Significance
For proteins to be able to have context-specific activities, they can adopt context-specific conformations that enhance or restrict their activity. For transcriptional regulatory factors, such a context-specific signal is provided by the sequence of the DNA response element to which it binds. Here we show how one signal, an alternative splicing event, rewires a transcriptional regulatory protein to respond differently to a second signal, the DNA sequence to which it binds, by changing the functional interplay between protein domains. Together, our findings argue that bidirectional allosteric signaling between the DNA:protein interface and other regulatory domains fine tunes the activity of transcriptional regulatory factors toward individual target genes.
Keywords: alternative splicing, steroid hormone receptor, sequence motifs
Abstract
In addition to guiding proteins to defined genomic loci, DNA can act as an allosteric ligand that influences protein structure and activity. Here we compared genome-wide binding, transcriptional regulation, and, using NMR, the conformation of two glucocorticoid receptor (GR) isoforms that differ by a single amino acid insertion in the lever arm, a domain that adopts DNA sequence-specific conformations. We show that these isoforms differentially regulate gene expression levels through two mechanisms: differential DNA binding and altered communication between GR domains. Our studies suggest a versatile role for DNA in both modulating GR activity and also in directing the use of GR isoforms. We propose that the lever arm is a ”fulcrum” for bidirectional allosteric signaling, conferring conformational changes in the DNA reading head that influence DNA sequence selectivity, as well as conferring changes in the dimerization domain that connect functionally with remote regulatory surfaces, thereby influencing which genes are regulated and the magnitude of their regulation.
The glucocorticoid receptor (GR) is a nuclear hormone receptor (NR) that integrates multiple cellular signals to regulate the expression of cell-type–specific target genes. Hormone binding to the GR ligand binding domain (LBD) induces conformational changes that facilitate interactions with cofactors and translocation to the nucleus, where it associates with specific genomic sequences via the DNA binding domain (DBD) (1). The majority of transcriptional cofactor interactions have thus far been mapped to activation functions 1 and 2 (AF1 and AF2) domains, which lie N terminal and C terminal to the DBD, respectively (2). The combinatorial assembly of different regulatory complexes (3) determines the magnitude and even the direction (activation or repression) of GR’s activity at individual target genes.
The GR binding sequence (GBS) is an unexpected source of regulation. Previously, DNA was thought to only attract GR to genomic loci. However, sequence variations within the GBS confer additional information to the receptor by altering specific or nonspecific contacts with DNA (4, 5) that change the conformation and activity of associated proteins. This indicates a role for binding sequence beyond affinity (5, 6). The changes in activity can be profound, as studies (7–9) suggest that distinct recognition sequences can instruct GR to act as either a transcriptional activator or a repressor. Sequence-specific structural changes are apparent in a region of the DBD termed “the lever arm” and in the DBD dimer interface (5, 7, 10). How sequence-specific structural changes in the DBD are transmitted to GR activation domains to specify GR activity is largely unknown.
Nonetheless, allosteric communication between receptor domains has been demonstrated in GR and other NRs (11–15). Notably, depending on the precise DNA sequences bound by retinoid X receptor (RXR)–vitamin D receptor (VDR) heterodimers, distinct domains outside of their DBDs adopt alternative conformations or changed dynamics, indicative of long-range allosteric communication (11). Further, ligand binding to the LBD of either NR in the heterodimer influenced the hydrogen/deuterium exchange (HDX) of the VDR DBD, indicating that allosteric communication between LBD and DBD can occur in both directions and that conformational changes can be propagated between dimer partners (10, 11). These observations highlight how several inputs are structurally integrated between distinct functional domains to define the function of multidomain transcriptional regulatory factors.
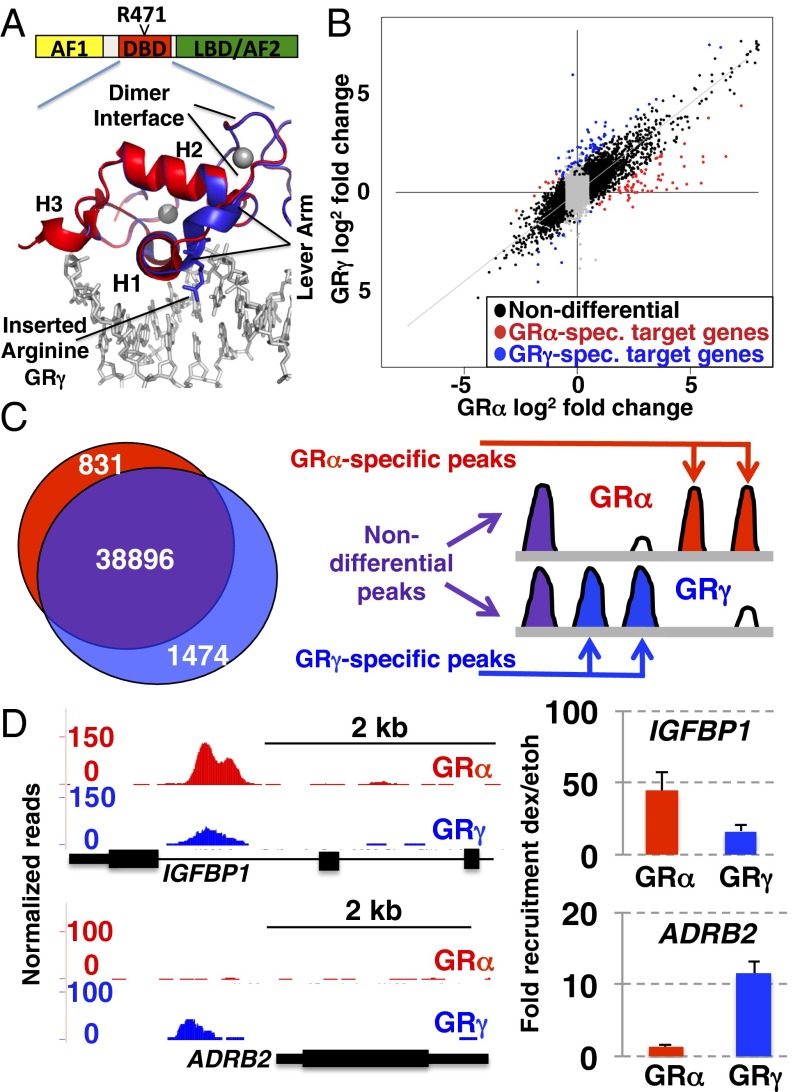
To understand how DNA sequence-specific structural changes originating in the DBD might define the gene-specific function of GR, we focused on the GR lever arm (residues 469–474; all numbering refers to rat GR), which links the DNA binding surface of GR to the dimerization interface. We compared two naturally occurring isoforms, GRα and GRγ, which differ by a single arginine insertion after position 471 in the lever arm (Fig. 1A). GRγ is present at 5–10% of GRα levels and is conserved in all mammals examined including platypus (16), which is estimated to have diverged from humans more than 200 million y ago (17). Although it has been correlated with glucocorticoid resistance (18), the physiological role of GRγ is unclear. The additional amino acid is a consequence of alternative splicing in which either one of two overlapping 5′ splice donor sites is used, resulting in the facultative inclusion of an arginine in the lever arm (Fig. 1A) (16). Previous studies have shown that the insertion does not prevent DNA binding or radically change the overall fold of the DBD, but instead specifically alters the lever arm’s ability to influence GR activity in a context-dependent manner (5).
Fig. 1.
Comparison of gene regulation and genomic binding by GRα and GRγ. (A) Domain structure highlighting the DNA binding domain and the inserted arginine in the lever arm of GRγ. Overlay of structures of GRα [red, Protein Data Bank (PDB) ID code 3G6U] and GRγ (blue, PDB ID code 3G6T) is shown. (B) Cells were treated for 3 h with 1 μM dexamethasone (dex) and isolated RNA was hybridized to microarrays. Shown are the log2 fold changes upon dex treatment for probes that are either GRα specific: red, GRγ specific: blue, or nondifferential: black. Gray points correspond to probes that did not reach both of the chosen thresholds for significance (log2 fold change >1.5 and adjusted P value <0.05). (C, Left) Venn diagram showing overlap between GRα and GRγ binding regions identified by ChIP-seq. (Right) Schematic diagram depicting the three classes of genomic binding: nondifferential (purple), GRα specific (red), and GRγ specific (blue). (D) Isoform-specific genomic binding. U2OS cells stably expressing GRα or GRγ were treated with dex (100 nM) for 90 min. Chromosomal GR binding for (Upper) IGFBP1, and (Lower) ADRB2 is shown as UCSC genome browser screenshot for the ChIP-seq data (Left) or as quantified by ChIP and RT-qPCR (Right). Fold enrichment relative to ethanol vehicle control ±SEM is shown (n = 3).
We reasoned that the structural changes induced by the arginine insertion in the lever arm would allow us to study its role in transmitting signals from the DNA to the rest of the protein. Here, we compared the transcriptional programs induced by these two isoforms using microarrays and the genomic locations by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq). Further, a combination of structural (NMR) and reporter assays was used to study how information is transmitted from the DNA:protein interface to remote domains of GR to shape the transcriptional responses of individual target genes.
Results
Transcriptional Regulation and Genomic Binding by GRα and GRγ.
Glucocorticoid-induced gene regulation was measured by human exonic evidence-based oligonucleotide (HEEBO) microarrays from clonal lines expressing either GRα or GRγ (5). The genes regulated by GRα and GRγ fell into three classes: similarly regulated, the largest subset with 94% of genes (2,261); GRα specific (71 genes, 3%); and GRγ specific (67 genes, 3%, Fig. 1B and Dataset S1). The latter two categories include genes that were either isoform specific or regulated by both isoforms but significantly stronger for one (Fig. 1B). We confirmed the isoform-specific gene regulation of a subset of genes by quantitative real-time PCR (qPCR) (Fig. S1A and Table S1) (5). For each of the genes presented, isoform-specific regulation was confirmed in additional clones derived from the same line.
To determine whether insertion of a single amino acid had an effect on GR occupancy, we performed ChIP-seq of GRα and GRγ in the same U2OS osteosarcoma cell lines. Consistent with our gene regulation observations, GR binding regions (GBRs) for GRα and GRγ were remarkably similar (nondifferential: 38,896/41,201; 94%), although a small portion of isoform-specific binding was observed (831/41,201; 2% GRα specific and 1,474/41,201; 4% GRγ specific, Fig.1C). We defined differential binding as regions with quantitatively different peak heights for both isoforms (Fig. 1C). Binding sites classified as isoform specific were typically observed as significantly differently enriched (Fig. S1B and Fig. 1D, IGFBP1), rather than exclusively binding one of the isoforms (Fig. 1D, ADRB2).
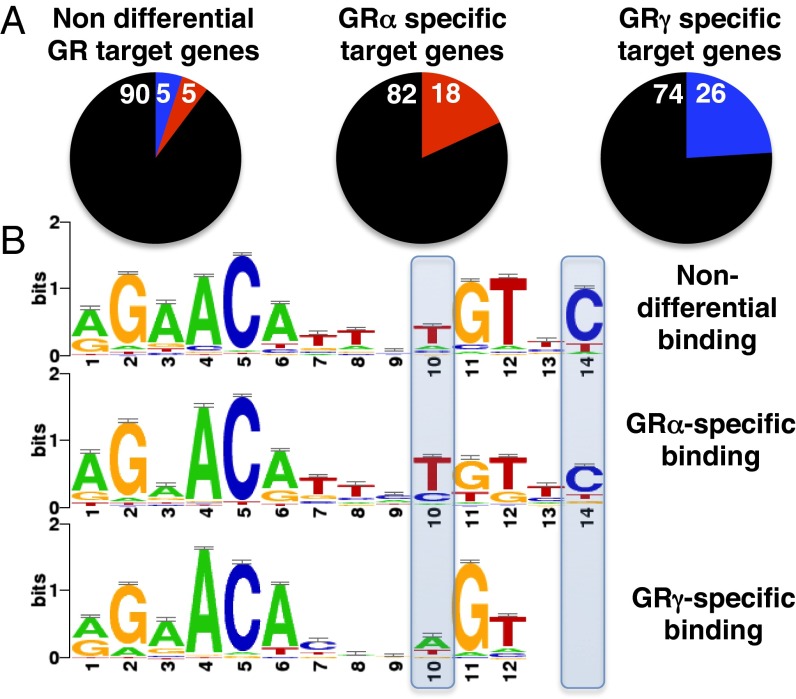
To assess whether differential occupancy could explain the isoform-specific transcriptional regulation, we compared the overlap between transcriptional regulation and occupancy and categorized genes as nondifferential (no significant difference between isoforms), GRα specific, and GRγ specific. Within each group, we looked for GBRs in a window of 20 kb centered on the transcriptional start site (TSS) of each gene and found peaks for 39% of nondifferentially regulated genes and for 47% and 38% of GRα- and GRγ-specific target genes, respectively. For nondifferential target genes at which GR was bound, we found similar levels of occupancy for GRα and GRγ for the majority of genes (815/908 genes: 90%) and some isoform-specific occupancy (Fig. 2A). Similarly, for most GRα-specific genes we found nondifferential occupancy (27/33 genes: 82%), but compared with the nondifferentially regulated genes, a larger fraction of genes had GRα-specific peaks (6/33 genes: 18% versus 47/908 genes: 5%). Notably, for this class of genes, no instances of GRγ-specific occupancy were observed. Similarly, for GRγ-specific regulated genes we found that most peaks were nondifferential (19/25 genes: 74%), with the remaining 26% (6/25) GRγ specific.
Fig. 2.
Overlap between gene regulation and genomic binding by GRα and GRγ. (A) Isoform-specific (deviated ≥2 SD, SI Materials and Methods) and nondifferential genomic binding by GRα and GRγ for the three classes of GR target genes: (Left) Non differential target genes, (Center) GRα specific target genes, (Right) GRγ specific target genes (black, nondifferential; red, GRα specific; and blue, GRγ specific). Numbers refer to the percentage of each group. (B) Motifs for the three classes of binding sites. Motifs were identified using peak motifs and clustered into familial consensus motifs using STAMP.
These data demonstrate that the gamma insertion influences GR activity toward its target genes by two distinct mechanisms: first by affecting GBR selection and second, by influencing events downstream from GBR binding.
Lever Arm Influences DNA Binding.
For those genes that rely on isoform-specific binding for differential regulation, we examined whether the gamma insertion altered the sequence preference of GR. We used peak motifs (19) to identify sequence motifs underlying the three classes of binding sites: nondifferential binding sites, GRα-specific and GRγ-specific binding sites. Because peak motifs intrinsically run multiple motif discovery algorithms, we clustered the identified motifs into three familial consensus binding motifs with STAMP (Fig. 2B) (20). Although all three motifs look similar, the most pronounced differences between the familial motifs were found for the motif derived from GRγ-specific GBRs. First, the GRγ motif preferentially has an A at position 10 (Fig. 2B), whereas the nonisoform-specific and GRα-specific motifs show a preference for a T. Second, the GRγ-specific motif is more degenerate, with one-half of the binding site being more loosely defined, lacking the C at position 14 and a reduced sequence preference for the spacer (positions 7–9). The observed preference for GRγ at position 10 and lack of preference at position 14 also diverges from the GR consensus motif identified in other studies (21–23).
To test the role of different DNA binding affinities in directing differential genomic binding of GRα and GRγ, we compared the binding of the DBD of both isoforms to several GBSs using electrophoretic mobility shift assays. We found that the in vitro affinity of the DBD of GRγ is higher than that of GRα for all GBSs tested (Fig. S2B) including one, the inositol 1,4,5-triphosphate receptor interacting protein gene (ITPRIP), that resembles the GRγ-specific motif by having an A at position 10 and missing a C at position 14. In contrast, the nonspecific affinity for DNA was comparable for both isoforms (Fig. S2A). This indicates that selective GRα occupancy is not due to higher intrinsic GBS affinity and instead may reflect other contributions such as flanking sequences and/or cofactors.
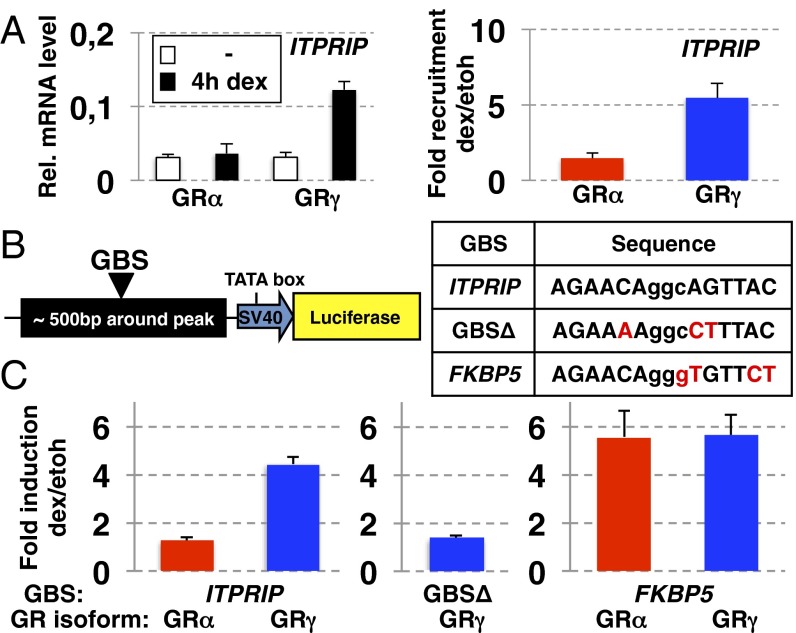
To study the role of the GBS in directing GRγ-specific binding and regulation, we constructed a reporter, containing a GRγ-specific peak (an approximately 500-bp region centered on the peak) with a GBS resembling the more degenerate GRγ-specific motif. The ITPRIP gene from which this reporter was derived is regulated by GRγ specifically and has a GRγ-specific GBR located ∼5 kb upstream of its transcriptional start site. The reporter recapitulated the isoform-specific regulation (Fig. 3 A and C), and deletion of the GRγ-specific GBS rendered the plasmid unresponsive to GRγ (Fig. 3C).
Fig. 3.
Isoform-specific binding and regulation by GRγ. (A) Regulation (Left) and genomic binding (Right) for ITPRIP by GRα and GRγ was determined as for Fig. S1A and Fig. 1D, respectively. (B) Genomic fragment centered on ITPRIP binding site with GBS variants as indicated were cloned upstream of a minimal SV40 promoter driving luciferase. (C) Fold induction (treated vs. nontreated) by GR isoform as indicated for the ITPRIP reporters with (Left) ITPRIP GBS, or (Center) with a deleted GBS, or (Right) with a FKBP5 GBS ±SEM (n = 3) are shown.
To test the influence of the GBS in directing the observed isoform-specific regulation by GRγ, we mutated the ITPRIP GBS to resemble the motif for nondifferential GBSs (Fig. 3 B and C). GRα and GRγ now activated transcription equally well, indicating a role for the GBS in directing GRγ-specific transcription in this reporter context. This suggests that isoform-specific regulation by GRγ can be explained for some genes (approximately 25%, Fig. 2A) by differential binding of GRγ to specific sequence motifs.
GRγ Lever Arm Insertion Rewires the Pattern of Domain Utilization.
For most GR-regulated genes, isoform-specific genomic occupancy does not explain the observed isoform-specific gene regulation (Fig. 2A). We previously showed that GBSs induce structural changes in the lever arm and hypothesized that these might be relayed to other domains to modulate GR activity (5, 10). To test if the differential transcriptional regulation by GRα and GRγ involves interplay between receptor domains, we compared the effect of point mutations in three functional domains (24) in the context of GRα and GRγ in stably transfected U2OS cell lines (Fig. 4 and Fig. S3A).
Fig. 4.
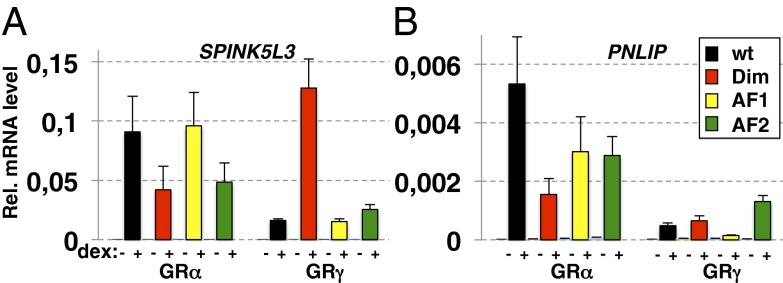
GR isoform-specific patterns of domain requirements. Effect of point mutations in functional domains on transcriptional regulation differs for GRα and GRγ. U2OS cells stably expressing GR variant as indicated were treated with 100 nM dex for 4 h and relative expression levels for treated and untreated cells were analyzed by RT-qPCR for (A) SPINK5L3, and for (B) PNLIP. Averages ± SEM are shown (n = 3).
Similar to the observations made for GBS variants (5), we found that GRγ used different patterns of functional domains than GRα at two GRα-specific target genes that are also regulated by GRγ (activation ∼80–90% lower than for GRα): pancreatic lipase (PNLIP) and serine protease inhibitor Kazal-type 5-like 3 (SPINK5L3) (Fig. 4). The effect of mutating the AF1 domain was similar for both isoforms with reduced activity at PNLIP but no obvious effect at SPINK5L3, whereas for both genes, the AF2 domain is essential for full activity of GRα, but not of GRγ. Moreover, mutating the dimer interface renders GRα less active at both the PNLIP and SPINK5L3 genes. In contrast, this mutation has little effect on PNLIP activation by GRγ and actually restores SPINK5L3 activation by GRγ to the level observed for GRα (an approximately eightfold increase). Thus, the effects of isoform-specific lever arms (which we shall henceforth denote as domains Lα and Lγ) and mutations in additional receptor domains were not simply additive. This indicates communication between these domains in controlling the activity of GR and is consistent with a functional connection between the lever arm and other GR domains. Additionally, we tested the effects of mutations in the AF1, AF2, or dimerization domains of GRα or GRγ by cotransfecting mutant receptors into U2OS cells with various transcriptional reporter plasmids (Fig. S3B). The CGT reporter contains a single GBS and is activated more strongly by GRα than GRγ (5). Two other reporters contained ∼500-bp genomic regions centered on GBRs associated with the GRγ-specifically regulated genes: kallikrein-related peptidase 3 (KLK3) and junctophilin 2 (JPH2). Similar to the findings with endogenous target genes, we found that the effects of perturbing the lever arm and mutations in additional receptor domains were not simply additive. For example, for the CGT reporter mutating the AF1 domain for GRα, or insertion of the arginine in the lever arm for GRγ, both resulted in reduced GR activity (Fig. S3B). However, the effect of combining the AF1 mutation and perturbing the lever arm did not result in a greater loss of activity than the effect of disrupting either single domain. Finally, we found that the dim mutation in the context of GRγ produced a dramatic approximately ninefold increase in activity from the CGT reporter (Fig. S3B). Furthermore, we found a different pattern of domain utilization at the KLK3 reporter, which recapitulates the GRγ-specific regulation of the KLK3 gene (Fig. S3C). For instance, although AF1 and AF2 and an intact dimer interface were all required for full activity of GRγ, mutation of any of those domains did not compromise regulation by GRα; in fact the dimerization interface mutation resulted in increased activation by GRα. In contrast, the patterns of domain utilization by GRα and GRγ were quite similar at the JPH2 reporter (Fig. S3D). This reporter is derived from a GBR where GRα and GRγ occupancy was indistinguishable. Hence, in this context, the lever arm affects GR function neither by altering binding to DNA nor by changing the pattern of functional domain utilization. Conceivably, the Lγ domain may communicate with other GR surfaces, or it might itself provide a functional surface to interact with cofactors directly.
Together, these data show that the effects of switching lever arm domains and mutation of other functional GR domains are not simply additive, indicating that the lever arm and other GR domains are functionally connected determinants of GR activity.
Structural Changes Induced by GRγ Lever Arm Insertion.
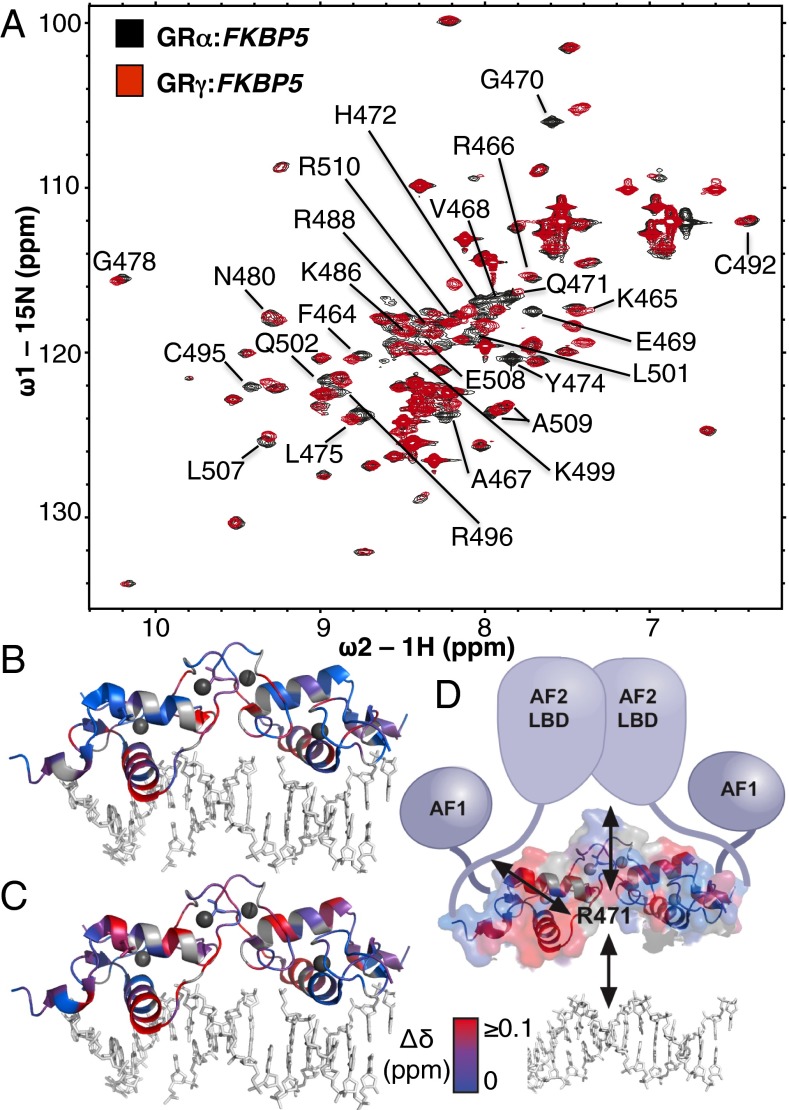
To understand the effects of the lever arm insertion and bound DNA sequence on DBD structure and activity, we used NMR. Previous comparison by X-ray crystallography of DNA-bound GRα or GRγ indicated that the structural changes induced by GBS sequence variants were restricted to the lever arm (5) but did not have sufficient resolution to detect changes elsewhere. 15N-labeled GRα-DBD and GRγ-DBD proteins were purified and incubated with GBSs from FKBP5, which resembles the nondifferential GBS motif, and ITPRIP, which resembles the GRγ-specific motif. 1H-15N HSQC peaks were assigned to amino acid residues for DNA-bound GRα-DBD (10) and assignments were transferred to GRα:FKBP5 and GRα:ITPRIP spectra (Fig. 5A and Fig. S4). Conformational shifts between GRα and GRγ were inferred from chemical shift difference analysis by calculating the distance between each peak in the GRα spectrum and the nearest peak in the corresponding GRγ spectrum. Notably, several residues within the lever arm of GRα and GRγ display peak splitting, indicating slow conformational exchange between two distinct lever arm conformations, as we previously observed for other GR–GBSs complexes (10).
Fig. 5.
Comparison of conformational changes induced by GBS or arginine insertion in the lever arm. (A) Overlay of 1H-15N HSQC spectra of GRα (black) and GRγ (red) bound to FKBP5 GBS. The chemical shift difference (Δδ) was calculated as the weighted sum of 1H and 15N peaks for GRα and GRγ. GRα peaks with a Δδ greater than the mean Δδ of all peaks are labeled by residue. (B) The weighted sum of 1H and chemical shift differences induced by either changing GBS sequence (ITPRIP versus FKBP5) or (C) arginine insertion in the lever arm (GRα versus GRγ bound to ITPRIP) are colored red onto the crystal structure of GRγ (PDB 3G6T). Blue represents residues without significant shifts. Gray represents unassigned residues. (D) Model indicating how the lever arm integrates signals from GBS variants and other functional domains of GR to influence regulatory complex composition and activity.
As expected, when we compared GRα and GRγ, the most pronounced changes in chemical shift mapped to the lever arm, at least in part reflecting the effects of the primary sequence difference between Lα and Lγ (e.g., E469 and G470) (Fig. 5A and Fig. S4). Importantly, the changes were not restricted to the lever arm but also mapped to residues that orient the dimerization interface (G478, N480, and R488). This indicates a structural interplay between these two subdomains, which parallels the functional interplay we observed in transcriptional regulation (Fig. 4). Additional changes were observed in the recognition helix (F464, R466, A467, and V468), helix 2 (C495, R496, L501, and C502), and near helix 3 (L507–R510), indicating that conformational shifts in the lever arm impact the structure of remote regions within the DBD (Fig. 5 and Fig S4).
To rationalize the chemical shifts we observed by NMR, we looked for structural clues by comparing the crystal structures of GRα and GRγ bound to the FKBP5 GBS (5). Indeed, the chemical shifts in the dimerization interface are consistent with conformational changes we observed by crystallography for residues in this domain (L475, I487, and R488) when we compare GRα and GRγ (Fig. S5).
To investigate possible mechanisms contributing to context-specific differential activity of GRα and GRγ (Fig. 3), we compared conformational shifts associated with Lα and Lγ at the isoform-indifferent (FKBP5) and the GRγ-specific GBS (ITPRIP) (Fig. S4D). There is extensive overlap between residues that are shifted by different GBS sequences and those that are shifted by the lever arm insertion (compare gray bars from Fig. S4D, Middle and Bottom). Interestingly, the few GBS-specific chemical shift differences for GRγ mainly map to the dimerization interface (I483, I484, and R488). Taken together with the reporter data showing GRγ-specific ITPRIP activation (Fig. 3), this suggests that the ITPRIP GBS induces rearrangement of the dimerization interface that limits activation in the Lα context but is compensated for by the structural changes induced specifically in Lγ.
Finally, we compared the chemical shift differences induced by GBS sequence changes with those induced by the lever arm isoform and found extensive overlap (Fig. 5 B and C). This indicates that the lever arm may be a key determinant of GR’s interpretation of DNA sequence. The widespread conformational shifts induced by the Lα–Lγ switch, together with the effects of the switch on transcriptional regulation, reinforce the idea that the lever arm is structurally and functionally linked to other GR surfaces (Fig. 5D).
Discussion
Transcription factor function is affected by signals that direct specific action, depending on cellular conditions. These signals are not encountered singly, but simultaneously and combinatorially, resulting in a fine tuning of transcription factor function. Here we show how one signal, an alternative splicing event, rewires GR to respond differently to a second signal, the DNA sequence to which it binds. This potentially regulatable signal can cause differential responses by three mechanisms: changing the affinity and selectivity of GR for DNA sequences to direct it to different genes; altering occupancy at equivalent affinity sites; and perhaps most interestingly, changing how GR functions at sites that are occupied.
Alternative splicing events that involve tandem 5′ and 3′ splice sites are widespread (25) and the resulting isoforms can have distinct functions. This was demonstrated in mice for the transcription factor WT1 (26), where the selective deletion of either one of two isoforms, that differ by a three-amino-acid insertion, showed nonoverlapping phenotypes. For GR, it is unclear if the GRα and GRγ isoforms serve distinct functions or if they are a simple consequence of erroneous splicing that is tolerated. However, the fact that these isoforms are conserved in mammalian genomes argues for a functional role. Furthermore, we found that the two GR isoforms have different activities, and enrichment analysis of the functional classifications of the isoform-specific target genes (27) showed that different functional categories were enriched (Dataset S1), indicating that alternative splicing of GR could allow GR to have context-specific activities. Because GRγ is naturally expressed at low levels (5–10%) compared with GRα (16), we also tested how having an excess of GRα influenced the ability of GRγ to regulate transcription of a reporter construct containing the GBR for the GRγ-specific gene, JPH2. We found that a 10-fold excess of GRα blunted the activation by GRγ, but that a 2-fold induction of JPH2 was still observed (Fig. S6). This indicates that GRγ might regulate genes even in the presence of excess amounts of GRα, either by forming GRγ homodimers or because GRγ function is dominant in GRγ:GRα heterodimers.
The lever arm links the DNA binding surface of GR to the dimerization interface, and provides both a structural readout and functional surface that is required for transmission of DNA sequence information to other domains within GR. We therefore hypothesized that perturbation of the lever arm by differential splicing might change how DNA sequence is interpreted by GR. The lever arm insertion indeed altered transcriptional regulation by GR in a context-specific manner. For most genes, we did not detect differential regulation, but for some genes, activity was reduced, whereas for others, activity increased. Together, this indicates that the Lα–Lγ lever arm switch can affect GR activity in either direction.
Role of the Lever Arm in Modulating DNA Binding.
The differential regulation could be explained, in part, by altering the DNA binding specificity of GR. Indeed, the consensus motif for sites specifically bound by GRγ was more degenerate than the nondifferential and GRα-specific motifs. In vitro, GRγ bound with an overall higher affinity to all GBSs tested, perhaps allowing GRγ to bind to these more degenerate sequences, whereas GRα, with its lower affinity, cannot bind. Comparison of the crystal structures showed that R471 of GRγ might form a weak hydrogen bond to the DNA backbone at one of the half-sites (Fig. S7), possibly contributing to the higher affinity observed for GRγ. Further, the NMR studies indicated changes in the DNA recognition helix 1, which could reflect altered DNA interactions for GRγ that result in the higher affinity observed. Additionally, introducing a residue in the lever arm might relieve strain on the dimer interface that spans the spacer to stabilize the interaction with DNA. This is supported by the relatively modest difference in affinity observed for the Pal binding site (the GBS with the narrowest spacer) as opposed to the biggest difference for FKBP5 (the GBS with the widest spacer, Fig. S2B).
The GRα motif did not show striking differences compared with the isoform-nondifferential binding site raising the question: Why does GRα bind to these regions more strongly than GRγ in vivo? One possible explanation is that although the intrinsic affinity of GRγ for these GBSs is higher, the interaction for GRα is stabilized by cofactors and that the GRγ lever arm disfavors these stabilizing interactions. Accordingly, for most of the isoform-specifically regulated genes, events downstream of binding may account for differential activities.
The Lever Arm Modulates GR Activity Downstream of DNA Binding.
Genetic epistasis refers to interactions where the phenotype of a mutation in one gene is masked by a mutation in another gene, thus placing them in the same genetic pathway. We used this type of analysis to study the interplay between the lever arm and other functional domains of GR by mutating domains individually or in combination. Previous studies comparing GBS variants showed that depending on the sequence of the binding site, different domains of the receptor and different cofactors are required for full activity, which indicates that events at the DNA:protein interface influence the function of other GR domains (5). Here we describe cases where GR transcriptional activity is impaired by either the GRγ isoform or a domain mutation, but where the effect of GRγ and the mutation together is not additive (Fig. 4 and Fig. S3). This epistasis indicates that the lever arm is in the same pathway as the mutated domain and raises the question how these domains are connected. Possibly, the lever arm and other functional domains together form a functional surface that allows the receptor to interact with cofactors involved in transcriptional regulation. The conformational rearrangements in the dimer interface induced by the GRγ insertion perhaps affects the cooperative formation of the GR dimer or changes the relative positioning of other functional domains of GR that might contact this surface. Either of these cases could influence GR activity by affecting binding affinity or global GR structure. Previous studies have shown that cofactors interacting with the DBD can propagate structural changes to other domains (28). Similarly, our results suggest that structural changes at the DNA–protein interface might be propagated within GR to change the structure and function of remote domains. The mechanism for GR interdomain signaling may be through long-range modulation of the conformation of other GR functional surfaces, analogous to findings with RXR/VDR NR heterodimers, demonstrating that DNA binding sequence affects the rate of hydrogen/deuterium exchange in the AF2 domain (11).
The structural and functional link between the lever arm and the dimerization interface illustrate how this communication might work. First, we observed that the conformation of the dimer interface conformation changed when we altered the conformation of the lever arm in much the same way that varying the DNA binding site changed the lever arm and the dimer interfaces (10). Importantly, this connectivity was confirmed when mutation of the dimer interface rescued the transcriptional defect imposed by the arginine lever arm insertion. This suggests that the dimer interface is part of an allosteric pathway between the lever arm and other functional surfaces of GR.
The structural changes induced by perturbing the lever arm showed extensive overlap with a chain of structurally connected residues in the DBD that shift in response to changes in GBS sequence (Fig. 5). In the context of full-length GR, the perturbed dimerization interface residues might propagate specific structural changes induced by DNA sequence to the other receptor domains (Fig. 5D). For GRγ, this allosteric pathway is altered, possibly explaining the changes in transcriptional activity induced by perturbation of the lever arm.
For proteins to be able to have context-specific activities, a likely mechanism is to adopt context-specific conformations that enhance or restrict activity. Studies with NRs related to GR have shown that sequence information within the DNA response element can be relayed to other domains structurally (11) and functionally (29). Interestingly, the flow of information can also occur in the opposite direction, as ligand binding induced structural changes in the DBD of VDR (11). We propose that the integration of various inputs acting on distinct domains of proteins shapes the structure and dynamics of transcriptional regulatory factors, thus allowing them to have context-specific activities. Alternative splicing can generate proteins in which the integration of these signals is altered by rewiring the connections between protein domains, thus allowing different responses to the same signal inputs. We suggest that the lever arm of GR mediates bidirectional allosteric signaling between the DNA:protein interface and other regulatory domains to specify such context-specific activities in gene regulation.
Materials and Methods
SI Materials and Methods includes information on cell lines, plasmids, proteins, qPCR, microarrays, transient transfections, EMSAs, ChIP, ChIP-seq, NMR, and computational analysis. Constructs and cell lines used in this study are available on request. Please contact Katja Borzym, borzym@molgen.mpg.de.
Supplementary Material
Acknowledgments
We thank Mike Love for suggestions regarding the statistical analysis of the data, Edda Einfeldt for help with protein purifications, and Matthew Knuesel and Kirk Ehmsen for reviewing the manuscript. This work was supported by National Institutes of Health Grant CA020535 (to K.R.Y.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316235110/-/DCSupplemental.
References
- 1.Tsai MJ, O’Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Thakur MK, Paramanik V. Role of steroid hormone coregulators in health and disease. Horm Res. 2009;71(4):194–200. doi: 10.1159/000201107. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20(3):560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 4.Rohs R, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meijsing SH, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia HG, et al. Operator sequence alters gene expression independently of transcription factor occupancy in bacteria. Cell Rep. 2012;2(1):150–161. doi: 10.1016/j.celrep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20(1):53–58. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Surjit M, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145(2):224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 9.Sakai DD, et al. Hormone-mediated repression: A negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2(9):1144–1154. doi: 10.1101/gad.2.9.1144. [DOI] [PubMed] [Google Scholar]
- 10.Watson LC, et al. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol. 2013;20(7):876–883. doi: 10.1038/nsmb.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18(5):556–563. doi: 10.1038/nsmb.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra V, et al. Structure of the intact PPAR-gamma-RXR-nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, et al. Interdomain signaling in a two-domain fragment of the human glucocorticoid receptor. J Biol Chem. 1999;274(35):24737–24741. doi: 10.1074/jbc.274.35.24737. [DOI] [PubMed] [Google Scholar]
- 14.Helsen C, et al. Evidence for DNA-binding domain—ligand-binding domain communications in the androgen receptor. Mol Cell Biol. 2012;32(15):3033–3043. doi: 10.1128/MCB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandra V, et al. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. 2013;495(7441):394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rivers C, et al. Characterization of conserved tandem donor sites and intronic motifs required for alternative splicing in corticosteroid receptor genes. Endocrinology. 2009;150(11):4958–4967. doi: 10.1210/en.2009-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rheede T, et al. The platypus is in its place: Nuclear genes and indels confirm the sister group relation of monotremes and Therians. Mol Biol Evol. 2006;23(3):587–597. doi: 10.1093/molbev/msj064. [DOI] [PubMed] [Google Scholar]
- 18.Haarman EG, Kaspers GJ, Pieters R, Rottier MM, Veerman AJ. Glucocorticoid receptor alpha, beta and gamma expression vs in vitro glucocorticod resistance in childhood leukemia. Leukemia. 2004;18(3):530–537. doi: 10.1038/sj.leu.2403225. [DOI] [PubMed] [Google Scholar]
- 19. Thomas-Chollier M, et al. (2011) RSAT 2011: Regulatory sequence analysis tools. Nucleic Acids Res 39(Web Server issue):W86–W91. [DOI] [PMC free article] [PubMed]
- 20. Mahony S, Benos PV (2007) STAMP: A web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35(Web Server issue):W253–W258. [DOI] [PMC free article] [PubMed]
- 21.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3(6):e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19(12):2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogatsky I, et al. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2003;100(24):13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller M, Platzer M. Widespread and subtle: Alternative splicing at short-distance tandem sites. Trends Genet. 2008;24(5):246–255. doi: 10.1016/j.tig.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Hammes A, et al. Two splice variants of the Wilms’ tumor 1 gene have distinct functions during sex determination and nephron formation. Cell. 2001;106(3):319–329. doi: 10.1016/s0092-8674(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 27. D’Andrea D, Grassi L, Mazzapioda M, Tramontano A (2013) FIDEA: A server for the functional interpretation of differential expression analysis. Nucleic Acids Res 41(Web Server issue):W84–W88. [DOI] [PMC free article] [PubMed]
- 28.Garza AS, Khan SH, Moure CM, Edwards DP, Kumar R. Binding-folding induced regulation of AF1 transactivation domain of the glucocorticoid receptor by a cofactor that binds to its DNA binding domain. PLoS ONE. 2011;6(10):e25875. doi: 10.1371/journal.pone.0025875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall JM, McDonnell DP, Korach KS. Allosteric regulation of estrogen receptor structure, function, and coactivator recruitment by different estrogen response elements. Mol Endocrinol. 2002;16(3):469–486. doi: 10.1210/mend.16.3.0814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.