Significance
Cuticular wax on plant leaves plays an important role in preventing water loss from drought stress. However, the genetic control of wax deposition under drought stress conditions has not been characterized. In this paper, a very large Drought-Induced Wax Accumulation 1 (DWA1) protein (2,391 aa in length) controlling the drought-induced wax load on leaves was characterized in rice, one of the most important crops. The DWA1 protein contains a multienzyme-like structure, including a prokaryotic nonribosomal peptide synthetase (NRPS)-like module and is conserved in vascular plants. Interestingly, the NRPS-like module with an AMP-binding domain can activate long-chain fatty acids for wax biosynthesis. This finding may have significant implications for improving the drought resistance of crop varieties through modifying wax biosynthesis.
Keywords: abiotic stress, NRPS, Oryza sativa
Abstract
Drought stress is a major limiting factor for crop production. Cuticular wax plays an important role in preventing water loss from drought stress. However, the genetic control of cuticular wax deposition under drought stress conditions has not been characterized. Here, we identified a rice gene Drought-Induced Wax Accumulation 1 (DWA1) encoding a very large protein (2,391 aa in length) containing multiple enzymatic structures, including an oxidoreductase-like domain; a prokaryotic nonribosomal peptide synthetase-like module, including an AMP-binding domain; and an allene oxide synthase-like domain. This previously unreported putative megaenzyme is conserved in vascular plants. A dwa1 KO mutant was highly sensitive to drought stress relative to the WT. DWA1 was preferentially expressed in vascular tissues and epidermal layers and strongly induced by drought stress. The dwa1 mutant was impaired in cuticular wax accumulation under drought stress, which significantly altered the cuticular wax composition of the plant, resulting in increased drought sensitivity. The mutant had reduced levels of very-long-chain fatty acids, and plants overexpressing DWA1 showed elevated levels of very-long-chain fatty acids relative to the WT. The expression of many wax-related genes was significantly suppressed in dwa1 under drought conditions. The AMP-binding domain exhibited in vitro enzymatic activity in activating long-chain fatty acids to form acyl-CoA. Our results suggest that DWA1 controls drought resistance by regulating drought-induced cuticular wax deposition in rice. This finding may have significant implications for improving the drought resistance of crop varieties.
Environmental stresses, such as drought, negatively affect plant growth and production. Plants have evolved diverse strategies to cope with these environmental stresses. Among these strategies, cuticular wax, the outermost hydrophobic layer covering the aerial parts of land plants, provides an essential barrier to limit nonstomatal water loss during periodic drying and drought stress, and it also plays important roles in protecting plants from other abiotic and biotic stresses (1, 2).
Cuticular wax is a complex mixture consisting mainly of very-long-chain fatty acids (VLCFAs) and derivatives, such as alkanes, alcohols, aldehydes, ketones, and wax esters (3–5). Progress has been made in understanding the major steps of wax biosynthesis for plant growth and development. Most of the biosynthetic steps were revealed by identifying wax-related genes of the eceriferum (cer) and glossy (gl) mutants in Arabidopsis and maize, respectively (3, 4, 6). These genes encode diverse proteins, including biosynthetic enzymes (CER6/CUT1, CER8/LACS1, and GL8) (7–9), wax transporters (CER5/ABCG12) (10), and transcription factors (e.g., WAX INDUCER1/SHINE1 [WIN1/SHN1], MYB30, MYB96) (11–13). To date, only a few genes involved in cuticular wax biosynthesis have been identified in rice. A mutant of wax-deficient anther1 (Wda1), a homolog of maize GL1 in rice, displayed reduced wax components and was defective in the biosynthesis of VLCFA in the anther wall (14). A wax deposition defect was also found in the leaf cuticle of the GL1-2 and GL1-1/WSL2 mutants (15, 16). Additionally, a mutation in a putative ketoacyl CoA synthase [wax crystal-sparse leaf1 (WSL1)] resulted in reduced wax load and crystals on mutant leaves (17).
Drought stress has a significant effect on cuticular wax biosynthesis and composition in plants (18, 19). However, few genes linking cuticular wax accumulation with drought stress have been reported. Overexpression of WXP1 and CER1 resulted in enhanced wax production and drought adaptability with decreased water loss and cuticle permeability (20, 21). Overexpression of SHN1/WIN1 caused elevated wax deposition and drought tolerance but unexpectedly increased cuticle permeability and water loss (22). Modulating the expression of rice SHN1/WIN1 homolog wax synthesis regulator 1 (OsWR1) altered wax production and drought tolerance (23). It is unknown how cuticular wax biosynthesis is regulated under drought stress; however, one recent report has shown that MYB96 regulates some wax-biosynthetic genes in Arabidopsis, suggesting it may be required for drought-induced cuticular wax biosynthesis (13).
Rice is one of the most important staple crops and a model plant for crop research. Mainly due to the increasing shortage of water resources and unpredictable climate changes, drought has become the major limiting factor for rice growth and yield stability (24, 25). Developing drought-resistant varieties has been one of the major goals in many breeding programs. Here, we identified a gene, DWA1, that plays an essential role in drought resistance in rice. The dwa1 mutant is defective in drought-induced wax accumulation and showed dramatic alteration in cuticular wax composition under drought stress conditions, which resulted in increased drought sensitivity. This gene encodes a multidomain protein that contains a prokaryotic nonribosomal peptide synthetase (NRPS)-like module and is conserved in vascular plants.
Results
Identification of a Drought-Sensitive Mutant dwa1.
To identify critical genes for drought resistance in rice, we screened transfer DNA (T-DNA) insertion mutants of japonica rice Zhonghua11 [collected from the Rice Mutant Database (26)] in the drought-prone field. One of the mutants, designated as dwa1, showed a highly drought-sensitive phenotype. The isolated flanking sequence analysis revealed an insertion of T-DNA in the sixth exon of a predicted gene (LOC_Os04g39780) in the rice genome (Fig. 1A). Transcription of this gene was verified using RNA samples from both homozygous mutant and WT plants. The DWA1 transcript was not detected in the mutant (Fig. 1B and Fig. S1B), indicating that dwa1 is a loss-of-function mutant.
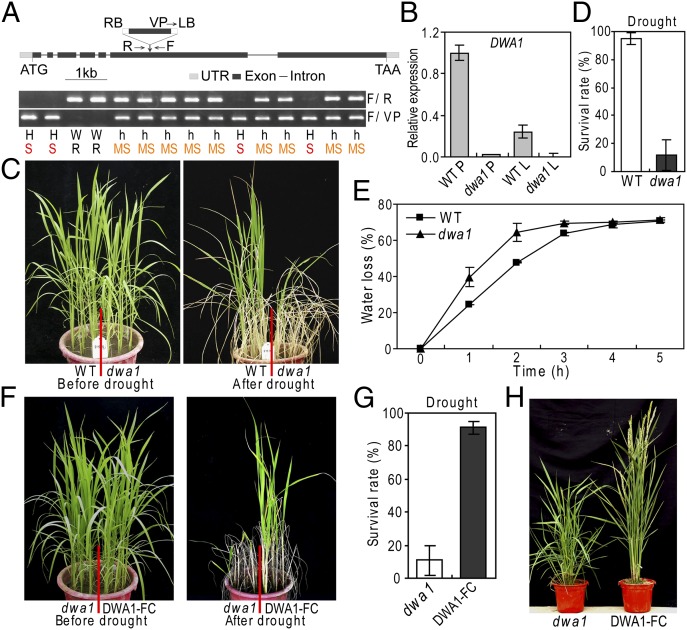
Fig. 1.
Identification of the drought-hypersensitive dwa1 mutant in rice, in which functional complementation of dwa1 restored the drought resistance. (A) Schematic diagram of DWA1 gene structure with the T-DNA insertion and genotyping of the mutant. h, heterozygote; H, homozygote; LB, left border; F and R, forward and reverse genomic primers; MS, moderate sensitivity; R, resistant to drought; RB, right border; S, sensitive to drought; VP, vector primer. (B) RT-qPCR expression analysis of DWA1 in WT and mutant tissues. L, leaf tissue; P, panicle. (C) Increased drought sensitivity of the dwa1 mutant at the seedling stage. (D) Survival rate comparisons of WT and mutant plants after drought stress. (E) Water loss rate of WT and mutant plants. (F) DWA1-FC functional complementation plants restored drought resistance. (G) Survival rate of DWA1-FC and dwa1 plants after drought stress treatment. (H) Recovery of DWA1-FC plants from reduced plant height of dwa1 at the reproductive stage.
The dwa1 mutant is the result of a single-copy T-DNA insertion (Fig. S1A). Under normal growth conditions, the homozygous dwa1 plants showed no phenotypic differences compared with the WT plants at vegetative stages. To confirm the drought-sensitive phenotype identified during the screening, dwa1 and WT seedlings were subjected to drought stress. During the course of drought treatment, the mutant wilted and exhibited leaf rolling earlier than the WT. After severe drought treatment followed by rewatering, more than 90% of the WT plants recovered, whereas the mutant plants exhibited a very low survival rate (almost all the plants were dead) (Fig. 1 C and D). The drought-sensitive phenotype cosegregated with the T-DNA insertion in the DWA1 gene (Fig. 1A).
In agreement with the leaf rolling phenotype, the leaf relative water content in the dwa1 mutant dropped significantly faster than in WT during the drought stress (Fig. 1E and Fig. S1C). An electrolyte leakage assay, a commonly used method for evaluating membrane damage, showed that the membrane system of dwa1, with a relative conductivity of 98.3%, was almost completely destroyed in the rolled leaves exposed to drought stress, whereas the conductivity of the WT was only 43.1% under the same stress conditions (Fig. S1D). The drought sensitivity of the dwa1 mutant was also tested at the reproductive stage in a paddy field and in PVC pots. After drought treatment, the mutant exhibited a significantly lower relative biomass and spikelet fertility than the WT (Fig. S1 E and F).
The phenotypic defects of the dwa1 mutant were further confirmed by a functional complementation test. The full-length genomic fragment of DWA1 was introduced into the dwa1 mutant. When subjected to drought stress conditions at the seedling stage, the drought-resistance phenotype was restored in the dwa1-complemented lines (designated as DWA1-functional complementation [FC]) (Fig. 1 F and G). Plant height at the reproductive stage (Fig. 1H) was also restored in the DWA1-FC lines. The results confirmed that the drought hypersensitivity of this mutant was caused by the DWA1 mutation.
DWA1 Encodes an Enzymatic Megaprotein Conserved in Vascular Plants.
The genomic sequence of DWA1 consists of seven exons (Fig. 1A). The open reading fame (7,176 bp) encodes a protein consisting of 2,391 aa in japonica rice, Nipponbare, which is one of the 30 megaproteins (>2,300 aa) predicted in the rice genome. A sequence search of DWA1 against the Rice Genome Annotation Database (http://rice.plantbiology.msu.edu/) found no other homologs in the rice genome. Because the full-length cDNA sequence is not available in the database, we confirmed the exon/intron structure of DWA1 by six RT-PCRs, with each primer pair located at two adjacent exons (Fig. S2 A–C), followed by sequencing. A Protein BLAST search against the National Center for Biotechnology Information protein database revealed that one homolog of DWA1 exists in most higher plant species and even in Selaginella moellendorffii, an ancient vascular plant (Table S1). The sequence identity of the homologs to DWA1 ranges from 42% (Selaginella) to >80% (Poaceae) (Fig. 2).
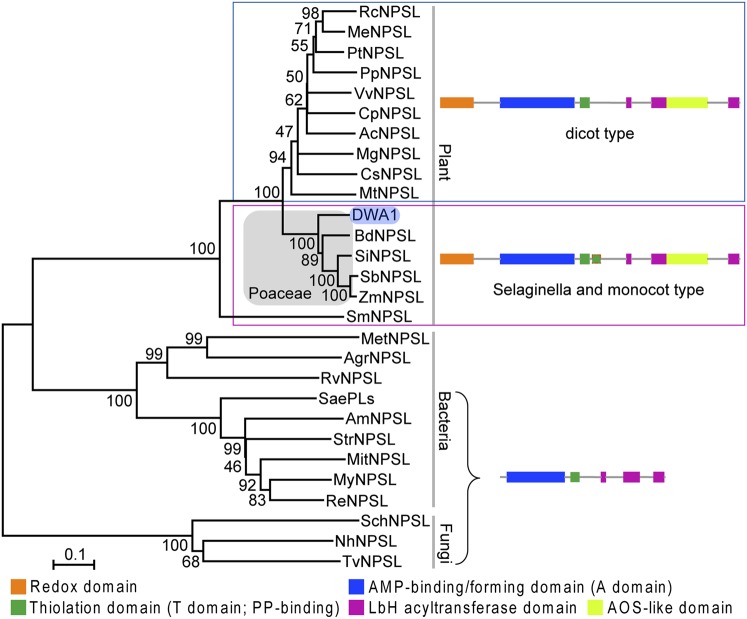
Fig. 2.
Unrooted phylogenetic tree and domain architecture of DWA1 protein orthologs in vascular plants and microbial relatives. Amino acid sequences of DWA1 orthologous proteins were analyzed using the neighbor-joining method with genetic distance calculated by MEGA4 www.megasoftware.net/. The numbers at the nodes represent percentage bootstrap values based on 1,000 replications. The length of the branches is proportional to the expected number of amino acid substitutions per site, with a scale provided at the bottom of the tree. Plant and microbial protein sequences are shown in Table S1. NPSL, NRPS-like protein; PP, phosphopantetheine.
The conserved domain analysis suggested that DWA1 may be a megaenzyme harboring at least five domains (27) (Fig. 2). Interestingly, each domain almost matches with a single enzyme-like structure. The N terminus is an oxidoreductase-like domain, which is followed by a core AMP-binding domain (A domain). The A domain shares significant homology with acyl-CoA synthases, and the AMP-binding domain enzymes consist of an AMP-binding protein (AMPBP) superfamily (28). Two repeats of a phosphopantetheine-binding subdomain follow the A domain and feature a thiolation domain (T domain). An allene oxide synthase (AOS)-like domain, showing high similarity to catalase, is located between the second and third repeats of the left-handed β-helix (LbH) domain at the C-terminal region, which may possess acyltransferase activity. Based on transmembrane prediction using hidden Markov models analysis, DWA1 features an integral membrane protein with eight transmembrane helices located around the LbH and AOS domains.
Possible relatives of DWA1 in nonplant organisms were also investigated. About 120 homologous sequences were retrieved. Interestingly, all of them were from microorganisms. They have very similar domain architectures to DWA1, except for the lack of the oxidoreductase-like and AOS-like domains (Fig. 2). All of them are annotated as putative NRPSs, which is a major branch of the AMP-binding enzyme superfamily. An NRPS is a multifunctional megaenzyme found in microorganisms directing nonribosomal peptide synthesis through a multienzyme thiotemplate mechanism (29). DWA1 and its plant homologs possess almost all the characteristics of an NRPS module, including a typical adenylation domain, followed by a thiolation domain catalyzing the activation and thiolation reaction of an amino acid instead of a carboxyl substrate for acyl-CoA synthetase (Fig. 2). These features suggest that DWA1 and its plant homologs may be derived from a prokaryotic NRPS rather than the acyl-CoA synthetase family, which possesses only the A domain.
Tempospatial and Stress-Responsive Expression of DWA1.
To unveil the function of DWA1, we first investigated the expression pattern of DWA1 by quantitative PCR (qPCR). The results showed that the DWA1 transcript was almost undetectable at the vegetative stages but was relatively high at the reproductive stages, especially in inflorescence tissues (Fig. S3A). The expression data of this gene obtained from microarrays (30) supported this pattern. DWA1 expression was further examined using a GUS reporter gene driven by a DWA1 promoter. Consistent with the qPCR result, the GUS signal was weak in most vegetative tissues. Relatively high levels of GUS expression were detected in the immature organs, such as young leaves and developing panicles at the reproductive stage. In the mature organs, GUS expression was detected mainly in specific regions, such as leaf veins, nerves of palea and lemma, the joint of stamen filament and anther, the pistil style, the base of the ovule, and pedicels of spikelets (Fig. S3B). Closer observation by SEM showed that the GUS staining in leaves was mainly distributed along the silica-cork cell lines in the cuticle (Fig. S3B). Cross-sections and semithin sections indicate that DWA1 is expressed mainly in vascular tissues and epidermal cell layers (Fig. S3B).
The increased drought sensitivity of dwa1 promoted us to examine the expression of DWA1 under abiotic stresses. DWA1 was dramatically up-regulated (23.6-fold) after 24 h of exposure to drought stress conditions and was slightly induced by cold stress (Fig. 3A and Fig. S3C). In addition, DWA1 expression was induced (2.3–4.9 fold) by treatment with various phytohormones, including abscisic acid, jasmonic acid, indole-3-acetic acid, and gibberellic acid (Fig. S3C). Promoter GUS staining also showed that DWA1 was induced under drought treatment (Fig. 3B). Many stress-related cis-elements are enriched in the DWA1 promoter (Table S2), which may be responsible for the stress-induced expression of DWA1.
Fig. 3.
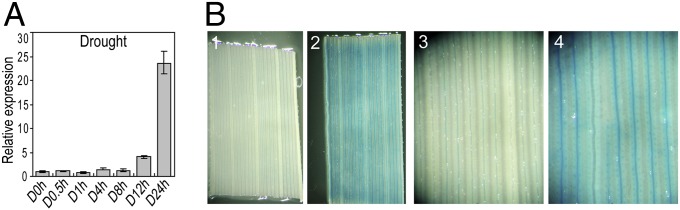
Drought-induced expression of DWA1. (A) Expression level of DWA1 under drought. (B) DWA1 promoter::GUS fusion expression response to drought in transgenic seedlings. GUS staining in leaf before (1 and 3) and after (2 and 4) drought stress is shown.
Defective Leaf Cuticle and Wax Deposition in dwa1 Under Drought Stress.
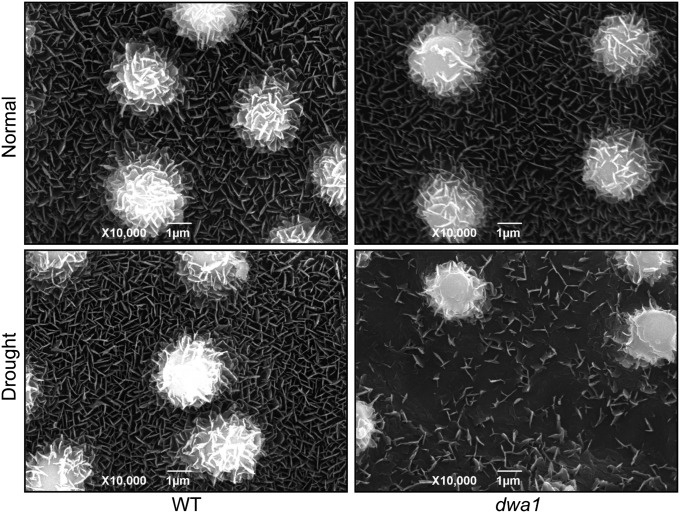
Even though the dwa1 mutant lost water at a faster rate than the WT under drought conditions, stomatal density and drought-triggered stomatal closure, two frequently documented factors related to water loss and drought sensitivity, were not significantly different in the dwa1 and the WT (Fig. S4A). DWA1 contains an AMP-binding domain that is commonly found in long-chain acyl-CoA synthetases (LACSs) involved in the biosynthesis of wax precursors (8), which prompted us to examine the cuticular wax of dwa1, because the wax layer on a leaf is another factor affecting water loss during exposure to drought stress. SEM observations suggested that the epicuticle properties had no difference under normal growth conditions. However, the epicuticular wax accumulation in the dwa1 mutant was strikingly impaired after prolonged drought treatment. The deposition of vertical plate-like wax crystals on the dwa1 leaf cuticle was severely reduced, and no wax crystals were observed in some areas, in contrast to the heavy deposition of wax crystals on the WT leaf surface (Fig. 4 and Fig. S4A). The reduced epicuticular wax deposition on the dwa1 leaves under drought stress may result in the increased water loss and hypersensitivity of the mutant to the stress. A chlorophyll leaching assay showed that the chlorophyll in the drought-stressed dwa1 leaves was leached much faster than in the WT leaves (Fig. S4B), suggesting that an increase in the permeability of the mutant leaf surface resulted from the reduced wax deposition.
Fig. 4.
SEM images of cuticle properties and wax crystals on leaf surface of dwa1 and WT seedlings under normal and drought conditions.
Wax Composition and Cellular Fatty Acids Were Altered in dwa1 Under Drought Stress.
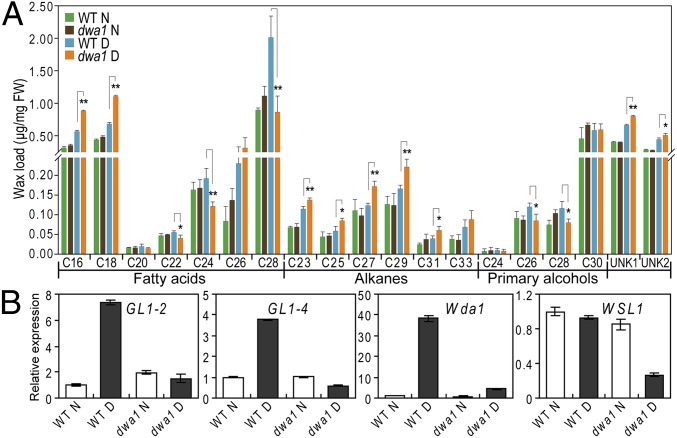
The leaf cuticular wax was quantified by GC/MS. Under normal growth conditions, no obvious difference was observed between the mutant and WT plants. Under drought stress conditions, the total wax load was reduced by 7.8% in the dwa1 mutant compared with that in the WT. The wax constituents were significantly changed in the mutant compared with the WT (Fig. S4C). The content of total fatty acids (FAs), including long-chain fatty acids (LCFAs) and VLCFAs, was about 11% less in the drought-induced dwa1 mutant than in the WT, whereas the total alkane content was 34% higher in the dwa1 mutant than in the WT under drought stress conditions. Obvious differences were also detected for the contents of individual FA components (Fig. 5A). Among them, VLCFAs, such as C20:0, C22:0, C24:0, and C28:0, were significantly reduced by 1.3- to 2.3-fold in the dwa1 mutant. On the contrary, the levels of LCFAs (e.g., C16:0, C18:0) were slightly higher (about 1.6-fold) in the dwa1 mutant (Fig. 5A). Almost all the alkane components (C21–C33) showed significantly increased levels in dwa1 (1.2- to 1.5-fold relative to the WT) after drought treatment (Fig. 5A). These results suggest that KO of DWA1 may result in a significant block in the biosynthesis of VLCFA and wax deposition on the leaf surface under drought stress and that the carbon flux may be shifted preferentially to the decarbonylation pathway for production of alkane constituents.
Fig. 5.
Cuticular wax composition analysis and the expression levels of wax-related rice genes in the dwa1 mutant and WT leaves under normal and drought conditions. (A) Cuticular wax composition and loads in the dwa1 mutant and WT leaves under both conditions. Error bars indicate SE of three biological repeats (t test: *P < 0.05; **P < 0.01). D, drought stress; N, normal growth; UNK, unknown metabolite. (B) Expression levels of wax-related genes in dwa1 and WT plants under drought stress.
Quantification of endogenous FAs revealed significant differences in the contents of C18:0 and C18:3 between the two genotypes under drought conditions (Fig. S4D). The content of C18:0 was reduced in the WT, but C18:0 was significantly higher in the dwa1 mutant. The content of C18:3 was the highest among the endogenous FAs, and its increase under drought stress was 38.3% higher in the dwa1 than in the WT. These differences suggest that KO of DWA1 may also disrupt the balance of endogenous FAs, some of which are precursors for epicuticular wax biosynthesis under drought conditions.
The DWA1-FC lines with the genotypes confirmed in Fig. S5A were also examined for wax deposition. SEM revealed that the vertical plate-like wax crystals on the leaf cuticle, which were diminished in the drought-stressed dwa1 mutant, were restored in the DWA1-FC lines exposed to drought stress (Fig. S5B). Wax composition analysis showed little difference between DWA1-FC and dwa1 grown under normal growth conditions. Under drought conditions, the block in VLCFA accumulation was alleviated in the DWA1-FC lines and the production of individual VLCFA (C20:0–C24:0, C28:0) was enhanced (Fig. S5C). Meanwhile, LCFAs (C16:0 and C18:0), which overaccumulated in the mutant, were significantly reduced in the DWA1-FC lines under drought conditions (Fig. S5C). These results further confirmed DWA1 as the gene responsible for the drought-induced wax deposition.
Expression of Wax-Related Genes Was Altered in dwa1 Under Drought Stress.
To understand the changes occurring in the cuticle wax of the dwa1 mutant better, we examined the expression levels of six cuticle and wax-related genes reported in rice, including GL1-2, GL1-4 (15), Wda1 (14), wilted dwarf and lethal 1 (WDL1) (31), WSL1 (17), and WR1 (23), as well as two LACS genes (LACS7, LOC_Os12g04990; LACS8, LOC_Os05g25310) (Table S3). The expression levels of these genes were not significantly different between the dwa1 and the WT under normal growth conditions; however, the gene expression levels were obviously different under drought treatment (Fig. 5B and Fig. S4E). Wda1 expression was strongly induced (38.4-fold) by drought stress in the WT, but it was only slightly induced in the mutant. GL1-2, GL1-4, and WR1 were also significantly induced by drought (3.4- to 7.4-fold) in the WT, whereas their expression levels decreased in the mutant under drought stress. Similar differences were observed for LACS7 and LACS8. Although the expression of WDL1 and WSL1 was not significantly affected by drought in the WT, their expression levels were significantly suppressed by drought in the mutant (only about 30% of that seen in the WT). The results suggest that loss of function of DWA1 affected the expression of important genes involved in cuticular wax accumulation under drought stress conditions.
The expression of the wax-related genes was also examined in the DWA1-FC lines (Fig. S5D). The expression of GL1-2, GL1-4, and Wda1, whose induction was largely suppressed in dwa1, was strongly induced by drought stress in the DWA1-FC lines. The expression levels of WDL1, WSL1, WR1, LACS7, and LACS8 were also restored in the DWA1-FC lines exposed to drought stress. These results further confirmed that KO of DWA1 resulted in the defect in drought-induced cuticular wax deposition.
Overexpression of DWA1 Enhanced VLCFA Production.
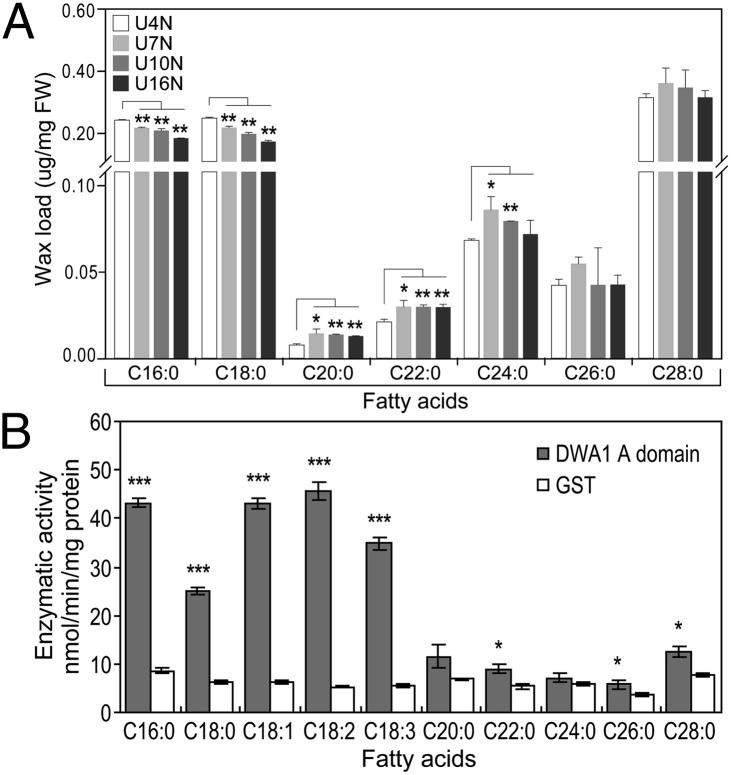
We further confirmed the role of DWA1 in epicuticular wax accumulation by examining transgenic rice overexpressing DWA1. Three independent overexpression plants and a negative control were selected for this analysis (Fig. S6A). Although the overexpression plants showed no visual differences in the epicuticular wax load compared with the control (Fig. S6B), the VLCFA constituents exhibited a significant increase in the overexpression lines compared with those in the WT (Fig. 6A). However, the contents of LCFAs (C16:0 and C18:0) were significantly reduced (by 10–30% relative to the control) in the overexpression plants (Fig. 6A). The alkanes and primary alcohols showed no significant changes (Fig. S6C). These results suggest that constitutive overexpression of DWA1 mainly affected LCFA and VLCFA production for cuticular wax. Under normal growth conditions, the expression levels of GL1-2, GL1-4, WR1, LACS8, and WSL1 were enhanced in the overexpression plants (Fig. S6D). Under drought stress conditions, however, only Wda1 showed a higher level of expression in the overexpression lines than in the control. Nevertheless, no obvious differences in drought resistance were observed between the overexpression and WT plants.
Fig. 6.
Changes in the FAs in the DWA1-overexpression rice and enzymatic activity of the DWA1 A domain. (A) Enhanced VLCFA and decreased LCFA constituents of leaf cuticular wax in DWA1-overexpression plants under normal conditions . U7, U10, and U16 are derived from three independent DWA1-overexpression plants. U4 is a transgenic negative control. All values are relative to fresh weight and represent the average of three independent experiments. Bars indicate SE of the mean (t test: *P < 0.05; **P < 0.01). FW, fresh weight. (B) Enzymatic activity of the DWA1 A domain in activating FAs to form acyl-CoA determined by a colorimetric method. Data are expressed as the mean ± SE with three replicates (t test: ***P < 0.001).
The A Domain of DWA1 Had Enzymatic Activity in Activating LCFAs.
The altered LCFA levels in the dwa1 mutant and DWA1-overexpression plants prompted us to examine if the A domain of DWA1 has enzymatic activity, because this domain has sequence similarity to LACS. The A domain, fused with a GST tag, was expressed in Escherichia coli, and enzymatic activity of the purified A domain protein was analyzed by an enzyme-coupled colorimetric method (32) using saturated free FAs with chain lengths from C16 to C28 and three unsaturated C18 FAs (C18:1/C18:2/C18:3) as substrates. As shown in Fig. 6B, the purified A domain of DWA1 exhibited high enzymatic activity for LCFAs (C16:0/C18:0) and the three unsaturated C18 FAs. The A domain showed slight enzymatic activity for VLCFAs (C20:0–C28:0) compared with the GST protein alone (negative control). These results are generally consistent with the changes of LCFAs and VLCFAs in the dwa1 mutant and DWA1-overexpression plants.
Discussion
DWA1 Encodes a Megaenzyme-Like Protein Highly Conserved in Vascular Plants.
Our results suggest that DWA1 plays vital roles in drought resistance and epicuticular wax deposition in rice. Compared with the reported genes for such traits in plants, DWA1 is a previously unknown gene, not only because it has never been described but because it encodes such a large protein with a complex composition of multiple domains, including an NRPS-like module that is likely derived from prokaryotic species. Among the multiple domains, the A (AMP-binding) domain shared by DWA1 and its homologs from other plants has a high sequence identity to LACSs (Fig. S7B), typical members of the AMPBP superfamily (28). LACSs function in activating free FAs with various chain lengths into acyl-CoAs as precursors for wax production and lipid metabolism. The recombinant protein of the AMP-binding domain of DWA1 can directly activate LCFAs similar to LACSs (Fig. 6B). However, the A domain and the T domain comprise the characteristic architecture of a monomodular NRPS, which also belongs to the AMPBP superfamily (29). Alignment of DWA1 with NRPS proteins showed that DWA1 possesses all the typical domains of an NRPS (Fig. 2). To date, almost all NRPS proteins that have been identified are from microorganisms, and some of them have been biochemically characterized with enzymatic activity using amino acids and their derivatives as substrates (33). However, no homolog of an NRPS has been documented in plants. The above evidence implies that DWA1 may have derived from a prokaryotic NRPS, and the actual origin and evolution of the DWA1 lineage in plants are very interesting issues for further study.
Homologous proteins with the same domain architecture as DWA1 ubiquitously exist in green vascular plants (Fig. 2). In the Phytozome database, DWA1 homologs were found in 16 genome-sequenced land plant species, ranging from the ancient Selaginella to angiosperm plants. Selaginella appears to be a primitive lycophyte, and the evidence suggests that the lycophyte lineage appeared shortly after land plants evolved vascular tissues (34). DWA1 homologs were not found in genome-sequenced nonvascular plants, such as moss (Physcomitrella patens) and green algaes (Chlamydomonas reinhardtii and Volvox carteri). This may suggest that the DWA1 ancestor appeared after the divergence of vascular plants from the nonvascular bryophyte lineage. Considering that DWA1 was strongly expressed in vascular tissues, the DWA1 ancestor may be related to the developmental evolution of vascular plants.
Role of DWA1 in Wax Accumulation Under Drought Stress.
Most plants have evolved the ability to increase cuticular wax production upon exposure to drought stress. The inducible wax accumulation is primarily due to an increase in the level of alkanes, which are the most abundant components of the cuticular wax in Arabidopsis. It has been proposed that cuticular alkanes confer greater resistance to water diffusion than VLCFAs (13, 19). However, total FAs are the major constituents of cuticular wax in rice (16, 23), and we obtained similar results in this study. Under drought stress, total FAs, including VLCFAs and LCFAs, were the major components contributing to the increase in the level of wax seen in the WT rice (Fig. 5A). In the dwa1 mutant, the total contents of VLCFAs were significantly reduced, but the contents of alkanes increased under exposure to drought stress. These results suggest that VLCFAs may play a more important role than alkanes in the drought adaptation of rice.
During the course of drought stress, wax crystal deposition on the leaf cuticle was abolished in the dwa1 mutant. Chemical analyses suggest that the defect in the epicuticular wax in the dwa1 mutant may have resulted from the alteration of wax composition rather than the reduction of wax production. KO of DWA1 resulted in a shift in the carbon flux for wax production from VLCFAs to alkanes. The mutation also affected the accumulation of LCFAs. In agreement with the change in wax components, the expression levels of some wax biosynthesis-related genes were also dramatically altered in the drought-stressed dwa1 mutant. These genes were up-regulated by drought stress, indicating their positive roles in drought-induced wax biosynthesis and deposition. The induction of three GL1-like genes (GL1-2, GL1-4, and Wda1), encoding the components of a wax biosynthetic complex, was largely suppressed in dwa1 during drought stress. Down-regulation of GL1-2 resulted in a reduction of wax crystals on the rice leaf cuticle (15). Wda1 has been proposed to function in the general process of VLCFA biosynthesis, and it is involved in an anther-specific FA elongation complex (14). WSL1, encoding a rate-limiting enzyme in the FA elongation multienzyme complex for the synthesis of VLCFAs (C20:0–C34:0) from LCFAs (C16:0/C18:0), was significantly suppressed in the dwa1 mutant. Loss of function of WSL1 led to reduced wax load, sparse wax crystals on the leaf cuticle, and a decrease in the levels of VLCFA precursors (17). The drought induction of LACS7 and LACS8, encoding LACSs for activation of free FAs with different chain lengths, was also suppressed in dwa1. The expression changes of these wax biosynthesis-related genes can largely explain the drought-induced alteration in wax composition, especially the LCFA overaccumulation and VLCFA reduction in dwa1 exposed to drought stress conditions. OsWR1, encoding a transcription factor involved in the regulation of wax production (23), was significantly suppressed in drought-stressed dwa1 plants. In addition, some of these genes (e.g., OsWR1, GL1-2, Wda1) showed significantly enhanced expression in the DWA1-overexpression lines under normal and/or drought stress conditions, and complementation of DWA1 successfully recovered the drought-induced expression of these genes in the dwa1 background. The recombinant protein of the DWA1 A domain exhibited strong activity in activating LCFAs but weak or no activity on VLCFAs. This was consistent with the overaccumulation of LCFAs and decreased VLCFAs for wax production in the dwa1 mutant compared with the WT under drought stress conditions. VLCFAs can serve as both the wax precursors for producing other wax constituents and the final products of epicuticular wax composition. Our results suggest that DWA1 may act as an essential enzyme in the activation of VLCFA biosynthesis for epicuticular wax production under drought stress conditions. Meanwhile, DWA1 may be involved in the control of the partition of FAs into different biosynthetic pathways, which remains largely unclear in plants (5). In addition, disruption of wax deposition by DWA1 loss of function may have feedback on the expression of wax biosynthesis-related genes.
In conclusion, DWA1 is a previously unreported megaenzyme conserved in green vascular plants and plays an essential role in drought-induced epicuticular wax deposition and drought resistance in rice.
Methods
General methods are presented here. Detailed information on methods is provided in SI Methods.
Plant Growth and Drought Treatments.
For phenotypic observation at the seedling stage, seeds of rice materials in this study were germinated and grown in sandy soil for 4 wk. The seedlings were then treated with drought stress (withholding water for 7 d), followed by recovery. For drought testing at the reproductive stage in PVC pots and in fields, rice plants at the booting stage were exposed to moderate or severe drought stress by following the protocol of Yue et al. (35) with minor modifications (SI Methods).
FA and Epicuticular Wax Quantification.
FA extraction was performed as described by Xia et al. (36). Wax extraction was conducted according to Lü et al. (8) with minor modifications. Quantification of FA and wax content was performed as described by Lü et al. (8) with some modifications (more details are provided in SI Methods).
Supplementary Material
Acknowledgments
We thank Z. L. Guo and K. Songyikhangsuthor for assistance in sampling, H. Zhang and L. K. Wang for help in heterologous expression and semithin sectioning, H. Du for providing phytohormone-treated RNA samples, R. J. Ye for kindly providing the pDX2181 vector, and F. C. Li and H. B. Liu for help in GC/MS analysis. This work was supported by the National Program on High Technology Development (Grant 2012AA10A303) and the National Program for Basic Research of China (Grant 2012CB114305).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316412110/-/DCSupplemental.
References
- 1.Jenks MA, Tuttle HA, Eigenbrode SD, Feldmann KA. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol. 1995;108(1):369–377. doi: 10.1104/pp.108.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xia Y, Nikolau BJ, Schnable PS. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell. 1996;8(8):1291–1304. doi: 10.1105/tpc.8.8.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Post-Beittenmiller D. Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47(1):405–430. doi: 10.1146/annurev.arplant.47.1.405. [DOI] [PubMed] [Google Scholar]
- 4.Kunst L, Samuels AL. Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res. 2003;42(1):51–80. doi: 10.1016/s0163-7827(02)00045-0. [DOI] [PubMed] [Google Scholar]
- 5.Samuels L, Kunst L, Jetter R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu Rev Plant Biol. 2008;59(1):683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- 6.Kunst L, Samuels L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr Opin Plant Biol. 2009;12(6):721–727. doi: 10.1016/j.pbi.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Fiebig A, et al. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell. 2000;12(10):2001–2008. doi: 10.1105/tpc.12.10.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lü S, et al. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009;59(4):553–564. doi: 10.1111/j.1365-313X.2009.03892.x. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich CR, et al. Characterization of two GL8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J. 2005;42(6):844–861. doi: 10.1111/j.1365-313X.2005.02418.x. [DOI] [PubMed] [Google Scholar]
- 10.Pighin JA, et al. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306(5696):702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- 11.Broun P, Poindexter P, Osborne E, Jiang C-Z, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA. 2004;101(13):4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raffaele S, et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008;20(3):752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seo PJ, et al. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell. 2011;23(3):1138–1152. doi: 10.1105/tpc.111.083485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung KH, et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell. 2006;18(11):3015–3032. doi: 10.1105/tpc.106.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam MA, Du H, Ning J, Ye H, Xiong L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol. 2009;70(4):443–456. doi: 10.1007/s11103-009-9483-0. [DOI] [PubMed] [Google Scholar]
- 16.Mao B, et al. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta. 2012;235(1):39–52. doi: 10.1007/s00425-011-1481-1. [DOI] [PubMed] [Google Scholar]
- 17.Yu D, et al. Wax Crystal-Sparse Leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta. 2008;228(4):675–685. doi: 10.1007/s00425-008-0770-9. [DOI] [PubMed] [Google Scholar]
- 18.Shepherd T, Wynne Griffiths D. The effects of stress on plant cuticular waxes. New Phytol. 2006;171(3):469–499. doi: 10.1111/j.1469-8137.2006.01826.x. [DOI] [PubMed] [Google Scholar]
- 19.Kosma DK, et al. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009;151(4):1918–1929. doi: 10.1104/pp.109.141911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J-Y, et al. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa) Plant J. 2005;42(5):689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- 21.Bourdenx B, et al. Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol. 2011;156(1):29–45. doi: 10.1104/pp.111.172320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aharoni A, et al. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16(9):2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol Biol. 2012;78(3):275–288. doi: 10.1007/s11103-011-9861-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q. Strategies for developing Green Super Rice. Proc Natl Acad Sci USA. 2007;104(42):16402–16409. doi: 10.1073/pnas.0708013104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukao T, Xiong L. Genetic mechanisms conferring adaptation to submergence and drought in rice: Simple or complex? Curr Opin Plant Biol. 2013;16(2):196–204. doi: 10.1016/j.pbi.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35(3):418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 27.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39(Database):D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shockey JM, Fulda MS, Browse JA. Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 2002;129(4):1710–1722. doi: 10.1104/pp.003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97(7):2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61(5):752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 31.Park J-J, et al. Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol Biol. 2010;74(1-2):91–103. doi: 10.1007/s11103-010-9656-x. [DOI] [PubMed] [Google Scholar]
- 32.Ichihara K, Shibasaki Y. An enzyme-coupled assay for acyl-CoA synthetase. J Lipid Res. 1991;32(10):1709–1712. [PubMed] [Google Scholar]
- 33.Yamanaka K, Maruyama C, Takagi H, Hamano Y. Epsilon-poly-L-lysine dispersity is controlled by a highly unusual nonribosomal peptide synthetase. Nat Chem Biol. 2008;4(12):766–772. doi: 10.1038/nchembio.125. [DOI] [PubMed] [Google Scholar]
- 34.Banks JA, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332(6032):960–963. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yue B, et al. Genetic basis of drought resistance at reproductive stage in rice: Separation of drought tolerance from drought avoidance. Genetics. 2006;172(2):1213–1228. doi: 10.1534/genetics.105.045062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia Y, et al. The glabra1 mutation affects cuticle formation and plant responses to microbes. Plant Physiol. 2010;154(2):833–846. doi: 10.1104/pp.110.161646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.