Abstract
Previous studies have shown that mammalian cells contain replicator sequences, which can determine where DNA replication initiates. However, the specific sequences that confer replicator activity were not identified. Here we report a detailed analysis of replicator sequences that dictate initiation of DNA replication from the human β-globin locus. This analysis suggests that the β-globin replication initiation region contains two adjacent, redundant replicators. Each replicator was capable of initiating DNA replication independently at ectopic sites. Within each of these two replicators, we identified short, discrete, nonredundant sequences, which cooperatively determine replicator activity. Experiments with somatic cell hybrids further demonstrated that the requirements for initiation at ectopic sites were similar to the requirements for initiation within native human chromosomes. The replicator clustering and redundancy exemplified in the human β-globin locus may account for the extreme difficulty in identifying replicator sequences in mammalian cells and suggest that mammalian replication initiation sites may be determined by cooperative sequence modules.
The initiation of DNA replication is a critical step in cell division. However, little is known about the cellular events that transmit signals from the cell cycle machinery to the DNA sequences at which DNA replication initiates. The replicon model of Jacob, Brenner, and Cuzin (28) proposed that all cells regulate DNA replication through the interaction of cis-acting DNA sequences called replicators with trans-acting initiation factors called initiators. Replicators are defined genetically as DNA sequences that contain information that allows them to confer initiation of DNA replication. Replication origins, or replication initiation sites, are determined biochemically and mark the regions from which replication forks emanate. Replicators often colocalize with chromosomal initiation sites in bacteria and in yeast (14). In mammalian cells, DNA sequences meeting the biochemical criteria of replication origins typically do not replicate extrachromosomally, unless they are linked to viral replicons or nuclear retention signals (14). Various biochemical strategies designed to identify initiation regions (IRs) in mammalian cells have sometimes generated conflicting data (8, 14). This led to controversy regarding whether replicators exist in mammals and whether initiation of replication in mammalian cells necessitates specific DNA sequences.
Mapping of replication initiation profiles in β-globin loci provided important clues regarding the roles of specific DNA sequences in directing initiation. The human β-globin locus resides on chromosome 11 and includes five genes that encode the β-subunit of hemoglobin. Kitsberg et al. (35) demonstrated that replication at this locus initiated from a single IR located between the two adult β-like globin genes. The naturally occurring Lepore deletion, which removed the IR, resulted in passive replication of the locus from an outside origin. These data suggested that replication required genetic information uniquely supplied by the deleted region. Gene transfer experiments, designed to circumvent the need for extrachromosomal replication assays (1), demonstrated that the IR can initiate DNA replication when transferred to genomic regions that lack inherent replicator activity. To control for position effects that may affect initiation capacity, these experiments were performed at fixed chromosomal locations using recombinase-mediated gene transfer (1). The IR was also used in human artificial chromosomes that can replicate extrachromosomally in human cells (25). These observations suggested that the initiation of DNA replication in mammalian cells requires specific replicator sequences and that the β-globin IR is such a replicator. Similar gene transfer experiments have recently identified mammalian replicators in other loci. For example, the replication IR from the human myc locus can initiate DNA replication at ectopic sites (30, 39), and one of the redundant replication origins residing within an expanded replication IR in the Chinese hamster dihydrofolate reductase (DHFR) locus (17) exhibits replicator activity (5).
The next challenge lies in determining which sequences are essential for replicator activity. Replication initiation sites, including the β-globin replicator, share some common features (22), but unlike genomic replicators in budding yeast, they do not exhibit a clear consensus sequence. These initiation sites tend to contain AT-rich regions, asymmetric purine:pyrimidine stretches, matrix/scaffolding attachment regions, potential cruciform structures, and regions that resemble yeast origin recognition complex (ORC) binding sites (8, 9, 44). Such sequence features are present within the β-globin IR; however, similar regions are also found in sequence elements that do not initiate DNA replication. The relevance of these elements to replicator activity is unclear.
We used the gene transfer approach to identify sequence features that are required for initiation of DNA replication from the globin IR to initiate DNA replication at constant ectopic chromosomal sites. A limited deletion analysis suggested that the β-globin replicator includes a core element that is essential, but not sufficient, for initiation of DNA replication (1). However, the specific sequences that confer the ability to initiate replication were not determined. Here we report a detailed analysis of the human β-globin replication IR. Using ectopic initiation assays, our data revealed that this region contained two redundant, independent, nonoverlapping replicators, each of which was sufficient to initiate replication at ectopic sites. Within each replicator, initiation of DNA replication required cooperation between at least two unique, nonredundant sequences. IR deletions that did not allow initiation at ectopic sites also failed to initiate replication within the intact native globin locus. These observations may account for the extreme difficulty of identifying replicator sequences in mammalian cells and suggest that mammalian replication initiation sites are determined by specific cooperative sequence modules.
MATERIALS AND METHODS
Cells and culture conditions.
Simian CV-1 E25B4 (B4) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and antibiotics as needed. DNA fragments to be tested for replicator activity were inserted downstream of the FLP recombination target (FRT) site in the shuttle vector pSFV (2). We used site-specific recombination to insert the replicator candidates into the B4 cell line. Insertion of the putative replicator fragments disrupted transcription from the lacZ marker, as described previously (1). Colonies were selected for hygromycin resistance and screened for insertion at the B4 site by staining for β-galactosidase activity, followed by PCR and hybridization analyses to verify that single copies of clones had been inserted.
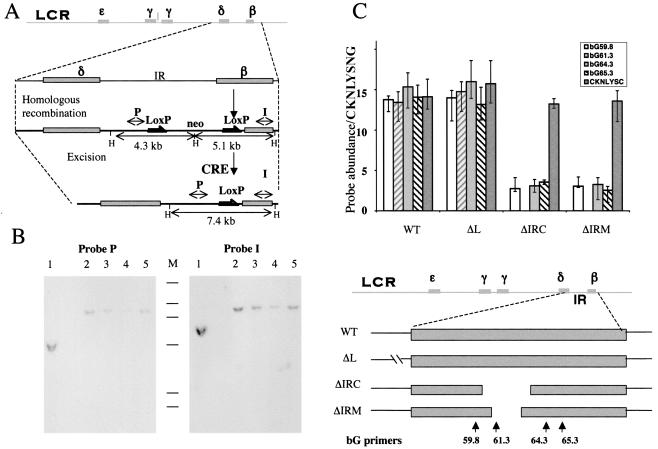
Chicken DT40 cells harboring human chromosome 11 have been described previously (16). Homologous recombination was used to replace sequences within the β-globin locus with the neomycin marker flanked by LoxP sites as described previously (16). The intervening neomycin marker was excised by CRE-mediated recombination, and the structure of the recombinants was verified by Southern hybridization analyses. The region between the PmeI and MfeI sites (coordinates, 61111 to 62805) was deleted in the clone designated ΔIRC; the region between the SnaBI and BspMI sites (coordinates, 61872 to 62267) was deleted in the clone designated ΔIRM, and the region between two PmlI sites 5′ of the IR (coordinates, 46122 to 55216) was deleted in the clone designated ΔL. All coordinates are for the compiled human β-globin locus (GenBank sequence accession no. U01317.1).
Replication initiation analyses.
The probes and primers used in this study are listed in Table 1. The IR fragments tested for replicator activities are listed in Table 2. Genomic DNA and nascent-strand DNA were prepared as described previously (4). Briefly, DNA was collected from asynchronous cultures and denatured by boiling followed by rapid cooling, and short DNA strands were size fractionated on neutral sucrose gradients. DNA strands ranging in size from 0.6 to 2.5 kb were collected and treated with λ exonuclease, as described previously (7, 36). Nascent strands were further size fractionated by gel electrophoresis and amplified by real-time PCR in an ABI 7900 thermocycler using a series of probe-primer combinations surrounding the inserted replicator and adjacent sequences. The amount of DNA in each sample was quantified by pico-green analysis (Molecular Probes, Eugene, Ore.). Genomic DNA that was not treated with exonuclease was used as a standard for calculating the number of molecules in the template. Genomic DNA from simian CV-1 cells was used as a nontemplate control to verify that primers used in the study were specific for the inserted DNA. The LacZ primer-probe combination, which lies nearly 5 kb away from the inserted replicator candidates, was used as a standard control for sequences that did not initiate DNA replication (1). Data from three PCRs for each primer-probe combination were used to calculate the amount of sequence-specific nascent strands, as described previously (4). All the experiments were performed at least three times, and a representative PCR analysis from a single experiment is shown.
TABLE 1.
Primers and probes used in the study
| Name | Acc no.a | Globin locus coord.b | Forward primer | Reverse primer | Probe |
|---|---|---|---|---|---|
| bG59.8 | U01317.1 | 59861-59834 | TGGAAAAGCAACCCCTGC | AACTATGGATCCTTCTCTTGTGTTGG | GCTGCAGATACCATCATCCTGGCTTCAA |
| bG61.3 | U01317.1 | 61274-61301 | ACAGAGGCTTTTTGTTCCCCC | GGTAATCAGTGGTGTCAAATAGGAGG | GACACTCTTGCAGATTAGTCCAGGCAGA |
| bG63.5 | U01317.1 | 63581-63608 | GGACAGCAAGAAAGCGAGCT | TCAGAAAGTGGTGGCTGGTG | GCTAATGCCCTGGCCCACAAGTATCACT |
| bG63.7 | U01317.1 | 63702-63740 | TTTATCTTATTTCTAATACTTTCCCTAATCTCTTT | CCTTAACCCAGAAATTATCACTGTTATTC | TCAGGGCAATAATGATACAATGTATCATGCCTCT |
| bG64.1 | U01317.1 | 64157-64131 | AGTGGCCAGGCGAGGAG | ACCACCTTTCCCCTGAAGTGT | AACCATCTCGCCGTAAAACATGGAAGG |
| bG64.3 | U01317.1 | 64257-64278 | GGCATGGTTTGACTGTCCTGT | TGCAGATTCCGGGTCACTG | CCCTTCTTCCCTGCCTCCCCCA |
| bG65.3 | U01317.1 | 65356-65332 | TGAGTAATAGTTTCCTGATTCTCCCA | AAAGTCACTCTCATGGAAACAGACA | CCCCAACCCCTGGAAACCATACCTC |
| Hygromycin resistance | AGCACGAGATTCTTCGCCC | TGATCGAAAAGTTCGACAGCG | CCGAGAGCTGCATCAGGTCGGAG | ||
| LacZ | GATGTGCGGCGAGTTGC | ACCGACATCGCAGGCTTCT | AGTTTCTTTATGGCAGGGTGAAACGCAGG | ||
| CKNLYZC | AF410481 | AGATCAACAGCCGCTGGTG | CACGGGATGTTGCACAGGT | ATGGCAGGACCCCAGGCTCCAG | |
| CKNLYZNG | AF410481 | TCCAGCAGGCCAAAGAGTCT | GGATACCATCTTACCTTGTTGAGAAAT | CTGAATGTTGTGTTGCCGGAGACCTG |
GenBank sequence accession number.
Coordinates of globin locus.
TABLE 2.
Fragments from the globin IR
| Name | Positiona | Restriction sites |
|---|---|---|
| bGRep-P | 59590-62187 | HindIII-Ncol |
| bGRep-I | 62188-65422 | NcoI-XbaI |
| ΔB | 59590-59882, 60677-62187 | HIII-BI, BI-NcoI |
| ΔH | 59590-60696, 61323-62187 | HIII-HII, HII-NcoI |
| ΔS | 59590-61503, 61870-62187 | HIII-Sphl, SnaBl-Ncol |
| ΔS-N | 59590-61870 | HIII-SnaBl |
| S-N | 61870-62187 | SnaBl-Ncol |
| ΔAT | 61557-61620 | |
| LI(AT) | 61557-61620 | |
| ΔAG | 62074-62118 | |
| ΔCCAAT | 62061-62065 | |
| ΔNP | 62611-67378 | PmII-HIII |
| ΔNE | 63532-65422 | EcoRI-Xbal |
| ΔNS | 63753-65422 | Swal-XbaI |
| ΔP | 626117-64302, 64561-67378 | PmII-Pstl, Pstl-HIII |
GenBank sequence accession no. U01317.1.
Mutagenesis.
The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to construct replicator mutants. Briefly, primer annealing was followed by extension with PfuTurbo DNA polymerase and incubation with DpnI, which digests the dam-methylated parental DNA template and selects for mutation-containing newly synthesized DNA. All mutants were verified by DNA sequencing (National Cancer Institute core sequencing facility).
RESULTS
The IR is composed of two independent replicators.
In a previous study, we suggested that the IR contains a central sequence of 2.3 kb, spanning from the promoter through the second intron of the β-globin coding sequence, and that this region is essential, but not sufficient, for initiation to occur. Initiation of DNA replication required interaction of this central sequence with other elements residing on either side of the IR. These data could imply that the central region contains unique information required for initiation, which could be complemented by redundant information at each of the two adjacent regions. Alternatively, the data could be interpreted to suggest that the IR contains two independent, redundant replicators that are both disrupted when the central region is deleted. To distinguish between these two hypotheses, we examined whether it is possible to dissect the IR into two DNA fragments that can direct independent initiation at ectopic sites.
We used the nascent-strand abundance assay to determine whether initiation of DNA replication occurred from a series of clones inserted at a fixed site in the E25B4 simian cells (B4 site), as described previously (1). This method involves isolation of nascent DNA strands, which are small (600 to 2,500 bases), RNA-primed DNA fragments, from cells containing replicator candidates (see Materials and Methods for details). Insertion of all the putative replicator candidates into a fixed site in the simian genome was performed using site-specific recombination to neutralize position effects, which are known to affect initiation capacity. To measure whether DNA replication initiated within the inserted DNA fragments, we probed the isolated nascent DNA strands for sequences derived from the inserted replicator. To facilitate the analysis of initiation of DNA replication from multiple clones inserted at ectopic sites, we modified our previous procedure to incorporate real-time PCR for measuring nascent-strand abundance. A high abundance of sequences from the inserted sequence, relative to distal fragments that do not initiate replication, indicated that replication initiated within the inserted fragment. This method allows rapid analysis of multiple clones in a consistent and reliable manner (4). It is important to note that probe abundance and amplification efficiency can vary up to twofold between independent nascent-strand preparations derived from the same cell line. This sample variation limits the use of the nascent-strand abundance method in fine mapping of initiation sites and does not allow detection of less-than-twofold variations in origin usage. Nevertheless, the method is very sensitive and reproducible in determining whether replication initiated from a specific known sequence. Combined with genetic analysis, this is therefore the method of choice for determining whether a fragmented or altered replicator retained its ability to initiate replication. An example of the nascent-strand abundance assay coupled with real-time PCR analysis is shown in Fig. 1.
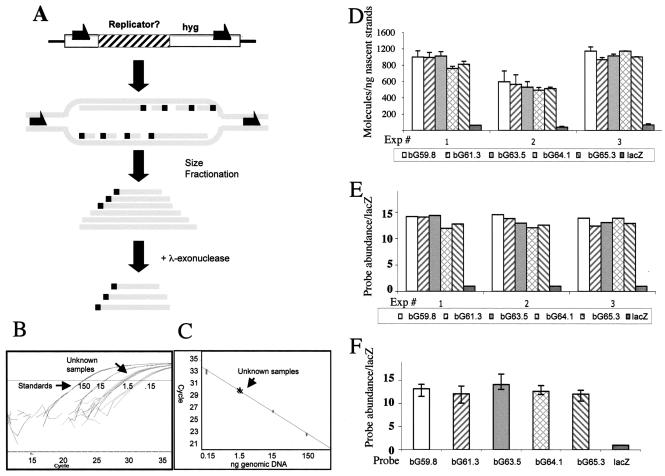
FIG. 1.
Replication initiation assays. (A) Schematic illustration of the methodology used for nascent-strand abundance assay. DNA strands are depicted as solid gray lines, and 5′ primer RNAs are shown as solid black boxes. Newly replicated origin-proximal DNA was selected by size (600 to 2,500 bp) and by resistance to lambda exonuclease (an enzyme that digests DNA with a 5′ DNA tail but not DNA with a 5′ RNA tail). Nascent strands isolated in this way were then subjected to real-time PCR with primers encompassing the locus of interest. (B) An example of real-time PCR output. Standards of genomic DNA at fixed concentrations were used in a PCR along with an unknown sample. The abundance of the PCR products in nascent DNA was calculated based on the cycle in which fluorescence from the real-time PCR crossed the manually set threshold. (C) A calibration curve based on the data shown in panel B. (D to F) An example of data processing. (D) PCR amplification of three independent preparations of nascent strands from CV-1 E25B4 cells containing the β-globin IR at the B4 site. Histogram bars show the average amplification efficiency of PCR products from a series of probes (for probe sequences and locations, see Table 1). Error bars indicate standard deviations. (E) The same data shown in panel D, represented as the ratio of the abundance of specific sequences to the abundance of sequences amplified by lacZ primers. (F) A histogram depicting the average abundance of specific sequences in nascent strands, represented as the average of the measurements shown in panel E from three independent nascent-strand preparations. Error bars indicate the upper and lower ranges of the measured ratios.
A series of nonoverlapping fragments of the IR were inserted into a fixed site within the simian genome, using site-specific recombination. Figure 2 shows a nascent-strand abundance assay with such simian cells containing two fragments that included a portion of the IR central core region. This region was previously determined to be necessary, but not sufficient, for initiation of DNA replication (Fig. 2A). The 5′ fragment, running from the HindIII site at position 59590 to the NcoI site at position 62187, contained a portion of the central core region and a region located 5′ to the central region that was unable to initiate replication but could complement the entire 2.3-kb central region in facilitating initiation. This fragment includes the promoter of the β-globin gene. Similarly, the 3′ fragment from the NcoI site at position 62188 to the XbaI site at position 65422 contained the complementary portion of the core and a sequence located 3′ to the central region, which could complement the 2.3-kb central region in facilitating initiation. This fragment includes the large intron of the β-globin gene.
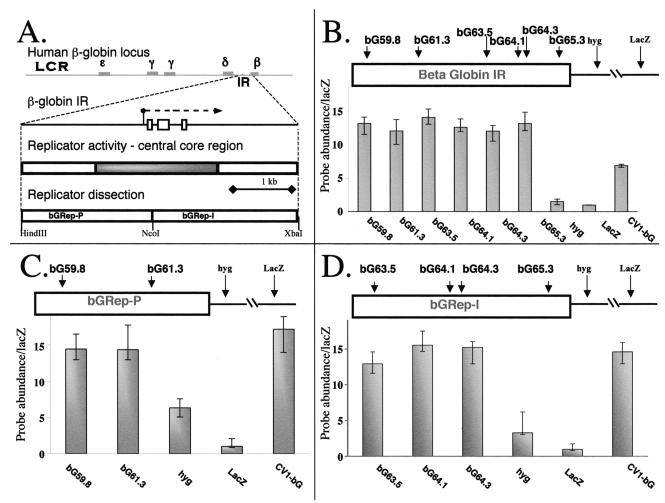
FIG. 2.
Identification of two nonoverlapping, independent replicators within the human β-globin IR. (A) A schematic representation of the IR from the human globin locus. Replication initiates from the region between the two adult β-globin genes (top line). The IR (IR) encompasses the promoter and the majority of the β-like-globin gene (second line); preliminary analysis identified a core central region within the IR, which was essential but not sufficient for initiation (third line) (1). The present analysis divided the IR into two fragments designated bGRep-P and bGRep-I (bottom line). The gray boxes on the top line represent the globin genes; white boxes in the second line represent exons, while the dashed line represents the direction of transcription. (B to D) DNA fragments originating from the human β-globin IR were inserted into the FRT site in CV-1 E25B4 cells as described. The abundance of IR-derived DNA sequences in short, exonuclease-resistant, newly replicated DNA was determined by real-time quantitative PCR and quantified as shown in panel F. A dissection of the locus at the NcoI site (coordinate 62187) preserved the ability to initiate DNA replication in both fragments when each fragment was inserted at the ectopic site, suggesting that both fragments can function as independent nonoverlapping replicators. Real-time PCR analysis of the entire IR (B), analysis of bGRep-P, (C), and analysis of bGRep-I (D) are shown. Data are represented as the number of molecules amplified from RNA-primed nascent strands divided by the number of molecules amplified from the same preparation by the lacZ primers. Each histogram bar depicts the average of three independent measurements. Error bars represent the range of measured ratios.
We performed real-time PCR on a preparation of newly replicated DNA strands to determine whether initiation of DNA replication occurred within the inserted fragments. We used a series of primers located within the replicator fragments, as well as primers amplifying sequences from the hygromycin resistance gene (hyg) and the lacZ marker, which is located 5 kb away from the inserted fragments and serves as a negative control. Sequences from the African green monkey β-globin region (CV-1 bG) served as positive controls. (Primers from the β-globin locus are labeled “bG” followed by a number that designates the map location of the amplified sequence in kilobases [Fig. 2B to D]. For primer and probe sequences, see Table 1). Figure 2B to D shows analyses of nascent strands derived from cells harboring either the entire, previously identified IR or each of the two fragments described above. Sequences from each of these fragments were abundant in newly replicated, short exonuclease-resistant DNA strands and exhibited amplification similar to that of the entire IR. These data suggest that each of the two nonoverlapping fragments was able to confer initiation at an ectopic location to the same extent as the entire IR. Hence, two redundant elements within the human β-globin IR contain information that is required for replicator activity. Of these two newly identified replicators, the upstream sequence, which contains the promoter of the β-like β-globin gene, was labeled β-globin replicator P (bGRep-P), whereas the adjacent downstream sequence, which includes the large intron within the β-like β-globin gene, was labeled β-globin replicator I (bGRep-I).
It should be noted that sequences derived from the hygromycin resistance marker were abundant in nascent strands from cells containing Rep-P, whereas these sequences were not abundant in nascent strands from cells containing Rep-I (for example, compare Fig. 2C with 2B and 2D). Since nascent strands collected in all experiments were of uniform size, these consistent variations indicate the occurrence of initiation events within the region extending from the center to the 3′ end of Rep-P, resulting in inclusion of sequences from the hygromycin resistance marker in nascent strands. The low abundance of hyg sequences in nascent strands from cells containing Rep-I suggests that initiation events are not frequent at the 3′ end of this replicator.
Activity requirements within the two replicators.
In order to determine which sequences are essential for the initiation of DNA replication, we created a series of cell lines harboring fragments from each of the replicators or containing point mutations in sequences we wished to test for replicator activity. All these mutants were inserted into a constant site in E25B4 cells (B4 site) and tested for replicator activity using the nascent-strand abundance assay.
An evolutionarily conserved AT-rich sequence was not required for initiation from replicator P.
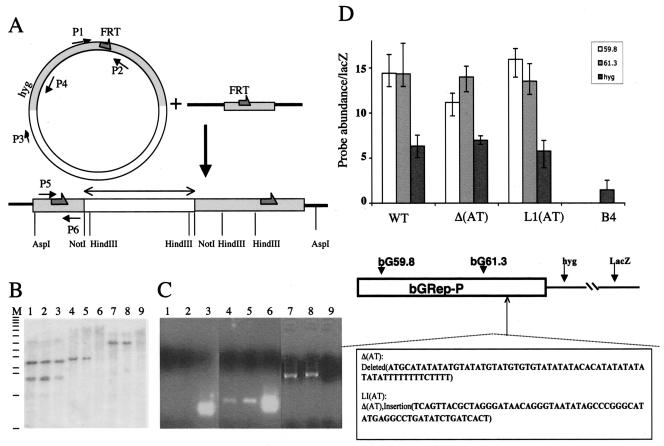
Evolutionary analyses had identified a conserved, AT-rich, alternate purine-pyrimidine sequence within the Rep-P region (21). Since this region was not essential for transcription, it was suggested that this conserved sequence might play a role in replication initiation. To test this hypothesis, we created a cell line harboring a modified Rep-P replicator in which the AT-rich sequence ATGCATATATATGTATATGTATGTGTGTATATATACACATATATATATATATTTTTTTTCTTTT was deleted. Then we created another cell line in which a non-AT rich sequence, TCAGTTACGCTAGGGATAACAGGGTAATATAGCCCGGGCATATGAGCCTGATATCTGATCACT, replaced the AT-rich sequence above. As shown in Fig. 3, fragments harboring the deletion of the AT-rich sequence [Δ(AT)] initiated DNA replication at the ectopic site, similar to results for the wild-type sequences. The deletion/linker insertion [L1(AT)] exhibited a similar behavior. These data suggest that the evolutionarily conserved AT-rich sequence was not essential for replicator activity.
FIG. 3.
An evolutionarily conserved alternate AT-rich stretch is not essential for replicator activity within the β-globin Rep-P replicator. (A) Insertion of the IR Rep-P fragment into an FRT-containing acceptor site in the simian genome using site-specific recombination. The insertion vector, used to clone putative replicator candidates, contains an FRT and a hygromycin resistance marker (hyg). The acceptor site has an identical FRT sequence inserted into the simian genome. Transfection of the vector into cells containing the target in the presence of excess FLP recombinase leads to frequent integrations of the entire insertion vector into the target. Integration disrupts the expression of the lacZ marker. Recombinant clones are selected based on hygromycin resistance and lack of lacZ expression, and Southern hybridization and PCR analyses are then used to verify that recombinant colonies contained single copies of the insertion vector in the acceptor sites. The filled grey arrows represent FRT sites; the double-headed arrow represents the location of the probe used in panel B; and the arrows designated P1 to P6 represent the locations of the primers used in panel C. (B) Analysis of the structure of recombinant clones using Southern hybridization. Lanes 1, 4, and 7 contain DNA from a colony with a single-copy integration of Rep-P at the target site; lanes 2, 5, and 8 contain DNA from a second colony containing the same insert exhibiting a similar structure; lanes 3, 6, and 9 contain DNA from a colony that exhibits a rearrangement of the inserted Rep-P fragment. DNAs were digested with HindIII (lanes 1 to 3), NotI (lanes 4 to 6), and AspI (lanes 7 to 9) and probed with a NotI fragment from the β-globin locus (double-headed arrow in panel A). Lane M shows molecular size markers ranging from 0.5 to 12.2 kb (Invitrogen, Ready-load 1 kb ladder). (C) PCR analysis of recombinant clones containing Rep-P. Lanes 1, 4, and 7 show the analysis of DNA from a colony with a single copy of Rep-P integrated at the target site, lanes 2, 5, and 8 show the analysis of DNA from a similar colony, and lanes 3, 6, and 9 show the analysis of DNA from a colony exhibiting integration at another genomic site. Lanes 1 to 3 show amplification products using primer pair P1 and P2; lanes 4 to 6 show amplification products using primer pair P3 and P4, and lanes 7 to 9 show amplification products using primer pair P5 and P6. Integration into the acceptor site will prevent amplification using primer pairs P1 and P2 but allow amplification using primer pairs P3 and P4 and P5 and P6. (D) DNA fragments containing Rep-P were mutated in vitro as indicated, inserted into the FRT site in CV-1 E25B4 cells, and tested for initiation activity as described in the legends to Fig. 1 and 2. The wild-type (WT) histogram bars show nascent-strand abundance data from the unaltered Rep-P. The B4 bar represents the nascent-strand abundance of a hygromycin marker at the FRT site in the absence of sequences from the β-globin replicator. Deletion of the AT-rich region, or replacement of this region with a non-AT-rich linker, did not affect initiation capacity.
Two separate sequence elements are required for initiation from replicator P.
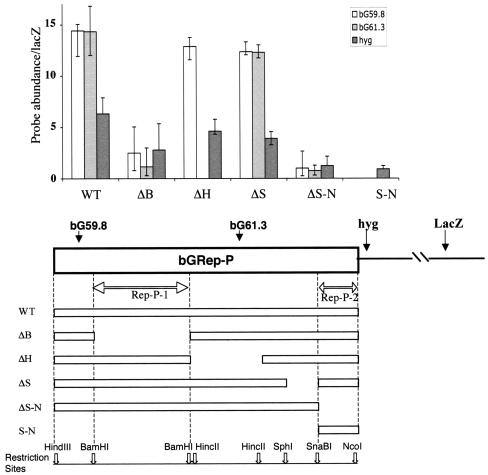
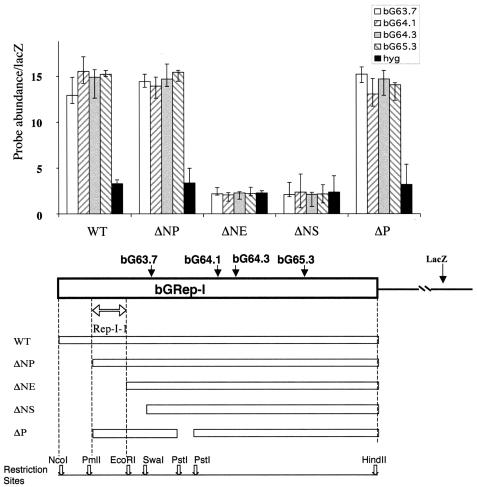
We next created a series of deletions in Rep-P in order to identify regions within this replicator that dictate initiation of DNA replication. An analysis of a series of such deletions is shown in Fig. 4. When the entire Rep-P is inserted, sequences from this replicator (wild type) are abundant in nascent strands. A deletion of 795 bp between the two BamHI sites (ΔB) dramatically diminished the ability of the fragment to initiate DNA replication, while a deletion of 627 bp between the two HincII sites (ΔH) did not affect initiation of DNA replication. Similarly, a deletion of 348 bp between the SphI site and the SnaBI site (ΔS) resulted in a slightly higher abundance of replicator sequences in nascent strands. However, this increase was within normal sample variation (the ratio between the amplification of replicator markers, such as the bG61.3 primers, and that of the distal lacZ marker was 14.2 for the wild-type insert versus 16.24 for the ΔS insert) and suggested that this deletion had no effect on replicator activity. In contrast, a deletion of 314 bp between the SnaBI site and the NcoI site (ΔS-N) dramatically diminished initiation of replication. The 314 bp between the SnaBI site and the NcoI site (S-N) could not initiate DNA replication when inserted independently into the B4 site. Therefore, two regions seemed to cooperate to initiate DNA replication from Rep-P: a region of 795 bp between the two BamHI sites, designated Rep-P-1, and a region of 314 bp between the SnaBI site and the NcoI site, designated Rep-P-2.
FIG. 4.
Genomic regions required for replicator activity within Rep-P. DNA fragments originating from bGRep-P (see Fig. 2) were inserted into the FRT site in CV-1 E25B4 cells and tested for initiation activity using the nascent-strand abundance assay described in the legends to Fig. 1 and 2. Deletion of a 300-bp fragment extending from the SnaBI site to the NcoI site (ΔS-N, coordinates 61870 to 62187) and deletion of an 800-bp fragment between the two BamHI sites (ΔB, coordinates 59882 to 60677) greatly diminished the representation of sequences from the inserted human β-globin fragment in nascent strands. Other deletions did not affect replicator activity, suggesting that the two 300- and 800-bp sequences (Rep-P-1 and Rep-P-2, outlined by double arrows) cooperate to confer the ability to initiate DNA replication at the ectopic site. WT, wild type.
A 921-bp region encompassing the large intron is essential for initiation from replicator I.
We next made a series of deletions in Rep-I, as shown in Fig. 5. Similar to the analysis of the Rep-P replicator, we found that several deletions had no effect on replicator capacity. For example, a deletion of 423 bp between the NcoI site and the PmlI (ΔNP) site did not affect initiation. In contrast, a deletion of 1,344 bp between the NcoI site and the EcoRI site (ΔNE), as well as a larger deletion of 1,665 bp between the NcoI site and the SwaI site (ΔNS), prevented initiation. A PstI fragment that lies 3′ to the EcoRI site (ΔP) was not required for initiation. Since deletion of the region between the NcoI site and the PmlI sites initiated replication at the ectopic site, whereas a larger deletion that also deleted the PmlI-EcoRI fragment could not initiate, we concluded that a region of 921 bp between the PmlI site and the EcoRI site was essential for replicator activity. This region, which was deleted from both the ΔNE and the ΔNS constructs, was designated Rep-I-1. Interestingly, the Rep-I-1 region contains a putative ORC binding site (8) and a putative cell cycle-dependent DNase-hypersensitive region (9, 10, 18, 19).
FIG. 5.
Genomic regions required for replicator activity within Rep-I. DNA fragments originating from Rep-I (see Fig. 2) were inserted into the FRT site in CV-1 E25B4 cells and tested for initiation activity using the nascent-strand abundance assay as described in the legends to Fig. 1 and 2. Deletion of DNA fragments, including a region extending from the PmlI site to the EcoRI site designated Rep-I-1 and outlined by a double arrow, did not allow replication. These data suggested that this region was essential for the initiation of DNA replication. WT, wild type.
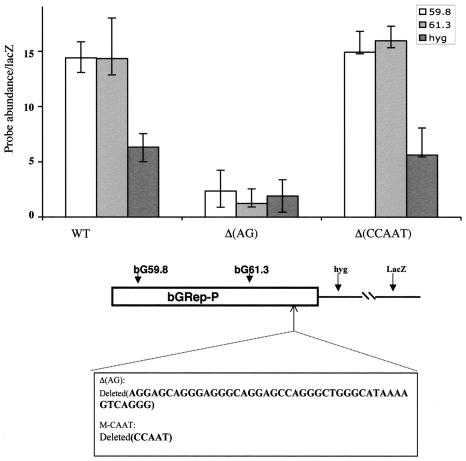
A purine:pyrimidine-rich domain was required for initiation of DNA replication from replicator P.
The sequence designated Rep-P-1 (Fig. 4), which was essential for initiation of DNA replication from Rep-P, contains an asymmetric, purine:pyrimidine stretch (purines on one strand and pyrimidines on the other strand; 78% AG), AGGAGCAGGGAGGGCAGGAGCCAGGGCTGGGTCAGGG. We used in vitro mutagenesis to delete this region from the 5′ replicator and inserted a fragment containing this deletion into the B4 site [Fig. 6, Δ(AG)]. The nascent-strand analyses shown in Fig. 6 suggested that this deletion blocked initiation of DNA replication from Rep-P, implicating this region as essential for replicator activity.
FIG. 6.
An asymmetric purine:pyrimidine stretch is required for replicator activity within Rep-P. DNA fragments containing Rep-P were mutated in vitro as indicated, inserted into the FRT site in CV-1 E25B4 cells, and tested for initiation activity as described in the legends to Fig. 1 and 2. Deletion of the CAAT box did not affect initiation capacity, whereas a deletion of the asymmetric purine:pyrimidine stretch (AG) blocked initiation from Rep-P sequences at the ectopic site. WT, wild type.
Deletion of the CCAAT box of the β-globin promoter had no effect on initiation.
The Rep-P replicator colocalizes with the β-globin promoter. Although replication initiates from the region between the two adult globin genes even in the absence of transcription, it was interesting to determine whether transcription may have a role in determining initiation of DNA replication from the independent Rep-P. Therefore, we next determined whether the CCAAT box, which is essential for transcription from the β-globin promoter, has a role in initiating DNA replication from Rep-P. As shown in Fig. 6 [Δ(CCAAT)], DNA replication initiated from Rep-P regardless of the presence or absence of the deletion of the CCAAT box. Nascent strands amplified from cells containing the CCAAT deletion fragments apparently exhibited stronger amplification than nascent strands from cells containing the wild-type fragments, but this variation was not significant, since the ratios between the amplification efficiency of sequences within the inserted replicators and that of the distal lacZ marker varied less than twofold. These data suggest that the deletion of the CCAAT box did not affect initiation efficiency, implying that the transcription potential of the β-globin promoter does not correlate with the initiation potential of the Rep-P replicator.
The intact β-globin locus and ectopic sites exhibit similar requirements for replicator activity.
The experiments outlined above provided new insight into the nature of replicator sequences that are required for initiation of DNA replication at ectopic locations. However, these studies did not establish whether these sequences are essential for initiation at the native locus. Although the Lepore deletion does not initiate replication from a region adjacent to the deleted IR, it remains to be determined whether this was due to deletion of the IR or to another defect in the diseased chromosome. We next directly addressed the question of whether the deletion of putative replicator sequences will prevent initiation in the context of the entire β-globin locus. To do this, we exploited the fact that the DNA replication initiation profile was conserved when the human chromosome containing the β-globin locus (chromosomes 11) was transferred into cells originating from other vertebrates, such as murine-human and chicken-human somatic cell hybrids (3). Figure 7 shows an analysis of initiation of DNA replication in β-globin loci contained within human chromosomes in chicken-human somatic cell hybrids. Initiation was measured in hybrids containing the entire human chromosome 11 and two derivatives in which parts of the IR were deleted, ΔIRC and ΔIRM. Each deletion removed parts of both the 5′ and the 3′ replicators, Rep-P and Rep-I, which were identified as essential through ectopic analyses. As shown in Fig. 7, the transferred wild-type human chromosome initiated replication. In contrast, replication failed to initiate in the region between the two adult genes in the ΔIRC and ΔIRM mutants, in which parts of the two replicators had been deleted. As a control, we included a fourth chromosome (ΔL), in which an extended region 5′ to the IR was removed. This chromosome initiated DNA replication from the IR, confirming that the process of creating the deletions did not interfere with the ability to initiate DNA replication.
FIG. 7.
Requirements for replicator activity within the human β-globin locus in the context of an intact human chromosome 11. The entire human chromosome 11 was transferred to the chicken DT40 cell line, as described previously (16). The indicated regions were deleted from the human β-globin locus: ΔIRC, deletion of a 1.7-kb fragment between the PmeI and MfeI sites; ΔIRM, deletion of a 500-bp fragment between the SnaBI and BspMI sites; and ΔL, deletion of a fragment between two PmlI sites 5′ of the IR. (A) Illustration of the homologous recombination followed by CRE-mediated excision of the IR fragments (for details, see reference 16). The top line shows a schematic illustration of the entire β-globin locus; the β-like globin genes are shown as shaded boxes. The second line shows a magnification of the IR region between the δ and the β genes. The third line shows the structure of a recombinant in which a neomycin resistance gene (neo) flanked by LoxP sites (grey arrows) replaced a part of the IR by homologous recombination. The fourth line shows the structure of a chromosome created by Cre-mediated excision, which deleted a part of the IR, leaving a LoxP site. Double-headed arrows designated P or I represent the location of probes from the human β-globin locus that straddle the inserted LoxP sites. These probes were used in the hybridization experiments shown in panel B. Sites marked by the letter H represent HindIII restriction sites. (B) Southern blot analysis of the structure of ΔIRC. Genomic DNA (10 μg) was digested with HindIII, fractionated on a 0.6% agarose gel, immobilized on a nylon membrane, and hybridized with probes designated I and P as indicated. Lanes 1 contain DNA from a homologous recombinant (third line from the top in panel A); lanes 2 to 5 contain DNA from three different clones resulting from CRE-mediated deletion. Lane 3 contains DNA from the chosen clone ΔIRC. M, molecular size markers derived from 32P-labeled HindIII digestion of bacteriophage λ DNA ranging from 2.3 to 23 kb. The insertion of the neo marker into the globin locus introduced a HindIII site. The two probes, P and I, identified two separate HindIII fragments in the homologous recombinant (4.3 kb and 5.1 kb). After excision, one of the HindIII sites was eliminated and both probes identified a 7.4-kb fragment. Excision was also verified by lack of hybridization to a probe containing the neomycin gene (data not shown). (C) Initiation of DNA replication was measured by the nascent-strand abundance assay as described in the legends to Fig. 1 and 2, using primers from the human β-globin locus except for the CKNLYSC and the CKNLYSNG primers, which were derived from the chicken lysozyme locus (see Table 1 for sequence information). The primer pair CKNLYSNG, which is not abundant in nascent strands from chicken cells (45), served as a standard. Primer locations are illustrated below the histograms. The ΔIRC and ΔIRM chromosomes did not initiate DNA replication, whereas the unaltered chromosomes and the ΔL chromosomes initiated DNA replication from the IR. WT, wild type.
DISCUSSION
Replicator activity at ectopic sites and native loci.
We measured replicator activity as defined by intrachromosomal initiation at ectopic sites. This approach is based on earlier observations suggesting that replication origins included within lambda phages and cosmid clones can initiate replication when integrated into the mammalian genome (12, 13, 23), on our previous observation that the human β-globin IR can initiate replication at ectopic sites (1), and on subsequent ectopic initiations observed for the DHFR (5) and the c-myc (30, 39) loci. These studies indicate that replicator activity is a feature of mammalian replication initiation sites and suggest that ectopic initiation assays can be used to identify DNA sequences that dictate the initiation of DNA replication. The ectopic initiation analyses reported here identified two redundant replicators within the human β-globin IR and demonstrated that initiation activity within these replicators was dictated by short sequences. Such short sequences could be mutated to produce nonfunctional replicators. Analyses using intact human chromosomes in human-chicken hybrids further revealed that deletions that produced nonfunctional replicators at the simian ectopic sites failed to initiate replication in the native chromosome. These observations demonstrate that the same sequences dictate initiation of DNA replication at both ectopic and native loci and validate the use of ectopic replicator assays to study the sequence requirements for initiation in mammalian cells. However, our data also imply that replicator clustering can mask the sequence requirements for replication initiation and complicate the interpretation of ectopic or in situ initiation data. For example, deletion of sequences essential for initiation in the context of a single replicator will not prohibit initiation in the context of the entire globin IR, since the other replicator stays intact. Replication clustering may, therefore, underlie the apparent lack of sequence specificity observed in some replication IRs in mammalian cells.
While the studies presented here helped identify DNA sequence elements required for initiation, they did not determine the minimal requirement for replicator function. Moreover, studies at native loci suggest that sequences required for replicator activity may localize at a considerable distance from replication origins. For example, in the human β-globin locus, deletion of the locus control region and upstream sequences in Hispanic thalassemia prevents replication initiation from the IR (2). By contrast, as shown here and in previous studies (1), the IR and fragments of the IR can initiate replication at simian ectopic sites in the absence of the LCR. These data suggest that other sequences may substitute for the LCR in providing an environment permissive for initiation. Similarly, distal sequences are required for initiation of DNA replication in the Chinese hamster DHFR locus (29, 42). Therefore, it is important to note that while the ectopic assays reported here were useful for identification of sequences required for replicator function, the question of whether specific combinations of these essential sequences are sufficient for initiation of DNA replication awaits further studies.
Sequence requirements for initiation of DNA replication in the human β-globin locus.
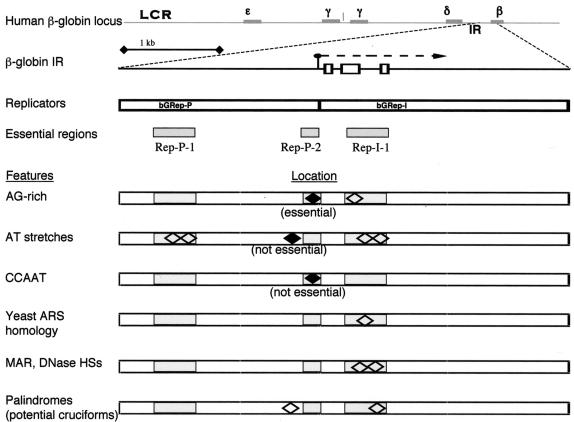
Unraveling the two replicators allowed us to determine which of the sequence elements abundant in replication origins were critical for dictating replicator activity within the globin IR. As summarized in Fig. 8, DNA sequences essential for initiation from Rep-P and Rep-I contain some AT-rich stretches and a region of asymmetric purines:pyrimidines, which was essential for initiation in Rep-P. However, the distances between these sequence features are not conserved, and the regions essential for replicator activities do not share significant sequence homology beyond the AT-rich region and the asymmetric purine:primidine stretches. Moreover, these essential regions do not exhibit easily identifiable homology to other mammalian replicators and replication origins identified to date (8). These observations may imply that although replicator activity is determined genetically, the sequence requirements for initiation are unique for each replicator. Alternatively, the data may suggest that replicator activity is determined by a combination of sequence features, such as AT-rich and asymmetric purine:pyrmidine elements, irrespective of distance or adjoining sequences. The studies presented here suggest that a combination of AT-rich and asymmetric purine:pyrimidine sequences have a role in facilitating initiation but had not determined whether these sequences were the sole requirements for initiation at ectopic sites. As shown in Fig. 8, our data do not support a role for other sequence features commonly observed in replication origins, such as palindromes, cruciforms, and bent-DNA regions, in determining initiation activity within the β-globin replicators.
FIG. 8.
Summary of sequence features implicated in replication IRs and their locations within the two β-globin replicators. Diamond shapes indicate the locations of sequence features within the β-globin IR relative to the locations of the two replicators, bGRep-P and bGRep-I, and the essential regions within the replicators. Sequences whose roles in initiating DNA replication were tested directly by in vitro mutagenesis are depicted as shaded diamonds and designated essential or not essential.
The role of AT-rich sequences.
In budding yeast (Saccharomyces cerevisiae), most replicators are comprised of a clearly identified ORC binding site, which is very AT rich, and certain auxiliary sequences (8, 32). However, in some cases, multiple copies of less than perfect ORC binding sites can be sufficient for initiation (52). In other organisms, including other yeast species, such as Schizosaccharomyces pombe, the requirements for initiation are not as well characterized, and the common features that seem to be present in replication initiation sites are mainly AT-rich tracts (8, 17, 33, 34, 39, 41-43, 47, 55, 56). This specificity reflects the affinity of the fission yeast ORC4, which contains AT hooks, to AT-rich sequences (37). At the molecular level, two AT-rich ORC binding sites cooperate for efficient formation of preinitiation complexes and initiation of DNA replication (50). Mammalian ORC can initiate replication in a sequence-independent manner in Xenopus egg extracts and does not exhibit sequence specificity beyond a strong preference for AT-rich sequences (53). Two of the regions essential for replicator activity, Rep-P-1 in Rep-P and Rep-I-1 in Rep-I, contain long series of AT-rich nucleotide stretches. These elements are necessary but not sufficient for initiation (Fig. 8) (1) and may represent preferred ORC binding sites or function as DNA-unwinding elements (26, 38). Our data also reveal that an evolutionarily conserved alternate (AT) sequence, which was speculated to play a role in replicator activity (21), was not essential for initiation at Rep-P.
The requirement for an asymmetric purine:pyrimidine sequence.
Analysis of the 5′ replicator, Rep-P, revealed that replicator activity depended on the purine:pyrimidine (AG-rich) tract at position 62074. This element was essential, but not sufficient, for replicator activity. An identical element is not present in Rep-I. However, a different AG-rich region (>80% AG) is present in Rep-I-1 (Fig. 8) which is necessary for initiation (starting at position 62695 in the globin locus [GenBank accession no. U01317.1]). This sequence lies just upstream of the AT-rich stretch. Similar, but not identical, AG-rich stretches (>75% AG) are present in Chinese hamster DHFR Ori-β and in the human Lamin B replication origins (starting at positions 4294 in the sequence under GenBank sequence accession no. 1731964 and 4277 in the sequence under GenBank sequence accession no. 186920, respectively). Functional analyses of these replication origins will be required to assess whether these other AG-rich sequences are indeed essential for initiation of DNA replication.
Role of transcriptional regulatory elements.
Replication initiation sites are preferentially localized to intergenic regions in yeast (11), and some mammalian replication IRs were also mapped in intergenic (24, 55) or promoter (6, 8, 35) regions (for a review, see references 8 and 22). The onset of transcription coincides with specification of replication origins in Xenopus development (27), and the transcription status of developmentally regulated loci may influence origin choice in flies (40) and mammals (56). Transcriptional regulatory elements may act as replication enhancers in viral systems (15), while transcription may inhibit plasmid replication in bacteria (49) and in yeast (48), and protection from transcription dictates the positions of initiation sites in yeast chromosomes (11, 48). Here, we found that although Rep-P colocalizes with the promoter of the β-like β-globin gene, deletion of the transcriptional regulatory CCAAT element did not affect the ability to initiate DNA replication. Moreover, Rep-I is located within the β-globin transcription unit. These observations, along with previous data for the CAD (31), RPS14 (51), and lamin B (6, 8, 35) replication origins, suggest that replicator activity in mammalian cells can localize in transcribed regions. These data are consistent with the observation that replication initiates from the human β-globin IR regardless of the transcriptional status of the locus and the identity of the gene that is being transcribed (3, 35).
Redundant modular replicators in the initiation of DNA replication.
Redundancy in replication initiation sites seems to be a prevalent feature in eukaryotes. In S. cerevisiae, most initiation sites are defined by distinct replicator sequences, which consist of well-defined sequence modules (8). However, some complex replication initiation sites contain redundant replicators (52). A similar clustering of replicators is observed in the replication initiation site of S. pombe ura4, which is adjacent to a “replication enhancer” that contains short stretches of AT-rich sequences (34). These enhancer sequences exhibit functional redundancy with sequences from within the replication origin. In mammalian cells, replication within the Chinese hamster DHFR gene initiates from a broad zone of potential initiation sites, in which some regions initiate replication at higher frequencies than others (17, 36). One of these frequent initiation sites, Ori-β, can act as a replicator in ectopic assays (5). However, unlike the case with the β-globin IR (35), deletion of Ori-β and other initiation sites from the native locus does not lead to loss of initiation activity in the remaining region (29, 42). At the human c-myc IR, each of a series of deletions decreases the initiation potential of the replicator at an ectopic site (39), suggesting that this initiation site also contains multiple elements essential for initiation. The human β-globin IR is an example of a replication origin that displays replicator activity both at ectopic sites and within the native locus. Here, we show that the IR is composed of two elements that can each satisfy the criteria for a replicator. Within these replicators, initiation requires cooperation between nonredundant sequence elements. Our data suggest that the two replicators that comprise the IR are truly redundant, in that each can direct initiation of DNA replication regardless of the presence of the other replicator. These data are consistent with previous analyses of replication fork direction. These studies observed replication forks emanating from the IR located between the two adult β-like globin genes but did not detect a strong preference for any direction within the IR (3, 35). Sequences from both replicators are represented in nascent strands, in agreement with previous observations in ectopic (1) and native (3) loci. Hence, unlike situations in yeast, in which initiation from some replicators inhibits initiation from adjacent potential replicators (20, 54), replication initiates from both replicators at the β-globin IR.
One possible role for the existence of redundant replicators in mammalian cells may be the need to modulate initiation site preferences to coordinate replication with transcription and chromatin remodeling. Examples for such preferences include the specification of initiation sites after mid-blastula in the developing Xenopus laevis embryo, in which replication initiates from random sites earlier in development (27), and the replication of puff II/9A of Sciara coprophila (40), which initiates from an 8-kb initiation zone in mitotic embryonic cells but specifies a contracted 1.2- to 2-kb replication origin during the amplification stage of fly development. Origin usage also changes during mammalian B-cell development, where a shift in replication timing of the IgH region is accompanied by the activation of a novel replication origin (55). Other examples in which replication origin usage was altered in a tissue-specific manner have also been reported (8). In all these cases, altered origin preferences correspond to significant changes in transcriptional activity. Although the β-globin locus undergoes significant chromatin remodeling and transcriptional activation during erythroid differentiation, vertebrate β-globin loci studied to date did not exhibit alterations in origin usage (3, 4, 35, 46). However, since all the data were obtained with somatic cell lines, we are unable to exclude a specific role for each of these replicators at specific times during development. Further studies are necessary to further investigate this question and to elucidate the functions of specific sequence modules in the initiation of DNA replication.
Acknowledgments
We thankfully acknowledge Mel DePamphilis for critical reading of the manuscript and for numerous helpful suggestions. We are grateful to Geoffrey M. Wahl, Yves G. Pommier, Kurt W. Kohn, Tsutomo Shimura, Haiqing Fu, Elsa Bronze Da Rocha, and Fred E. Indig for helpful suggestions. We thank Luo-Wei Rodewald for assistance in making the deletion mutants shown in Fig. 1 and Agnes Telling for assistance in making the DT-40 cells containing human chromosome 11 and deletions in the human β-globin locus in these cells.
Work in M.I.A.'s laboratory is supported by the NIH's intramural program. M.G. is supported by NIH grants DK44746 and HL57620.
REFERENCES
- 1.Aladjem, M., L.-W. Rodewald, J. L. Kolman, and G. M. Wahl. 1998. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science 281:1005-1009. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem, M., and G. M. Wahl. 1997. Mapping replication origins by leading strand analysis in the absence of protein synthesis. Methods Companion Methods Enzymol. 13:281-292. [DOI] [PubMed] [Google Scholar]
- 3.Aladjem, M. I., M. Groudine, L. L. Brody, E. S. Dieken, R. E. K. Fournier, G. M. Wahl, and E. M. Epner. 1995. Participation of the human beta globin locus control region in initiation of DNA replication. Science 270:815-819. [DOI] [PubMed] [Google Scholar]
- 4.Aladjem, M. I., L. W. Rodewald, C. M. Lin, S. Bowman, D. M. Cimbora, L. L. Brody, E. M. Epner, M. Groudine, and G. M. Wahl. 2002. Replication initiation patterns in the beta-globin loci of totipotent and differentiated murine cells: evidence for multiple initiation regions. Mol. Cell. Biol. 22:442-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman, A. L., and E. Fanning. 2001. The Chinese hamster dihydrofolate reductase replication origin beta is active at multiple ectopic chromosomal locations and requires specific DNA sequence elements for activity. Mol. Cell. Biol. 21:1098-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biamonti, G., G. Perini, F. Weighardt, S. Riva, M. Giacca, P. Norio, L. Zentilin, S. Diviacco, D. Dimitrova, and A. Falaschi. 1992. A human DNA replication origin: localization and transcriptional characterization. Chromosoma 102(1 Suppl.):S24-S31. [DOI] [PubMed] [Google Scholar]
- 7.Bielinsky, A. K., and S. A. Gerbi. 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279:95-98. [DOI] [PubMed] [Google Scholar]
- 8.Bogan, J. A., D. A. Natale, and M. L. Depamphilis. 2000. Initiation of eukaryotic DNA replication: conservative or liberal? J. Cell. Physiol. 184:139-150. [DOI] [PubMed] [Google Scholar]
- 9.Boulikas, T. 1996. Common structural features of replication origins in all life forms. J. Cell. Biochem. 60:297-316. [DOI] [PubMed] [Google Scholar]
- 10.Boulikas, T. 1993. Homeodomain protein binding sites, inverted repeats, and nuclear matrix attachment regions along the human beta-globin gene complex. J. Cell. Biochem. 52:23-36. [DOI] [PubMed] [Google Scholar]
- 11.Brewer, B. J. 1994. Intergenic DNA and the sequence requirements for replication initiation in eukaryotes. Curr. Opin. Genet. Dev. 4:196-202. [DOI] [PubMed] [Google Scholar]
- 12.Burhans, W. C., L. T. Vassilev, M. S. Caddle, N. H. Heintz, and M. L. DePamphilis. 1990. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell 62:955-965. [DOI] [PubMed] [Google Scholar]
- 13.Carroll, S. M., P. Gaudray, R. M. De, J. F. Emery, J. L. Meinkoth, E. Nakkim, M. Subler, H. D. Von, and G. M. Wahl. 1987. Characterization of an episome produced in hamster cells that amplify a transfected CAD gene at high frequency: functional evidence for a mammalian replication origin. Mol. Cell. Biol. 7:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DePamphilis, M. L. 1997. The search for origins of DNA replication. Methods 13:211-219. [DOI] [PubMed] [Google Scholar]
- 15.DePamphilis, M. L. 1988. Transcriptional elements as components of eukaryotic origins of DNA replication. Cell 52:635-638. [DOI] [PubMed] [Google Scholar]
- 16.Dieken, E. S., E. M. Epner, S. Fiering, R. E. Fournier, and M. Groudine. 1996. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat. Genet. 12:174-182. [DOI] [PubMed] [Google Scholar]
- 17.Dijkwel, P. A., S. Wang, and J. L. Hamlin. 2002. Initiation sites are distributed at frequent intervals in the Chinese hamster dihydrofolate reductase origin of replication but are used with very different efficiencies. Mol. Cell. Biol. 22:3053-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djeliova, V., G. Russev, and B. Anachkova. 2002. DNase I sensitive site in the core region of the human beta-globin origin of replication. J. Cell. Biochem. 87:279-283. [DOI] [PubMed] [Google Scholar]
- 19.Djeliova, V., G. Russev, and B. Anachkova. 2001. Dynamics of association of origins of DNA replication with the nuclear matrix during the cell cycle. Nucleic Acids Res. 29:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, K. L., J. D. Diller, B. M. Ferguson, S. V. Nyland, B. J. Brewer, and W. L. Fangman. 1996. Multiple determinants controlling activation of yeast replication origins late in S phase. Genes Dev. 10:1595-1607. [DOI] [PubMed] [Google Scholar]
- 21.Fullerton, S. M., J. Bond, J. A. Schneider, B. Hamilton, R. M. Harding, A. J. Boyce, and J. B. Clegg. 2000. Polymorphism and divergence in the beta-globin replication origin initiation region. Mol. Biol. Evol. 17:179-188. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert, D. M. 2001. Making sense of eukaryotic DNA replication origins. Science 294:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handeli, S., A. Klar, M. Meuth, and H. Cedar. 1989. Mapping replication units in animal cells. Cell 57:909-920. [DOI] [PubMed] [Google Scholar]
- 24.Heintz, N. H., J. D. Milbrantdt, K. S. Greisen, and J. L. Hamlin. 1983. Cloning of the initiation region of a mammalian chromosomal replicon. Nature 302:439-441. [DOI] [PubMed] [Google Scholar]
- 25.Henning, K. A., E. A. Novotny, S. T. Compton, X. Y. Guan, P. P. Liu, and M. A. Ashlock. 1999. Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl. Acad. Sci. USA 96:592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, R.-Y., and D. Kowalski. 1993. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J. 12:4521-4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyrien, O., C. Maric, and M. Mechali. 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270:994-997. [DOI] [PubMed] [Google Scholar]
- 28.Jacob, F., J. Brenner, and F. Cuzin. 1964. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp. Quant. Biol. 288:329. [Google Scholar]
- 29.Kalejta, R. F., X. Li, L. D. Mesner, P. A. Dijkwel, H. B. Lin, and J. L. Hamlin. 1998. Distal sequences, but not ori-beta/OBR-1, are essential for initiation of DNA replication in the Chinese hamster DHFR origin. Mol. Cell 2:797-806. [DOI] [PubMed] [Google Scholar]
- 30.Kamath, S., and M. Leffak. 2001. Multiple sites of replication initiation in the human beta-globin gene locus. Nucleic Acids Res. 29:809-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly, R. E., M. L. DeRose, B. W. Draper, and G. M. Wahl. 1995. Identification of an origin of bidirectional DNA replication in the ubiquitously expressed mammalian CAD gene. Mol. Cell. Biol. 15:4136-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly, T. J., and G. W. Brown. 2000. Regulation of chromosome replication. Annu. Rev. Biochem. 69:829-880. [DOI] [PubMed] [Google Scholar]
- 33.Kim, S. M., and J. A. Huberman. 1998. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol. Cell. Biol. 18:7294-7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S. M., D. Y. Zhang, and J. A. Huberman. 2001. Multiple redundant sequence elements within the fission yeast ura4 replication origin enhancer. BMC Mol. Biol. 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitsberg, D., S. Selig, I. Keshet, and H. Cedar. 1993. Replication structure of the human beta-globin gene domain. Nature 366:588-590. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi, T., T. Rein, and M. L. DePamphilis. 1998. Identification of primary initiation sites for DNA replication in the hamster dihydrofolate reductase gene initiation zone. Mol. Cell. Biol. 18:3266-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong, D., and M. L. DePamphilis. 2002. Site-specific ORC binding, prereplication complex assembly and DNA synthesis at Schizosaccharomyces pombe replication origins. EMBO J. 21:5567-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowalski, D., and M. J. Eddy. 1989. The DNA unwinding element: a novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 8:4335-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, G., M. Malott, and M. Leffak. 2003. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 23:1832-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lunyak, V. V., M. Ezrokhi, H. S. Smith, and S. A. Gerbi. 2002. Developmental changes in the Sciara II/9A initiation zone for DNA replication. Mol. Cell. Biol. 22:8426-8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesner, L. D., J. L. Hamlin, and P. A. Dijkwel. 2003. The matrix attachment region in the Chinese hamster dihydrofolate reductase origin of replication may be required for local chromatid separation. Proc. Natl. Acad. Sci. USA 100:3281-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesner, L. D., X. Li, P. A. Dijkwel, and J. L. Hamlin. 2003. The dihydrofolate reductase origin of replication does not contain any nonredundant genetic elements required for origin activity. Mol. Cell. Biol. 23:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavlov, Y. I., C. S. Newlon, and T. A. Kunkel. 2002. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell 10:207-213. [DOI] [PubMed] [Google Scholar]
- 44.Pearson, C. E., H. Zorbas, G. B. Price, and M. Zannis-Hadjopoulos. 1996. Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication. J. Cell. Biochem. 63:1-22. [DOI] [PubMed] [Google Scholar]
- 45.Phi-van, L., and W. H. Stratling. 1999. An origin of bidirectional DNA replication is located within a CpG island at the 3" end of the chicken lysozyme gene. Nucleic Acids Res. 27:3009-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prioleau, M. N., M. C. Gendron, and O. Hyrien. 2003. Replication of the chicken β-globin locus: early-firing origins at the 5′ HS4 insulator and the ρ- and βA-globin genes show opposite epigenetic modifications. Mol. Cell. Biol. 23:3536-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma, K., M. Weinberger, and J. A. Huberman. 2001. Roles for internal and flanking sequences in regulating the activity of mating-type-silencer-associated replication origins in Saccharomyces cerevisiae. Genetics 159:35-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snyder, M., R. J. Sapolsky, and R. W. Davis. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:2184-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stueber, D., and H. Bujard. 1982. Transcription from efficient promoters can interfere with plasmid replication and diminish expression of plasmid specified genes. EMBO J. 1:1399-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi, T., E. Ohara, H. Nishitani, and H. Masukata. 2003. Multiple ORC-binding sites are required for efficient MCM loading and origin firing in fission yeast. EMBO J. 22:964-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tasheva, E. S., and D. J. Roufa. 1994. A mammalian origin of bidirectional DNA replication within the Chinese hamster RPS14 locus. Mol. Cell. Biol. 14:5628-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Theis, J. F., and C. S. Newlon. 2001. Two compound replication origins in Saccharomyces cerevisiae contain redundant origin recognition complex binding sites. Mol. Cell. Biol. 21:2790-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vujcic, M., C. A. Miller, and D. Kowalski. 1999. Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol. Cell. Biol. 19:6098-6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou, J., N. Ashouian, M. Delepine, F. Matsuda, C. Chevillard, R. Riblet, C. L. Schildkraut, and B. K. Birshtein. 2002. The origin of a developmentally regulated Igh replicon is located near the border of regulatory domains for Igh replication and expression. Proc. Natl. Acad. Sci. USA 99:13693-13698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, J., O. V. Ermakova, R. Riblet, B. K. Birshtein, and C. L. Schildkraut. 2002. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 22:4876-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]