 7 and 10) domains closer together (Fig. 7C). In the inward-facing conformation helices 5 and 10 move away from the center, likely dislocating Q137 and W357 from the glucose-binding site, and thus opening the cavity toward the cytoplasm (Fig. 7D). Glucose is released (Fig. 7E), followed by the H+ release (Fig. 7F), which returns D22 and R102 to their salt bridge form, opening wide the cavity. In this scheme, our structure is shown in Fig. 7E, and the liganded XylE structure (13) is shown in Fig. 7C. Glucose translocation for most human GLUTs skips the steps shown in Fig. 7 A and F, because the residue corresponding to D22 is Asn. The absence of these steps in human GLUTs makes biological sense considering the environmental conditions, i.e., the relatively high and stable concentration of glucose in animal cells versus the limiting conditions of the bacterial milieu. Therefore, bacterial glucose transporters generally have lower Km values for glucose, have higher transport activity than human GLUTs, and can move glucose uphill at the expense of ∆µ̃H+.
Fig. 7.
Proposed mechanism of glucose/H+ symport; see text for details. S, carbohydrate substrate (glucose) of the transporter; H+, proton.
How do bacterial H+ symporters achieve these improvements in transport? We propose that an H+-binding site decreases the energetic barrier mediating different conformations. Our model predicts that GLUT10 (which has a Glu at the D22 position from GlcPSe) may be able to function as a H+ glucose symporter and that mutation of N29 of GLUT1 (corresponding to D22 in GlcPSe) to Asp can transform GLUT1 into a glucose/H+ symporter. Most human GLUTs likely lost their ability to couple H+ transport to glucose transport because they are exposed to a steady, relatively high concentration of glucose.
Materials and Methods
Protein Expression and Purification.
The GlcPSe gene was cloned into the pET-15b vector (Novagen) and was expressed in E. coli C41 cells (36). Cells were grown at 37 °C, using Terrific Broth or Luria Broth medium supplemented with 1% (vol/vol) glycerol. Cells were disrupted by sonication. The membrane fraction was collected by ultracentrifugation and then was solubilized with 1% (wt/vol) dodecyl-β-d-maltopyranoside (DDM; EMD Chemicals) or other detergents at 4 °C. Protein was purified with Talon metal affinity resin (Clontech). The N-terminal poly-His tag was removed by thrombin (BioPharm Laboratories). The buffer of the purified protein was exchanged to 20 mM Tris (pH 7.5), 0.02% DDM, and the protein was brought to a concentration of 20 mg/mL by ultrafiltration using Amicon Ultra concentrator (Millipore). More details are given in SI Materials and Methods.
Crystallization of GlcPSe and Data Collection.
Crystallization by hanging-drop vapor diffusion was set up by combining 1 µL of 10 mg/mL purified GlcPSe with 1 µL of precipitating solution. Rod-like crystals of 0.05 × 0.05 × 0.2 mm appeared within 2–3 d at 22–26% PEG 400, 0.1 M calcium acetate, 0.1 M NaCl, 0.1 M MOPS (pH 7.0), at 18 °C. Crystals were dehydrated to improve their diffraction. Data were collected at Advanced Photon Sources at Argonne National Laboratory. Further details are given in SI Materials and Methods.
Structural Determination.
Initial phases were obtained from a Hg-derivative crystal by single anomalous diffraction using the program ShelxC/D/E (37). The manual tracing of the 12 transmembrane helices was done with the program COOT (38) (Fig. S6). The initially built model was subjected to molecular replacement to native data. The model was built using COOT (38) and XtalView (39) and was refined with Phenix (40) and Refmac (41). Details are given in SI Materials and Methods.
Transport Assay.
For whole-cell assays, the glucose transporter-deficient bacterial strains JM1100 (Yale E. coli Genetic Stock Center) contained GlcPSe cloned in the pBAD vector (Invitrogen). The RSO vesicles of E. coli JM1100 cells were prepared as described by Kaback and colleagues (42, 43). Transport assay was initiated by the addition of 14C-radiolabeled glucose or other sugars to 50 μL of cells or RSO vesicles (at an OD600 of 2.0) containing 20 mM ascorbate and 0.2 mM phenazine methosulfate in 0.1 M KPi [pH 7.5 (or pH 5.5 when the effect of pH was examined)], 10 mM MgSO4. The transport was stopped by the ice-chilled quench buffer [0.1 M KPi (pH 5.5), 0.1 M LiCl] after 1 min unless otherwise specified. The solution was filtered with a 0.4-µm cellulose nitrate membrane filter (Whatman), and the filter was washed twice with the quench buffer. The membrane filter was placed into a vial filled with BioSafe II scintillation liquid (Research Products International Corp.), and radioactivity was quantified. Kinetic parameters were determined by nonlinear algorithm plots supplied by Prism (GraphPad Software). More details are given in SI Materials and Methods.
Calculation of H+/Glucose Stoichiometry.
The volume of intravascular fluid per milligram of membrane protein (for the internal concentration of solutes accumulated by the RSO vesicles) is 2.2 μL (44, 45). A concentration gradient of ∼10-fold was determined from ratio of internal to external glucose concentrations at steady state. At pH 7.5, the membrane potential is the only component of the proton electrochemical gradient, and it is around −75 mV (46). Because the glucose concentration gradient is ∼10-fold (Fig. 1A), the stoichiometry of H+/glucose transport is ∼1:1 for GlcPSe.
Entrance Counterflow Transport Assay.
Proteoliposomes of GlcPSe were prepared as described previously (47). Two microliters of liposomes or proteoliposomes (OD600 = 25), preloaded with 10 mM cold glucose, were diluted into 100 μL of assay buffer (50 mM KPi) at different pH (5.5, 7.5, or 8.5) containing 4.3 μM 14C-glucose. Details are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staffs of the General Medicine and Cancer Institutes Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory, and the Canadian Macromolecular Crystallography Facility, Canadian Light Source for support and extensive screening time; Prof. Peter Henderson for suggesting the H+-binding location; Dr. Andrey Lebedev for help with data processing; and Drs. Carl Correll, Marc Glucksman, David Mueller, Kenneth Neet, and Kyoung Joon Oh for comments on the manuscript. This work was supported by National Institutes of Health Grant 1R01DK091754 and American Cancer Society Grant ILLBASIC00050068 (to J.-y.C.) and by United Nations Educational, Scientific and Cultural Organization-L’OREAL International Fellowships (to J.Z.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank (PDB), www.pdb.org (PDB ID code 4LDS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311485110/-/DCSupplemental.
References
- 1.Uldry M, Thorens B. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch. 2004;447(5):480–489. doi: 10.1007/s00424-003-1085-0. [DOI] [PubMed] [Google Scholar]
- 2.Thorens B, Mueckler M. Glucose transporters in the 21st Century. Am J Physiol Endocrinol Metab. 2010;298(2):E141–E145. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klepper J, Fischbarg J, Vera JC, Wang D, De Vivo DC. GLUT1-deficiency: Barbiturates potentiate haploinsufficiency in vitro. Pediatr Res. 1999;46(6):677–683. doi: 10.1203/00006450-199912000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Santer R, et al. Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat Genet. 1997;17(3):324–326. doi: 10.1038/ng1197-324. [DOI] [PubMed] [Google Scholar]
- 5.Medina RA, Owen GI. Glucose transporters: Expression, regulation and cancer. Biol Res. 2002;35(1):9–26. doi: 10.4067/s0716-97602002000100004. [DOI] [PubMed] [Google Scholar]
- 6.Airley RE, Mobasheri A. Hypoxic regulation of glucose transport, anaerobic metabolism and angiogenesis in cancer: Novel pathways and targets for anticancer therapeutics. Chemotherapy. 2007;53(4):233–256. doi: 10.1159/000104457. [DOI] [PubMed] [Google Scholar]
- 7.Watson RT, Pessin JE. Intracellular organization of insulin signaling and GLUT4 translocation. Recent Prog Horm Res. 2001;56:175–193. doi: 10.1210/rp.56.1.175. [DOI] [PubMed] [Google Scholar]
- 8.Uldry M, et al. Identification of a mammalian H(+)-myo-inositol symporter expressed predominantly in the brain. EMBO J. 2001;20(16):4467–4477. doi: 10.1093/emboj/20.16.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson-O’Brien AL, Patron N, Rogers S. Evolutionary ancestry and novel functions of the mammalian glucose transporter (GLUT) family. BMC Evol Biol. 2010;10:152. doi: 10.1186/1471-2148-10-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kruckeberg AL, Dickinson JR. Carbon metabolism. In: Dickinson JR, Schweizer M, editors. Metabolism and Molecular Physiology of Saccharomyces cerevisiae. 2nd Ed. London: CRC; 2004. pp. 42–103. [Google Scholar]
- 11.Williams LE, Lemoine R, Sauer N. Sugar transporters in higher plants—a diversity of roles and complex regulation. Trends Plant Sci. 2000;5(7):283–290. doi: 10.1016/s1360-1385(00)01681-2. [DOI] [PubMed] [Google Scholar]
- 12.Slewinski TL. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: A physiological perspective. Mol Plant. 2011;4(4):641–662. doi: 10.1093/mp/ssr051. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490(7420):361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 14.Hirai T, et al. Three-dimensional structure of a bacterial oxalate transporter. Nat Struct Biol. 2002;9(8):597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- 15.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Lemieux MJ, Song J, Auer M, Wang D-N. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301(5633):616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312(5774):741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467(7316):734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 19.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30(2):417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497(7451):647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan H, et al. Structure and mechanism of a nitrate transporter. Cell Rep. 2013;3(3):716–723. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quistgaard EM, Löw C, Moberg P, Trésaugues L, Nordlund P. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nat Struct Mol Biol. 2013;20(6):766–768. doi: 10.1038/nsmb.2569. [DOI] [PubMed] [Google Scholar]
- 24.Martin GE, et al. Forskolin specifically inhibits the bacterial galactose-H+ transport protein, GalP. J Biol Chem. 1994;269(40):24870–24877. [PubMed] [Google Scholar]
- 25.Sanderson NM, Qi D, Steel A, Henderson PJ. Effect of the D32N and N300F mutations on the activity of the bacterial sugar transport protein, GalP. Biochem Soc Trans. 1998;26(3):S306. doi: 10.1042/bst026s306. [DOI] [PubMed] [Google Scholar]
- 26.Konings WN, Barnes EM, Jr, Kaback HR. Mechanisms of active transport in isolated membrane vesicles. 2. The coupling of reduced phenazine methosulfate to the concentrative uptake of beta-galactosides and amino acids. J Biol Chem. 1971;246(19):5857–5861. [PubMed] [Google Scholar]
- 27.Ramos S, Schuldiner S, Kaback HR. The electrochemical gradient of protons and its relationship to active transport in Escherichia coli membrane vesicles. Proc Natl Acad Sci USA. 1976;73(6):1892–1896. doi: 10.1073/pnas.73.6.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kasahara T, Kasahara M. Expression of the rat GLUT1 glucose transporter in the yeast Saccharomyces cerevisiae. Biochem J. 1996;315(Pt 1):177–182. doi: 10.1042/bj3150177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sergeant S, Kim HD. Inhibition of 3-O-methylglucose transport in human erythrocytes by forskolin. J Biol Chem. 1985;260(27):14677–14682. [PubMed] [Google Scholar]
- 30.Seyfang A, Duszenko M. Specificity of glucose transport in Trypanosoma brucei. Effective inhibition by phloretin and cytochalasin B. Eur J Biochem. 1991;202(1):191–196. doi: 10.1111/j.1432-1033.1991.tb16362.x. [DOI] [PubMed] [Google Scholar]
- 31.Mueckler M, Makepeace C. Model of the exofacial substrate-binding site and helical folding of the human Glut1 glucose transporter based on scanning mutagenesis. Biochemistry. 2009;48(25):5934–5942. doi: 10.1021/bi900521n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashiramoto M, et al. Site-directed mutagenesis of GLUT1 in helix 7 residue 282 results in perturbation of exofacial ligand binding. J Biol Chem. 1992;267(25):17502–17507. [PubMed] [Google Scholar]
- 33.Mueckler M, Weng W, Kruse M. Glutamine 161 of Glut1 glucose transporter is critical for transport activity and exofacial ligand binding. J Biol Chem. 1994;269(32):20533–20538. [PubMed] [Google Scholar]
- 34.Carrasco N, et al. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989;28(6):2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- 35.Bloch R. Inhibition of glucose transport in the human erythrocyte by cytochalasin B. Biochemistry. 1973;12(23):4799–4801. doi: 10.1021/bi00747a036. [DOI] [PubMed] [Google Scholar]
- 36.Arechaga I, et al. Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett. 2000;482(3):215–219. doi: 10.1016/s0014-5793(00)02054-8. [DOI] [PubMed] [Google Scholar]
- 37.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McRee DE, Israel M. XtalView, protein structure solution and protein graphics, a short history. J Struct Biol. 2008;163(3):208–213. doi: 10.1016/j.jsb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Mustyakimov M, Afonine PV, Langan P. Generalized X-ray and neutron crystallographic analysis: More accurate and complete structures for biological macromolecules. Acta Crystallogr D Biol Crystallogr. 2009;65(Pt 6):567–573. doi: 10.1107/S0907444909011548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaback HR. Bacterial membranes. Methods Enzymol. 1971;22:99–120. [Google Scholar]
- 43.Short SA, Kaback HR, Kohn LD. Localization of D-lactate dehydrogenase in native and reconstituted Escherichia coli membrane vesicles. J Biol Chem. 1975;250(11):4291–4296. [PubMed] [Google Scholar]
- 44.Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 1. Effect of pH on efflux, exchange, and counterflow. Biochemistry. 1979;18(17):3691–3697. doi: 10.1021/bi00584a009. [DOI] [PubMed] [Google Scholar]
- 45.Kaback HR, Barnes EM., Jr Mechanisms of active transport in isolated membrane vesicles. II. The mechanism of energy coupling between D-lactic dehydrogenase and beta-galactoside transport in membrane preparations from Escherichia coli. J Biol Chem. 1971;246(17):5523–5531. [PubMed] [Google Scholar]
- 46.Ramos S, Kaback HR. The electrochemical proton gradient in Escherichia coli membrane vesicles. Biochemistry. 1977;16(5):848–854. doi: 10.1021/bi00624a006. [DOI] [PubMed] [Google Scholar]
- 47.Geertsma ER, Nik Mahmood NAB, Schuurman-Wolters GK, Poolman B. Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc. 2008;3(2):256–266. doi: 10.1038/nprot.2007.519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.