Significance
Temporal regulation of a typical developmental control gene depends on multiple enhancers, which makes it important to understand how enhancers coordinate their activities in time. We describe a mechanism for such coordination in Drosophila oogenesis, where the expression of the transcription factor Broad depends on sequential activities of two enhancers. The early enhancer is essential for activity of the late one. To explain this requirement, we propose a mechanism based on a network with feedforward and feedback loops. This network interprets and modulates the epidermal growth factor receptor (EGFR) signaling gradient that controls both enhancers. Experiments and computational modeling show that understanding Broad expression is impossible without simultaneously considering two enhancers, a dynamic gradient, and a network with extracellular and intracellular components.
Keywords: morphogen gradients, mathematical modeling
Abstract
Although it is widely appreciated that a typical developmental control gene is regulated by multiple enhancers, coordination of enhancer activities remains poorly understood. We propose a mechanism for such coordination in Drosophila oogenesis, when the expression of the transcription factor Broad (BR) evolves from a uniform to a two-domain pattern that prefigures the formation of two respiratory eggshell appendages. This change reflects sequential activities of two enhancers of the br gene, early and late, both of which are controlled by the epidermal growth factor receptor (EGFR) pathway. The late enhancer controls br in the appendage-producing cells, but the function of the early enhancer remained unclear. We found that the early enhancer is essential for the activity of the late enhancer and induction of eggshell appendages. This requirement can be explained by a mechanism whereby the BR protein produced by the early enhancer protects the late enhancer from EGFR-dependent repression. We illustrate this complex mechanism using a computational model that correctly predicts the wild-type dynamics of BR expression and its response to genetic perturbations.
A typical developmental control gene is controlled by multiple enhancers (1). A canonical example is provided by even skipped (eve), a pair rule gene in Drosophila. In the blastoderm embryo, eve is expressed in a striped pattern. This pattern reflects simultaneous activities of four enhancers, which are active in different regions of the embryo and respond to different signals (2). The striped pattern is succeeded by a pattern with expression limited to a subset of the heart precursor cells, which is driven by a different enhancer and a different set of signals (3). Enhancers controlling a gene at different times may respond to the same inductive signal, which makes it important to study how they coordinate their activities over time. Here, we present a temporal coordination strategy that involves two enhancers, early and late, which coordinate their actions through feedback modulation of their common inductive cue. The early enhancer prevents ectopic repression of the other enhancer, which is activated later in time and is required for the formation of a specific morphological structure.
The experimental system is Drosophila oogenesis, during the stages when the epidermal growth factor receptor (EGFR) patterns the eggshell, a structure derived from the follicular epithelium, a cell sheet that envelops the developing oocyte (4). EGFR is expressed in all follicle cells and is activated by Gurken (GRK), a ligand secreted from the dorsal cortex of the oocyte. The resulting gradient of EGFR activity defines the dorsoventral axis of the future embryo and induces the formation of dorsal eggshell structures, including a pair of respiratory appendages. Induction of eggshell appendages depends on broad (br), which encodes a Zn-finger transcription factor and is expressed in a dynamic pattern (5, 6). Initially, BR is expressed throughout the follicular epithelium. In response to GRK, it is repressed in cells exposed to the maximal levels of EGFR activation. Subsequently, BR is up-regulated in the cells that form appendages and down-regulated in the rest of the epithelium.
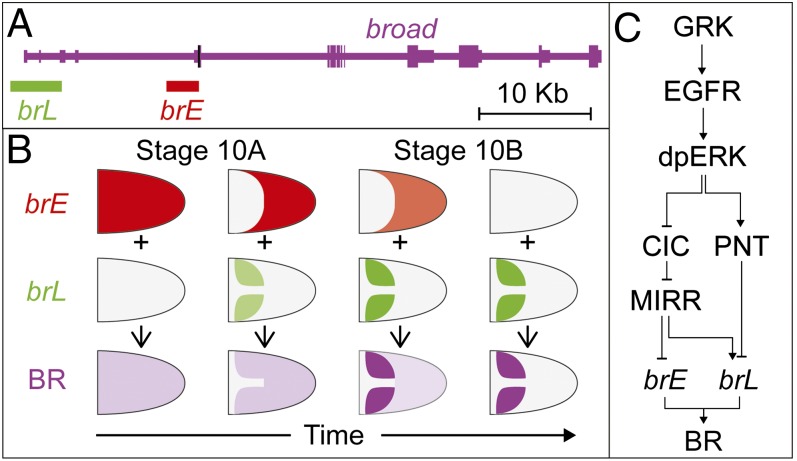
These changes result from activities of two enhancers of the br gene (Fig. 1A) (7). The early enhancer, brE, is first active in all follicle cells and then repressed by EGFR signaling. The brL enhancer is activated later, in a two-domain pattern (Fig. 1B). To generate this pattern, EGFR induces both brL and its repressor, which requires a higher level of EGFR activation. Repression of brL depends on the ETS-family transcription factor Pointed (PNT) (Fig. 1C) (8). The late enhancer controls br in the appendage-producing cells, but the function of the early enhancer remained unclear. We show that brE is essential for the two-domain pattern of brL and for appendage morphogenesis. Based on experiments with genetic mosaics and disruption of enhancer activities, we propose that brE prevents ectopic repression of brL. Our experimental results can be summarized using a predictive computational model that accounts for two different enhancers of the br gene and their interactions with the EGFR network.
Fig. 1.
The spatiotemporal pattern of broad is generated by the superposition of two regulatory regions. (A) The pattern of br is controlled by two enhancers, brE (red) and brL (green) within the 5′-UTR of the br locus (start site marked in black). (B) The dynamics of BR (magenta) expression reflects the dynamic activities of two enhancers. (C) The EGFR pathway regulates BR through the transcription factors PNT, Capicua (CIC), and MIRR.
Results
Both Enhancers of br Are Required for Dorsal Appendage Formation.
BR controls multiple aspects of egg development, including patterning of eggshell appendages during midoogenesis and chorion gene amplification at early stages (9). Clear differences in the activity patterns of the brL and brE enhancers suggest that they are responsible for BR regulation and function at different stages of egg development. In the simplest case, brL could control appendage patterning, whereas brE, active in an earlier and wider pattern, could control the production of chorion proteins.
To test this idea, we used the GAL4-UAS system to interfere with the activities of the brL and brE enhancers. We generated transgenic flies in which these enhancers drive the expression of the GAL4 protein (10). These lines were then used to activate the expression of a UAS-br-RNAi transgene. To avoid interference with the activities of brL and brE in other stages of development, we also expressed the temperature-sensitive GAL80ts protein under the control of the αTub84B promoter. We raised larvae at 18 °C, where the GAL80ts protein binds to GAL4 and represses its transcriptional activity. We then transferred the adults to 29 °C, to derepress GAL4 and start br-RNAi transcription.
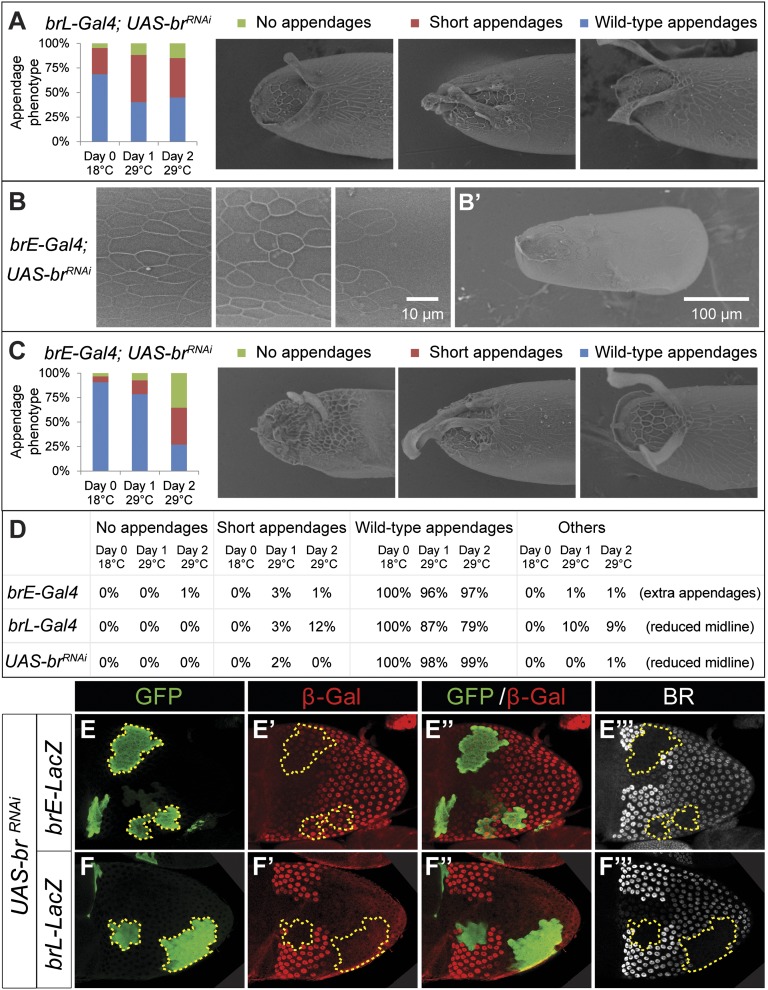
We first inspected eggs laid by females expressing br-RNAi under the control of brL. Upon transfer to 29 °C, there was a clear increase in the percentage of eggshells with shortened or absent appendages (Fig. 2A). Other features of the eggshell, such as the cell imprints on the chorion, were unaffected. These observations are consistent with the notion that brL is required mainly for appendage formation. A similar analysis with the brE enhancer led to defects throughout the eggshell. We recorded multiple instances of eggshell thinning and loss of chorion cell imprints (Fig. 2B). After 2 d at 29 °C, there were clear signs of loss of eggshell integrity (Fig. 2B′). An earlier study attributed this phenotype to reduced levels of chorion proteins (11). Our results suggest that this process is controlled by the brE enhancer.
Fig. 2.
Both brL and brE are required for dorsal appendage formation. (A–C) The effects of brL and brE on eggshell morphogenesis were assayed using RNAi-mediated down-regulation of br transcript. The bars summarize appendage phenotypes scored before and after temperature shift. (A) Down-regulation of BR by brL only compromised appendage formation. Upon transfer to 29 °C, the percentage of eggshells with shortened or absent dorsal appendages increased from 31%, to 60% and 55% after 1 and 2 d, respectively (n = 560 eggs). (B and B′) Down-regulation of BR by brE produced loss of chorionic imprints in the posterior and ventral regions of the eggshell (three samples with different degrees of severity shown), and eventually loss of eggshell integrity after 2 d at 29 °C. (C) Down-regulation of BR by brE also compromised appendages formation, with an increase from 8%, to 21% and 73% after 1 and 2 d at 29 °C, respectively (n = 2,508 eggs). (D) The described eggshell malformations were clearly different from those produced when the brE-Gal4, brL-Gal4, and UAS-br-RNAi (each in combination with tub-Gal80ts) were assayed on their own (n = 1,464, 1,183, 913, respectively). (E–F′′′) Immunostaining for GFP (green), β-GAL (red), and BR (gray) of stage 10B egg chambers carrying either brE-LacZ or brL-LacZ reporters. Expression of br-RNAi is marked by GFP. The yellow outlines highlight clone boundaries. (E–E′′′) Loss of BR does not affect brE expression (n = 26 egg chambers). (F–F′′′) Loss of BR down-regulates brL (n = 25 egg chambers).
Surprisingly, disruption of BR expression caused by the brE enhancer also resulted in a large percentage of eggshells with dorsal appendage malformations (Fig. 2C). Control experiments, where the driver or responder lines were subject to the same treatment, did not lead to the same phenotypes (Fig. 2D), demonstrating that the effect is specific to the knockdown of BR. It is possible that this reflects reduced secretion of chorion proteins due to premature BR down-regulation. However, some of the eggshells with no appendages had well-defined imprints on the dorsal side of the eggshell (Fig. 2C). Based on this, we argue that loss of appendages cannot be attributed to a simple lack of protein secretion and is caused by defects in patterning.
BR Protein Is Required for Induction of the brL Enhancer.
Our results suggest that both brE, which is repressed in the appendage-producing cells, and brL, which is activated in these cells, are required for patterning of eggshell appendages. Because BR protein produced by brE is still present in the dorsal domain at the onset of brL expression, we asked whether BR is required for brL induction. To test this idea, we used the mosaic analysis with repressive cell marker (MARCM) system to generate clones of cells expressing the br-RNAi transgene (12). We used the CY2-GAL4 driver, which is active throughout the follicular epithelium beginning at stage 8 of oogenesis. This allowed us to circumvent the early requirements of BR for cell viability in the follicular epithelium.
This system allowed us to induce clones with normal-sized nuclei, unlike the small nuclei produced by clones of the brnpr-3 null allele (13, 14). This shows that we had indeed bypassed the early requirement for BR in oogenesis. In general, there was a good overlap between the GFP marker and loss of BR protein (Fig. 2 E, F, E′′′, and F′′′), confirming the efficacy of this approach. Testing both brE-LacZ and brL-lacZ reporter lines in BR knockdown clones shows that BR is essential for expression of brL in the appendage-forming domain, but does not affect brE (Fig. 2 E–F′′′). Thus, the activity of brE is essential for the later activity of brL.
Regulation of brL by BR Depends on PNT.
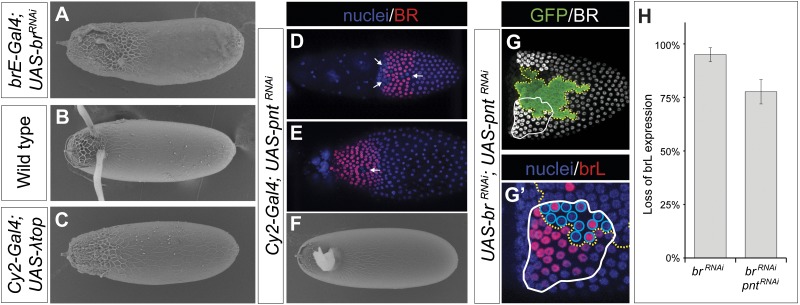
Many eggshells derived from egg chambers in which br-RNAi was driven by the brE enhancer had an increased operculum, a part of the eggshell that provides a site for larva hatching (Fig. 3A). In the wild-type eggs, the operculum is located anteriorly and in between the appendages, and can be recognized by the rough appearance of the chorion (Fig. 3B). Formation of the operculum depends on the transcription factor PNT, which is induced by maximal levels of EGFR activation and splits the pattern of brL into two smaller domains located on either side of the dorsal midline (8).
Fig. 3.
Loss of BR expression leads to PNT-mediated down-regulation of brL. (A) Some eggshells produced by follicle cells expressing br-RNAi under the control of brE-Gal4 produced operculum-like chorion imprints in the dorsal domain. (B) In wild-type eggshells, the operculum is formed by follicle cells exposed to high levels of EGFR signaling in the dorsal midline and is characterized by a rough chorion imprints. (C) Operculum-like imprints through the anterior of the eggshell can be generated by a constitutively active EGFR transgene (λ-top) under the control of the CY2-Gal4 driver. (D and E) Expression of pnt-RNAi leads to ectopic BR in the dorsal midline cells in stages 10B and 11 egg chambers. The incomplete ectopic expression in the midline suggests that PNT down-regulation was incomplete (white arrows, n = 14 of 17 egg chambers). (F) pnt-RNAi expression generates eggshells with fused dorsal appendages (100%, n = 290 eggs). (G) Clones with BR and PNT double knockdown (yellow dotted line) spanning the appendage primordium (white line) were assayed for loss of brL expression. (G′) Magnified view of lower left area in G showing expression of brL. The proportion of cells (blue circles) both in the appendage primordium and in double-knockdown clones expressing brL (red) was quantified by visual inspection. (H) Loss of brL expression in BR and PNT double-knockdown cells (78 ± 6% SD, n = 36 egg chambers) was significantly lower (P < 0.01) than in BR knockdown cells (95 ± 3% SD, n = 25 egg chambers). P value calculated from a t test of empirical bootstrap distributions.
The eggshell phenotype of BR knockdown produced by the brE enhancer resembles the operculum expansion induced by overactivation of EGFR (Fig. 3C) (15). Based on this, we hypothesized that loss of appendages in our experiments results from ectopic repression of brL by PNT. We tested this idea using a UAS-pnt-RNAi transgene. When expressed uniformly in the follicular epithelium under the control of the CY2-Gal4 driver, pnt-RNAi produced eggshells with a single wide appendage (Fig. 3F). This phenocopies eggshells observed in experiments with a null allele of pnt (8) and correlates with the ectopic expression of BR in the dorsal midline of the follicular epithelium (Fig. 3 D and E).
To test whether loss of brL activity in the BR knockdown cells depended on PNT, we induced GFP-marked clones of cells depleted for both BR and PNT. We quantified the number of cells downregulating brL in BR and PNT double knockdown clones. Although the BR single knockdown down-regulated brL-LacZ expression in 95 ± 3% of the cells that landed within the brL expression domain, the double knockdown of both PNT and BR down-regulated brL-LacZ in only 78 ± 6% of such cells (Fig. 3 G and H). The rescue of the effect of BR on brL when PNT was down-regulated was statistically significant (P < 0.01). This supports the idea that loss of brL activity in the absence of BR is PNT dependent. The incomplete rescue can be attributed to the incomplete knockdown of PNT by the RNAi construct (Materials and Methods).
Proposed Mechanism for Interaction Between br Enhancers.
In the wild type, PNT represses BR only in cells exposed to the maximal levels of EGFR activation (7, 16, 17). This is consistent with a model where BR is induced by intermediate levels of EGFR signaling, whereas PNT, which represses BR, has a high threshold. Our data suggest that, in the absence of BR, PNT represses the brL enhancer in cells with high and intermediate levels of EGFR activation. Importantly, this happens without changes in the level of the oocyte-derived GRK. Based on the increase in the region where BR is repressed, we suggest that disruption of early expression of BR results in ectopic high levels of EGFR activation and ectopic BR repression by PNT.
To explain how loss of BR can increase the levels of EGFR activation, we noted that EGFR activation in the follicle cells depends on both GRK, produced by the oocyte, and Spitz (SPI), produced by the follicle cells (18–20). Production of SPI requires rhomboid (rho), a gene encoding a protease that processes a uniformly expressed inactive precursor of SPI. In response to GRK, rho is expressed in a wide dorsal domain of the follicular epithelium. With time, expression is down-regulated in cells expressing high levels of BR (where brL is active) (20). The down-regulation of rho in the appendage primordia reflects its repression by BR. This was suggested based on experiments with an enhancer of rho that recapitulates the late, L-shaped pattern of the gene and is ectopically expressed in br mutant cells (13).
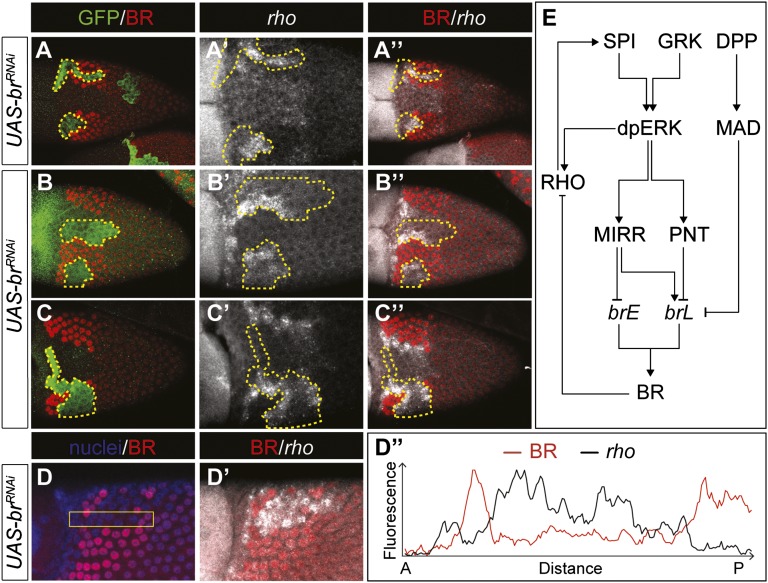
We confirmed BR-dependent repression of rho by visualizing the pattern of rho transcripts in egg chambers with marked clones of cells expressing br-RNAi (Fig. 4 A–D″). We also found that stage 10B mosaic egg chambers show premature, ectopic rho inside BR knockdown clones that span the appendage primordium. Ectopic expression could be observed before the L-shaped pattern of rho was visible (Fig. 4A″). Later, when the endogenous L-shaped pattern became visible, ectopic expression could be observed throughout the clone (Fig. 4 B–B″ and D–D″) and then only at the clone borders, next to cells with high levels of BR (Fig. 4 C–C″). These results confirm that BR regulates both the early (dorsal) and late (L-shaped) patterns of rho expression.
Fig. 4.
BR modulates EGFR signaling through repression of rhomboid. (A–C″) Immunostaining for GFP (green) and BR (red), and FISH for rho transcript (gray) in stage 10B egg chamber. BR knockdown clones spanning the dorsal domain show ectopic expression of rho (n = 23 egg chambers). (A–A″) rho is prematurely expressed in BR knockdown clones. (B–B″) Late stage 10B egg chamber shows ectopic rho in clones, in addition to the later L-shaped wild-type pattern. (C–C″) Later, the level of rho expression is high in the cells at the boundaries, and low in the center of the clone. (D and D′) Large clones spanning most of the appendage confirm rho expression throughout the clone. (D″) Quantification along the outlined area in D shows that rho is up-regulated both along the boundaries and in the center of the clone. (E) Proposed regulation of brE and brL by the EGFR and DPP pathways.
Because rho expression is sufficient to induce high levels of EGFR activation, our results suggest that BR knockdown leads to ectopic and premature activation of high levels of EGFR signaling even in cells exposed to intermediate levels of GRK. Based on these observations, rho expression dynamics can be related to the activity patterns of the br enhancers: The early pattern of rho matches the dorsal repression of brE, which relieves rho repression in the dorsal domain. Later on, brL reestablishes BR in the appendage primordia, leading to lateral repression of rho. According to this scenario, induction of rho requires repression of brE in the dorsal follicle cells. In the absence of BR or in the absence of brE activity, rho is expressed prematurely, leading to earlier and ectopic production of SPI, which amplifies EGFR signaling to the levels that are high enough to repress the brL enhancer. The observed effects are essentially cell autonomous, consistent with the short range of SPI (21).
Computational Modeling of the Mechanism.
Our mechanism for temporal coordination of enhancer activities is based on an inductive signal that is amplified by positive feedback and controls two different enhancers, which in turn affect the pattern of EGFR activation. To explore the feasibility of this complex mechanism, we analyzed it computationally, using earlier models of the GRK gradient and br regulation (22, 23). These earlier models did not account for rho-dependent amplification of EGFR signaling. We extended these models to incorporate the effects of the early enhancer and positive feedback. We did not model the later, spatially restricted phase of rho expression. Because this pattern appears after the pattern of brL is established (Fig. 4 A–C″), we do not expect this simplification to affect our results. The GRK gradient in our model has a fixed shape and amplitude that peaks over time (Fig. 5 A and A′). This reflects the initiation of GRK signaling and the later synthesis of the vitelline membrane, which is believed to separate the oocyte and the follicle cells (18, 20). We also included the effect of Decapentaplegic (DPP), a secreted ligand, which represses BR in the anterior cells (6) (Fig. 5 B and B′) (24).
Fig. 5.
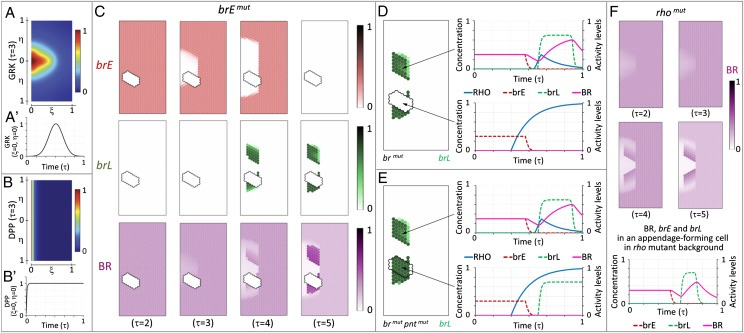
Mathematical model of the follicular epithelium patterning. (A) Spatial distribution of GRK. (A′) Production of GRK is modeled with a temporal Gaussian distribution, resulting in a dynamic profile. (B) Spatial distribution of DPP. (B′) Influx of DPP from the anterior boundary is constant, producing a constant concentration profile. (C) Dynamics of brE, brL, and BR expression in a clone with inactivated brE spanning the lower appendage primordium. (D) Simulation shows loss of brL in clones where BR production has been inactivated. Dynamics of RHO, BR, brE, and brL expression in a wild-type (Upper), and a clone cell (Lower). (E) Simulation shows no loss of brL in clones where both BR and PNT production have been inactivated. Dynamics of RHO, BR, brE, and brL in a wild-type (Upper), and a clone cell (Lower). (F) Computational modeling of rho-mutant epithelium. Loss of rho can fine-tune the dynamics of brE and brL (compare the temporal pattern with that of the wild type in D), but does not prevent the formation of two domains with high levels of BR expression.
We first implemented a one-dimensional version of the model to select parameters consistent with the temporal patterns of brE, brL, BR, and rho expression (SI Text). We then used these parameters in a 2D implementation of the model, and kept only those parameters for which the activity of the late enhancer was abolished by inactivating the early enhancer. Results obtained with a representative set of parameters are shown in Fig. 5C. In these simulations, we predicted pattern formation in a genetic mosaic where the brE enhancer was inactivated in a part of the follicular epithelium spanning one of the two appendage primordia. Consistent with the proposed mechanism, inactivation of brE abolishes the activity of brL. This effect results from PNT-dependent repression of brL and is triggered by high levels of EGFR activation.
Our model correctly predicts the requirement of BR protein for the activity of the brL enhancer. In the absence of BR, cells exposed to intermediate levels of GRK express rho, have high levels of EGFR signaling, and repress brL (Fig. 5D), as shown in our clones of follicle cells expressing the br-RNAi transgene (Figs. 2 F–F″ and 4 A–C″). Furthermore, the model recapitulates the rescue of brL expression in clones where both BR and PNT are down-regulated (Figs. 3G′ and 5E). Cells in such clones experience elevated levels of rho expression and EGFR signaling, but they are insensitive to the PNT-dependent down-regulation of brL.
Previous studies established that loss of rho does not affect the number of dorsal appendages (8, 25). Fig. 5F shows numerical solution of the model where rho has been removed, while keeping all other parameters the same. Consistent with published data, the number of BR expression domains and their separation is not affected. Thus, removal of rho does not prevent the formation of two domains of high BR expression. However, disruption of the brE activity leads to ectopic activation of rho-dependent feedback. This produces ectopic regions of cells with high levels of EGFR signaling and represses the brL enhancer, resulting in the loss of high levels of BR in cells that would normally form appendages (Fig. 5D). Therefore, rho-dependent feedback does not significantly affect brL activity in the wild type, but this feedback represses the brL enhancer when the early phase of BR expression is disrupted.
Discussion
Temporal control of transcription can be provided by changes in the levels of inductive signals, by cross-regulatory interactions between genes, and by dynamic use of different enhancers. For example, the dynamic expression of rho in the early Drosophila embryo results from sequential activities of two different rho enhancers, responding to two different inductive cues (26). In another control strategy, the early enhancer initiates expression, and the late enhancer maintains it through positive autoregulation. This mechanism controls Krox20 during the hindbrain segmentation in vertebrates (27). Both of these scenarios are different from the mechanism that coordinates br enhancers in Drosophila oogenesis. First, both the early and late enhancers respond to the same inductive signal. Second, the early enhancer is needed not to the initiate the expression of the late enhancer, but to protect it from ectopic and premature repression.
In the wild-type egg chamber, brL is repressed only in cells exposed to the maximal levels of GRK. In the absence of brE, signaling levels sufficient for repression are realized in the appendage primordia, due to amplification of EGFR activation resulting from ectopic expression of rho. This model is supported by eggshell defects induced by the RNAi-based disruption of BR expression by brE, and by ectopic expression of rho mRNA and PNT-dependent loss of brL activity in the absence of BR. We tested the requirement for the rho-dependent amplification of EGFR signaling computationally, by analyzing a simplified model in which BR and PNT repress each other directly, without feedback by rho (SI Text). Our extensive exploration of the parameter space in this model could not identify a set of parameters that would be consistent with both the wild-type expression of brL and its response to genetic perturbations. Based on this, we argue that amplification of EGFR signaling by rho is essential for explaining our results.
Going beyond br and rho, we note that dozens of genes regulated by GRK are expressed in dynamic patterns (28). Some of these patterns may be explained using the proposed computational model based on the interplay of multiple enhancers and dynamic signals. Although these models are more complex than existing models of developmental patterning, their analysis is essential for understanding temporal control of gene expression in development.
Materials and Methods
Fly Stocks, Clonal Analysis, and Eggshell Examination.
The following Drosophila stocks were used: UAS -br-RNAi HMS00042 and UAS-pnt-RNAi HMS01452 [Transgenic RNAi Project (TRiP)], tubP-GAL80ts on the second and third chromosomes [Bloomington Drosophila Stock Center (BDSC)], and brE-LacZ and brL-LacZ (7). Both the RNAi lines target the sequence of the gene common to all their isoforms.
Eggshells with increased operculum (Fig. 3C) were produced by females expressing the constitutively active EGFR transgene UAS-λ-top under the control of the CY2-Gal4 driver (16).
The MARCM system was used to generate gain-of-function clones. The hsFLP,tubP-Gal80,FRT19A and FRT19A stocks (BDSC) were used to generate knockdown clones marked by the expression of GFP. The CY2-Gal4 and UAS-2xEGFP-AH2 (BDSC) transgenes were recombined onto the same chromosome, and used to drive transgene expression and to mark clones. Mitotic recombination was induced by subjecting flies to a 37 °C heat shock for 5 h for 2 consecutive days, 4–5 d before dissection.
For comparison of brL down-regulation in BR, and BR and PNT double knockdowns, both genotypes were heat shocked, dissected, and imaged in parallel. Combined results of four experiments are shown in Fig. 3H. Error bars indicate 2 SDs for each genotype.
Eggshells from females expressing UAS-pnt-RNAi controlled by the CY2-Gal4 driver were raised at 18 °C until adulthood. All eggshells showed a single, wide dorsal appendage, even though ectopic expression of BR in the dorsal domain egg chambers was incomplete (arrows in Fig. 3D). The proportion of cells expressing high levels of BR in the midline increased after stage 11 (arrow in Fig. 3E), possibly due to reduced activity of the driver or cell rearrangement during morphogenesis.
Generation of brE-Gal4 and brL-Gal4 Drivers.
The genomic sequences of brE and brL were amplified by PCR, and cloned upstream of Gal4 under the control of a minimal hsp70-promoter in the vector pGal4AttB. The vector was generated from placZattB (a gift from K. Basler, University of Zurich, Zurich, Switzerland), by replacing the lacZ cassette with a hsp70-Gal4 fragment from the vector phsp70Gal4 (a gift from W. J. Gehring, University of Basel, Basel, Switzerland), and inserting a Gateway attP1-ccdB-CmR-attP2 cassette (Invitrogen) upstream of the hsp70-Gal4 cassette. Both constructs were inserted by PhiC31-mediated integration into chromosomal position 22A3 of the VK37 line by BestGene.
Immunostaining, FISH, and Microscopy.
Immunostaining of ovaries was performed as described elsewhere (7). Primary antibodies included mouse anti-BR-Core [1:100; Developmental Studies Hybridoma Bank (DSHB)], rabbit anti–β-GAL (1:500; MP Biomedicals; antibody was preabsorbed with fixed histone-GFP Drosophila embryos to reduce background), and sheep anti-GFP (1:1,000; AbD Serotec). FISH was performed as described elsewhere. Primary antibodies used in FISH included mouse anti–BR-Core (1:50; DSHB), sheep anti-DIG (3:500; Roche), and Alexa Fluor 488-conjugated rabbit anti-GFP (1:100; Invitrogen). Alexa Fluor-conjugated secondary antibodies (1:500; Molecular Probes) were used.
Egg chambers were imaged using a Leica SP5 confocal microscope, and processed with ImageJ 1.45s (National Institutes of Health). Eggshell images were acquired with a Hitachi TM-1000 tabletop scanning electron microscope (SEM) and processed with Photoshop CS5 (Adobe Systems).
Computational Modeling.
Details on the 1D and 2D model implementation are provided in the SI Text. Briefly, we solved the set of 11 ordinary differential equations representing each component of the network shown in Fig. 4E using Comsol Multiphysics 4.3 (Spatial Corporation). We modeled a single appendage primordium. In reporting the results of computational clonal analysis, the wild-type image is presented in a mirrored format, for illustrative purposes.
Supplementary Material
Acknowledgments
We are indebted to Trudi Schüpbach, Miriam Osterfield, Nir Yakoby, and Jeremiah Zartman for helpful discussions and comments on the manuscript. We thank Eric Wieschaus for the use of SEM and Miriam Osterfield for help with eggshell images. We thank the TRiP at Harvard Medical School (National Institutes of Health/National Institute of General Medical Sciences Grant R01-GM084947) for providing the transgenic RNAi fly stocks used in this study. This work was supported by Human Frontiers Science Program Grant RGP0052/2009-C and by National Institutes of Health Grant P50-GM071508.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304753110/-/DCSupplemental.
References
- 1.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20(17):R754–R763. doi: 10.1016/j.cub.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papatsenko D. Stripe formation in the early fly embryo: Principles, models, and networks. Bioessays. 2009;31(11):1172–1180. doi: 10.1002/bies.200900096. [DOI] [PubMed] [Google Scholar]
- 3.Halfon MS, et al. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103(1):63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- 4.Cheung LS, Schüpbach T, Shvartsman SY. Pattern formation by receptor tyrosine kinases: Analysis of the Gurken gradient in Drosophila oogenesis. Curr Opin Genet Dev. 2011;21(6):719–725. doi: 10.1016/j.gde.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng WM, Bownes M. Two signalling pathways specify localised expression of the Broad-Complex in Drosophila eggshell patterning and morphogenesis. Development. 1997;124(22):4639–4647. doi: 10.1242/dev.124.22.4639. [DOI] [PubMed] [Google Scholar]
- 6.Yakoby N, Lembong J, Schüpbach T, Shvartsman SY. Drosophila eggshell is patterned by sequential action of feedforward and feedback loops. Development. 2008;135(2):343–351. doi: 10.1242/dev.008920. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs A, Cheung LS, Charbonnier E, Shvartsman SY, Pyrowolakis G. Transcriptional interpretation of the EGF receptor signaling gradient. Proc Natl Acad Sci USA. 2012;109(5):1572–1577. doi: 10.1073/pnas.1115190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boisclair Lachance JF, Fregoso Lomas M, Eleiche A, Bouchard Kerr P, Nilson LA. Graded Egfr activity patterns the Drosophila eggshell independently of autocrine feedback. Development. 2009;136(17):2893–2902. doi: 10.1242/dev.036103. [DOI] [PubMed] [Google Scholar]
- 9.Tzolovsky G, Deng WM, Schlitt T, Bownes M. The function of the broad-complex during Drosophila melanogaster oogenesis. Genetics. 1999;153(3):1371–1383. doi: 10.1093/genetics/153.3.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 11.Hackney JF, Pucci C, Naes E, Dobens L. Ras signaling modulates activity of the ecdysone receptor EcR during cell migration in the Drosophila ovary. Dev Dyn. 2007;236(5):1213–1226. doi: 10.1002/dvdy.21140. [DOI] [PubMed] [Google Scholar]
- 12.Wu JS, Luo LQ. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1(6):2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- 13.Ward EJ, Zhou XF, Riddiford LM, Berg CA, Ruohola-Baker H. Border of Notch activity establishes a boundary between the two dorsal appendage tube cell types. Dev Biol. 2006;297(2):461–470. doi: 10.1016/j.ydbio.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Zartman JJ, et al. Cad74A is regulated by BR and is required for robust dorsal appendage formation in Drosophila oogenesis. Dev Biol. 2008;322(2):289–301. doi: 10.1016/j.ydbio.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neuman-Silberberg FS, Schupbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120(9):2457–2463. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- 16.Queenan AM, Ghabrial A, Schüpbach T. Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development. 1997;124(19):3871–3880. doi: 10.1242/dev.124.19.3871. [DOI] [PubMed] [Google Scholar]
- 17.Clifford R, Schüpbach T. Molecular analysis of the Drosophila EGF receptor homolog reveals that several genetically defined classes of alleles cluster in subdomains of the receptor protein. Genetics. 1994;137(2):531–550. doi: 10.1093/genetics/137.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95(3):355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- 19.Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125(2):191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- 20.Peri F, Bökel C, Roth S. Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev. 1999;81(1–2):75–88. doi: 10.1016/s0925-4773(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 21.Peri F, Technau M, Roth S. Mechanisms of Gurken-dependent pipe regulation and the robustness of dorsoventral patterning in Drosophila. Development. 2002;129(12):2965–2975. doi: 10.1242/dev.129.12.2965. [DOI] [PubMed] [Google Scholar]
- 22.Simakov DS, Cheung LS, Pismen LM, Shvartsman SY. EGFR-dependent network interactions that pattern Drosophila eggshell appendages. Development. 2012;139(15):2814–2820. doi: 10.1242/dev.077669. [DOI] [PubMed] [Google Scholar]
- 23.Zartman JJ, et al. Pattern formation by a moving morphogen source. Phys Biol. 2011;8(4):045003. doi: 10.1088/1478-3975/8/4/045003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lembong J, Yakoby N, Shvartsman SY. Spatial regulation of BMP signaling by patterned receptor expression. Tissue Eng Part A. 2008;14(9):1469–1477. doi: 10.1089/ten.tea.2008.0098. [DOI] [PubMed] [Google Scholar]
- 25.Zartman JJ, Kanodia JS, Cheung LS, Shvartsman SY. Feedback control of the EGFR signaling gradient: Superposition of domain-splitting events in Drosophila oogenesis. Development. 2009;136(17):2903–2911. doi: 10.1242/dev.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ip YT, Park RE, Kosman D, Bier E, Levine M. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev. 1992;6(9):1728–1739. doi: 10.1101/gad.6.9.1728. [DOI] [PubMed] [Google Scholar]
- 27.Labalette C, et al. Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development. 2011;138(2):317–326. doi: 10.1242/dev.057299. [DOI] [PubMed] [Google Scholar]
- 28.Yakoby N, et al. A combinatorial code for pattern formation in Drosophila oogenesis. Dev Cell. 2008;15(5):725–737. doi: 10.1016/j.devcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.