Murphy’s Law, “anything that can happen, will happen,” constitutes a reasonable working philosophy for biologists. A corollary is that seemingly minor processes can have major consequences. In exploring how the immune system monitors cells for abnormal gene expression, Apcher et al. (1) provide solid evidence supporting two highly controversial processes: first, protein translation in the nucleus, and second, compartmentalized translation by ribosome subclasses (immunoribosomes) to facilitate immunosurveillance.
Jawed vertebrates evolved a remarkable system to monitor cellular gene expression to detect tumors, viruses, and other intracellular pathogens. CD8+ T cells express clonally restricted antigen receptors that recognize small peptides (typically 8- to 13-mers) bound noncovalently to MHC class I molecules (MHC I). Nearly all nucleated cells constitutively express MHC I. Extended oligopeptides are continuously generated in all cells by the action of proteasomes and other cytosolic proteases and transported from the cytosol to nascent class I molecules in the endoplasmic reticulum by TAP (transporter associated with antigen processing), the dedicated oligopeptide transporter. Peptides derive both from “retirees” (proteins exhibit a characteristic lifespan that typically follow first-order degradation kinetics) and defective ribosomal products (DRiPs) (2). DRiPs are translation products that for myriad reasons (Murphy’s Law redux) fail to achieve a native, stable structure and are shunted for degradation. DRiPs likely provide the bulk of constitutively presented peptides and are particularly important for detecting viruses, because their tight linkage with translation (3) enables CD8+ T-cell killing of infected cells before progeny viruses can be released (Fig. 1).
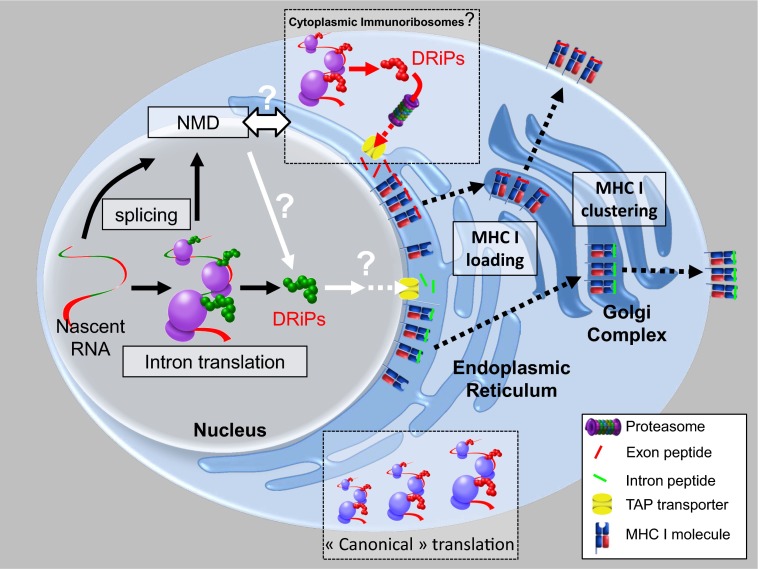
Fig. 1.
Nuclear intron translation and other possible examples of compartmentalized DRiP antigen processing. Apcher et al. (1) show that immunosurveillance begins in the nucleus, when introns are translated in situ to generate DRiPs that feed the class I antigen processing pathway. Intron encoded peptides (green) reach ER TAP via an uncertain route, where they are loaded onto MHC I. NMD pioneer translation of spliced message also contributes peptides (9, 10), although the site of translation (nucleus vs. cytoplasm) is uncertain. Both pathways are consistent with the immunoribosome hypothesis that translation is compartmentalized to facilitate presentation of peptides from low abundance mRNA (8). Peptides (red) also arise from translation of standard mRNA in the cytoplasm. This may be through DRiPs arising from canonical translation. Alternatively, dedicated DRiPs may be synthesized by cytoplasmic immunoribosomes. Longer DRiPs are initially processed by proteasomes into peptide fragments that are transported by TAP (transporter associated with antigen processing) into the ER. Peptide loading may be compartmentalized (6) to generate clusters of MHC I with the same peptide (20), clusters are retained trafficking through the Golgi complex to the cell surface, where clustered pMHC increases CD8+ T-cell sensitivity (20).
The abundance and nature of DRiPs remain a major question for immunology and cell biology. Recent evidence (4) supports original findings that a large fraction of translation products are rapidly degraded (t1/2 ∼ 7 min) (2). Whatever the fraction, it appears that only a subset of rapidly degraded proteins is efficiently targeted to TAP for immunosurveillance (5), consistent with the concept that proteasomes differ in their capacity to generate immunosurveillance relevant peptides. Proteasome substrate selectivity is probably key to an immunosurveillance system that is not overwhelmed by peptides from the most abundantly translated gene products. Peptide competition studies suggest that antigen processing is compartmentalized in a manner that distinguishes peptides derived from DRiPs from cytosolic peptides generated by other means (6). Nonstoichiometric representation of antigenic information may have evolved to enable immunosurveillance of tumors, where tumor-specific CD8+ T cells often recognize extremely low abundance nontraditional translation products (7). There are several reports describing CD8+ T-cell recognition of intronic peptides, but translation from prespliced mRNA itself was ruled out or uncertain.
Translating immature RNA is an attractive possibility for immunosurveillance. Misspliced nascent RNAs are degraded based on detection of premature termination codons in a proposed pioneer round of translation. Such nonsense mediated decay (NMD)-related translation was proposed as a possible example of immunoribosome function, i.e., ribosomes whose translation products are selectively processed to enable immunosurveillance based on mRNA quality and not simply quantity (8). Elegant evidence for NMD in immunosurveillance comes from the Fahraeus and Shastri laboratories (9, 10).
Apcher et al. (1) now look at peptide generations from introns in prespliced mRNA by inserting a model peptide into an intron and measuring T-cell activation under a variety of circumstances. They find that peptide presentation is equivalent whether the peptide is expressed intronically vs. exonically. Even more remarkably, blocking mRNA export from the nucleus selectively inhibits presentation of the exon-encoded peptide while enhancing presentation of the intron-encoded peptide.
These findings raise the highly controversial possibility of nuclear translation. Translation in the nucleoplasm and nucleolus (the site of ribosomal preassembly) was reported 60 y ago but fell out of vogue. Nuclear translation was resurrected by Iborra et al. in 2001, only to be buried by criticism that skirted Iborra’s actual findings (11).
New technology reliably provides a fresh angle on old controversies. David et al. (12) jumped into the fray with their ribopuromycylation method (RPM), which uses an antibody specific for puromycin to localize, via standard immunofluorescence, ribosome catalyzed puromycylated nascent chains tethered to ribosomes in cultured cells. RPM revealed a considerable level of translation in the nucleus, particularly the nucleolus, echoing original findings from the 1950s.
Apcher et al. directly support nuclear translation by localizing an intron-coded peptide in the nucleus using an antipeptide antibody and showing that peptide levels were controlled by proteasomes (abundant in nuclei). The peptide colocalized with RPM-labeled ribosomes, and the intimate relationship between peptide and puromycin or ribosomes themselves was supported using an immunoproximity assay, in which secondary antibodies generate a signal when attached aptamers are sufficiently close to interact. Nuclear translation was also recently reported by Al Jubran et al. (13), who used RPM to detect translation in the nucleoli and active transcription sites in Drosophila cell nuclei and further elegantly showed that assembled 80S ribosomes are present at these sites using bimolecular fluorescence complementation between ribosomal proteins on 60S and 40S ribosomal subunits.
Together, these studies robustly support the existence of nuclear translation and raise many questions.
• What is the nature of intron-translating ribosomes? Are they immature ribosomes (ribosome assembly is completed in the cytosol) or are cytosolic ribosomes reimported into the nucleus?
• Are all preRNAs scanned for immunosurveillance? What other functions do intron-encoded peptides possess?
• What is the nature of nuclear/nucleolar translation? Apcher et al. describe noncanonical translation independent of the cap-binding translation factor eIF4E. What other types of translation can be distinguished in the nucleoplasm and nucleolus?
• What is being translated? Application of new high-throughput methods for identifying nascent proteins or translating mRNAs (14, 15) should provide a detailed accounting of the variety proteins/peptides being translated by ribosomes isolated from various nuclear subdomains.
• What fraction of nuclear translation is devoted to antigen presentation? Dolan et al. (16) reported possible nuclear translation contributing to the generation of influenza A virus peptides. Does this represent another type of nuclear translation that contributes to immunosurveillance?
• How are nuclear peptides transported to class I molecules? Because the inner nuclear membrane lacks TAP, do nuclear peptides reach TAP via the cytosol? Is there another route for accessing the ER from the nucleus?
• What are the other functions of nuclear translation? The demonstration of nuclear translation in invertebrate cells (11), which appear to lack any semblance of a class immunosurveillance system, strongly supports nonimmunological functions.
• Are there cytoplasmic immunoribosomes that contribute to immunosurveillance? More generally, echoing the ribosome filter hypothesis (17), how are ribosomes and the translation machinery specialized to translate individual mRNAs or classes of mRNAs? Does such specialization extend to recruiting modified aminoacyl tRNA synthetases to modify the genetic code on an mRNA by mRNA basis (18).
The broadest issue broached by Apcher et al. regards compartmentalization of cellular processes. Although we think in terms of membrane-bound organelles, it is likely that translation (19), antigen processing (6, 20), and many other cellular processes are chemiophysically organized to increase efficiency. Such interactions are obliterated once cells are lysed for biochemical procedures, providing yet another reason for studying natural phenomena in as natural a setting as experimentally feasible.
Acknowledgments
J.W.Y. is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID).
Footnotes
The authors declare no conflict of interest.
See companion article on page 17951.
References
- 1. Apcher S, Millot G, Daskalogianni C, Scherl A, Manoury B, Fåhraeus R (2013) Translation of pre-spliced RNAs in the nuclear compartment generates peptides for the MHC class I pathway. Proc Natl Acad Sci USA 110:17951–17956. [DOI] [PMC free article] [PubMed]
- 2.Yewdell JW. DRiPs solidify: Progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32(11):548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croft NP, et al. Kinetics of antigen expression and epitope presentation during virus infection. PLoS Pathog. 2013;9(1):e1003129. doi: 10.1371/journal.ppat.1003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol Cell. 2013;50(3):368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian SB, Princiotta MF, Bennink JR, Yewdell JW. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J Biol Chem. 2006;281(1):392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- 6.Lev A, et al. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc Natl Acad Sci USA. 2010;107(15):6964–6969. doi: 10.1073/pnas.0910997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starck SR, Shastri N. Non-conventional sources of peptides presented by MHC class I. Cell Mol Life Sci. 2011;68(9):1471–1479. doi: 10.1007/s00018-011-0655-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: Support, controversy, refinement and extension. Trends Immunol. 2006;27(8):368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Apcher S, et al. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci USA. 2011;108(28):11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt A. Nonsense Mediated Decay Associated Pioneer Round of Translation as Source for Peptides for Presentation by MHC Class I. Koln, Germany: PhD dissertation Univ of Koln; 2009. [Google Scholar]
- 11.Iborra FJ, Jackson DA, Cook PR. Coupled transcription and translation within nuclei of mammalian cells. Science. 2001;293(5532):1139–1142. doi: 10.1126/science.1061216. [DOI] [PubMed] [Google Scholar]
- 12.David A, et al. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol. 2012;197(1):45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Jubran K, et al. Visualization of the joining of ribosomal subunits reveals the presence of 80S ribosomes in the nucleus. RNA. 2013 doi: 10.1261/rna.038356.113. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147(4):789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aviner R, Geiger T, Elroy-Stein O. Novel proteomic approach (PUNCH-P) reveals cell cycle-specific fluctuations in mRNA translation. Genes Dev. 2013;27(16):1834–1844. doi: 10.1101/gad.219105.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolan BP, Knowlton JJ, David A, Bennink JR, Yewdell JW. RNA polymerase II inhibitors dissociate antigenic peptide generation from normal viral protein synthesis: A role for nuclear translation in defective ribosomal product synthesis? J Immunol. 2010;185(11):6728–6733. doi: 10.4049/jimmunol.1002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci USA. 2002;99(19):12031–12036. doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462(7272):522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stapulionis R, Deutscher MP. A channeled tRNA cycle during mammalian protein synthesis. Proc Natl Acad Sci USA. 1995;92(16):7158–7161. doi: 10.1073/pnas.92.16.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, et al. Endogenous viral antigen processing generates peptide-specific MHC class I cell-surface clusters. Proc Natl Acad Sci USA. 2012;109(38):15407–15412. doi: 10.1073/pnas.1208696109. [DOI] [PMC free article] [PubMed] [Google Scholar]