Significance
High-grade astrocytomas (HGAs), including glioblastomas (GBMs), are the most common human brain tumors, and they remain fatal with no effective treatment. The most prevalent form, primary GBM, presents clinically as advanced disease, thus providing no access to or understanding of early stages. We report a comprehensive study in the mouse that establishes causal relationships and an evolutionary etiology in HGA development. Events yielding disease, both engineered and spontaneous, indicate grade-specific roles culminating in the development of GBMs with characteristics of primary GBMs, including molecular alignment with the mesenchymal subclass, asymptomatic early disease, and rapid emergence of high-grade aggressive cancer. These genetically engineered models provide a path to basic understanding of disease etiology and a window into diagnostic and therapeutic discovery.
Keywords: mouse model preclinical, cancer progression, cancer initiation
Abstract
Glioblastoma (GBM), the most common brain malignancy, remains fatal with no effective treatment. Analyses of common aberrations in GBM suggest major regulatory pathways associated with disease etiology. However, 90% of GBMs are diagnosed at an advanced stage (primary GBMs), providing no access to early disease stages for assessing disease progression events. As such, both understanding of disease mechanisms and the development of biomarkers and therapeutics for effective disease management are limited. Here, we describe an adult-inducible astrocyte-specific system in genetically engineered mice that queries causation in disease evolution of regulatory networks perturbed in human GBM. Events yielding disease, both engineered and spontaneous, indicate ordered grade-specific perturbations that yield high-grade astrocytomas (anaplastic astrocytomas and GBMs). Impaired retinoblastoma protein RB tumor suppression yields grade II histopathology. Additional activation of v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) network drives progression to grade III disease, and further inactivation of phosphatase and tensin homolog (PTEN) yields GBM. Spontaneous missense mutation of tumor suppressor Trp53 arises subsequent to KRAS activation, but before grade III progression. The stochastic appearance of mutations identical to those observed in humans, particularly the same spectrum of p53 amino acid changes, supports the validity of engineered lesions and the ensuing interpretations of etiology. Absence of isocitrate dehydrogenase 1 (IDH1) mutation, asymptomatic low grade disease, and rapid emergence of GBM combined with a mesenchymal transcriptome signature reflect characteristics of primary GBM and provide insight into causal relationships.
Astrocytomas are the most common malignant brain tumors. Overall survival for patients with diffuse astrocytoma [A2; World Health Organization (WHO); grade II], anaplastic astrocytoma (AA; WHO grade III), and glioblastoma (GBM; WHO grade IV) has not significantly changed in the past 50 y and remains at 8–10 y, 2–3 y, and 9–12 mo, respectively (1). High-grade astrocytomas (AA and GBM, HGAs), for which there is no curative treatment (1), are characterized by aggressive diffuse brain invasion, cytological pleomorphism, and elevated mitotic activity, with GBM additionally showing microvascular proliferation (MVP) and/or necrosis. Although some GBMs arise subsequent to a previously diagnosed low-grade glioma (secondary GBM), 90% appear without prior clinical history (primary GBM), thus precluding insight into disease progression (2).
Most GBMs harbor aberrations in three major signaling networks, with tumor-specific lesions occurring at various known pathway points (2, 3). Seventy to 90% of GBMs carry lesions in CDKN2A/CDK4-CyclinD/RB, receptor tyrosine kinase (RTK)/RAS/PI3K/PTEN, and MDM2/CDKN2A(ARF)/p53 networks. However, based on global transcriptome studies, several distinct GBM molecular subtypes exist that may manifest in distinct clinical and/or therapeutic outcomes (2–6). Given the critical role of the microenvironment in cancer evolution, comprehensive examination of astrocytoma etiology requires assessment of both engineered and spontaneous molecular aberrations in vivo. Genetically engineered mouse (GEM) models provide an experimental format for such studies and may offer promise for preclinical development of therapeutics and diagnostics (7–9). Although several studies have reported combinations of genetic events that can predispose to GEM-HGA, specific contributions to disease stages and evolutionary patterns have yet to be elucidated.
Here, we dissected the relative roles of inactivating retinoblastoma protein (RB)-tumor suppression (TS), activating the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) network and inactivating phosphatase and tensin homolog (PTEN) in GBM etiology. We examined tumor evolution for evidence of spontaneous mutation of tumor suppressor Trp53 and/or PTEN deletion in conjunction with engineered events and report progression of events that yield HGAs possessing a spectrum of heterogeneous molecular and histological disease features typical of human astrocytomas. The study provides unique insights that distinguish contributions to initiation and progression in the transition from low- to high-grade disease.
Results
Adult-Inducible Targeting of Networks Commonly Perturbed In Human GBMs.
Based on the prominent networks frequently aberrant in GBMs, we examined contributions to disease etiology upon RB-TS and/or PTEN inactivation and/or KRAS activation singly and in all combinations. Events were targeted to adult astrocytes using 4OHT-directed activation of CreERT2 expressed from a human glial fibrillary acidic protein (hGFAP)-CreERT2 transgene (10) (Fig. S1). In the adult brain, GFAP-directed transgene expression is restricted to mature astrocytes and adult neural stem cells (10). RB-TS was conditionally abolished by Cre-dependent expression of T121, which dominantly inactivates RB and compensatory proteins p107 and p130 (11). Upon exposure to Cre, TgGZT121 transgenic mice switch from LacZ expression to T121 expression, also under GFAP regulatory control (12). KRAS, commonly hyperactivated in GBMs (5, 13), was activated via Cre-conditional endogenous-level induction of the KRAS+/lsl-G12D allele (14). Finally, we examined the impact of homozygous or heterozygous PTEN deletion.
Events were induced by 4OHT intraperitoneal (IP) injection of 3-mo-old animals (Fig. S1A). All genotypes post-4OHT treatment are abbreviated herein as shown in Fig. S1B: T = TgG(ΔZ4OHT)T121;hGFAP-CreERT2, R = KRASG12D;hGFAP-CreERT2, PL/L = PTENL/L; hGFAP-CreERT2, and PL/+ = PTENL/+;hGFAP-CreERT2. Cre activation was validated by allele-specific PCR of TR brain DNA (Fig. S2A and Table S1) and immunostaining for T121 (Fig. S2B). Deletion of loxP-flanked cassettes was readily apparent post induction (p.i.). By 2 wk p.i., T121-expressing cells (present only in induced brains) were also positive for GFAP (cytoplasmic astrocyte marker) and S100β (nuclear/cytoplasmic glial marker) (Fig. S2C).
Only RB-TS Inhibition Initiates Disease.
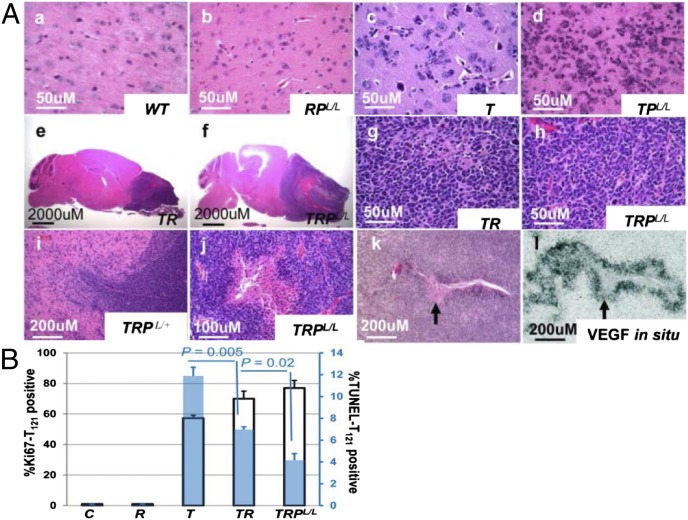
Of the three single events targeted to adult astrocytes, only loss of RB-TS function was sufficient to initiate astrocytomagenesis (Figs. 1A and 2A) based on the induction of grade II-like histopathology (Fig. 1 A, c). Neither KRASG12D activation nor PTEN inactivation, alone or in combination (Fig. 1 A, b), produced detectable brain pathology (Fig. 2A). In T mice, grade II pathology was detectable within 2 wk of induction and became more cellular with time (Fig. S2D). Atypical astrocytes proliferated aberrantly (Fig. 1B and Fig. S3A), had an increased apoptotic rate (Fig. 1B and Fig. S3A), and, as in the human disease, associated extensively with neurons and vasculature (perineuronal and perivascular satellitosis; secondary structures of Scherer) (Fig. S2 D, f and D, j). Two of 11 aged T mice (18%) at 455 and 537 d p.i. harbored diffuse grade III pathology based solely on mitotic activity. No tumor masses were present.
Fig. 1.
Characterization of GEM astrocytomas. (A) H&E-stained mouse brain and tumor sections of indicated genotypes as described in Results. Invasiveness (i) and pseudopalisading necrosis (j) were observed in GBMs. In situ hybridization detected VEGF mRNA expression in perinecrotic GBM cells (l, arrow) as shown in the H&E-stained serial section (k). (B) Quantification of Ki67 (left y axis, black-bordered bar) and TUNEL (right y axis, blue bar) in cortex of mice of indicated genotypes 2 wk p.i.. Data are presented as mean ± SEM.
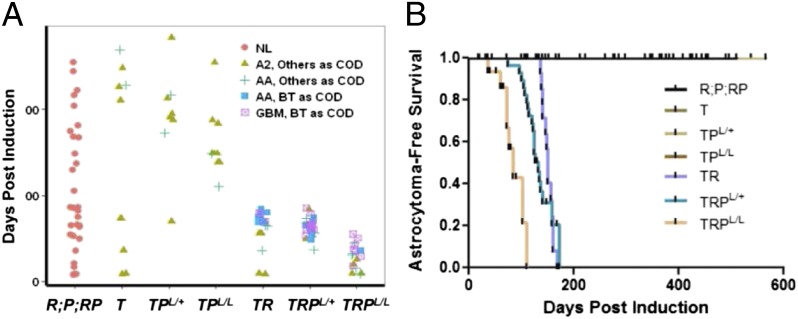
Fig. 2.
Analysis of genotype-associated astrocytoma phenotypes. (A) Brain histopathology of mice at the time of sacrifice p.i. (y axis) is shown (colored shapes). Mice were euthanized at several time points for analysis or when tumors were apparent. Brain tumor (BT) as cause of death (COD): mice were killed due to neurological or physical signs of CNS abnormality and a brain tumor was confirmed; COD other than BT: scheduled analysis time point with no sign of abnormality and mice with non-BT masses. The highest brain pathology grade present is indicated. Genotypes of mice induced with 4OHT at 3 mo of age are indicated on the x axis: T = TgG(ΔZ4OHT)T121;hGFAP-CreERT2+/−, R = KRASG12D;hGFAP-CreERT2+/−, PL/L = PTENL/L;hGFAP-CreERT2+/−, RPL/L = KRASG12D;PTENL/L;hGFAP-CreERT2+/−, TPL/+ = TgG(ΔZ4OHT)T121;PTENL/+;hGFAP-CreERT2+/−, TPL/L = TgG(ΔZ4OHT)T121;PTENL/L;hGFAP-CreERT2+/−, TR = TgG(ΔZ4OHT)T121;KRASG12D;hGFAP-CreERT2+/−, TRPL/+ = TgG(ΔZ4OHT)T121;KRASG12D;PTENL/+;hGFAP-CreERT2+/−, and TRPL/L = TgG(ΔZ4OHT)T121;KRASG12D;PTENL/L;hGFAP-CreERT2+/−. R, P, and RP mice were grouped as R;P;RP because none developed brain pathology. NL, normal. (B) Kaplan–Meier analysis of astrocytoma-specific tumor-free survival after 4OHT administration in GEMs. Median survival for TR, TRPL/+, and TRPL/L mice was 5, 4.4, and 2.8 mo, respectively. Mice with COD other than brain tumor or unknown COD were censored (tick marks). T, TPL/+, and TPL/L mice did not develop life-threatening disease although diffuse grade II astrocytoma pathology was consistently present. Numbers of mice analyzed: R;P;RP (n = 32), T (n = 11), TPL/+ (n = 8), TPL/L (n = 11), TR (n = 21), TRPL/+ (n = 28), and TRPL/L (n = 17). Littermate control mice (either hGFAP-CreERT2 negative or vehicle injected) were not plotted due to lack of phenotype.
Roles for KRAS Activation and PTEN Deletion in Progression to HGA.
To test whether KRAS activation and/or PTEN deletion or heterozygosity would accelerate and/or progress disease, these events were combined with RB-TS inactivation, singly and together. Neither AA nor GBM appeared in TPL/+ or TPL/L mice (Fig. 2A) although the cellularity of grade II astrocytoma was accelerated in TPL/L brains (Fig. 1 A, d), consistent with previous results in which PTEN haploinsufficiency (12) or focal PTEN deletion (15) reduced RB-TS inactivation-induced apoptosis. In contrast to PTEN deletion, the combination of RB-TS loss and KRASG12D expression (TR mice) dramatically shortened survival (median = 150 d p.i.) compared with RB-TS loss alone (no lethal brain malignancy up to 537 d) (Fig. 2). In TR mice, lethal masses consistently progressed from the background of widespread grade II pathology (Fig. 1 A, e and A, g). Ten of 13 TR mice (77%) developed lethal AA based on the presence of masses with extensive mitoses (Table S2). The remaining masses (3 of 13; 23%) were GBMs based on the additional presence of small necrotic centers; these tumors likely reflect stochastic progression via additional oncogenic event(s) (see next paragraph). A single TR mouse requiring premature euthanasia due to thymic hyperplasia (see below and Table S2) had also developed diffuse grade III astrocytoma.
Unlike the combination of Rb-TS inactivation and PTEN deletion, impaired PTEN function together with RB-TS loss and KRASG12D expression in TRPL/+ and TRPL/L astrocytes led to reduced median survival (132 d and 84 d p.i., respectively) compared with TR mice (150 d) (Fig. 2). Sixty percent (12 of 20) of TRPL/+ and 88% (7 of 8) of TRPL/L animals developed lethal necrotic GBM (Table S2 and Fig. 1 A, j). For each genotype, tumor development occurred within a reproducible time p.i.. As in humans, GEM-GBMs were invasive as evidenced by infiltrative interfaces (Fig. 1 A, i) and the frequent presence of necrosis (Fig. 1 A, j). By RNA in situ hybridization, VEGF expression was evident in perinecrotic tumor cells (Fig. 1 A, k and A, l). GBMs were astrocytic as shown by GFAP, nestin, and olig2 positivity and synaptophysin (a neuronal marker) negativity (Fig. S2E). Skin and thymic lesions arose in some TRPL/+ and TRPL/L mice due to GFAP–driven recombination in these tissues (16, 17), thus precluding assessment of terminal brain disease (Fig. S3 D and E and Table S2). However, all brains minimally contained significant diffuse astrocytoma pathology consistent with the age at termination.
Astrocyte inactivation of RB-TS in adult central nervous system (CNS) led to aberrant astrocyte proliferation and apoptosis (Fig. 1B and Fig. S3A). We assessed acute effects on astrocyte proliferation and apoptosis to determine relative acute contributions of progression events, KRASG12D expression (TR brain at 2 wk p.i.) and subsequent PTEN deletion (TRPL/L brain at 2 wk p.i.). Compared with T mice, astrocyte apoptosis was significantly decreased upon KRASG12D expression and was further reduced with PTEN deletion (Fig. 1B and Fig. S3A). There were no significant increases in proliferation. This observation is consistent with the hypothesis that pAkt downstream of PI3K may be the effector of increased survival because its activity can be induced downstream of RAS-triggered PI3K activation and further enhanced upon loss of PI3K negative regulator PTEN. By this mechanism, apoptosis provides a selective pressure for loss of PTEN function during GBM evolution. Indeed, KRASG12D activation is required in this model to facilitate stochastic loss of PTEN (see Spontaneous PTEN Loss and Trp53 Mutations Validate Evolutionary HGA Progression and Identity with Human Disease).
HGA Development Is Dictated by Genetic Lesions.
In the above studies, HGA development could reflect true cause–effect relationships in human disease progression; however, it was possible that the widespread astrocyte abnormality created by diffuse induction of genetic events had artificially contributed to disease. To distinguish these possibilities, we focally induced the genetic lesions that predisposed to GEM-GBM (astrocyte-specific RB-TS inactivation, KRASG12D activation, and PTEN loss or heterozygosity) by stereotactic introduction of self-deleting Cre-lentivirus into the frontal cortex of TRPL/+ and TRPL/L mice without the hGFAP-CreERT2 allele (Fig. S3B). Focal induction led to GBM development with histopathology identical to that developed in broadly induced brains in 3/5 TRPL/+ and 3/5 TRPL/L mice. Thus, GBM was caused by the engineered and associated evolving lesions and not as a secondary result of widespread gliosis. As expected, no skin or thymic lesions developed when genetic lesions were induced focally in brain. These results confirm the interpretation of cause/effect relationships derived from diffuse astrocyte induction studies described above.
Spontaneous PTEN Loss and Trp53 Mutations Validate Evolutionary HGA Progression and Identity with Human Disease.
Although significant molecular and biological aspects of human HGA are represented in these GEM models, it was possible that engineered events inappropriately produced such similarity but did not reflect the natural evolution of human disease. Thus, we assessed the stochastic loss of wild-type PTEN alleles in GBMs of TR (two wild-type PTEN alleles) and TRPL/+ (one wild-type PTEN allele) mice. As described above, engineered PTEN deletion in the context of RB-TS loss and KRAS activation (TRPL/L mice) drives GBM progression, and PTEN deletion is frequent in human GBMs. Quantitative PCR assessment of tumor DNA showed that, among 8 samples from 6 TR-derived tumors, 5 (63%) had lost one allele, and 2 (25%) had lost both PTEN alleles (Table S3). Of 12 samples from 9 TRPL/+ tumors, 7 (58%) had lost the WT PTEN allele. Of the 5 tumors (2 TR and 3 TRPL/+) that were analyzed from 2 distinct regions, 3 tumors (2 TR and 1 TRPL/+) showed intratumoral heterogeneity of wild-type PTEN allele loss. Three TRPL/+ samples appeared to retain both PTEN alleles, indicating either that recombination had not occurred in that region or that PTEN wild-type astrocytes had assumed tumor-cell properties within the tumor microenvironment (18). By similar analyses, grade II astrocytomas retained both PTEN alleles, consistent with KRASG12D expression being required, directly or indirectly, to establish the pressure for PTEN inactivation selection.
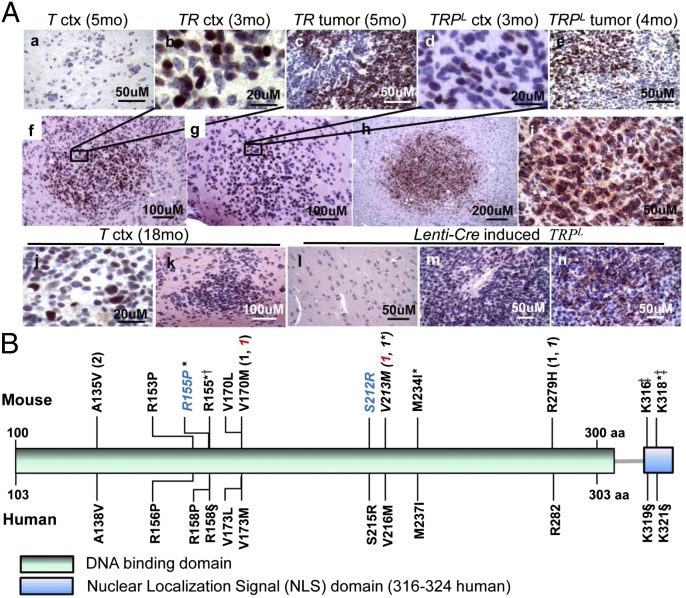
Eighty-seven percent of human GBMs harbor p53 network aberrations, with 42% of these having p53 missense mutations and only 1% with p53 homozygous allele loss (5). We assessed GEM-GBMs by IHC for p53 accumulation (indicative of mutation) and subcellular localization. Tumors, but not adjacent regions, expressed high p53 levels (Fig. 3 A, c and A, e). DNA sequencing of 18 GBMs revealed that 10 (56%) harbored Trp53 missense mutations, 1 (∼6%) carried a frame-shift mutation, and 1 (∼6%) a splice site acceptor mutation at the −1 position of Trp53 exon 5 (Fig. 3B and Table S4). Most of the missense mutations were analogous to those observed in human GBMs, and all have been observed in human cancers (http://p53.iarc.fr/; IARC TP53 database R16) (19). In addition, of the 6 GBMs that contained wild-type p53 coding exons, p53 was mislocalized to the cytoplasm in 4 (67%) (Fig. 3 A, h and A, i and Table S4). Of unknown significance, the 2 tumors that expressed nuclear wild-type p53 carried intronic base changes not represented in SNP databases (Table S4). GBMs induced focally via lentivirus-driven Cre injection also expressed p53 mutants (Fig. 3 A, m and A, n). Furthermore, HGAs induced both by systemic and focal induction methods appeared to be heterogeneous for p53 changes based on the observation that not all tumor cells within a given mass had accumulated p53 (e.g., Fig. 3 A, c and A, e). As with PTEN allele loss, this observation could indicate that wild-type cells within the tumor mass may assume abnormal properties due to paracrine mechanisms.
Fig. 3.
p53 status in TR and TRPL tumors and cortex. (A) Representative images of p53 IHC staining at different p.i. time points in systemic-induced T, TR, and TRPL brains (a–k) and lenti-Cre induced TRPL brain (l–n). Accumulation of p53 was evident in nucleus (b–g, j, k, and m) or cytoplasm (h, i, and n). p53 accumulation appeared in early clusters/nodules (b and f; d and g; and h and i). ctx, cortex; mo, months p.i.. p53 was negative in the remaining brain of lenti-Cre induced brain (l). (B) Trp53 missense mutations and frame shifts identified in TR, TRPL/+, and TRPL/L HGAs (Upper) and corresponding human TP53 missense mutations present in human GBMs (Lower). All samples were from fresh frozen HGA tissues except italicized samples, which were from primary cells generated from HGA or cortex region. Same color of blue or red indicates that tissue samples were taken from the same animal but different regions (one from tumor mass and the other from non-tumor mass cortex region). All tumors were diagnosed as GBM except asterisk-labeled (AA). †In frame insertion; ‡Frame shift; §Corresponding frame shift found in human GBMs; parentheses, number of tumors that had the same missense mutation.
To determine the stage(s) in tumor progression in which stochastic p53 mutation/mislocalization arose, we assessed p53 immunopositivity in brains of T, TR, TRPL/+, and TRPL/L mice at various ages (Table S5). Early p53-positive nodules consistently emerged within 2–3 mo p.i. only in mice predisposed to HGA (TR, TRPL/+, and TRPL/L mice). Nodules appeared before mass formation and expressed abundant p53 in either the nucleus or cytoplasm (Fig. 3 A, b; A, d; and A, f–i). Indeed, based on sequencing, 5 of 8 (62.5%) AA (GIII) tumors harbored spontaneous Trp53 mutations, 2 (25%) had wild-type Trp53, and 1 carried intronic base changes (Fig. 3B and Table S4). Despite active grade II (GII) disease, T brains did not detectably express p53. The presence of a small number of p53 positive cells detected in 36% (5 of 14) of 12- to 23-mo-p.i. brains may indicate the accumulation of multiple stochastic changes reflecting early progression (Fig. 3 A, j and A, k and Table S5).
Human Disease Properties in GEM-GBM.
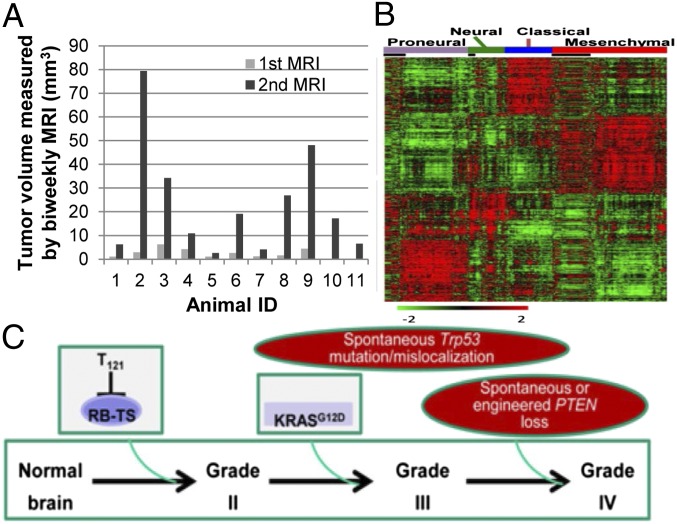
In primary GBM, the first clinical symptoms occur after advanced disease is present. Similarly, overt physical signs of low-grade disease were not observed in GEMs until HGA was present in TR, TRPL/+, and TRPL/L mice. To determine how quickly HGA developed, we monitored tumor development by serial magnetic resonance images (MRI) in TRPL/+ mice. Masses were readily detectable grossly (Fig. S3 C, a) or with MRI (Fig. S3 C, d), and, as in human (20), computer-enhanced vascular reconstruction analysis revealed extensive vessel tortuosity both intrinsic and adjacent to tumor masses (Fig. S3 C, b). To monitor tumor development, scans were performed biweekly starting at 14 wk p.i., and tumor volumes were calculated using ImageJ software (Fig. 4A). Lesions were detectable in all TRPL/+ mice between 16 and 23 wk p.i.. Within a 2-wk interval, 9 of 11 animals (82%) transitioned from a tiny detectable lesion (1.037–6.194 mm3) to a large life-threatening mass (2.609–79.355 mm3) (Fig. 4A) (animal ID 1–9). In the time frame of this experiment, two mice (18%) had a detectable lesion only at the terminal imaging session (Fig. 4A) (animal IDs 10 and 11; 17.235 mm3 and 6.5505 mm3, respectively). Mice were moribund within 1 wk after detecting a mass, with only one exception (Fig. 4A) (animal ID 1; within 2 wk). Thus, tumor growth subsequent to major progression event(s) follows a rapid lethal course, possibly explaining symptomless early disease associated with primary GBM in humans.
Fig. 4.
Disease development, molecular characterization, and model for GEM-GBM etiology. (A) Tumor volume was measured by biweekly T2-weighted magnetic resonance images (MRI) in TRPL/+ mice starting at 14 wk p.i.. (B) Microarray analysis of RNA from 44 GBMs (TR, TRPL/+, and TRPL/L animals) was clustered with TCGA human GBM data using 840 classifiers identified by Verhaak et al. (21). Genes were ordered based on predictive subtypes. Mouse tumor data are denoted with black boxes and align as follows: Proneural (n = 12), Neural (n = 2), Classical (n = 0), and Mesenchymal (n = 30). (C) A model of astrocytoma molecular pathogenesis based on study results. RB-TS inhibition, modeled here through expression of T121, is necessary and sufficient to initiate the neoplastic process (grade II disease). Constitutively activated KRASG12D is not sufficient for astrocytoma initiation but rather is required for progression to high-grade tumors, in part by imposing selection for spontaneous Trp53 mutation/mislocalization that occurs before grade III mass formation. PTEN loss (spontaneous or engineered) contributes to grade IV (GBM) progression only after RAS activation and p53 alteration.
To determine whether GEM-GBMs recapitulated specific molecular subtypes of human primary GBMs, we performed transcriptome analysis using the orthologous 840-gene classifier set identified by Verhaak et al. (21). Thirty of 44 GBMs (68%) clustered with the mesenchymal subclass, the most aggressive of the human classes, 12 (27%) with the proneural, and 2 (5%) with the neural subgroups (Fig. 4B). No tumors clustered with the classical subtype. The predominance of mesenchymal-like GEM-GBM is consistent with the observation that this human subtype correlates with a high rate of mutation in RB1 (13%), NF1 (37%, indicative of KRAS activation), PTEN (32%), and TP53 (32%) compared with other subtypes (21). However, the clustering of molecular subtypes does not align with specific genotypes of GEMs, indicating some heterogeneity in GBM development and possible association with stochastic progression events. Recently, murine GBMs induced via stereotactic introduction of a lentivirus expressing HRAS and shRNA suppressing p53 function were also shown to harbor a mesenchymal transcriptome signature (22), further supporting alignment of this class with aberrant RAS and p53 functions.
Somatic isocitrate dehydrogenase 1 (IDH1) mutations are present in 73–85% of secondary and in only 3.7–4.9% of primary GBMs (23, 24). Among the primary GBMs, the mesenchymal subtype was not associated with IDH1 mutation, where 30% of the proneural subtype was shown to harbor IDH1 mutations (21). Consistent with the majority of GEM-GBMs harboring a mesenchymal phenotype, no IDH1 mutations were detected by DNA sequencing in GEM-grade II (n = 5), GEM-AA (n = 18), or GEM-GBM (n = 15) tissues. Together, the GEM-GBM phenotype, absence of IDH1 mutations, predominance of the mesenchymal transcriptome subclass, and rapid asymptomatic tumor development findings indicate that the GEM-GBMs are most characteristic of human primary disease.
Discussion
We used combinations of engineered alleles along with analyses of stochastic genetic changes to probe the etiology of high-grade astrocytomas (HGAs) in GEMs. Three major networks perturbed in most human GBMs (RTK/RAS/PI3K/PTEN, p53, and RB signaling) contributed to distinct disease stages. RB tumor suppression (TS) inhibition was required to initiate disease, with constitutive perturbation of KRAS signaling followed by stochastic p53 aberration driving the transition from low- to high-grade disease and yielding grade III tumors (AA). Additional PTEN loss (engineered or stochastic) drove progression to grade IV (GBM) (summarized in Fig. 4C). These conclusions were possible based on the following logic. Of all single events and double combinations, only RB-TS inactivation initiated disease and resulted in GII pathology. Of double combinations, only the combination of Rb-TS inactivation and KRASG12D induction predisposed to GIII tumors. Finally, either stochastic loss of PTEN from GIII tumors or engineered PTEN deletion along with Rb-TS inactivation and KRASG12D induction drove GBM (GIV) progression.
Astrocytoma Initiation.
Recent studies have reported event combinations within the same major GBM-perturbed networks that predispose to GEM-GBM, wherein engineered events were insufficient for initiation, thus limiting analysis of disease evolution. In this study, RB-TS inactivation acutely induced low-grade disease. That no brain pathology was caused by KRASG12D induction or PTEN deletion or both combined is consistent with other recent reports examining adult brain (25–28). The RB network is perturbed in most human GBMs. Although CDKN2A locus [CDKN2A (p16/INK4A) and CDKN2B (p15/INK4B)] deletions are the most frequent events (50–55%), aberrations can occur at multiple points, including CDKN2C (p18/INK4C) deletion (2%), CDK gene amplification (CDK4, 18%; CDK6, 1%), or RB1 deletion/mutation (11–12%) (5, 6). In mouse, deletion of INK4A does not elicit brain pathology (29, 30). There is clear redundancy among the CDKNs in mouse tissues (31) as there is among RB family members. Directly modeling RB-TS inactivation in mouse astrocytes, as well as in most other cell types, requires inhibition of multiple RB family proteins (32–34). Thus, use of a single allele to disable RB, p107, and p130 via the dominant activity of T121 was key to exploring role(s) of RB-mediated TS, and thus GBM initiation. In Chow et al. (25), HGA predisposed by adult astrocyte PTEN and p53 deletion often carried copy-number alterations in genes acting upstream of RB (e.g., CDK4, CDK6, CCND1, CCND2, and CCND3 amplification), supporting a role for stochastic perturbation of this network. Although RB1 deletion alone did not yield disease, it could reduce the latency of HGAs predisposed by codeletion of PTEN and p53, and several resulting tumors had also amplified the E2F3 gene (25).
HGA Progression: A Model for Disease Evolution.
KRASG12D induction was required for progression to HGA. Although stochastic KRAS mutation is rare in human GBM (∼2%) (5), KRAS regulators are frequently aberrant (see Introduction). Causal roles for RAS network aberrations in GEM-HGA have been confirmed in several engineered mouse studies. Unique to the current study is placement of the RAS activation impact at the transition from low- to high-grade disease and before GBM formation in the context of RB network perturbation. Engineered PTEN deletion alone could not drive HGA from low-grade astrocytoma (induced by RB-TS inactivation); however, engineered or stochastic PTEN loss was associated with progression of AA to GBM. Thus, KRASG12D-driven pathway(s) and/or subsequent stochastic events provide selective pressure(s) for PTEN inactivation in the evolution from GIII to GIV disease. Consistent with results herein and with previous observations is the possibility that the PI3K network, activated by KRAS and negatively regulated by PTEN, is involved. Consistent with the hypothesis that RAS network perturbation is required for driving high-grade astrocytoma, Chow et al. also observed numerous stochastic RTK (MET, EGFR, and PDGFR) amplifications in engineered p53/PTEN-deficient GEM-HGAs (25).
Stochastic p53 Missense Mutation in HGA Evolution.
In previous reports, Trp53 and PTEN deletion in embryonic radial glial cells (35) or adult astrocytes (25, 36) predisposed mice to GEM-HGA. Relative contributions to disease could not be assessed. The predominant p53 aberration in human GBMs, missense mutation (5, 35), wasn’t previously examined. Here, we chose not to engineer p53 mutations, but rather to assess emerging tumors for the presence of stochastic p53 mutation. This approach facilitated assessment of p53 mutation requirement in HGA evolution, of spontaneously selected mutation type(s), and of the validity in modeling HGA. Strikingly, as in humans, most GEM-GBMs expressed missense mutant p53, with mutations analogous to those found in human GBMs. Total p53 loss was not observed although cytoplasmic mislocalization of wild-type p53 occurred in several cases. p53 accumulation first appeared in early nodules predisposed by the combination of RB-TS inhibition and KRAS activation, with or without PTEN inactivation. Thus, KRAS directly or indirectly positively selected for p53 missense mutant or mislocalized cells. Recent assessment of high-grade gliomas derived from cultured mouse PTEN−/− ;INK4a/ARF−/− neurospheres orthotopically injected into immunocompromised mice also detected two Trp53 missense mutations (37).
Primary vs. Secondary GBM.
Primary and secondary GBMs are defined by clinical presentation, with the former presenting as HGA and the latter appearing subsequent to previously diagnosed low-grade disease. Some tumor characteristics are more prominent in one class compared with the other, but these divisions are not absolute. For example, although IDH1 mutations are present in 73–85% of secondary GBMs, they also appear in 4–5% of primary GBMs. Although GBMs with a mesenchymal transcriptome signature are dominated by the primary class, secondary GBMs can also harbor this signature. The current study has produced relevant models of low- and high-grade disease, with evident tumor heterogeneity and the majority of GBMs having transcriptional similarity to the aggressive human mesenchymal subtype (4, 5, 38). Because tumors did not carry IDH1 mutations and arose rapidly without prior evidence of symptoms, they most closely resemble primary GBMs. Given that the models may represent etiology of a primary GBM subset, they could provide the first insight into possible evolutionary mechanisms and early disease stages. Overlap in tumor evolution mechanisms among some secondary and primary GBMs is likely, with the determining features of clinical presentation lying outside the actual mechanism(s) of evolution. The use of relevant disease models for discovery of early disease biomarkers and mechanisms of etiology may help elucidate clinically relevant features amenable to disease prevention, intervention, and management.
Materials and Methods
Genetically Engineered Mice and Systemic Induction.
TgGZT121 conditional transgenic mice (12) were maintained by crossing to BDF1 mice, and KRASG12D (39), PTENL/L (40), and hGFAP-CreERT2 (10) mice to C57BL/6J mice. Genetic events were induced by injecting freshly prepared 4-OH-tamoxifen (4OHT; Sigma) intraperitoneally into adult mice (mean 3.5 ± 0.6 mo) with 1 mg per mouse per day for 5 consecutive days (see SI Materials and Methods for 4OHT preparation and genotyping). Methods described herein have been approved by the Institutional Animal Care and Use Committees at the University of North Carolina-Chapel Hill and at the National Cancer Institute-Frederick.
Microarray Transcriptome Analysis.
Total RNA was extracted from frozen tumor tissues using Qiagen RNeasy. Affymetrix mouse 430.2 array or Custom Agilent SurePrint microarrays were performed following standard protocols by the LMT (Science Applications International Corporation-Frederick) and the Institute for Systems Biology microarray core. Microarray data were processed and analyzed with R/Bioconductor packages and Gaggle software platform (41). Each gene in the mouse tumor data was median-centered and divided by its SD. The transformed data were clustered together with human TCGA GBM data using the 840 classifiers identified by Verhaak et al. (21). All raw data are publicly available (GEO26069).
Trp53 and IDH1 Sequencing.
Genomic DNA was prepared from HGA tumor tissues, cortex, or primary GEM-GBM cells. All 11 exons including UTRs of Trp53 and all coding exons for IDH1 were PCR amplified and Sanger sequenced by the LMT.
Additional Methods.
More detailed methods are shown in SI Materials and Methods, and antibody information is shown in Table S6.
Supplementary Material
Acknowledgments
We thank A. Wolthusen, D. Roth, S. Sweeney, S. Henderson, S. Rausch, and D. Swing for excellent animal care; the Laboratory Animal Medicine and the Histopathology Core Facility at the University of North Carolina-Chapel Hill and the transgenic core at the National Cancer Institute (NCI)-Frederick and Pathology/Histotechnology Laboratory at Science Applications International Corporation (SAIC)-Frederick for mouse husbandry and histopathological service; S. Lockett for confocal images; L. Ileva for assistance with MRI; the Laboratory of Molecular Technology (LMT) at SAIC-Frederick for sequencing; and LMT and the Institute for Systems Biology microarray core for microarray studies. We thank S. Jansen, P. Martin, L. Cleveland, L. Lu, A. Adhikari, T. Sullivan, N. K. Collins, D. Householder, T. Guerin, H. Duong, and O. Pacheco-Velez for technical assistance; P. Duke for providing a plasmid containing VEGF-A165 cDNA; and G. Merlino for critical evaluation of the manuscript. This research was supported by the Intramural Research Program of the NCI-Center for Cancer Research, NCI Grants CA084314 and CA046283, the Goldhirsh Foundation, the Brain Tumor Society, and the Waxman Cancer Research Foundation (T.V.D.), NCI CA124608 (E.B.), National Institutes of Health NS020212 and NS033938 (K.D.M.), and the Luxembourg Centre for Systems Biomedicine and the University of Luxembourg (B.K. and L.H.). C.R.M. is a Damon Runyon-Genentech Clinical Investigator (CI-45-09).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray raw data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GEO26069).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317026110/-/DCSupplemental.
References
- 1. Louis DN, Ohgaki H, Weistler OD, Cavenee WK, eds (2007) WHO Classification of Tumours of the Central Nervous System (IARC Press, Lyon) [DOI] [PMC free article] [PubMed]
- 2.Brennan C. Genomic profiles of glioma. Curr Neurol Neurosci Rep. 2011;11(3):291–297. doi: 10.1007/s11910-011-0198-7. [DOI] [PubMed] [Google Scholar]
- 3.Vitucci M, Hayes DN, Miller CR. Gene expression profiling of gliomas: Merging genomic and histopathological classification for personalised therapy. Br J Cancer. 2011;104(4):545–553. doi: 10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries NA, Beijnen JH, van Tellingen O. High-grade glioma mouse models and their applicability for preclinical testing. Cancer Treat Rev. 2009;35(8):714–723. doi: 10.1016/j.ctrv.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Furnari FB, et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 9.Hambardzumyan D, Parada LF, Holland EC, Charest A. Genetic modeling of gliomas in mice: New tools to tackle old problems. Glia. 2011;59(8):1155–1168. doi: 10.1002/glia.21142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casper KB, Jones K, McCarthy KD. Characterization of astrocyte-specific conditional knockouts. Genesis. 2007;45(5):292–299. doi: 10.1002/dvg.20287. [DOI] [PubMed] [Google Scholar]
- 11.Stubdal H, et al. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17(9):4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao A, Wu H, Pandolfi PP, Louis DN, Van Dyke T. Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell. 2002;1(2):157–168. doi: 10.1016/s1535-6108(02)00029-6. [DOI] [PubMed] [Google Scholar]
- 13.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 14.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 15.Xiao A, et al. Somatic induction of Pten loss in a preclinical astrocytoma model reveals major roles in disease progression and avenues for target discovery and validation. Cancer Res. 2005;65(12):5172–5180. doi: 10.1158/0008-5472.CAN-04-3902. [DOI] [PubMed] [Google Scholar]
- 16.Danielyan L, et al. Colocalization of glial fibrillary acidic protein, metallothionein, and MHC II in human, rat, NOD/SCID, and nude mouse skin keratinocytes and fibroblasts. J Invest Dermatol. 2007;127(3):555–563. doi: 10.1038/sj.jid.5700575. [DOI] [PubMed] [Google Scholar]
- 17.Mentlein R, Kendall MD. The brain and thymus have much in common: A functional analysis of their microenvironments. Immunol Today. 2000;21(3):133–140. doi: 10.1016/s0167-5699(99)01557-1. [DOI] [PubMed] [Google Scholar]
- 18.Inda MM, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24(16):1731–1745. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petitjean A, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 20.Parikh AH, Smith JK, Ewend MG, Bullitt E. Correlation of MR perfusion imaging and vessel tortuosity parameters in assessment of intracranial neoplasms. Technol Cancer Res Treat. 2004;3(6):585–590. doi: 10.1177/153303460400300608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niola F, et al. Mesenchymal high-grade glioma is maintained by the ID-RAP1 axis. J Clin Invest. 2013;123(1):405–417. doi: 10.1172/JCI63811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715. [DOI] [PubMed] [Google Scholar]
- 24.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–316. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasheed BK, et al. PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer Res. 1997;57(19):4187–4190. [PubMed] [Google Scholar]
- 27.Marumoto T, et al. Development of a novel mouse glioma model using lentiviral vectors. Nat Med. 2009;15(1):110–116. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holland EC, et al. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet. 2000;25(1):55–57. doi: 10.1038/75596. [DOI] [PubMed] [Google Scholar]
- 29.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 30.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413(6851):83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 31.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602(1):73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 32.Robanus-Maandag E, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12(11):1599–1609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14(23):3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sage J, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14(23):3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacques TS, et al. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 2010;29(1):222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim HS, et al. (2012) Gliomagenesis arising from Pten- and Ink4a/Arf-deficient neural progenitor cells is mediated by the p53-Fbxw7/Cdc4 pathway, which controls c-Myc. Cancer Res 72(22):6065–6075. [DOI] [PubMed]
- 38.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki A, et al. T cell-specific loss of Pten leads to defects in central and peripheral tolerance. Immunity. 2001;14(5):523–534. doi: 10.1016/s1074-7613(01)00134-0. [DOI] [PubMed] [Google Scholar]
- 41.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: An open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.