Significance
Microorganisms in the gastrointestinal tract interact with their host in many ways. Lipid metabolism by gastrointestinal microbes generates multiple fatty acid species that can affect host health. In the representative gut bacterium Lactobacillus plantarum, we revealed a fatty acid metabolism, saturation metabolism of polyunsaturated fatty acid, that generates hydroxy fatty acids, oxo fatty acids, conjugated fatty acids, and partially saturated trans-fatty acids as intermediates. Furthermore, fatty acid analysis in mice suggests that the fatty acid metabolism by gastrointestinal microbes modifies fatty acid composition of the host. Therefore, functional investigations of lipid metabolisms of gastrointestinal microbes may provide new methods for improving our health by altering lipid metabolism related to the onset of metabolic syndrome.
Keywords: biohydrogenation, hydratase, fatty acid isomerase, conjugated linoleic acid, lipid nutrition
Abstract
In the representative gut bacterium Lactobacillus plantarum, we identified genes encoding the enzymes involved in a saturation metabolism of polyunsaturated fatty acids and revealed in detail the metabolic pathway that generates hydroxy fatty acids, oxo fatty acids, conjugated fatty acids, and partially saturated trans-fatty acids as intermediates. Furthermore, we observed these intermediates, especially hydroxy fatty acids, in host organs. Levels of hydroxy fatty acids were much higher in specific pathogen-free mice than in germ-free mice, indicating that these fatty acids are generated through polyunsaturated fatty acids metabolism of gastrointestinal microorganisms. These findings suggested that lipid metabolism by gastrointestinal microbes affects the health of the host by modifying fatty acid composition.
Dietary fats are metabolized not only by humans but also by microbes in our gastrointestinal tracts. Microorganisms in the gastrointestinal tract interact with their host in many ways and contribute significantly to the maintenance of host health (1). Lipid metabolism by gastrointestinal microbes generates multiple fatty acid species, such as conjugated fatty acids and trans-fatty acids, that can affect host lipid metabolism (2). However, lipid metabolism by gastrointestinal microbes has not been explored in detail. Saturation metabolism of polyunsaturated fatty acids, a representative mode of lipid metabolism by gastrointestinal microbes, is a detoxifying metabolism of anaerobic bacteria, such as lactic acid bacteria, that reside in colon and intestine. This process transforms growth-inhibiting free polyunsaturated fatty acids into less toxic free saturated fatty acids (3). This saturation metabolism generates characteristic fatty acids (e.g., conjugated fatty acids and trans-fatty acids, which are well known to present in ruminant-derived foods and exert various physiological activities).
“Conjugated fatty acid” is a collective term for positional and geometric isomers of fatty acids with conjugated double bonds. In particular, conjugated linoleic acids (CLAs), such as cis-9,trans-11-CLA and trans-10,cis-12-CLA, reduce carcinogenesis (4), atherosclerosis (5), and body fat (6). With regard to lipid metabolism, CLA is a potent peroxisome proliferator-activated receptor (PPAR)α agonist (7), and treatment with CLA increases the catabolism of lipids in the liver of rodents (8). Based on these findings, CLA is now commercialized as a functional food for control of body weight, especially in the United States and European countries.
On the other hand, consumption of trans-fatty acids increases the risk of coronary heart disease by increasing LDL and reducing HDL cholesterol levels (9). Consequently, trans-fatty acids are considered to be harmful for health, and nutritional authorities have recommended that consumption of trans-fatty acids be reduced to trace amounts (10). Therefore, it is important to control fatty acid saturation processes that generate these fatty acids (11); however, the precise metabolic pathway and enzymes involved have not been clearly identified.
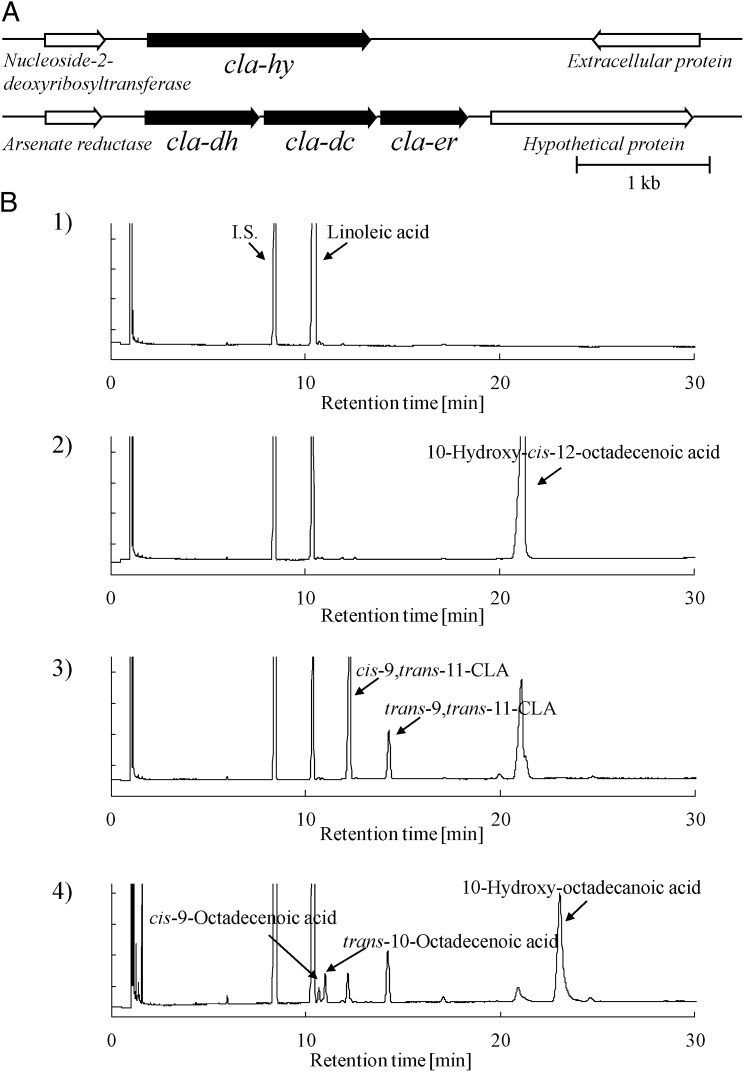
Our analyses on conjugated fatty acid synthesis in representative gut bacteria, the lactic acid bacteria (12–15), demonstrated that Lactobacillus plantarum AKU 1009a (AKU Culture Collection, Faculty of Agriculture, Kyoto University) can transform the cis-9,cis-12 diene structure of C18 fatty acids such as linoleic acid, α-linolenic acid, and γ-linolenic acid into the conjugated diene structures cis-9,trans-11 and trans-9,trans-11 (16–21). In addition, this strain can saturate these conjugated dienes into the trans-10 monoene. Our subsequent metabolic analysis indicated that 10-hydroxy-12-octadecenoic acid is an intermediate of CLA synthesis, and further investigations of hydroxy fatty acid metabolism by lactic acid bacteria revealed that CLA is produced from hydroxy fatty acids such as ricinoleic acid in castor oil (22–25). In cell-free extracts from this strain, we identified the enzymes involved in CLA synthesis (26). Three enzymes, CLA-HY, CLA-DH, and CLA-DC, are necessary for synthesis of conjugated fatty acids such as CLA. Only the combined action of these three enzymes can generate CLA from linoleic acid, with 10-hydroxy-cis-12-octadecenoic acid arising as an intermediate (Fig. 1B, 3). The reactions catalyzed by each enzyme, however, were not revealed in those studies. Through genomic analysis in L. plantarum WCFS1, we found that cla-dh (GenBank accession no. NC_004567; region: 59613-60473) and cla-dc (GenBank accession no. NC_004567; region: 60505-61350) are located in a cluster with another gene, cla-er (GenBank accession no. NC_004567; region: 61378-62031) (Fig. 1A). In light of this, we tried to identify the function of the gene product (CLA-ER) together with those of CLA-HY, CLA-DH, and CLA-DC.
Fig. 1.
Gene clusters for polyunsaturated fatty acid metabolism and GC chromatograms. (A) Gene clusters for fatty acid metabolic enzymes in L. plantarum. (B) GC chromatograms of substrate (1); reaction with CLA-HY (2); reaction with CLA-HY, CLA-DH, and CLA-DC together with FAD and NADH (3); and reaction with CLA-HY, CLA-DH, CLA-DC, and CLA-ER together with FAD and NADH (4). I.S., internal standard.
Results and Discussion
Based on the sequence of the L. plantarum WCFS1 genome, we designed primers to amplify the CLA-ER gene from L. plantarum AKU 1009a genomic DNA. The PCR-amplified product, cla-er (GenBank accession no. AB812091), consisted of 654 bp and encoded a protein whose amino acid sequence is 99.8% identical to the homologous sequence from L. plantarum WCFS1. We transformed the resulting plasmid containing cla-er from L. plantarum AKU 1009a, pCLA-ER, into Escherichia coli Rosetta2 (DE3) to generate E. coli Rosetta/pCLA-ER. The gene product, CLA-ER, had a molecular mass of ∼25 kDa (including a His tag) and could be detected in soluble cell-free extracts of E. coli Rosetta/pCLA-ER. We examined the function of CLA-ER in fatty acid metabolism using purified CLA-ER in combination with other purified enzymes (CLA-HY, CLA-DH, and CLA-DC) from the corresponding transformants (E. coli Rosetta/pCLA-HY, E. coli Rosetta/pCLA-DH, and E. coli Rosetta/pCLA-DC).
In a reaction with these four enzymes (CLA-HY, CLA-DH, CLA-DC, and CLA-ER) as catalysts, cis-9,trans-11-CLA (CLA1), trans-9,trans-11-CLA (CLA2), 10-hydroxy-cis-12-octadecenoic acid, trans-10-octadecenoic acid, and cis-9-octadecenoic acid (oleic acid) were generated from linoleic acid (Fig. 1B, 4). Thus, the combined action of these four enzymes generated saturated products of oleic acid and trans-10-octadecenoic acid from linoleic acid (i.e., these four enzymes catalyzed the saturation of an polyunsaturated fatty acid).
In our previous studies, we revealed that linoleic acid could be converted into 10-hydroxy-cis-12-octadecenoic acid by CLA-HY (Fig. 1B, 2), as well as into CLA1 and CLA2 along with 10-hydroxy-cis-12-octadecenoic acid by CLA-HY, CLA-DH, and CLA-DC (Fig. 1B, 3). We purified the resulting 10-hydroxy-cis-12-octadecenoic acid by HPLC and used it as a substrate for reactions containing each enzyme together with the oxidoreduction cofactors (i.e., FAD, NADH, or NADPH) that enhanced CLA synthesis by CLA-HY, CLA-DH, and CLA-DC (14). 10-Hydroxy-cis-12-octadecenoic acid was converted into linoleic acid and trans-10,cis-12-octadecadienoic acid by CLA-HY in the presence of FAD and NADH (SI Appendix, Fig. S1). The same substrate was converted into 10-oxo-cis-12-octadecenoic acid by CLA-DH in the presence of NAD+ (SI Appendix, Fig. S2). As before, we purified the resulting trans-10,cis-12-octadecadienoic acid and 10-oxo-cis-12-octadecenoic acid by HPLC and used them as substrates in subsequent reactions.
None of the enzymes converted trans-10,cis-12-octadecadienoic acid under any conditions tested. By contrast, 10-oxo-cis-12-octadecenoic acid was converted into 10-oxo-trans-11-octadecenoic acid by CLA-DC in the absence of cofactors (SI Appendix, Fig. S3). HPLC-purified 10-oxo-trans-11-octadecenoic acid was converted into 10-oxo-octadecanoic acid by CLA-ER in the presence of FAD/FMN and NADH (SI Appendix, Fig. S4) and was converted into 10-hydroxy-trans-11-octadecenoic acid by CLA-DH in the presence of NADH (SI Appendix, Fig. S5). Purified 10-oxo-octadecanoic acid was converted into 10-hydroxy-octadecanoic acid by CLA-DH in the presence of NADH (SI Appendix, Fig. S6), and this product was, in turn, converted into cis-9-octadecenoic acid and trans-10-octadecenoic acid by CLA-HY in the presence of FAD and NADH (SI Appendix, Fig. S7). The 10-hydroxy-trans-11-octadecenoic acid could also be converted into cis-9,trans-11-CLA and trans-9,trans-11-CLA by CLA-HY in the presence of FAD and NADH (SI Appendix, Fig. S8).
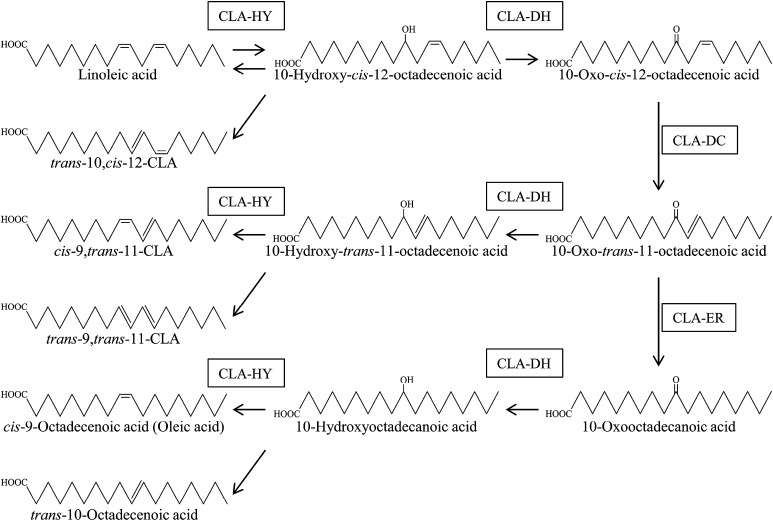
These results demonstrate that the linoleic acid-saturation metabolism of L. plantarum consists of multiple reactions. The first reaction of linoleic acid metabolism is hydration of the carbon–carbon double bond at the Δ9 position, catalyzed by CLA-HY, to generate 10-hydroxy fatty acid. The second reaction is dehydrogenation of the hydroxy group at C10, catalyzed by CLA-DH, to generate 10-oxo fatty acid. The third reaction is isomerization of the carbon–carbon double bond at Δ12, catalyzed by CLA-DC, to generate the conjugated enone structure, 10-oxo-trans-11-fatty acid. The fourth reaction is hydrogenation of the carbon–carbon double bond at Δ11, catalyzed by CLA-ER, to generate the carbon–carbon single bond. The fifth reaction is hydrogenation of the oxo group at C10, catalyzed by CLA-DH, to generate 10-hydroxy fatty acid. The last reaction is dehydration of hydroxy group at C10, catalyzed by CLA-HY, to generate cis-9 and trans-10 monoenoic fatty acids. Through a branch of the saturation-metabolism pathway, conjugated fatty acids are generated by the combined actions of three of the enzymes, CLA-HY, CLA-DH, and CLA-DC. The branched pathway starts with hydrogenation of the oxo group at C10 in 10-oxo-trans-11-fatty acid, catalyzed by CLA-DH, to generate 10-hydroxy-trans-11-fatty acid; the final reaction is dehydration of hydroxy group at C10 in 10-hydroxy-trans-11-fatty acid, catalyzed by CLA-HY, to generate cis-9,trans-11– and trans-9,trans-11–conjugated fatty acids (Fig. 2). As we reported previously, C18 fatty acids with Δ9 and Δ12 diene systems such as α-linolenic acid, γ-linolenic acid, and stearidonic acid undergo the same transformations in L. plantarum AKU 1009a (20), indicating that the corresponding intermediates (hydroxy, oxo, conjugated, and partially saturated trans-fatty acids) are produced by the combined actions of these enzymes. The revealed fatty acid-saturation metabolism consists of similar reactions in known fatty acid biosynthesis and degradation pathway; however, it is a pathway that uses only free fatty acids as substrates but not CoA- or acyl carrier protein-activated fatty acids.
Fig. 2.
Polyunsaturated fatty acid-metabolism pathway.
In the experiments described above, we revealed the pathway of unsaturated fatty acid metabolism in L. plantarum and the enzymes involved in this metabolism. These enzymes function as catalysts of hydration/dehydration (CLA-HY), oxidation of hydroxy groups/reduction of oxo groups (CLA-DH), migration of carbon–carbon double bonds (CLA-DC), and saturation of carbon–carbon double bonds (CLA-ER). The genes that encode CLA-DH, CLA-DC, and CLA-ER form a gene cluster in L. plantarum. When we searched for gene clusters containing cla-dh, cla-dc, and cla-er in other microorganisms using the Kyoto Encyclopedia of Genes and Genomes database (www.genome.jp/kegg), we found that Lactobacillus casei and Lactobacillus rhamnosus have the same gene cluster, as well as the CLA-HY gene. Furthermore, many species of lactic acid bacteria have one or more of these four genes. For example, Lactobacillus salivarius has cla-dc, cla-er, and cla-hy; and Lactobacillus amylovorus, Lactobacillus johnsonii, Lactobacillus helveticus, and Lactobacillus crispatus have cla-dh, cla-er, and cla-hy. Therefore, acting in concert, these species may mediate the polyunsaturated fatty acid-saturation metabolism in gastrointestinal tract. The in vivo distributions of these strains in relation to the fatty acid profiles and the health conditions of host organisms are also of interest.
The reactions and enzymes we identified will be useful for modifying the properties of fatty acids in foods. The apparent isomerization reaction catalyzed by the combined activities of CLA-HY, CLA-DH, and CLA-DC will be useful for production of cis-9,trans-11– and trans-9,trans-11–conjugated fatty acids. The other isomerization reaction catalyzed by CLA-HY will be useful for production of trans-10,cis-12–conjugated fatty acids. Furthermore, the dehydration reaction catalyzed by CLA-HY might determine the ratio of trans to cis-fatty acids. In other words, enhancing cis-dehydration by CLA-HY could be useful for reducing the amounts of trans-fatty acid in foods. This might be possible as we reported previously that the CLA1/CLA2 ratio (cis/trans ratio) can be controlled by optimizing the reaction conditions (14, 16). Furthermore, not only in the food industry but also in the chemical industry, the reactions found in the fatty acid-saturation metabolism are useful (e.g., enzymatic production of hydroxy fatty acids for polymer industry).
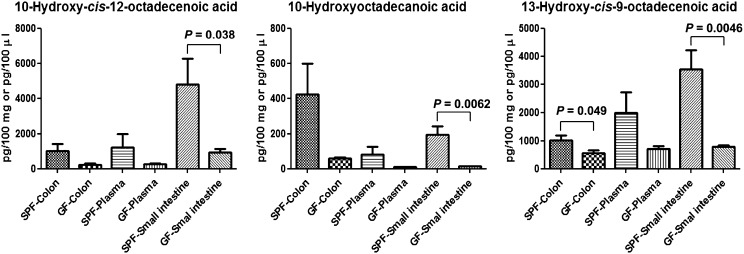
To evaluate the effects of gastrointestinal bacteria on the profile of fatty acids in host tissues, we monitored endogenous formation of the fatty acid intermediates of polyunsaturated fatty acid-saturation metabolism (i.e., hydroxy and oxo fatty acids) in mice bred in either germ-free or specific pathogen-free (SPF) conditions. In the colon, small intestine, and plasma of both groups of mice, we detected 10-hydroxy-cis-12-octadecenoic acid, 10-hydroxyoctadecanoic acid, and 10-oxooctadecanoic acid. By contrast, we did not detect 10-oxo-cis-12-octadecenoic acid or 10-oxo-trans-11-octadecenoic acid derived from linoleic acid or metabolites derived from α-linolenic acid and γ-linolenic acid. There were significant differences in the levels of hydroxy fatty acids between SPF and germ-free mice: in particular, we observed higher levels of 10-hydroxy fatty acids derived from linoleic acid, oleic acid, or both in SPF mice than in germ-free mice (Fig. 3). We also detected a structurally related hydroxy fatty acid, 13-hydroxy-9-cis-octadecenoic acid, which is produced by lactic acid bacteria (27), at higher levels in SPF mice than in germ-free mice (Fig. 3). These differences in the levels of hydroxy fatty acids could be clearly observed in the small intestine, the primary site of fatty acid absorption. These results indicate that gastrointestinal microbes play roles in modifying the fatty acid profiles of their host mice, in particular, by increasing the levels of hydroxy fatty acids that are initial intermediates of polyunsaturated fatty acid-saturation metabolism. The investigations using monocolonize germ-free mice with L. plantarum or a strain deficient in one or more of the fatty acid saturating genes to see whether the host accumulates intermediates are also future interests.
Fig. 3.
Detection and quantitative analyses of polyunsaturated fatty acid-saturation metabolism intermediates in mice. Lipids extracted from colon (100 mg), intestine (100 mg), or plasma (100 μL) of SPF or germ-free (GF) mice were analyzed by LC-MS/MS-based lipidomics as described in Materials and Methods. Data are presented as means ± SEM (n = 8). The statistical significance between mean values was determined by unpaired t test with Welch’s correction.
The intermediates found in the polyunsaturated fatty acid-saturation metabolism described here are predicted to exert specific physiological functions. For example, hydroxy and oxo fatty acids act as ligands for PPARγ (28); oxo-fatty acids found in tomato juice are potent PPARα activators and decrease the amount of triacylglycerol in obese diabetic mice (29). Therefore, functional investigations of these fatty acid intermediates of the polyunsaturated fatty acid-saturation metabolism may provide new methods for improving our health by altering lipid metabolism related to the onset of the metabolic syndrome. Exploration of the lipid metabolism of gastrointestinal microorganisms at the enzymatic and genetic levels, and integration of these findings with metagenomic information, might enable us to promote health by controlling intestinal lipid metabolism.
Materials and Methods
Chemicals.
Standard samples of cis-9,trans-11-CLA, trans-9,trans-11-CLA, and 10-hydroxy-cis-12-octadecenoic acid were prepared as described previously (11, 13). Linoleic acid and fatty acid-free (<0.02%) BSA were purchased from Sigma. All other chemicals were of analytical grade and were obtained commercially.
Cloning and Expression of Recombinant CLA-ER Proteins in E. coli.
Primers were designed to amplify the cla-er sequence from L. plantarum AKU 1009a genomic DNA. The PCR-amplified product was ligated into the pET101/D-TOPO expression vector (Invitrogen), according to the manufacturer’s instructions. The resulting plasmid, pCLA-ER, was transformed into E. coli Rosetta2 (DE3) (Novagen) to generate E. coli Rosetta/pCLA-ER. The integrity of the cloned gene was verified by DNA sequencing using a Beckman-Coulter CEQ8000.
Expression of Recombinant Proteins in E. coli.
Transformants (E. coli Rosetta/pCLA-HY, E. coli Rosetta/pCLA-DH, E. coli Rosetta/pCLA-DC, and E. coli Rosetta/pCLA-ER) were cultured in Luria–Bertani medium at 37 °C for 1 h with shaking at 300 rpm, and then isopropyl-β-thiogalactopyranoside (IPTG) was added to a final concentration of 1.0 mM. After addition of IPTG, the transformed cells were cultivated at 20 °C for 6 h with shaking at 300 rpm. The cells were harvested by centrifugation (12,000 × g; 10 min), washed twice with 0.85% NaCl, and stored at −20 °C until further use.
Purification of Enzymes from Transformants.
To determine the approximate concentration of protein eluted in chromatography, effluents were monitored by UV detection at 280 nm. Enzymes were purified using a fast protein liquid chromatography system (Amersham Pharmacia Biotech) equilibrated with binding buffer [20 mM potassium phosphate buffer (KPB), 50 mM imidazole; pH 7.4] or standard buffer (20 mM KPB, 1 mM DTT, 10% (vol/vol) ethylene glycol; pH 6.5). Fractions with enzymatic activity were collected and concentrated using an Amicon Ultra YM-10 (Millipore). All procedures were carried out at 4 °C.
CLA-HY and CLA-ER were purified as His-tagged proteins. E. coli Rosetta/pCLA-HY and E. coli Rosetta/pCLA-ER cells (8 g) in 1.5 L of culture broth were suspended in binding buffer and disrupted with an Insonator 201M ultrasonic oscillator (Kubota). After ultracentrifugation (100,000 × g; 60 min) of the cell lysate, the resulting supernatant containing His-tagged CLA-HY or His-tagged CLA-ER was loaded onto a Ni-Sepharose column (His Trap HP; GE Healthcare) preequilibrated with binding buffer. After washing, the bound proteins were eluted with elution buffer (20 mM KPB, 250 mM imidazole; pH 7.4). Active fractions were collected and concentrated with a Centriprep YM-3 (Millipore) and applied to a Hi-load 26/60 Superdex 200 prep-grade column (GE Healthcare) equilibrated with 50 mM KPB (pH 6.5). Active fractions were collected, dialyzed with 50 mM KPB (pH 6.5) including 50% (vol/vol) glycerol, and stored at −20 °C until use.
For purification of CLA-DH, E. coli Rosetta/pCLA-DH cells (8 g) in 1.5 L of culture broth were suspended in BugBuster Master Mix (Merck) (30 mL) and incubated for 20 min at room temperature. After ultracentrifugation (100,000 × g; 60 min) of the cell lysate, the resulting supernatant was concentrated with a Centriprep YM-3 and applied to a HiLoad 26/60 Superdex 200 prep-grade column that had been equilibrated with standard buffer and eluted. CLA-DH was purified further using a Mono Q 10/100 GL column and a Superdex 200 10/300 GL column (GE Healthcare). The purified CLA-DH was dialyzed with 50 mM KPB (pH 6.5) including 50% (vol/vol) glycerol and stored at −20 °C until use.
For purification of CLA-DC, E. coli Rosetta/pCLA-DC cells (8 g) in 1.5 L of culture broth were suspended in standard buffer and disrupted with an Insonator 201M ultrasonic oscillator. After centrifugation (20,000 × g; 30 min) of the cell lysate, solid sulfate was added to the resulting supernatant to 50–80% saturation. The precipitate was recovered by centrifugation, dissolved in 10 mL of standard buffer, and then dialyzed three times against 2 L of standard buffer for 8 h. CLA-DC was purified further using a Phenyl Superose HR 10/10 column and a Mono Q 5/50 GL column (GE Healthcare). Purified CLA-DC was dialyzed with 50 mM KPB (pH 6.5) including 50% (vol/vol) glycerol and stored at −20 °C until use.
Reaction Conditions.
Reactions were performed in test tubes (16.5 × 125 mm) that contained 1 mL of reaction mixture (20 mM KPB; pH 6.5) with 0.1% (wt/vol) fatty acid complexed with BSA [0.02% (wt/vol)] as the substrate and purified enzymes (CLA-HY, CLA-DH, CLA-DC, and CLA-ER) in various combinations. The reactions were performed with 5 mM NADH, 5 mM NAD+, 0.1 mM FMN, or 0.1 mM FAD under microaerobic conditions in a sealed chamber with an O2 absorber (Anaeropack Kenki; Mitsubishi Gas Chemical) and gently shaken (120 strokes per minute) at 37 °C for 12 h. The oxygen concentration under microaerobic conditions was maintained below 0.1% (<1,000 ppm) and monitored with an oxygen indicator (Mitsubishi Gas Chemical). All experiments were performed in triplicate, and the averages of three separate experiments that were reproducible within ±10% are presented in the figures.
Lipid Analyses.
Before lipid extraction, n-heptadecanoic acid was added to the reaction mixture as an internal standard. Lipids were extracted from 1 mL of the reaction mixture with 5 mL of chloroform/methanol/1.5% KCl in H2O (2:2:1, by volume), according to the procedure of Bligh-Dyer, and concentrated by evaporation under reduced pressure (30). The resulting lipids were dissolved in 2 mL of methanol and 3 mL of benzene and then methylated with 300 μL of 1% trimethylsilyldiazomethane at 28 °C for 30 min. After methylation, the resulting fatty acid methyl esters were concentrated by evaporation under reduced pressure. The resulting fatty acid methyl esters were analyzed by gas–liquid chromatography (GC) using a Shimadzu GC-1700 gas chromatograph equipped with a flame-ionization detector and split-injection system, fitted with a capillary column (SPB-1; 30 m length × 0.25 mm i.d.; Supelco). The initial column temperature was 180 °C for 30 min but was subsequently increased to 210 °C at a rate of 60 °C/min and then maintained at that temperature for 29.5 min. The injector and detector were operated at 250 °C. Helium was used as a carrier gas at a flow rate of 1.4 mL/min. The fatty acid peaks were identified by comparing retention times to known standards.
Isolation and Identification of Reaction Products.
Reaction products were separated by reverse-phase HPLC using a Shimadzu LC-10A system equipped with a Cosmosil column (5C18-AR; 20 × 250 mm; Nacalai Tesque). The mobile phase was acetonitrile-H2O (8:2, by volume) at a flow rate of 3.0 mL/min, and the effluent was monitored by UV detection (205 and 233 nm). The methyl esters of purified fatty acids were transformed to the pyrrolidide and trimethylsilyl (TMS) derivatives. Pyrrolidide derivatives were prepared by direct treatment of the purified fatty acid methyl esters with pyrrolidine-acetic acid [10:1 (vol/vol)] in screw-cap tubes for 1 h at 115 °C, followed by extraction with dichloromethane. The organic extract was washed with water and dried over anhydrous Na2SO4, and then the solvent was removed under vacuum in a rotary evaporator. The TMS derivatives were prepared by direct treatment of the purified fatty acid methyl esters with a mixture of TMS agent (pyridine/hexamethyldisilazane/trimethylchlorosilane; 9:3:1, by volume) in screw-cap tubes for 30 min at 60 °C, followed by extraction with chloroform. The chemical structures of purified fatty acid methyl esters, pyrrolidide derivatives, and TMS derivatives were determined by mass spectroscopy (MS), and the chemical structures of purified free fatty acids were determined by 2D proton NMR (1H-NMR) techniques including 1H-1H double-quantum–filtered chemical-shift correlation spectroscopy and 2D nuclear Overhauser effect spectroscopy, as described previously (20).
Mice.
SPF and germ-free BALB/c mice (9 wk; female) were obtained from CLEA Japan and maintained under SPF and germ-free conditions with a sterile diet (CL-2; CLEA Japan), respectively, at the Experimental Animal Facility, Institute of Medical Science, The University of Tokyo. Isolated tissues were immediately frozen by liquid nitrogen and always kept at −80 °C before fatty acid analysis. All experiments were approved by the Animal Care and Use Committee of the University of Tokyo and conducted in accordance with their guidelines.
Fatty Acid Analysis in Mice.
Liquid chromatography–tandem MS (LC-MS/MS)-based lipidomics was performed as described (31). Briefly, samples were subjected to solid-phase extraction using a Sep-Pak C18 cartridge (Waters) with a deuterium-labeled internal standard (arachidonic acid-d8, leukotriene B4-d4, 15-hydroxyeicosatetraenoic acid-d8, prostaglandin E2-d4). Lipidomic analyses were performed using an HPLC system (Waters UPLC) with a linear ion-trap quadrupole mass spectrometer (QTRAP 5500; AB SCIEX) equipped with an Acquity UPLC BEH C18 column (1.0 mm × 150 mm × 1.7 µm; Waters). Samples were eluted with a mobile phase consisting of water/acetate [100:0.1 (vol/vol)] and acetonitrile/methanol [4:1 (vol/vol)] (73:27) for 5 min; increased to 30:70 after 15 min, increased to 20:80 after 25 min, and held for 8 min; and then increased to 0:100 after 35 min and held for 10 min with flow rates of 70 µL/min (0–30 min), 80 µL/min (30–33 min), and 100 µL/min (33–45 min). MS/MS analyses were conducted in negative-ion mode, and fatty acid metabolites were identified and quantified by multiple-reaction monitoring. Quantitation was performed using calibration curves constructed for each compound, and recoveries were monitored using added deuterated internal standards.
Supplementary Material
Acknowledgments
We thank Y. Suzuki, E. Hashimoto, and R. Sumiya for technical assistance with animal experiments. This work was supported, in part, by the Industrial Technology Research Grant Program in 2007 (Grant 07A08005a to S.K.); the Project for Development of a Technological Infrastructure for Industrial Bioprocesses on Research and Development of New Industrial Science and Technology Frontiers (S.S.); the New Energy and Industrial Technology Development Organization of Japan; Scientific Research Grants in Aid 19780056 (to S.K.), 16688004 (to J.O.), 18208009 (to S.S.), and 23116506 (to J.K.); grants in aid for the Leading-edge Research Infrastructure Program (to J.K. and H.K.); the Centers of Excellence for Microbial-Process Development Pioneering Future Production Systems from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; the Bio-Oriented Technology Research Advancement Institution of Japan (J.O. and to J.K.); the Institute for Fermentation, Osaka, Japan; and grants from the Ministry of Health and Welfare of Japan (to J.K. and H.K.), the Yakult Bio-Science Foundation (to J.K.), and Core Research for Evolutional Science and Technology (to H.K.). S.K. received Research Fellowship 01985 from the Japan Society for the Promotion of Science for Young Scientists.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB812091).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312937110/-/DCSupplemental.
References
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griinari JM, Bauman DE. In: Advances in Conjugated Linoleic Acid Research. Yurawecz MP, et al., editors. Champaign, IL: American Oil Chemists’ Society Press; 1999. pp. 180–199. [Google Scholar]
- 3.Polan CE, McNeill JJ, Tove SB. Biohydrogenation of unsaturated fatty acids by rumen bacteria. J Bacteriol. 1964;88(4):1056–1064. doi: 10.1128/jb.88.4.1056-1064.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pariza MW, Ha YL. In: Antimutagenesis and Anticarcinogenesis Mechanisms II. Kuroda Y, Shankel D, Waters MD, editors. New York: Plenum; 1990. pp. 167–170. [Google Scholar]
- 5.Lee KN, Kritchevsky D, Pariza MW. Conjugated linoleic acid and atherosclerosis in rabbits. Atherosclerosis. 1994;108(1):19–25. doi: 10.1016/0021-9150(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 6.Park Y, et al. Effect of conjugated linoleic acid on body composition in mice. Lipids. 1997;32(8):853–858. doi: 10.1007/s11745-997-0109-x. [DOI] [PubMed] [Google Scholar]
- 7.Moya-Camarena SY, Vanden Heuvel JP, Blanchard SG, Leesnitzer LA, Belury MA. Conjugated linoleic acid is a potent naturally occurring ligand and activator of PPARalpha. J Lipid Res. 1999;40(8):1426–1433. [PubMed] [Google Scholar]
- 8.Gudbrandsen OA, et al. Trans-10, cis-12-conjugated linoleic acid reduces the hepatic triacylglycerol content and the leptin mRNA level in adipose tissue in obese Zucker fa/fa rats. Br J Nutr. 2009;102(6):803–815. doi: 10.1017/S0007114509297200. [DOI] [PubMed] [Google Scholar]
- 9.Food and Nutrition Board, Institute of Medicine . Dietary Reference Intakes for Energy, Carbohydrate, Fiber, Fat, Fatty acids, Cholesterol, Protein, and Amino Acids. Washington, DC: National Academies Press; 2005. pp. 422–541. [Google Scholar]
- 10.Brouwer IA, Wanders AJ, Katan MB. Effect of animal and industrial trans fatty acids on HDL and LDL cholesterol levels in humans—a quantitative review. PLoS ONE. 2010;5(3):e9434. doi: 10.1371/journal.pone.0009434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kepler CR, Hirons KP, McNeill JJ, Tove SB. Intermediates and products of the biohydrogenation of linoleic acid by Butyrinvibrio fibrisolvens. J Biol Chem. 1966;241(6):1350–1354. [PubMed] [Google Scholar]
- 12.Ogawa J, Matsumura K, Kishino S, Omura Y, Shimizu S. Conjugated linoleic acid accumulation via 10-hydroxy-12-octadecaenoic acid during microaerobic transformation of linoleic acid by Lactobacillus acidophilus. Appl Environ Microbiol. 2001;67(3):1246–1252. doi: 10.1128/AEM.67.3.1246-1252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishino S, Ogawa J, Omura Y, Matsumura K, Shimizu S. Conjugated linoleic acid production from linoleic acid by lactic acid bacteria. J Am Oil Chem Soc. 2002;79(2):159–163. [Google Scholar]
- 14.Kishino S, et al. Structural analysis of conjugated linoleic acid produced by Lactobacillus plantarum, and factors affecting isomer production. Biosci Biotechnol Biochem. 2003;67(1):179–182. doi: 10.1271/bbb.67.179. [DOI] [PubMed] [Google Scholar]
- 15.Kishino S, Ogawa J, Yokozeki K, Shimizu S. Linoleic acid isomerase in Lactobacillus plantarum AKU1009a proved to be a multi-component enzyme system requiring oxidoreduction cofactors. Biosci Biotechnol Biochem. 2011;75(2):318–322. doi: 10.1271/bbb.100699. [DOI] [PubMed] [Google Scholar]
- 16.Kishino S, Ogawa J, Ando A, Shimizu S. Conjugated α-linolenic acid by Lactobacillus plantarum AKU 1009a. Eur J Lipid Sci Technol. 2003;105(10):572–577. [Google Scholar]
- 17.Ogawa J, et al. Production of conjugated fatty acids by lactic acid bacteria. J Biosci Bioeng. 2005;100(4):355–364. doi: 10.1263/jbb.100.355. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa J, et al. Screening and industrial application of unique microbial reactions involved in nucleic acid and lipid metabolisms. Biosci Biotechnol Biochem. 2006;70(3):574–582. doi: 10.1271/bbb.70.574. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa J, Kishino S, Shimizu S. In: Biocatalysis and Biotechnology for Functional Foods and Industrial Products. Hou CT, Shaw JF, editors. New York: CRC; 2006. pp. 121–136. [Google Scholar]
- 20.Kishino S, Ogawa J, Yokozeki K, Shimizu S. Metabolic diversity in biohydrogenation of polyunsaturated fatty acids by lactic acid bacteria involving conjugated fatty acid production. Appl Microbiol Biotechnol. 2009;84(1):87–97. doi: 10.1007/s00253-009-1949-0. [DOI] [PubMed] [Google Scholar]
- 21.Kishino S, Ogawa J, Ando A, Yokozeki K, Shimizu S. Microbial production of conjugated γ-linolenic acid by Lactobacillus plantarum AKU 1009a. J Appl Microbiol. 2010;108(6):2012–2018. doi: 10.1111/j.1365-2672.2009.04609.x. [DOI] [PubMed] [Google Scholar]
- 22.Kishino S, Ogawa J, Ando A, Omura Y, Shimizu S. Ricinoleic acid and castor oil as substrates for conjugated linoleic acid production by washed cells of Lactobacillus plantarum. Biosci Biotechnol Biochem. 2002;66(10):2283–2286. doi: 10.1271/bbb.66.2283. [DOI] [PubMed] [Google Scholar]
- 23.Ando A, Ogawa J, Kishino S, Shimizu S. CLA production from ricinoleic acid by lactic acid bacteria. J Am Oil Chem Soc. 2003;80(9):889–894. [Google Scholar]
- 24.Ando A, Ogawa J, Kishino S, Shimizu S. Conjugated linoleic acid production from castor oil by Lactobacillus plantarum JCM 1551. Enzyme Microb Technol. 2004;35(1):40–45. [Google Scholar]
- 25.Kishino S, Ogawa J, Yokozeki K, Shimizu S. Microbial production of conjugated fatty acids. Lipid Technol. 2009;21(8-9):177–181. [Google Scholar]
- 26.Kishino S, et al. Novel multi-component enzyme machinery in lactic acid bacteria catalyzing C=C double bond migration useful for conjugated fatty acid synthesis. Biochem Biophys Res Commun. 2011;416(1-2):188–193. doi: 10.1016/j.bbrc.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi M, et al. Hydroxy fatty acid production by Pediococcus sp. Eur J Lipid Sci Technol. 2013;115(4):386–393. [Google Scholar]
- 28.Itoh T, et al. Structural basis for the activation of PPARγ by oxidized fatty acids. Nat Struct Mol Biol. 2008;15(9):924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YI, et al. Potent PPARα activator derived from tomato juice, 13-oxo-9,11-octadecadienoic acid, decreases plasma and hepatic triglyceride in obese diabetic mice. PLoS ONE. 2012;7(2):e31317. doi: 10.1371/journal.pone.0031317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 31.Arita M. Mediator lipidomics in acute inflammation and resolution. J Biochem. 2012;152(4):313–319. doi: 10.1093/jb/mvs092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.