Significance
Invasive species negatively impact both natural ecosystems and human society and are notoriously difficult to control once established. Thus, identifying potentially invasive taxa and preventing their dislocation is the most efficient management method. Darwin’s naturalization hypothesis, which predicts that the less closely related to native flora species are, the more likely they are to succeed as invaders, is tested here with an unprecedentedly thorough molecular phylogenetic approach, examining >100,000 phylogenies of the weed-rich thistle tribe Cardueae. Branch lengths between taxa were used as measures of evolutionary relatedness. Results show that invasive thistles are more closely related to natives than noninvasive introduced thistles, suggesting they share preadaptive traits with the natives that make them more likely to succeed as invaders.
Keywords: Asteraceae, exotic species, evolutionary relatedness
Abstract
Invasive species have great ecological and economic impacts and are difficult to control once established, making the ability to understand and predict invasive behavior highly desirable. Preemptive measures to prevent potential invasive species from reaching new habitats are the most economically and environmentally efficient form of management. Darwin’s naturalization hypothesis predicts that invaders less related to native flora are more likely to be successful than those that are closely related to natives. Here we test this hypothesis, using the weed-rich thistle tribe, Cardueae, in the California Floristic Province, a biodiversity hotspot, as our study system. An exhaustive molecular phylogenetic approach was used, generating and examining more than 100,000 likely phylogenies of the tribe based on nuclear and chloroplast DNA markers, representing the most in-depth reconstruction of the clade to date. Branch lengths separating invasive and noninvasive introduced taxa from native California taxa were used to represent phylogenetic distances between these groups and were compared at multiple biogeographical scales to ascertain whether invasive thistles are more or less closely related to natives than noninvasive introduced thistles are. Patterns within this highly supported clade show that not only are introduced thistles more closely related to natives more likely to be invasive, but these invasive species are also evolutionarily closer to native flora than by chance. This suggests that preadaptive traits are important in determining an invader’s success. Such rigorous molecular phylogenetic analyses may prove a fruitful means for furthering our understanding of biological invasions and developing predictive frameworks for screening potential invasive taxa.
Spread beyond their native ranges, some species have become numerically and ecologically dominant in new regions (1) and become of great interest and concern to scientists, policymakers, and the public. Such invasive species, sensu Colautti and MacIsaac (2), affect biodiversity, ecosystem function, and human health (3) and have ecological and economic impacts that cannot be ignored (4, 5), making the ability to understand and predict the invasiveness of species of great importance. Also, once exotic species become established in a new region, they are often extremely difficult to control (6, 7). Identifying and preventing new potentially invasive exotic species from reaching ground zero is, by far, the most economically and environmentally efficient management method (8). Hence, there is great need for early warning systems to determine the probability that a given species will become invasive (9–12).
The number of plant species introduced into the United States far exceeds that of other groups of organisms (13). However, although many case studies have illuminated various aspects of plant invasions (14–22), it has proven difficult to quantify and/or make generalizations about traits, characteristics, and circumstances that contribute to plant invasiveness across multiple geographic scales and ecological systems (10, 23–28). The difficulty of devising a framework to predict the behavior of exotic plants following dislocation and the challenges of designing effective control strategies for the ones that have become invasive result from the uniqueness of the organisms involved in each case, as well as the complexity of interactions between invaders and native communities (29). Few studies have provided a practical means of addressing these issues (30, 31).
Quantifiable measures that can provide robust predictions are therefore required (32), and phylogenetic relationships between native and introduced taxa may reveal patterns that invoke testable hypotheses that could not be derived from examining species traits alone (33). Distinct sets of traits evolve in response to environmental conditions, which in turn reflect past and present selection pressures and are therefore expected to differ not only among geographic regions and local communities (34), but also among evolutionary lineages. Hence species’ responses are not statistically independent from their shared evolutionary histories (35–37), and phylogeny may affect a species’ biotic interactions when it is introduced to a novel environment (38). As Darwin (39) observed, this results in a link between the evolutionary relatedness of organisms in a community, their characteristics, and the ecological processes that determine their distributions and abundance. Darwin’s naturalization hypothesis posits that invaders that are closely related to native taxa are less likely to be successful than those that are not. Assuming evolutionary relatedness is correlated with ecological similarity, such a pattern might emerge as a result of niche overlap and competitive exclusion between introduced taxa and their native relatives, in addition to being subject to the same predators and pathogens (40, 41). The enemy escape hypothesis also supports this view (42, 43). An alternative, opposing hypothesis is that relatedness to native taxa may convey degrees of preadaptation to the conditions of the invaded environment, rendering close relatives more likely to succeed once introduced (32, 39, 44).
Previous studies have been equivocal, finding evidence both for (30, 31, 38, 45–50), and against (24, 32, 41, 44, 51–55) Darwin’s hypothesis. However, few have used a strict phylogenetic approach based on evolutionary divergence, instead predominantly relying on taxonomic ranks (e.g., refs. 45 and 56, reviewed in refs. 57 and 58), which are highly subjective as measures of relatedness (59). In instances where phylogenetic trees were used, some have used supertrees compiled from multiple studies (46, 60, 61), with estimated branch lengths that may not accurately reflect the evolutionary distances between taxa. Other studies have been based on community phylogenetic trees (47, 60, 62). This approach may be problematic, as communities are not necessarily monophyletic groups, but collections of co-occurring species whatever their evolutionary relationships may be (63), and sampling in such studies is unlikely to include adequate representation of all lineages present, whereas phylogenetic analyses assume monophyly of the ingroup and their accuracy is dependent on sampling that is representative of the diversity within (64, 65).
This study sought to address theoretical and methodological issues that may have limited progress toward resolving Darwin’s naturalization conundrum (56) by using phylogenies based on molecular markers to assess the evolutionary distances between native and nonnative taxa in a strongly supported monophyletic group, the thistle tribe (Cardueae, Asteraceae), in a well-defined biogeographic area, the California Floristic Province (CAFP) (66, 67). The thistles of California offer an ideal opportunity to test Darwin’s hypothesis. The tribe, which boasts an impressive list of Mediterranean and temperate invaders, is most prolific in Mediterranean climate regions, which not only rank among the most biodiversity-rich biomes on the planet (68, 69), but among the most imperiled as well (70–74). The CAFP is one of such biodiversity hotspots, as defined by Conservation International, and is often recognized as a biogeological entity (75, 76). Also, programs and legislation regarding invasive taxa are usually state specific, making the study of invasive Cardueae in California both politically and biologically meaningful.
Phylogenies of Cardueae based on sequences from three genomic regions commonly used in phylogenetic studies of angiosperms were generated with taxon sampling representing the full lineage diversity of the tribe, including all species native and naturalized in the CAFP. Phylogenetic trees were constructed using parsimony, maximum likelihood, and Bayesian approaches. Phylogenetic distances of invasive and noninvasive introduced species from natives were compared using a comprehensive set of metrics and statistical tests to assess the utility of phylogenetic distance from natives as a predictor of invasive behavior. This study finds evidence contrary to Darwin’s hypothesis and demonstrates the robustness of such metrics, which should be more informative and meaningful than taxonomic groupings (30, 46, 77–79), especially when supported by a well-resolved molecular phylogeny.
Results
Phylogenetic trees of 202 species spanning the entire diversity of Cardueae, including the 51 species that occur in the CAFP, were constructed using Bayesian inference, maximum likelihood, and parsimony analyses based on two different combinations of three genomic regions: the internal transcribed spacer 1, 5.8S rRNA gene, and internal transcribed spacer 2 (nrDNA, ITS); the trnL intron, the 3′ trnL exon, and the intergenic spacer between trnL and trnF (cpDNA, trnL-trnF IGS); and maturase K (cpDNA, matK), per Sussanna et al. (80). Of the 73 recognized genera (81), 61 were represented. The issue of whether wider sampling of taxa is more important to accurate phylogenetic reconstructions than wider sampling of characters has been debated (82). Hence, two datasets, one with 165 taxa represented by three markers and another with 202 taxa represented by two markers, were examined. The chloroplast matK region was excluded in the two-marker dataset, as it contained the least number of variable sites.
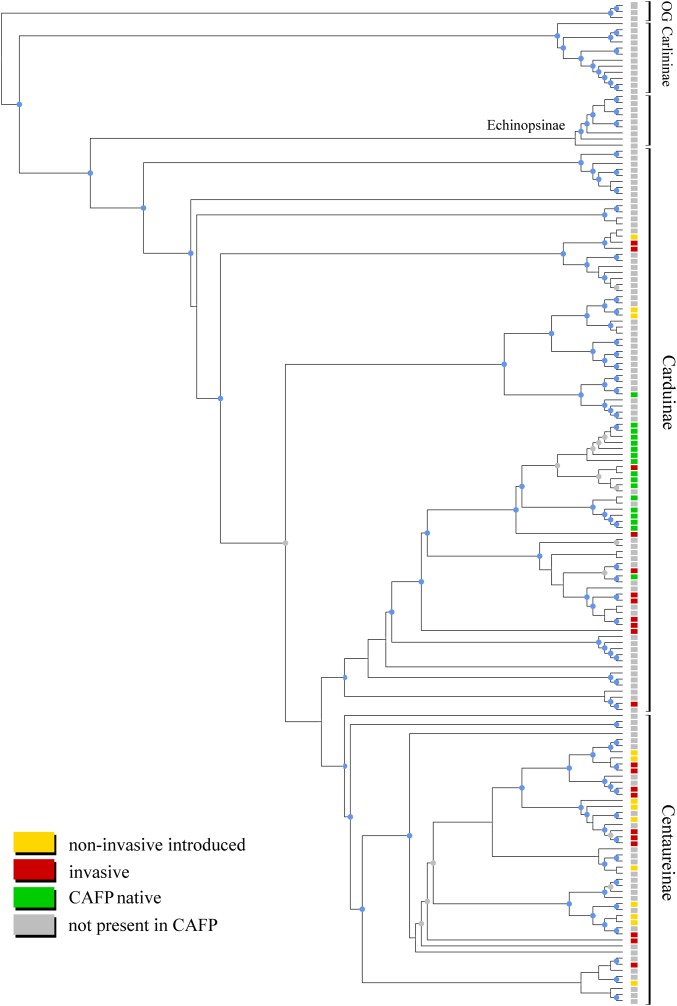
The total number of possible topologies examined for both datasets and three tree building methods combined was 112,626. These trees represent the most thoroughly sampled phylogenetic reconstructions of the tribe Cardueae to date. The Bayesian consensus tree in Fig. 1 illustrates the phylogenetic distribution of species of the four possible biogeographic and ecological categories used in this study (invasive, noninvasive exotic, native, and not present in CAFP), and is largely congruent with previous studies, including the paraphyly of subtribe Carduinae and strong support for the monophyly of Centaureinae as traditionally circumscribed (80, 83). Nonnative (introduced) species were classified as either invasive or noninvasive based on the California Invasive Plant Council invasive plant inventory (84).
Fig. 1.
Bayesian majority rule consensus tree of Cardueae (ITS+matK+trnL-trnF). Tips represent species, which are color coded according to their invasive status. Blue circles on the nodes indicate posterior probabilities of 0.9 and higher; gray circles represent posterior probabilities of 0.75 and higher. Major subtribes are labeled, and OG represents the outgroup.
The mean phylogenetic distance between each introduced species and all native species (MPD) and that between each introduced species and its nearest native relative (MNND) were calculated separately for invasive and noninvasive introduced species, and the results were compared for all 112,626 trees. Invasive species were significantly more closely related to the native community than were noninvasive exotic species in all topologies examined (P < 0.05). Invasive species also tended to have significantly closer nearest native relatives than noninvasive exotics in 99% of the trees tested (Table 1). Also, Cardueae species not occurring in CA were found to be significantly less related to the native CA thistle community than were invasive nonnative CA Cardueae in all cases (Table S1).
Table 1.
Proportion of evolutionary topologies contrary to Darwin’s naturalization hypothesis
| Maximum parsimony |
Bayesian inference |
|||||
| Bioregion | Statistic | ITS+matK+trnL-F (n = 13,528) | ITS+trnL-F (n = 19,092) | ITS+matK+trnL-F (n = 40,002) | ITS+trnL-F (n = 40,002) | Sum of all trees (n = 112,626) |
| CAFP | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 100.00 | 100.00 | 99.70 | 96.81 | 98.76 | |
| NRI | 100.00 | 100.00 | 99.99 | 99.21 | 99.72 | |
| NTI | 100.00 | 100.00 | 99.86 | 90.01 | 96.40 | |
| CaR | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 100.00 | 100.00 | 91.12 | 91.79 | 93.93 | |
| NRI | 100.00 | 98.82 | 84.55 | 35.28 | 71.32 | |
| NTI | 100.00 | 99.41 | 83.03 | 35.11 | 70.83 | |
| CW | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | |
| NRI | 100.00 | 100.00 | 99.94 | 94.41 | 97.99 | |
| NTI | 100.00 | 100.00 | 99.93 | 94.20 | 97.91 | |
| GV | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 100.00 | 0.31 | 95.58 | 42.08 | 60.96 | |
| NRI | 18.13 | 0.00 | 2.21 | 0.02 | 2.97 | |
| NTI | 19.20 | 0.00 | 2.21 | 0.02 | 3.10 | |
| NW | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 0.00 | 0.00 | 90.61 | 10.19 | 35.80 | |
| NRI | 100.00 | 100.00 | 99.12 | 58.85 | 85.07 | |
| NTI | 96.69 | 12.48 | 93.07 | 17.33 | 52.94 | |
| SN | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 100.00 | 100.00 | 99.99 | 68.89 | 88.95 | |
| NRI | 100.00 | 100.00 | 99.98 | 90.92 | 96.77 | |
| NTI | 100.00 | 100.00 | 99.97 | 89.66 | 96.32 | |
| SW | MPD | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| MNND | 0.00 | 100.00 | 3.52 | 100.00 | 53.72 | |
| NRI | 100.00 | 100.00 | 100.00 | 99.97 | 99.99 | |
| NTI | 100.00 | 100.00 | 100.00 | 99.98 | 99.99 | |
Values indicate the percentage of P values that were <0.05, when investigating whether invasive taxa are significantly more closely related to natives than are noninvasive exotic taxa (MPD, MNND), or more closely related to natives than by random assembly (NTI, NRI). Bioregions are abbreviated as follows: Cascade Ranges (CaR), Central Western California (CW), Great Central Valley (GV), Northwest California (NW), Sierra Nevada (SN), Southwestern California (SW).
To further examine the nature of the nonnative community of thistles in CA, the net relatedness index (NRI), a standardized measure that quantifies the degree of clustering of taxa over the entire phylogeny, and nearest taxon index (NTI), a measure of nearest taxon distance that quantifies the clustering of the terminal nodes (85, 86), were calculated as follows:
 |
where MPDobs and MNNDobs are the mean phylogenetic distance and mean nearest neighbor distance between introduced species and native species of the observed original data and MPDrndm and MNNDrndm are those of 10,000 randomly generated communities. SdsMPDrndm and SdsMNNDrndm refer to the SDs of the MPDs and MNNDs of the random assemblages. P values for the calculated NRI/NTIs were obtained by dividing the number of random assemblages with means greater than or equal to observed by the number of randomizations + 1 (85). Thus, positive NRI/NTI values indicate phylogenetic clustering, and negative values indicate overdispersion, relative to the native community. Positive values were retrieved in all instances for invasive taxa, indicating that invasive taxa were more closely related to their respective nearest native relatives, as well as to the native thistle community as a whole, than would be expected by chance. Results were statistically significant (P < 0.05) in nearly all of the topologies investigated (Table 1). In contrast, noninvasive exotic taxa were not significantly closer to native species than by chance.
To examine Darwin’s naturalization hypothesis at a local and possibly more ecologically meaningful scale (57), the aforementioned analyses were replicated at the level of individual bioregions, using the phytogeographic boundaries as defined in The Jepson Manual (Table 1) (87). Again, the majority of the investigated evolutionary topologies resolved invasive species as more closely related to the natives than are noninvasive exotic taxa. Despite smaller sample sizes, in most bioregions, invasive species were significantly more closely related to native species than by random chance as well. However, these patterns were not as well supported in the Great Central Valley as in the other bioregions, which generally represent coastal and mountainous areas of California.
These results suggest that, within the thistle tribe, introduced taxa more closely related to the native community and/or have closer native relatives are more likely to become invasive than those that are more distantly related to natives. The robustness of this pattern is demonstrated by the fact that it was upheld at multiple geographic scales, across various tree-building approaches.
Discussion
In the midst of a plethora of conflicting results and methodologies, some have argued that predicting which species will be invasive is nigh impossible (11); however, there may exist an optimal scale and scope of analysis at which the prediction of invasiveness is feasible, focusing on meaningfully circumscribed taxonomic and biogeographic entities (88). The phylogenetic scope of this study, encompassing a highly supported clade of 202 species within 64 genera, allows for circumvention of some of the problems that might occur in similar studies aimed at testing Darwin’s naturalization hypothesis at lesser or greater taxonomic scales. Examining the relationships between introduced species and native species where the community is composed of taxa classified in multiple families (47) and possibly separated by over a hundred million years of evolution (89), may not be ecologically meaningful, as it becomes difficult to assume ecological similarities between such taxa are the result of shared evolutionary history. Also, at such a large scale, it is difficult to construct well sampled, robust phylogenies of an all-encompassing monophyletic group. At the other extreme, examining the relationships among species within a smaller clade, as might be represented by a single genus, would likely provide results whose applicability to other situations would be quite limited. The scale used here provides a reasonable middle ground where taxa maintain a relatively high level of genetic and structural uniformity, yet are diverse enough to display a wide range of ecological adaptations.
The reliability of such comparative analyses depends on whether the phylogeny used accurately reflects the true evolutionary history of the taxa involved. However, phylogenetic reconstructions are prone to error and uncertainty, and most studies currently use one or few working phylogenies (90). Here, by examining a large number of likely evolutionary scenarios generated by a range of methods, it was possible to deal, to an extent, with the problem of phylogenetic uncertainty.
Results of this study suggest that introduced species with highly negative ecological impacts are phylogenetically closer to their native counterparts than are largely benign, noninvasive introduced species. Together with the finding that these invasive species were more closely related to native taxa than by chance, this suggests that preadaptive advantages, evolved over shared history and shared through common ancestry, may outweigh the importance of enemy escape or competitive exclusion, at least in certain stages of biological invasions. This trend mirrors those found in studies using taxonomic ranks and supertrees at large continental scales (44, 55, 91, 92) but has rarely been observed at local community scales (61).
Although invasive species tended to be closer to both the native community (MPD) and their closest native relatives (MNND) than were noninvasive introduced species, higher support was found for the former in most of the cases analyzed here. Distances to the nearest native relatives may reflect similarity to, and competition with, a single species, the one likely to hold the closest ecological niche, and therefore may be a metric that better reflects Darwin’s original rationalizations than mean distance to the entire native community (46). However, given that the CAFP native taxa are highly clustered phylogenetically, the slightly lower support for each invasive thistle having a closer native relative than each noninvasive introduced thistle is likely an artifact of small sample size.
Although the same patterns were found at the bioregion level, the results were not as highly supported across all phylogenies. This finding is not unexpected, as the sample sizes were considerably smaller (n < 30), and the results should be interpreted with caution. Indeed, the fact that these patterns were upheld across bioregions and phylogenies to this degree, although surprising, suggests that preadaptations continue to be important to species’ success at local scales and is a testament to the robustness of using phylogenetic relatedness to gauge the success of invading thistles. Interestingly, further support was found when examining the species compositions of natural reserves in the University of California Natural Reserve System for which species lists are publically available (93). Of the 14 reserves in which both native and invasive thistle species co-occur, 12 had at least one invasive species in the same genus with a native, supporting the patterns found at larger scales. No noninvasive introduced thistles were found in the reserve system. However, the Great Central Valley of California seems to be an exception; although this region is far from being the smallest, or most thistle-free bioregion, little support was found for phylogenetic relatedness determining the success of introduced taxa. This result is likely due to the fact that the Central Valley is predominantly farmland and grazing land for livestock and is almost entirely in private ownership (94). Very little land is left unmanaged, and remaining natural habitats are mostly small and fragmented. It may be impossible to observe ecological patterns underlying biological invasions in such areas.
Only a small fraction of the many exotics that have been introduced to alien habitats over the years have become highly invasive (93). Clearly, the net outcome of a plethora of opportunities and obstacles introduced plants are faced with will determine whether their populations ultimately thrive against, or are suppressed by, the native community (2). As demonstrated here, a phylogenetic framework can represent such interactions by addressing the shared evolutionary history of species. Thus, the evolutionary relatedness of exotic taxa to natives can provide information for identifying threats to native communities, as well as extending our understanding of why certain introduced species prove to be more invasive than others. This study suggests that monitoring and regulating exotic species that are closely related to native taxa but not yet introduced or escaped should be a priority.
Methods
Study System.
The Asteraceae, the largest eudicot family and possibly the largest angiosperm family, represents 8% of all flowering plant species and, although most prominent in drier, Mediterranean climates, can be found on all continents except Antarctica (95). While comprising many important crop and horticultural species, the family also includes a disproportionately high global representation of invasive species (96, 97). Of specific interest is the thistle tribe, Cardueae, one of the largest tribes of Asteraceae, with ∼2,500 species. Previous molecular studies have unanimously confirmed Cardueae as monophyletic (80, 95, 98–101). The tribe comprises roughly half of the invasive Asteraceae species in California, as well as many natives and endemics (102). Native and nonnative introduced taxa were delimited according to the second edition of The Jepson Manual (87).
Collecting Materials.
Sampling was based on previous systematics studies (80, 83, 101) to represent most of the genera and major clades of the tribe Cardueae. Plant material was collected both from the field and herbarium specimens. A small amount of leaf tissue was removed from each specimen and dried in silica gel and/or frozen for DNA extraction. Collected specimens were pressed and deposited at the University of California–Davis Center for Plant Diversity. Relevant sequences were also downloaded from GenBank. Species were chosen to represent the entire diversity of the tribe and achieve sufficient depth of taxon sampling within that monophyletic group. To minimize inaccurate placements of taxa, sampling of hybrids was avoided. The origins of the samples and their GenBank accession numbers are listed in Table S2.
DNA Extraction, Amplification, and Sequencing.
DNA extractions were performed using Qiagen miniprep kits. The ITS region was amplified and sequenced using primers ITS6 and ITS9 (103) and separately using combinations of ITS2 and ITS5, and ITS3 and ITS4, respectively (104). The trnL intron, the 3′ trnL exon, and the intergenic spacer between trnL and trnF were amplified and sequenced together. The universal primers trnL-c and trnL-f were used for amplifying the trnL-trnF IGS region. In some cases, combinations of trnL-d and trnL-e were used together as well (105). The first 1,000 bp of the 5′ end of the gene maturase K (matK), which account for most of the variability found in the gene (106), were amplified with the primers trnK-710 F (107) and AST-1R (101). In some cases, combinations of the primers matK1F, matK1R, matK2F, and matK2R were used as well (95). PCR products were run on a 1% agarose gel. Identifiable bands were cut out and purified with Qiagen Gel Extraction Kits. Sanger sequencing of the purified PCR products was performed on ABI 3730 Capillary Electrophoresis Genetic Analyzers at the University of California–Davis College of Biological Sciences Sequencing Facility, with the same primers used for amplification.
Phylogenetic Analyses.
The ITS region, as well as the chloroplast matK gene and the trnL-F region, was chosen not only because of their widespread use in phylogenetics, but also because the combination of relatively fast-evolving regions and slower ones was necessary to provide the required deep and shallow level resolution for performing phylogenetic analyses at the species level. The combination of these markers has been used in previous studies of the group (80, 83), and Shimodaira-Hasegawa tests (108) were performed in PAUP (109) to confirm lack of significant conflict in phylogenetic signal between nuclear and plastid regions (Table S3). Sequences of each region were aligned independently with ClustalX (110), edited further by hand using MEGA5 (111), and combined into a single matrix. Multiple phylogenies of the tribe Cardueae were estimated based on two combinations of the three aforementioned markers: ITS and trnL-F (n = 202) and ITS, trnL-F, and matK (n = 165). To minimize error and bias in taxon placement and branch length calculations, all taxa in the phylogenies were represented by all markers, with only 0.15% missing data. In addition, to investigate a full breadth of possible evolutionary scenarios, three different tree building methods were used as described below.
Garli 2.0 (112) was run on the Cipres Science Gateway server to generate maximum likelihood (ML) phylogenies. The general time reversible (GTR + Γ + I) (113) model was used with default settings. Four parallel runs were performed to ensure that the resulting tree was not lodged on a local optimum. Maximum parsimony analyses were conducted with PAUPRat (114) through the Cipres Science Gateway. Analyses were run with default settings with TBR branch swapping for 43,000 Ratchet repetitions and 200 Ratchet iterations for the ITS + trnLF dataset and 50,000 repetitions and 200 iterations for the ITS + trnL-F + matK dataset, yielding 19,092 and 13,528 most parsimonious trees, respectively. Maximum likelihood trees and bootstrap values are presented in Figs. S1 and S2. Bayesian inference analyses were carried out using MrBayes 3.2.1 (115). Models of molecular evolution were evaluated with JMODELTEST (116); the best fit models were GTR + Γ + I for ITS and GTR + Γ for plastid regions, based on both the modified Akaike information criterion and Bayesian inference criterion. Analyses were run under default settings for 13 million generations, sampling two Markov chain Monte Carlo (MCMC) chains every 500 generations. A total of 40,002 trees were saved for each dataset after discarding 23% as burn-in.
Investigating Darwin’s Naturalization Hypothesis.
To assess whether exotic invaders tend to be more closely related to their native relatives, the MPDs from each nonnative taxon to all native taxa, as well as the distance from each nonnative taxon to its nearest native relative, i.e., the MNNDs, were calculated using the ICOMDIST function in phylocom (85). These two metrics were compared using a t test in R (117). To determine the degree to which variance in tree topologies affects these results, this process was repeated for all of the phylogenies generated by the aforementioned methods using a custom Perl script, for a total of > 100,000 trees. Identical analyses were also conducted at the bioregion level as defined by The Jepson Manual (87), to seek a more focused view of species interactions.
Custom R scripts were used to calculate NRI and NTIs across all phylogenies. In accordance to the independent swap null model, nonnative taxa were randomly sampled to generate random exotic assemblages of equal species richness as observed communities (118). A total of 10,000 random assemblages were generated per tree.
Supplementary Material
Acknowledgments
We thank E. Dean and J. Shepard of the University of California–Davis Center for Plant Diversity for assistance with sampling herbarium material for DNA analyses; L. Celesti, D. Viciani, E. Badalamenti, and S. Peccenini for invaluable help collecting in Italy; I. Korf and K. Battenberg for instructive discussions on Perl programming; and D.S.P.’s dissertation committee members for insightful comments on the project and manuscript. This work was supported by funding from Davis Botanical Society and National Science Foundation Grant DEB1210526.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KC969472–KC969614).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309948110/-/DCSupplemental.
References
- 1.Crooks JA. Characterizing ecosystem-level consequences of biological invasions: The role of ecosystem engineers. Oikos. 2002;97(2):153–166. [Google Scholar]
- 2.Colautti RI, MacIsaac HJ. A neutral terminology to define 'invasive' species. Divers Distrib. 2004;10(2):135–141. [Google Scholar]
- 3.Crowl TA, Crist TO, Parmenter RR, Belovsky G, Lugo AE. The spread of invasive species and infectious disease as drivers of ecosystem change. Front Ecol Environ. 2008;6(5):238–246. [Google Scholar]
- 4.Mack RN, Lonsdale WM. Humans as global plant dispersers: Getting more than we bargained for. Bioscience. 2001;51(2):95–102. [Google Scholar]
- 5.Pimentel D. In: Invasive Plants: Their Role in Species Extinctions and Economic Losses to Agriculture in the USA. Management of Invasive Weeds, Invading Nature - Springer Series in Invasion Ecology. Inderjit, editor. Vol 5. The Netherlands: Springer, Dordrecht; 2009. pp. 1–7. [Google Scholar]
- 6.Rejmánek M, Richardson DM, Higgins SI, Pitcairn M, Grotkopp E. Ecology of invasive plants: State of the art. In: Monney HA, McNeely JA, Neville L, Schei PJ, Waage J, editors. Invasive Alien Species: A New Synthesis. Washington, DC: Island Press; 2005. pp. 104–161. [Google Scholar]
- 7.Duncan RP, Blackburn TM, Sol D. The ecology of bird introductions. Annu Rev Ecol Evol Syst. 2003;34:71–98. [Google Scholar]
- 8.Wittenberg R, Cock MJW, editors. Invasive Alien Species: A Toolkit of Best Prevention and Management Practices (CAB International. Wallingford, Oxon, UK: 2001. pp. xvii–228. [Google Scholar]
- 9.Andreu J, Vila M. Risk analysis of potential invasive plants in Spain. J Nat Conserv. 2010;18(1):34–44. [Google Scholar]
- 10.Kolar CS, Lodge DM. Progress in invasion biology: Predicting invaders. Trends Ecol. Evolut. 2001;16(4):199–204. doi: 10.1016/s0169-5347(01)02101-2. [DOI] [PubMed] [Google Scholar]
- 11.Kolar CS, Lodge DM. Ecological predictions and risk assessment for alien fishes in North America. Science. 2002;298(5596):1233–1236. doi: 10.1126/science.1075753. [DOI] [PubMed] [Google Scholar]
- 12.Forsyth DM, Duncan RP, Bomford M, Moore G. Climatic suitability, life-history traits, introduction effort, and the establishment and spread of introduced mammals in Australia. Conserv Biol. 2004;18(2):557–569. [Google Scholar]
- 13.Pimentel D, Zuniga R, Morrison D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ. 2005;52(3):273–288. [Google Scholar]
- 14. Baker J, Bomford M (2009) Opening the climate modelling envelope. Plant Protect Qtly 24(3 Special Issues SI):88–91.
- 15.Mack RN. Assessing the extent, status, and dynamism of plant invasions: Current and emerging approaches. In: Mooney HA, Hobbs RJ, editors. Invasive species in a changing world. Washington, DC: Island Press; 2000. pp. 141–168. [Google Scholar]
- 16.Reaser JK, Meyerson LA, Von Holle B. Saving camels from straws: How propagule pressure-based prevention policies can reduce the risk of biological invasion. Biol Invasions. 2008;10(7):1085–1098. [Google Scholar]
- 17.Mack RN, et al. Biotic invasions: Causes, epidemiology, global consequences, and control. Ecol Appl. 2000;10(3):689–710. [Google Scholar]
- 18. Simberloff D (2009) The role of propagule pressure in biological invasions. Annual Review of Ecology Evolution and Systematics (Annual Reviews, Palo Alto, CA), Vol 40, pp 81–102.
- 19.Richardson DM, Allsopp N, D'Antonio CM, Milton SJ, Rejmanek M. Plant Invasions: The Role of Mutualisms. Cambridge, UK: Cambridge Univ Press; 2000. pp. 65–93. [DOI] [PubMed] [Google Scholar]
- 20.Pyšek P, Richardson DM. Traits Associated with Invasiveness in Alien Plants: Where Do we Stand? In: Nentwig W, editor. Biological Invasions, Ecological Studies. Vol 193. Berlin: Springer; 2007. pp. 97–125. [Google Scholar]
- 21.Catford JA, Jansson R, Nilsson C. Reducing redundancy in invasion ecology by integrating hypotheses into a single theoretical framework. Divers Distrib. 2009;15(1):22–40. [Google Scholar]
- 22.Rejmánek M, Richardson DM. What attributes make some plant species more invasive? Ecology. 1996;77(6):1655–1661. [Google Scholar]
- 23.Rejmanek M. A theory of seed plant invasiveness: The first sketch. Biol Conserv. 1996;78(1-2):171–181. [Google Scholar]
- 24.Richardson DM, Allsopp N, D’Antonio CM, Milton SJ, Rejmánek M. Plant invasions—the role of mutualisms. Biol Rev Camb Philos Soc. 2000;75(1):65–93. doi: 10.1017/s0006323199005435. [DOI] [PubMed] [Google Scholar]
- 25.Grotkopp E, Rejmánek M, Rost TL. Toward a causal explanation of plant invasiveness: Seedling growth and life-history strategies of 29 pine (Pinus) species. Am Nat. 2002;159(4):396–419. doi: 10.1086/338995. [DOI] [PubMed] [Google Scholar]
- 26.Hallett SG. Dislocation from coevolved relationships: A unifying theory for plant invasion and naturalization? Weed Sci. 2006;54(2):282–290. [Google Scholar]
- 27.Moles AT, Gruber MAM, Bonser SP. A new framework for predicting invasive plant species. J Ecol. 2008;96(1):13–17. [Google Scholar]
- 28.Zuppinger-Dingley D, et al. In their native range, invasive plants are held in check by negative soil-feedbacks. Ecosphere. 2011;2(5):54. [Google Scholar]
- 29.Lodge DM. Biological invasions: Lessons for ecology. Trends Ecol Evol. 1993;8(4):133–137. doi: 10.1016/0169-5347(93)90025-K. [DOI] [PubMed] [Google Scholar]
- 30.van Wilgen NJ, Richardson DM. Is phylogenetic relatedness to native species important for the establishment of reptiles introduced to California and Florida? Divers Distrib. 2011;17(1):172–181. [Google Scholar]
- 31.Harvey KJ, Nipperess DA, Britton DR, Hughes L. Australian family ties: Does a lack of relatives help invasive plants escape natural enemies? Biol Invasions. 2012;14(11):2423–2434. [Google Scholar]
- 32.Ricciardi A, Mottiar M. Does Darwin’s naturalization hypothesis explain fish invasions? Biol Invasions. 2006;8(6):1403–1407. [Google Scholar]
- 33.Cadotte MW, et al. Phylogenetic patterns differ for native and exotic plant communities across a richness gradient in Northern California. Divers Distrib. 2010;16(6):892–901. [Google Scholar]
- 34.Griffiths ME, Lawes MJ. Biogeographic, environmental, and phylogenetic influences on reproductive traits in subtropical forest trees, South Africa. Ecography. 2006;29(4):614–622. [Google Scholar]
- 35.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- 36.Harvey PH. Pagel MD. The Comparative Methods in Evolutionary Biology. New York: Oxford Univ Press; 1991. p. 239. [Google Scholar]
- 37.Ives AR, Midford PE, Garland T., Jr Within-species variation and measurement error in phylogenetic comparative methods. Syst Biol. 2007;56(2):252–270. doi: 10.1080/10635150701313830. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell CE, et al. Biotic interactions and plant invasions. Ecol Lett. 2006;9(6):726–740. doi: 10.1111/j.1461-0248.2006.00908.x. [DOI] [PubMed] [Google Scholar]
- 39.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. p. 490. [PMC free article] [PubMed] [Google Scholar]
- 40. Elton CS (1958) The Ecology of Invasions by Animals and Plants (Wiley, New York)
- 41.Daehler CC. Darwin’s naturalization hypothesis revisited. Am Nat. 2001;158(3):324–330. doi: 10.1086/321316. [DOI] [PubMed] [Google Scholar]
- 42.Liu H, Stiling P. Testing the enemy release hypothesis: A review and meta-analysis. Biol Invasions. 2006;8(7):1535–1545. [Google Scholar]
- 43.Maron JL, Vilà M. When do herbivores affect plant invasion? Evidence for the natural enemies and biotic resistance hypotheses. Oikos. 2001;95(3):361–373. [Google Scholar]
- 44.Duncan RP, Williams PA. Ecology: Darwin’s naturalization hypothesis challenged. Nature. 2002;417(6889):608–609. doi: 10.1038/417608a. [DOI] [PubMed] [Google Scholar]
- 45.Agrawal AA, et al. Enemy release? An experiment with congeneric plant pairs and diverse above- and belowground enemies. Ecology. 2005;86(11):2979–2989. [Google Scholar]
- 46.Strauss SY, Webb CO, Salamin N. Exotic taxa less related to native species are more invasive. Proc Natl Acad Sci USA. 2006;103(15):5841–5845. doi: 10.1073/pnas.0508073103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer H, Hardy OJ, Silva L, Barraclough TG, Savolainen V. Testing Darwin’s naturalization hypothesis in the Azores. Ecol Lett. 2011;14(4):389–396. doi: 10.1111/j.1461-0248.2011.01600.x. [DOI] [PubMed] [Google Scholar]
- 48.Mack RN. Predicting the identity and fate of plant invaders: Emergent and emerging approaches. Biol Conserv. 1996;78(1-2):107–121. [Google Scholar]
- 49.Lockwood J, Simberloff D, McKinney M, Von Holle B. How many, and which, plants will invade natural areas? Biol Invasions. 2001;3(1):1–8. [Google Scholar]
- 50.Ricciardi A, Atkinson SK. Distinctiveness magnifies the impact of biological invaders in aquatic ecosystems. Ecol Lett. 2004;7(9):781–784. [Google Scholar]
- 51.Lambdon PW, Hulme PE. How strongly do interactions with closely-related native species influence plant invasions? Darwin's naturalization hypothesis assessed on Mediterranean islands. J Biogeogr. 2006;33(6):1116–1125. [Google Scholar]
- 52.Lambdon PW. Is invasiveness a legacy of evolution? Phylogenetic patterns in the alien flora of Mediterranean islands. J Ecol. 2008;96(1):46–57. [Google Scholar]
- 53.Ricotta C, Godefroid S, Rocchini D. Invasiveness of alien plants in Brussels is related to their phylogenetic similarity to native species. Divers Distrib. 2010;16(4):655–662. [Google Scholar]
- 54.Tingley R, Phillips BL, Shine R. Establishment success of introduced amphibians increases in the presence of congeneric species. Am Nat. 2011;177(3):382–388. doi: 10.1086/658342. [DOI] [PubMed] [Google Scholar]
- 55.Ferreira RB, Beard KH, Peterson SL, Poessel SA, Callahan CM. Establishment of introduced reptiles increases with the presence and richness of native congeners. Amphib-reptil. 2012;33(3-4):387–392. [Google Scholar]
- 56.Diez JM, Sullivan JJ, Hulme PE, Edwards G, Duncan RP. Darwin’s naturalization conundrum: Dissecting taxonomic patterns of species invasions. Ecol Lett. 2008;11(7):674–681. doi: 10.1111/j.1461-0248.2008.01178.x. [DOI] [PubMed] [Google Scholar]
- 57.Thuiller W, et al. Resolving Darwin’s naturalization conundrum: A quest for evidence. Divers Distrib. 2010;16(3):461–475. [Google Scholar]
- 58.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. [Google Scholar]
- 59.Hill S, Kotanen P. Phylogenetically structured damage to Asteraceae: Susceptibility of native and exotic species to foliar herbivores. Biol Invasions. 2010;12(9):3333–3342. [Google Scholar]
- 60.Hill SB, Kotanen PM. Evidence that phylogenetically novel non-indigenous plants experience less herbivory. Oecologia. 2009;161(3):581–590. doi: 10.1007/s00442-009-1403-0. [DOI] [PubMed] [Google Scholar]
- 61.Carboni M, et al. Darwin's naturalization hypothesis: scale matters in coastal plant communities. Ecography. 2013;36(5):560–568. doi: 10.1111/j.1600-0587.2012.07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davies KF, Cavender-Bares J, Deacon N. Native communities determine the identity of exotic invaders even at scales at which communities are unsaturated. Divers Distrib. 2011;17(1):35–42. [Google Scholar]
- 63.Grandcolas P. Phylogenetic analysis and the study of community structure. Oikos. 1998;82(2):397–400. [Google Scholar]
- 64. Henning W (1966) Phylogenetic Systematics (Univ of Illinois Press, Urbana, IL)
- 65.Hillis DM, Pollock DD, McGuire JA, Zwickl DJ. Is sparse taxon sampling a problem for phylogenetic inference? Syst Biol. 2003;52(1):124–126. doi: 10.1080/10635150390132911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Axelrod DI (1977) Terrestrial Vegetation of California, eds Barbour M, Major J (Wiley, New York), pp 139–194.
- 67.Rabeler RK. California flora. Science. 1993;262(5131):261. doi: 10.1126/science.262.5131.261. [DOI] [PubMed] [Google Scholar]
- 68.Cowling RM, Rundel PW, Lamont BB, Kalin Arroyo M, Arianoutsou M. Plant diversity in mediterranean-climate regions. Trends Ecol Evol. 1996;11(9):362–366. doi: 10.1016/0169-5347(96)10044-6. [DOI] [PubMed] [Google Scholar]
- 69.Sauquet H, et al. Contrasted patterns of hyperdiversification in Mediterranean hotspots. Proc Natl Acad Sci USA. 2009;106(1):221–225. doi: 10.1073/pnas.0805607106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greuter W. Botanical diversity, endemism, rarity, and extinction in the mediterranean area: An analysis based on the published volumes of med-checklist. Botanika Chronika. 1991;10:63–79. [Google Scholar]
- 71.Sala OE, et al. Global biodiversity scenarios for the year 2100. Science. 2000;287(5459):1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- 72.Seabloom EW, et al. Human impacts, plant invasion, and imperiled plant species in California. Ecol Appl. 2006;16(4):1338–1350. doi: 10.1890/1051-0761(2006)016[1338:hipiai]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 73.Underwood EC, Viers JH, Klausmeyer KR, Cox RL, Shaw MR. Threats and biodiversity in the mediterranean biome. Divers Distrib. 2009;15(2):188–197. [Google Scholar]
- 74.Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403(6772):853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 75.Quézel P. Definition of the Mediterranean region and the origin of its flora. In: Gómez-Campo C, editor. Plant Conservation in the Mediterranean Area. The Netherlands: Dr. W. Junk Publishers; 1985. pp. 9–24. [Google Scholar]
- 76.Médail F, Quezél P. Biodiversity hotspots in the Mediterranean basin: Setting global conservation priorities. Conserv Biol. 1999;13(6):1510–1513. [Google Scholar]
- 77.Vamosi SM, Heard SB, Vamosi JC, Webb CO. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol Ecol. 2009;18(4):572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- 78.Cadotte MW, Hamilton MA, Murray BR. Phylogenetic relatedness and plant invader success across two spatial scales. Divers Distrib. 2009;15(3):481–488. [Google Scholar]
- 79.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. The merging of community ecology and phylogenetic biology. Ecol Lett. 2009;12(7):693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 80.Susanna A, Garcia-Jacas N, Hidalgo O, Vilatersana R, Garnatje T. The Cardueae (Compositae) revisited: Insights from its, trnL-trnF, and matK nuclear and chloroplast DNA analysis. Ann Mo Bot Gard. 2006;93(1):150–171. [Google Scholar]
- 81.Susanna A, Garcia-Jacas N. Cardueae. In: Kadereit JW, Jeffrey C, editors. Flowering Plants, Eudicots, Asterales, The Families and Genera of Vascular Plants. Berlin: Springer; 2007. pp. 123–147. [Google Scholar]
- 82.Nabhan AR, Sarkar IN. The impact of taxon sampling on phylogenetic inference: A review of two decades of controversy. Brief Bioinform. 2012;13(1):122–134. doi: 10.1093/bib/bbr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Susanna A, et al. Lessons from Plectocephalus (Compositae, Cardueae-Centaureinae): ITS disorientation in annuals and Beringian dispersal as revealed by molecular analyses. Ann Bot (Lond) 2011;108(2):263–277. doi: 10.1093/aob/mcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cal-IPC . California Invasive Plant Inventory. Berkeley, CA: California Invasive Plant Council; 2006. [Google Scholar]
- 85.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24(18):2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 86.Webb CO. Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat. 2000;156(2):145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 87.Baldwin BG, et al., editors. The Jepson Manual: Vascular Plants of California. 2nd Ed. Berkeley: Univ of California Press; 2012. [Google Scholar]
- 88.Muth NZ, Pigliucci M. Traits of invasives reconsidered: Phenotypic comparisons of introduced invasive and introduced noninvasive plant species within two closely related clades. Am J Bot. 2006;93(2):188–196. doi: 10.3732/ajb.93.2.188. [DOI] [PubMed] [Google Scholar]
- 89.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97(8):1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- 90.Rezende EL, Diniz-Filho JAF. Phylogenetic Analyses: Comparing Species to Infer Adaptations and Physiological Mechanisms. Comprehensive Physiology. Wiley, New York; 2011. [DOI] [PubMed] [Google Scholar]
- 91.Diez JM, et al. Learning from failures: Testing broad taxonomic hypotheses about plant naturalization. Ecol Lett. 2009;12(11):1174–1183. doi: 10.1111/j.1461-0248.2009.01376.x. [DOI] [PubMed] [Google Scholar]
- 92.Escobedo VM, Aranda JE, Castro SA. Hipótesis de naturalización de Darwin evaluada en la flora exótica de Chile continental. Rev Chil Hist Nat. 2011;84:543–552. [Google Scholar]
- 93.Umbach KW. Sacramento, CA: California Research Bureau; 1998. A Statistical Tour of California's Great Central Valley. [Google Scholar]
- 94.Williamson M, Fitter A. The varying success of invaders. Ecology. 1996;77(6):1661–1666. [Google Scholar]
- 95.Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: Major clades of the Asteraceae revealed. Mol Phylogenet Evol. 2008;47(2):757–782. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 96.Daehler CC. The taxonomic distribution of invasive angiosperm plants: Ecological insights and comparison to agricultural weeds. Biol Conserv. 1998;84(2):167–180. [Google Scholar]
- 97.Pysek P. Is there a taxonomic pattern to plant invasions? Oikos. 1998;82(2):282–294. [Google Scholar]
- 98.Funk VA, et al. Everywhere but Antarctica: Using a supertree to understand the diversity and distribution of the Compositae. Kongelige Danske Videnskabernes Selskab Biologiske Skrifter. 2005;55:343–374. [Google Scholar]
- 99.Panero JL, Funk VA. Toward a phylogenetic subfamilial classification for the Compositae (Asteraceae) Proc Biol Soc Wash. 2002;115(4):909–922. [Google Scholar]
- 100.Susanna A, Garnatje T, Garcia-Jacas N, Vilatersana R. On the correct subtribal placement of the genera Syreitschikovia and Nikitinia (Asteraceae, Cardueae): Carduinae or Centaureinae? Bot J Linn Soc. 2002;140(3):313–319. [Google Scholar]
- 101.Garcia-Jacas N, Garnatje T, Susanna A, Vilatersana R. Tribal and subtribal delimitation and phylogeny of the Cardueae (Asteraceae): A combined nuclear and chloroplast DNA analysis. Mol Phylogenet Evol. 2002;22(1):51–64. doi: 10.1006/mpev.2001.1038. [DOI] [PubMed] [Google Scholar]
- 102.Kelch DG, Baldwin BG. Phylogeny and ecological radiation of New World thistles (Cirsium, Cardueae-Compositae) based on ITS and ETS rDNA sequence data. Mol Ecol. 2003;12(1):141–151. doi: 10.1046/j.1365-294x.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- 103.Potter D, et al. Phylogeny and classification of Rosaceae. Plant Syst Evol. 2007;266(1-2):5–43. [Google Scholar]
- 104.Baldwin BG, et al. The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard. 1995;82(2):247–277. [Google Scholar]
- 105.Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17(5):1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 106.Hilu K, Liang H. The matK gene: Sequence variation and application in plant systematics. Am J Bot. 1997;84(6):830. [PubMed] [Google Scholar]
- 107.Johnson LA, Soltis DE. Phylogenetic inference in Saxifragaceae sensu stricto and Gilia (Polemoniaceae) using matK sequences. Ann Mo Bot Gard. 1995;82(2):149–175. [Google Scholar]
- 108.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Molecular Biology and Evolution. 1999;16(8):1114–1116. [Google Scholar]
- 109.Swofford DL. Sunderland, MA: Sinauer Associates; 2003. PAUP*: phylogenetic analysis using parsimony, version 4.0b10. [Google Scholar]
- 110.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tamura K, et al. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zwickl DJ. Austin, TX: University of Texas at Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation. [Google Scholar]
- 113.Yang Z. Maximum-likelihood models for combined analyses of multiple sequence data. J Mol Evol. 1996;42(5):587–596. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- 114. Sikes DS, Lewis P (2001) PAUPRat: PAUP* Implementation of the Parsimony Ratchet (Department of Ecology and Evolutionary Biology, Univ of Connecticut, Storrs, CT)
- 115.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 116.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 117. R Core Team (2013) R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna)
- 118.Gotelli NJ, Entsminger GL. Swap and fill algorithms in null model analysis: rethinking the knight's tour. Oecologia. 2001;129(2):281–291. doi: 10.1007/s004420100717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.