Abstract
The general transcription factor TFIID is composed of TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs). TFIID mediates the transcriptional activation of a subset of eukaryotic promoters. The N-terminal domain (TAND) of TAF1 protein (Taf1p) inhibits TBP by binding to its concave and convex surfaces. This study examines the role of the TAND in transcriptional regulation and tests whether the TAND is an autonomous regulator of TBP. The TAND binds to and regulates TBP function when it is fused to the amino or carboxy terminus of Taf1p, the amino or carboxy terminus of Taf5p, or the amino terminus of Taf11p. However, a carboxy-terminal fusion of the TAND and Taf11p is not compatible with several other TAF proteins, including Taf1p, in the TFIID complex. These results indicate that there is no or minimal geometric constraint on the ability of the TAND to function normally in transcriptional regulation as long as TFIID assembly is secured.
Transcription of eukaryotic protein-coding genes is regulated by the concerted action of a number of distinct transcription factors (reviewed in references 34 and 38). These transcription factors are recruited to the promoter region in a precise gene-specific order (reviewed in reference 10). Transcription factor IID (TFIID) is a general transcription factor complex that is composed of the TATA-binding protein (TBP) and 14 TBP-associated factors (TAFs) (39, 41). It recognizes and binds to several core promoter elements and is thought to be a platform for the subsequent assembly of the preinitiation complex (reviewed in reference 3). TFIID also plays a role in transcriptional activation (reviewed in references 3 and 34). Extensive studies using in vitro reconstitution systems have shown that the involvement of the TAFs that make up TFIID in transcriptional activation can vary depending on the experimental conditions, e.g., whether mediator components are included and/or nucleosomal DNA is used as a template (16, 35, 43). In good agreement with these observations, genetic studies of yeast revealed that the requirement for TAFs in in vivo transcription depends on the promoter structures of the target genes (reviewed in reference 13).
Yeast promoters are classified as TAF dependent (TAFdep) or TAF independent (TAFind) (23, 25). In vivo cross-linking and chromatin immunoprecipitation analyses indicate that TAFs interact infrequently with TAFind promoters but frequently with TAFdep promoters at an approximately equimolar ratio to TBP. In the case of the TAFdep promoters of ribosomal protein (RP) genes, TAF recruitment is directed by upstream activating sequences (UAS) but not by the core promoter and it does not depend on the function of TBP and pol II (26, 32). In contrast, the TAFind UAS in the PGK1 and PYK1 promoters do not efficiently recruit TAFs to TAFdep or TAFind core promoters (32). These observations indicate that an activator(s) bound to a TAFdep UAS may specifically interact with TAF(s) and recruit TFIID onto the core promoter. In addition, there are some functional compatibilities between the UAS and the core promoter (5, 26, 32). For instance, while the TAFdep RPS5 UAS activates the TAFind ADH1 and CUP1 core promoters at the same levels as its own TAFdep RPS5 core promoter, the TAFind ADH1 and CUP1 UAS were largely unable to activate the TAFdep RPS5 core promoter (26).
Despite these intriguing observations, the question of how activators bound to TAFdep UAS can recruit TFIID to the core promoter remains unresolved. The unstable TFIID-core promoter complex formed during in vitro transcription is stabilized by transcriptional activators that induce a conformational change in TFIID (7, 27). This suggests that activators may act in two steps during this process, namely, they may first recruit TFIID to the core promoter by directly interacting with TAFs and then convert TFIID into an active form that is competent to initiate transcription.
We have previously proposed a two-step handoff model describing TAF- and TBP-dependent regulation of transcription. The model involves a pivotal triadic interplay between an activator, the TAF1 N-terminal domain (TAND), and TBP. The TAND is comprised of two functionally distinct subdomains, TAND1 and TAND2, which bind to the concave and convex surfaces of TBP, respectively, and thereby inhibit TBP (2, 18, 19, 22). An important observation is the fact that some activation domains (AD) are functionally interchangeable with TAND1 in vitro and in vivo (20). Thus, it is possible that TAND1 prevents TBP from binding to the TATA element during the initial stage of TFIID recruitment. However, once TFIID is recruited onto the DNA, TAND1 may be displaced sequentially by an AD and the TATA element. These steps are associated with conformational changes in TFIID that render it competent to initiate transcription (20).
An increasing amount of evidence suggests that the TAND participates in transcriptional activation even though it is not absolutely required (5, 6, 8, 14, 15, 29). For example, the TBP-inhibitory activity of human TAF1 can be reduced in vitro by the action of the AD (14, 15). In addition, deletion of TAND1 impairs the activating function of RPS5 UAS and twice-repeated synthetic GAL4 binding sites on the RPS5 core promoter (5). A microarray experiment also showed that deletion of TAND1 decreases the transcription of a subset of genes (8).
The proposed two-step handoff model suggests that the TAND modulates the interaction between TBP and the TATA element after TFIID is loaded onto the core promoter. The TAND may maintain TBP in an inactive state until it encounters the AD, which dissociates the TAND from TBP and allows activated TBP to bind to the TATA element. This model predicts that the TAND could function independently of other regions of TFIID; in other words, the TAND may be an autonomous regulator of TBP. This notion is supported by the well-known fact that some transcription factors have autonomous DNA-binding and activation domains (36). Autonomous domains function independently and can be readily swapped between heterologous transcription factors.
This study tests the autonomy of the TAND. The TAND was fused to the amino- or carboxy-terminal ends of TAFs that occupy different spatial positions in TFIID. The novel forms of TFIID with the TAND in different spatial positions were tested for the ability to complement temperature-sensitive TAND-deficient yeast cells and for transcriptional competence. When the TAND was fused to the carboxy terminus of Taf1p, the amino or carboxy terminus of Taf5p, or the amino terminus of Taf11p, the novel TFIID was transcriptionally competent. These findings indirectly support the proposed two-step handoff model and demonstrate that the TAND is a largely autonomous regulator of transcription.
MATERIALS AND METHODS
Yeast strains, media, and cultures.
Standard techniques were used for yeast growth, transformation, and tetrad dissection (1). The yeast strains used in this study are listed in Table 1. The details of the methods used to construct these strains are available upon request.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| Y22.1 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pYN1/TAF1 |
| YTK2741 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pM1169/TAF1 |
| YST3 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pM3315/TAF1 (Y19A,F57A) |
| YTK2233 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pM1001/TAF1(ΔTAND) |
| YST139 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pM3707/TAF1(ΔTAND)-TAND |
| YST142 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 pM3707/TAF1(ΔTAND)-TAND* |
| YTK6047 | MATaura3-52 trp1-63 leu2,3-112 Δtaf5 pYN31/TAF5 |
| YST136 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pRS315 |
| YST145 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pRS315 |
| YST148 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pM3681/TAF5 |
| YST151 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pM3682/TAND-TAF5 |
| YST154 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pM3683/TAND*-TAF5 |
| YST157 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pM3684/TAF5-TAND |
| YST160 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pYN31/TAF5 pM3685/TAF5-TAND* |
| YTK3991 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 |
| YST163 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pRS315 |
| YST166 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pM3681/TAF5 |
| YST169 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pM3682/TAND-TAF5 |
| YST172 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pM3683/TAND*-TAF5 |
| YST175 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pM3684/TAF5-TAND |
| YST178 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pYN31/TAF5 pM3685/TAF5-TAND* |
| YST181 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pM3681/TAF5 |
| YST184 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pM3682/TAND-TAF5 |
| YST187 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pM3683/TAND*-TAF5 |
| YST190 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pM3684/TAF5-TAND |
| YST193 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1169/TAF1 pM3685/TAF5-TAND* |
| YST196 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pM3681/TAF5 |
| YST199 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pM3682/TAND-TAF5 |
| YST202 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pM3683/TAND*-TAF5 |
| YST205 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pM3684/TAF5-TAND |
| YST208 | MATα ura3-52 trp1-63 leu2,3-112 Δtaf1 Δtaf5 pM1001/TAF1(ΔTAND) pM3685/TAF5-TAND* |
| Y13.2 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 pYN1/TAF1 |
| YTK6048 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pYN1/TAF1 pM2060/TAF11 |
| YST57 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11 |
| YST340 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11 pRS315 |
| YST343 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::His3pM1169/TAF1 pM2160/TAF11pM2093/TAF11 |
| YST346 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11pM2181/TAND-TAF11 |
| YST349 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11pM3566/TAND*-TAF11 |
| YST352 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11pM3567/TAF11-TAND |
| YST355 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2160/TAF11pM3568/TAF11-TAND* |
| YST60 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11 |
| YST358 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11 pRS315 |
| YST361 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11pM2093/TAF11 |
| YST364 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11pM2181/TAND-TAF11 |
| YST367 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11pM3566/TAND*-TAF11 |
| YST370 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11pM3567/TAF11-TAND |
| YST373 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2160/TAF11pM3568/TAF11-TAND* |
| YST376 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2093/TAF11 |
| YST379 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM2181/TAND-TAF11 |
| YST382 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3566/TAND*-TAF11 |
| YST391 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2093/TAF11 |
| YST394 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM2181/TAND-TAF11 |
| YST397 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3566/TAND*-TAF11 |
| YST406 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11 |
| YST409 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pRS315 |
| YST412 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pM2093/TAF11 |
| YST415 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pM2181/TAND-TAF11 |
| YST418 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pM3566/TAND*-TAF11 |
| YST421 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pM3567/TAF11-TAND |
| YST424 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1169/TAF1 pM3838/TAF11pM3568/TAF11-TAND* |
| YST427 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11 |
| YST430 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11 pRS315 |
| YST433 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11pM2093/TAF11 |
| YST436 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11pM2181/TAND-TAF11 |
| YST439 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11pM3566/TAND*-TAF11 |
| YST442 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11pM3567/TAF11-TAND |
| YST445 | MATα ura3-52 trp1-63 leu2,3-112 his3-609 Δtaf1 Δtaf11::HIS3pM1001/TAF1(ΔTAND) pM3838/TAF11pM3568/TAF11-TAND* |
The YTK2741, YST3, YTK2233, YST139, and YST142 strains were generated from Y22.1 (Δtaf1 strain) (19) by a plasmid shuffle technique. The YTK6047 strain was generated from H2440 (19) by targeted disruption of the TAF5 gene using a marker cassette that has a URA3 gene between duplicate copies of a Salmonella hisG gene segment. YTK6047 (Δtaf5 strain) was then crossed with YTK2233 (Δtaf1 strain) and dissected to obtain the haploid strain YTK3991, which carries double deletions of the TAF1 and TAF5 genes. As TAF1 and TAF5 are both essential genes, the growth of YTK3991 is supported by pYN31/TAF5 (URA3 marker) and pM1001/taf1ΔTAND (TRP1 marker). pM1001 of YTK3991 was replaced with pM1169/TAF1 (TRP1 marker) to create YST136. YTK3991 and YST136 were then transformed with plasmids listed in Table 1 to generate the strains used (see Fig. 2B). These strains were grown on 5-fluoroorotic acid (5-FOA) plates to generate other strains used (see Fig. 2C).
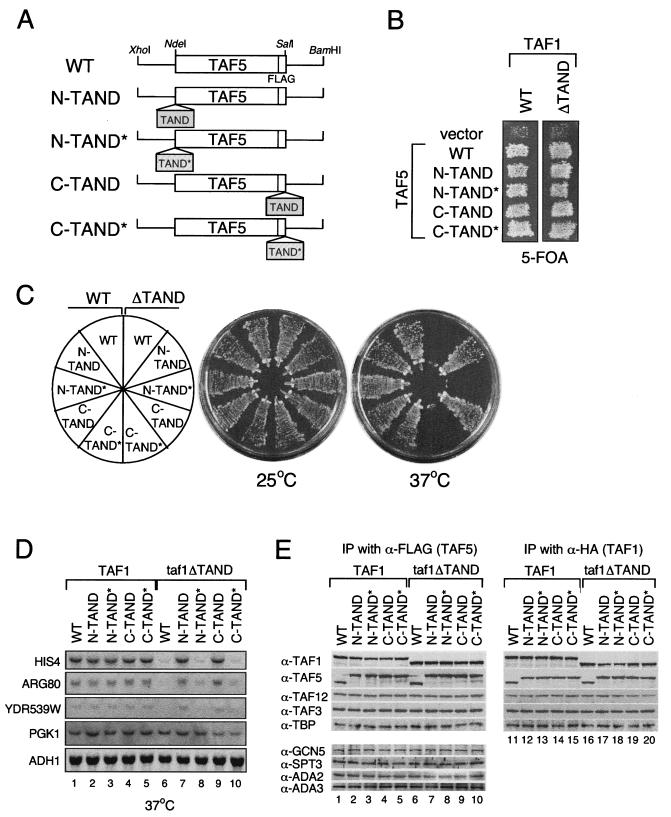
FIG. 2.
Characterization of amino- and carboxy-terminal TAND-Taf5p. (A) Schematic diagram of the plasmids encoding TAND-Taf5p derivatives with FLAG tags at their carboxy termini. (B) Strains expressing the indicated TAF5p derivatives were grown on 5-FOA plates at 30°C for 5 days to induce the loss of the plasmid encoding WT Taf5p. The host yeast strain is Δtaf1 Δtaf5 and expresses WT Taf1p or Taf1p-ΔTAND and WT Taf5p as indicated. (C) The strains were grown on YPD plates for 3 days at the indicated temperatures. (D) Transcription of TAND-dependent and TAND-independent genes. The expression of HIS4, ARG80, YDR539W, PGK1, and ADH1 was examined as described in the legend to Fig. 1E. (E) Immunoprecipitation (IP) of TFIID and SAGA complexes. Whole-cell extracts were prepared from the indicated strains and immunoprecipitated with anti-FLAG (α-FLAG; lanes 1 to 10) or anti-HA (lanes 11 to 20) monoclonal antibodies. Note that FLAG and HA epitope tags were added to Taf5p and Taf1p derivatives, respectively, in these strains. Proteins coprecipitating with Taf5p (lanes 1 to 10) or Taf1p (lanes 11 to 20) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with the indicated antibodies.
The YTK6048 strain was generated from Y13.2 (Δtaf1 strain) (19) by targeted disruption of the TAF11 gene using a PCR-based gene deletion method (30). A pair of plasmids, i.e., pYN1/TAF1 and pM2060/TAF11, carried by YTK6048 was replaced with four different sets of plasmids listed in Table 1 to generate strains YST57, YST60, YST406, and YST427. These strains were then transformed with the same set of six plasmids (i.e., pRS315 [40], pM2093, pM2181, pM3566, pM3567, and pM3568) to generate the 24 strains used (see Fig. 3B and 4B). Some of the strains obtained in this way were grown further on 5-FOA plates to generate other strains used (see Fig. 3C).
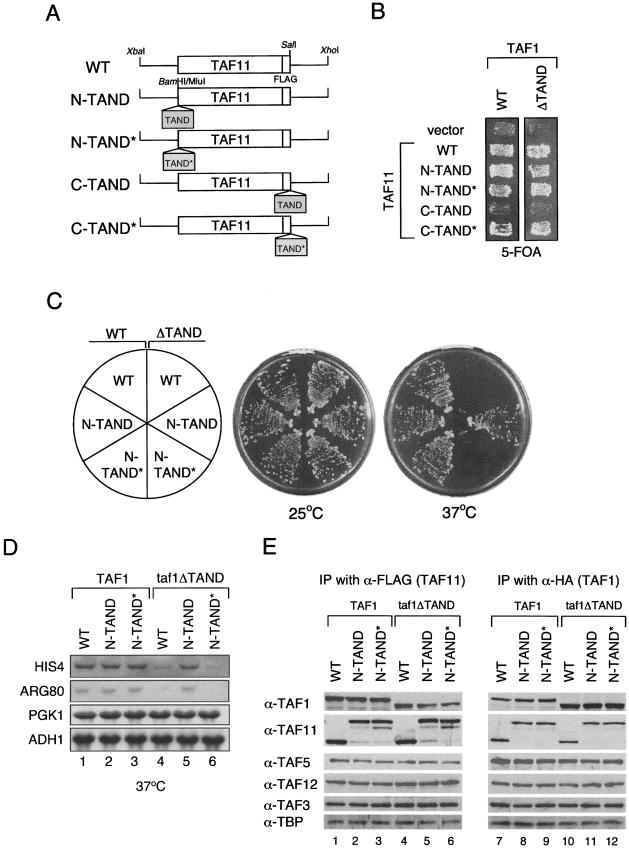
FIG. 3.
Characterization of amino- and carboxy-terminal TAND-Taf11p. (A to D) The TAF11 gene was manipulated as described in the legends to Fig. 2A to D. (E) Immunoprecipitation (IP) analyses of the TFIID complex in the presence of TAND-Taf11p derivatives. As described in the legend to Fig. 2E, whole-cell extracts were prepared from the indicated strains and immunoprecipitated with anti-FLAG (α-FLAG; lanes 1 to 6) or anti-HA (lanes 7 to 12) monoclonal antibodies that react with Taf11p and Taf1p, respectively. Proteins coprecipitating with Taf11p (lanes 1 to 6) or Taf1p (lanes 7 to 12) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with the indicated antibodies.
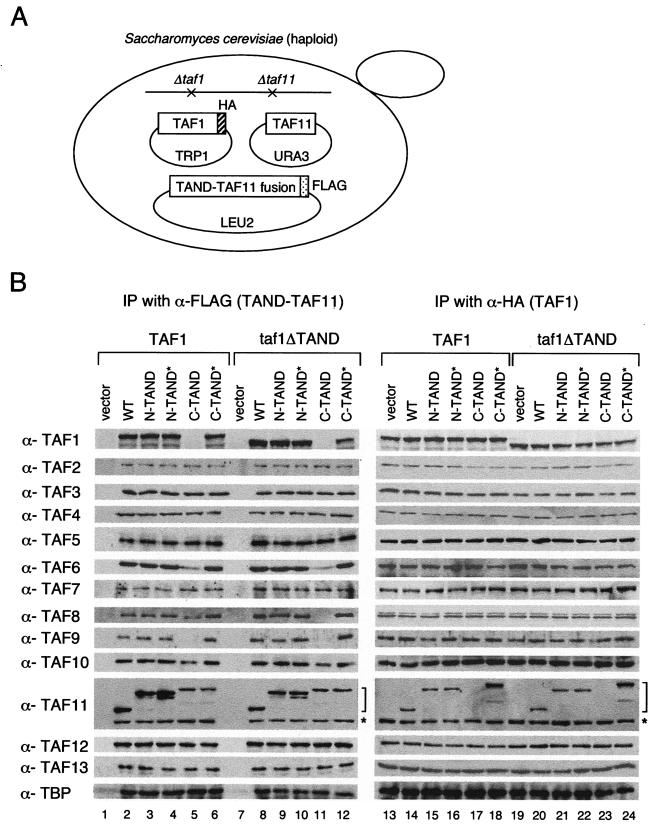
FIG. 4.
Taf11p bearing a carboxy-terminal TAND and Taf1p cannot be incorporated into the same TFIID complex. (A) Schematic representation of the strains used in this experiment. The WT Taf11p was not tagged with an epitope. (B) Whole-cell extracts (WCE) were prepared from the indicated strains expressing a TAF11p derivative and WT Taf1p or Taf1p lacking a TAND and untagged Taf11p. Aliquots of WCE were immunoprecipitated with anti-FLAG (α-FLAG; lanes 1 to 12) or HA (lanes 13 to 24) antibodies. Proteins coprecipitating with Taf11p (lanes 1 to 12) or with Taf1p (lanes 13 to 24) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with the indicated antibodies. The positions of FLAG-tagged Taf11p and untagged WT Taf11p are marked with brackets and asterisks, respectively.
Construction of plasmids encoding TAF1 genes.
The details of plasmid construction and oligonucleotide sequences are available upon request. Briefly, pM1169 and pM1001 were constructed by inserting a DNA fragment encoding four repeats of the hemagglutinin (HA) epitope tag at the carboxy terminus of Taf1p encoded by pM11 and pM10 (21), respectively. pM1169 was subjected to site-specific mutagenesis to create pM3315 by using oligonucleotides TK48 and TK311. These two oligonucleotides were used to create a mutant form of the TAND that we called TAND*. This form carries two mutations, Y19A and F57A, that abolish its TBP-binding activity.
pYN2 (19) was mutagenized to create pM384 (taf1ΔTAND/pRS314) using the oligonucleotides TK3, TK5, and T903. TK3 generates NheI and SphI sites at the carboxy terminus of Taf1p. The NheI-SphI DNA fragment encoding the TAND was subcloned into the NheI/SphI sites of pM384 to create pM877 (taf1ΔTAND-TAND/pRS314). pM877 was then mutagenized by oligonucleotides TK48 and TK311 to create pM3316. pM3707 and pM3708 were obtained from pM877 and pM3316, respectively, by adding four repeats of the HA epitope tag to the carboxy terminus of Taf1p.
Construction of plasmids encoding TAF5 genes.
pYN32 was constructed by ligating the 3.2-kb ClaI-PvuII TAF5 gene fragment into the ClaI/SmaI sites of pBluescript II KS(+) (Stratagene) and then subjected to site-specific mutagenesis to create pYN33 using oligonucleotides T612 and T614, which generate NdeI and XbaI sites at the amino and carboxy termini of Taf5p, respectively. pM3556 was constructed by ligating the 3.2-kb ClaI-BamHI TAF5 gene fragment of pYN33 into the ClaI/BamHI sites of pRS424 (9). pM3558 was then created from pM3556 by disrupting the internal BamHI site and instead creating the SalI site in juxtaposition with the XbaI site. After ligating the XhoI-SalI DNA fragment encoding three repeats of the FLAG epitope tag into the SalI site of pM3558, the 3.2-kb XhoI-BamHI TAF5 gene fragment was subcloned into the XhoI/BamHI sites of pRS315 to create pM3681. pM3682 and pM3683 were constructed by ligating the NdeI-NdeI DNA fragment encoding the TAND or TAND*, respectively, into the NdeI site of pM3681. Similarly, pM3684 and pM3685 were constructed by ligating the SalI-SalI DNA fragment encoding the TAND or TAND*, respectively, into the SalI site of pM3681.
Construction of plasmids encoding TAF11 genes.
The 2.2-kb XbaI-XhoI DNA fragment containing the TAF11 gene was amplified by PCR using the TK1481 and TK1484 primer pair and then subcloned into the XbaI/XhoI sites of pRS315 to create pM2060. After three repeats of the FLAG epitope tag were added to the amino terminus of Taf11p, the XbaI-XhoI TAF11 gene fragment was transferred into pRS316 to create pM2160. pM3838 was similarly constructed from pM2060 but without the FLAG epitope tag. pM2093 was obtained by adding three repeats of the FLAG epitope tag to the carboxy terminus of Taf11p in pM2060.
pM2060 was mutagenized by oligonucleotide TK1939 to create BamHI, BssHII, and MluI sites at the initiation codon of the TAF11 gene. After three repeats of the FLAG epitope tag were added to the carboxy terminus of Taf11p, the BamHI-MluI fragment encoding the TAND of pM1169 was ligated into the the BamHI/MluI sites to create pM2181. The BamHI-MluI fragment of pM2181 was replaced with the corresponding region of pM3315 to create pM3566. pM3567 and pM3568 were constructed by ligating the SalI-SalI fragments encoding the TANDs of pM3005 and pM3569, respectively, into the SalI site of pM2093.
GST pull-down assay.
pM1431 and pM3629 were constructed so that they would express glutathione S-transferase (GST)-TAND (amino acids [aa] 6 to 71) and GST-TAND* (aa 6 to 71) proteins, respectively, in Escherichia coli (DH5α) by ligating the 0.2-kb PCR-amplified BamHI-EcoRI fragment into the same sites of pGEX2T (Amersham Biosciences). Saccharomyces cerevisiae TBP was expressed as a histidine-tagged protein using a pET system (Novagen). The preparation of these proteins and GST pull-down experiments were conducted as described previously (21).
Immunoblot and coimmunoprecipitation analyses.
Immunoblot and coimmunoprecipitation analyses were conducted as described previously (21, 42). Polyclonal antibodies directed against TAF1, TAF3, TAF12, and TBP were prepared as described previously (17, 21, 42). Antibodies against ADA2, ADA3, GCN5, and HA epitopes were purchased from Santa Cruz Biotechnology, Inc. Antibodies against FLAG epitope were purchased from Sigma. Polyclonal antibodies directed against SPT3 (aa 1 to 200), TAF2 (aa 591 to 790), TAF4 (aa 1 to 200), TAF5 (aa 460 to 798), TAF6 (aa 1 to 200), TAF7 (aa 1 to 214), TAF8 (aa 1 to 200), TAF9 (aa 1 to 157), TAF10 (aa 1 to 206), TAF11 (aa 1 to 176), and TAF13 (aa 1 to 167) were raised in rabbits by using recombinant, gel-purified His-tagged polypeptides expressed in bacteria as the antigens.
Northern blot analyses.
Northern blot analyses were performed as described previously (42). To prepare the probes, DNA fragments surrounding the initiating methionine were amplified by PCR from yeast genomic DNA, purified, and 32P labeled using a random priming method.
RESULTS
Taf1p functions with the TAND at the amino or carboxy terminus.
The autonomy of the TAND was initially tested by comparing the functions of wild-type (WT) Taf1p, Taf1p with the TAND fused to the carboxy terminus instead of the amino-terminus, and a novel TAND mutant carrying Y19A and F57A amino acid substitutions. Previous studies showed that Y19A or F57A significantly reduces TBP binding of TAND1 and TAND2, respectively (20, 21). The double Y19A-F57A mutant, referred to here as TAND*, was used as a negative control for the domain-swapping experiments described below. GST pull-down assays showed that TAND* completely lacks TBP-binding activity (Fig. 1B).
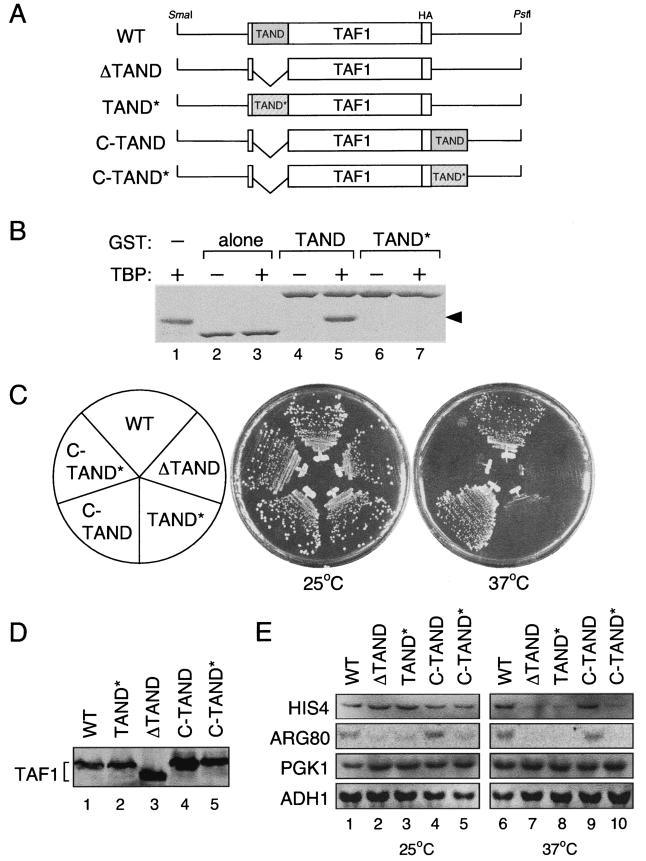
FIG. 1.
Comparison of WT Taf1p and carboxy-terminal TAND-Taf1p. TAND* designates a TAND double mutant bearing the Y19A and F57A substitutions. (A) Schematic diagram of plasmid constructs encoding Taf1p derivatives with HA tags at the carboxy termini. (B) GST pull-down assay using TBP and GST-TAND derivatives or GST. Negative controls (−) lack cell lysates with TBP (lanes 2, 4, and 6); the positive control is purified TBP (lane 1). TBP is indicated by the arrowhead. (C) Comparison of growth of several taf1 mutants at 25 and 37°C. Strains lacking the TAF1 gene but carrying one of several plasmids encoding Taf1p derivatives as indicated were grown on yeast-peptone-dextrose (YPD) plates for 3 days at the indicated temperatures. (D) Expression of Taf1p derivatives. Logarithmically grown cells in YPD were harvested 24 h after the temperature shift from 25 to 37°C. Aliquots of whole-cell extracts were electrophoresed on a sodium dodecyl sulfate-polyacrylamide gel and transferred to a nitrocellulose membrane. Taf1p derivatives were detected using anti-TAF1 polyclonal antibodies (bracketed on the left). (E) Transcription of TAND-dependent or -independent genes in taf1 mutants. Expression of HIS4 and ARG80 (TAND dependent) and PGK1 and ADH1 (TAND independent) was measured in the indicated strains by Northern blot analysis. Cultures were grown in YPD medium to log phase at 25°C and shifted to 37°C for 2 h (lanes 6 to 10) or maintained at 25°C (lanes 1 to 5).
The Taf1p derivatives used in these experiments are shown schematically in Fig. 1A. ΔTAND and TAND* lack the TAND region or TAND activity, respectively. C-TAND and C-TAND* express the TAND or TAND*, respectively, at the carboxy-terminal end of Taf1p. If the TAND is autonomous, C-TAND, but not C-TAND*, should rescue the deficiency of ΔTAND or TAND*.
TAND activity can be readily detected in vivo because TAND deficiency causes yeast strains to be temperature sensitive for growth at 37°C (Fig. 1C) (19). Alternatively, TAND activity can be detected by measuring the expression of TAND-dependent genes (17). In this study, the expression of the TAND-dependent genes HIS4, ARG80, and YDR539W were monitored in yeast strains expressing variant TAFs.
C-TAND and C-TAND* substituted effectively for WT Taf1p in a plasmid shuffle assay (data not shown), indicating that C-TAND or C-TAND* supports cell growth at the permissive temperature in the absence of WT Taf1p. However, only C-TAND rescued the temperature-sensitive phenotype of the ΔTAND or TAND* strains (Fig. 1C). This result suggests that the TBP-binding activity of the TAND is essential for growth at the restrictive temperature. Immunoblotting analysis demonstrated that similar levels of the Taf1p derivatives were expressed under all conditions, eliminating the possibility that differential expression of the TAF derivatives influenced the results (Fig. 1D). mRNA expression was also measured for several TAND-dependent genes at the permissive and restrictive temperatures (Fig. 1E). Expression of HIS4 mRNA decreased at 37°C and expression of ARG80 mRNA decreased at 25 and 37°C in strains expressing ΔTAND or TAND*. In contrast, expression of the TAND-independent genes PGK1 and ADH1 was not affected by TAND deficiency at 25 or 37°C. Furthermore, expression of HIS4 and ARG80 mRNA returned to the WT level in strains expressing C-TAND but not in strains expressing C-TAND* (Fig. 1E). These results are self-consistent (compare Fig. 1E and C) and support the conclusion that the TAND functions normally when it is fused to the carboxy terminus of Taf1p.
Taf5p functions with the TAND at the amino or carboxy terminus.
The autonomy of the TAND was also tested by fusing it to the amino or carboxy terminus of Taf5p, which is a subunit of TFIID and SAGA (TAND-Taf5p fusion proteins are shown schematically in Fig. 2A). The ability of TAND-Taf5p fusions to support normal growth was tested in a double-deletion Δtaf1 Δtaf5 yeast strain in which WT Taf5p was expressed from a selectable URA3 vector, a TAND-Taf5p variant was expressed from a selectable LEU2 vector (Fig. 2A), and WT Taf1p or Taf1p-ΔTAND was expressed from a selectable TRP vector (data not shown). The TAND-Taf5p fusion proteins did not have a dominant-negative phenotype in the presence of WT Taf5p and were also fully functional in vivo as a sole source of Taf5p (Fig. 2B). This suggests that an extraneous TAND or TAND* does not interfere with the function(s) of TFIID or SAGA. Indeed, SAGA-dependent activation of the GAL1 promoter was normal when these strains were grown on raffinose or galactose (data not shown). Furthermore, amino- or carboxy-terminal TAND-Taf5p fusion proteins suppressed the temperature sensitivity (Fig. 2C, right half circle) and transcriptional deficiency of HIS4, ARG80, and YDR539W in taf1ΔTAND yeast (Fig. 2D, lanes 6, 7, and 9). However, the transcriptional defect was not restored when amino- or carboxy-terminal TAND-Taf5p was replaced with amino- or carboxy-terminal TAND*-Taf5p (Fig. 2D, lanes 8 and 10). In addition, adverse effects on growth or transcription were not observed in strains expressing TAND-Taf5p and WT Taf1p (Fig. 2C, left half circle, and D). This result suggests that multiple TANDs can be present in TFIID without causing the loss of TFIID function.
Immunoprecipitation was used to determine if TAND-Taf5p altered the stability or composition of TFIID and/or SAGA. Anti-FLAG antibodies that cross-react with TAND-Taf5p were used to analyze protein complexes in whole-cell extracts. The results showed that TFIID-specific (Taf1p, Taf3p, and TBP), SAGA-specific (Gcn5p, Spt3p, Ada2p, and Ada3p), and shared (Taf12p) subunits coprecipitated in similar amounts from 10 different strains expressing TAND-Taf5p variants (Fig. 2E, left). Essentially the same results were obtained by a reciprocal immunoprecipitation experiment using anti-HA antibodies that cross-react with Taf1p (Fig. 2E, right). Thus, we conclude that amino- or carboxy-terminal TAND-Taf5p does not alter the composition or stability of TFIID or SAGA.
Taf11p functions with amino-terminal but not with carboxy-terminal TAND.
The TAND was also fused to Taf11p, a TFIID-specific subunit (TAND-Taf11p derivatives are shown schematically in Fig. 3A), but the results were different from the Taf1p and Taf5p TAND fusions. First, carboxy-terminal TAND-Taf11p did not support normal growth in the absence of WT Taf11p (Fig. 3B, C-TAND), but carboxy-terminal TAND*-Taf11p did. One possible explanation is that carboxy-terminal TAND-Taf11p forms a TAND-TBP complex that sterically interferes with the assembly or conformational flexibility of TFIID.
Amino-terminal TAND-Taf11p has different properties from carboxy-terminal TAND-Taf11p. For example, amino-terminal TAND-Taf11p suppressed the temperature-sensitive phenotype of ΔTAND (Fig. 3C), and the TAND-Taf11p, but not the TAND*-Taf11p, fusion protein restored the normal transcription of HIS4 and ARG80 in ΔTAND strains (Fig. 3D). In addition, Taf1p, Taf3p, Taf5p, Taf12p, TBP, and Taf11p derivatives coimmunoprecipitated in similar amounts using either anti-FLAG or HA antibodies that cross-react with Taf11p and Taf1p, respectively (Fig. 3E). Amino-terminal TANDs in both Taf1p and Taf11p are compatible with normal TAF functions and support normal growth (Fig. 3C, left half circle), transcription (Fig. 3D, lanes 1 to 3), and TFIID complex formation (Fig. 3E, lanes 1 to 3 and 7 to 9). Collectively, these observations indicate that the TAND is functionally competent when it is fused to the amino but not the carboxy terminus of Taf11p.
Taf1p and carboxy-terminal TAND-Taf11p are not compatible in the TFIID complex.
The composition of TFIID was examined in strains expressing WT Taf11p and carboxy-terminal TAND-Taf11p (Fig. 4A). In this experiment, WT Taf11p lacks a FLAG tag while carboxy-terminal TAND-Taf11p has a FLAG tag. Cells also expressed HA-tagged WT Taf1p or Taf1p lacking a TAND (TAF1 and taf1ΔTAND in Fig. 4B). Immunoblots using anti-TAF11 antibodies confirmed that WT Taf11p and TAND-Taf11p were expressed at similar levels in all strains, as were Taf1p, Taf2p, Taf3p, Taf4p, Taf5p, Taf6p, Taf7p, Taf8p, Taf9p, Taf10p, Taf12p, Taf13p, and TBP (data not shown). However, anti-FLAG antibodies coimmunoprecipitated a significant amount of WT Taf11p along with FLAG-tagged TAND-Taf11p (Fig. 4B, lanes 2 to 6 and 8 to 12). This result suggests that Taf11p exists as a dimer or multimer in TFIID. In addition, no Taf1p/Taf9p and less Taf6p/Taf8p were coprecipitated with FLAG-tagged carboxy-terminal TAND-Taf11p (Fig. 4B, lanes 5 and 11). Notably, the association of Taf8p depends on the TAND of Taf1p (Fig. 4B, lanes 5 and 11). In contrast, only slightly less Taf10p, which is an interaction partner of Taf8p via histone fold domains (12), was coprecipitated in either strain expressing WT Taf1p or Taf1p lacking a TAND (Fig. 4B, lanes 5 and 11). Thus, most of the Taf10p may be anchored to TFIID by its association with Taf3p, another histone fold domain-containing partner of Taf10p, or with other TAFs. A reciprocal coimmunoprecipitation assay revealed that FLAG-tagged carboxy-terminal TAND-Taf11p cannot be coprecipitated with HA-tagged Taf1p whether the latter protein carries the TAND or not (Fig. 4B, lanes 17 and 23). In contrast, Taf2p, Taf3p, Taf4p, Taf5p, Taf7p, Taf12p, Taf13p, and TBP coimmunoprecipitated at similar levels under all conditions (Fig. 4B, lanes 2 to 6, 8 to 12, and 13 to 24). These results could be explained if carboxy-terminal TAND-Taf11p and Taf1p/Taf9p/Taf6p/Taf8p cannot coexist or can coexist only poorly in a single TFIID complex. This possibility would also explain the functional deficiency of carboxy-terminal TAND-Taf11p (Fig. 3B).
DISCUSSION
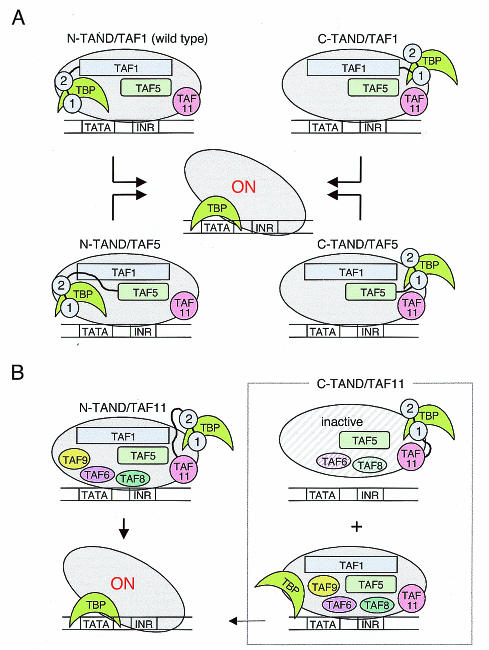
Here, we have studied the autonomy of the TAND at the carboxy terminus of Taf1p, the amino or carboxy terminus of Taf5p, and the amino terminus of Taf11p. Our preliminary experiments show that the TAND can also be functional at the amino termini of Taf2p, Taf4p, and Taf13p but not at the amino terminus of TFIIB (unpublished observations). Thus, it appears that the TAND is largely autonomous in that it functions similarly at the amino terminus of Taf1p and at the termini of several other subunits of TFIID but not at the termini of other components of the preinitiation complex. The study is summarized in Fig. 5. The ability of the TAND to bind TBP appears to be crucial to its ability to function in any position. These data suggest that the TAND modulates the TBP-TATA interaction independently of the other domains of TFIID.
FIG. 5.
Model for an autonomous TAND as a regulator of TFIID. (A) TFIID bearing the TAND at the amino or carboxy terminus of Taf1p or Taf5p can mediate normal levels of transcription in vivo. Activators may induce a conformational change in TFIID that stabilizes its binding to the core promoter. The activated form of TFIID is indicated as ON. The relative orientations of the TAFs within TFIID are arbitrarily depicted. INR, initiator. (B) TFIID bearing the TAND at the amino terminus of Taf11p can function in the same way as shown in panel A (left). However, two distinct forms of TFIID are produced in yeast cells that express carboxy-terminal TAND-Taf11p. One form contains carboxy-terminal TAND-Taf11p and less Taf6p/Taf8p and lacks Taf1p/Taf9p (top right), whereas the other form contains all TAF proteins, including WT Taf11p, but lacks carboxy-terminal TAND-Taf11p (bottom right). The former is inactive, but the latter is capable of normal function.
The two-step handoff model, in which a transcription factor AD triggers sequential events that displace TAND1 from TBP and facilitate the binding of TBP to the TATA element, was proposed previously (20). According to this model, if the AD contacts the TAND-TBP complex, wherever it may be situated in TFIID, it can displace the TAND and thereby activate transcription. In other words, the TAND may not be a simple repressor of TBP but rather a more sophisticated regulator that maintains TBP in an inactive state near the TATA element by anchoring to other TAFs until it encounters an activation signal from the AD. This model could explain why the TAND is largely autonomous in TFIID but is not functional outside TFIID, e.g., at the amino termini of TFIIB (unpublished observations).
This study also suggests that there are two or more molecules of Taf11p per TFIID complex (Fig. 4). This result was somewhat unexpected, because Sanders et al. showed that HA-tagged Taf11p does not self-associate in vivo and that highly purified TFIID contains approximately one molecule of Taf11p per molecule of Taf1p or TBP (39). The reason for the discrepancy between these two studies is not known, but it may relate to the use of different tag epitopes in different positions in the protein sequence or structure; the previous study used amino-terminally HA-tagged Taf11p, and this study used carboxy-terminally FLAG-tagged Taf11p. A recently constructed three-dimensional model of TFIID based on immunoelectron microscopy suggests that at least two molecules of Taf11p are in lobes A and B, presumably as a component of a Taf11p-Taf13p heterodimer (24). This microscopic model is consistent with the results of the present study.
Yeast strains expressing Taf11p with a carboxy-terminal TAND are nonviable, probably because this Taf11p variant prevents several TAFs, including Taf1p, from being stably incorporated into TFIID (Fig. 5B). Other studies have also reported partial TFIID complexes that lack Taf1p. For example, the TBP mutant K151L K156Y remains associated with Taf6p and Taf14p at the restrictive temperature despite the dissociation of Taf1p, Taf5p, Taf10p, and Taf12p (37). In addition, our study also showed that TBP associates stably with Taf2p, Taf3p, Taf4p, Taf5p, Taf7p, Taf11p, Taf12p, and Taf13p in the absence of Taf1p (Fig. 4B). These observations suggest that Taf1p may not be essential for the assembly of TFIID, which contradicts an earlier study that reported that Taf1p acts as a scaffold for TFIID assembly (4). Further studies are required to reveal the transcriptional defects displayed by such partial TFIID complexes that specifically lack several TAFs.
This study shows that TFIID retains normal function when there is more than one TAND in the complex (i.e., the TANDs at the amino terminus of Taf1p and at the amino or carboxy terminus of Taf5p or the amino terminus of Taf11p). TFIID may include two copies of Taf5p (39), so TFIID may actually contain three TANDs when the TAND is fused to Taf5p. The ratio of TBP to some TAFs may be constrained (Fig. 2E), and the number of TBP monomers per TFIID may be limited to one.
Studies of the basic mechanisms of transcription have rarely tested whether a functional domain of one factor retains its function when repositioned as a domain of another factor. In this regard, it is worth describing a previous study. Dove et al. showed that a carboxy-terminal portion of σ factor (region 4), which is a functional target for the bacteriophage λ cI protein (λcI), can function as a domain of the α subunit of prokaryotic RNA polymerase (11). Since λcI specifically stimulates transcription at the isomerization step, it probably induces a conformational change in RNA polymerase during activation via physical contact with σ region 4. This is remarkably analogous to the relationship between TFIID and eukaryotic activators. Furthermore, given that σ and TFIID both recognize core promoters, they can be regarded as functional counterparts. Thus, the fact that region 4 of σ factor and the TAND are largely autonomous may not be entirely surprising. These observations suggest that transcription is regulated by mechanisms that have been conserved from prokaryotes through to eukaryotes.
A recent model suggesting that the three-dimensional positions of σ region 1.1 in the holoenzyme and the open complex differ dramatically reminds us of another resemblance between σ and TFIID (31). It has been proposed that when σ region 1.1 is in a holoenzyme, it autoinhibits the DNA binding of RNA polymerase by occupying a channel for downstream duplex DNA but that it is ejected from the channel during open-complex formation (31, 33). Interestingly, the idea that σ region 1.1 may serve as a molecular mimic for DNA when it occupies the channel (31) is entirely consistent with the previous observations that TAND1 of Drosophila Taf1p mimics the three-dimensional structure of the TATA element when bound to TBP (28). Although the functional relationships between σ region 1.1 and/or 4 and the TAND remain elusive, further work on these domains will help elucidate the evolutionarily conserved mechanisms of transcriptional regulation.
Acknowledgments
We thank Y. Ohyama for constructing the Δtaf1Δtaf5 double-deletion strain and Y. Ohyama, Y. Tsukihashi, A. Kobayashi, H. Ohta, and E. Watanabe for sharing their antibodies. We also thank Y. Nakatani, A. G. Hinnebusch, and M. Longtine for yeast strains and plasmids and H. Iwasaki and other members of our laboratory and M. Ptashne for advice and comments on this work.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Mitsubishi Foundation.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Bai, Y., G. M. Perez, J. M. Beechem, and P. A. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAF(II)130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, J. X., M. Floer, P. Ononaji, G. Bryant, and M. Ptashne. 2002. Responses of four yeast genes to changes in the transcriptional machinery are determined by their promoters. Curr. Biol. 12:1828-1832. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J. X., J. Nevado, Z. Lu, and M. Ptashne. 2002. The TBP-inhibitory domain of TAF145 limits the effects of nonclassical transcriptional activators. Curr. Biol. 12:934-937. [DOI] [PubMed] [Google Scholar]
- 7.Chi, T., and M. Carey. 1996. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 10:2540-2550. [DOI] [PubMed] [Google Scholar]
- 8.Chitikila, C., K. L. Huisinga, J. D. Irvin, A. D. Basehoar, and B. F. Pugh. 2002. Interplay of TBP inhibitors in global transcriptional control. Mol. Cell 10:871-882. [DOI] [PubMed] [Google Scholar]
- 9.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 11.Dove, S. L., F. W. Huang, and A. Hochschild. 2000. Mechanism for a transcriptional activator that works at the isomerization step. Proc. Natl. Acad. Sci. USA 97:13215-13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gangloff, Y. G., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 13.Green, M. R. 2000. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 25:59-63. [DOI] [PubMed] [Google Scholar]
- 14.Guermah, M., S. Malik, and R. G. Roeder. 1998. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell. Biol. 18:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAF(II) functions that suggest a dynamic TFIID structure and elicit synergy with traps in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, Y.-J., S. Bjorklund, Y. Li, M. H. Sayre, and R. D. Kornberg. 1994. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77:599-608. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi, A., T. Miyake, M. Kawaichi, and T. Kokubo. 2003. Mutations in the histone fold domain of the TAF12 gene show synthetic lethality with the TAF1 gene lacking the TAF N-terminal domain (TAND) by different mechanisms from those in the SPT15 gene encoding the TATA box-binding protein (TBP). Nucleic Acids Res. 31:1261-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokubo, K., S. Yamashita, M. Horikoshi, R. G. Roeder, and Y. Nakatani. 1994. Interaction between the N-terminal domain of the 230-kDa subunit and the TATA box-binding subunit of TFIID negatively regulates TATA box-binding. Proc. Natl. Acad. Sci. USA 91:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokubo, T., M. J. Swanson, J. I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotani, T., K. Banno, M. Ikura, A. G. Hinnebusch, Y. Nakatani, M. Kawaichi, and T. Kokubo. 2000. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc. Natl. Acad. Sci. USA 97:7178-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotani, T., T. Miyake, Y. Tsukihashi, A. G. Hinnebusch, Y. Nakatani, M. Kawaichi, and T. Kokubo. 1998. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem. 273:32254-32264. [DOI] [PubMed] [Google Scholar]
- 22.Kou, H., J. D. Irvin, K. L. Huisinga, M. Mitra, and B. F. Pugh. 2003. Structural and functional analysis of mutations along the crystallographic dimer interface of the yeast TATA binding protein. Mol. Cell. Biol. 23:3186-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 24.Leurent, C., S. Sanders, C. Ruhlmann, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21:3424-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 26.Li, X. Y., S. R. Bhaumik, X. Zhu, L. Li, W. C. Shen, B. L. Dixit, and M. R. Green. 2002. Selective recruitment of TAFs by yeast upstream activating sequences. Implications for eukaryotic promoter structure. Curr. Biol. 12:1240-1244. [DOI] [PubMed] [Google Scholar]
- 27.Lieberman, P. M., and A. J. Berk. 1994. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 8:995-1006. [DOI] [PubMed] [Google Scholar]
- 28.Liu, D., R. Ishima, K. I. Tong, S. Bagby, T. Kokubo, D. R. Muhandiram, L. E. Kay, Y. Nakatani, and M. Ikura. 1998. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94:573-583. [DOI] [PubMed] [Google Scholar]
- 29.Lively, T. N., H. A. Ferguson, S. K. Galasinski, A. G. Seto, and J. A. Goodrich. 2001. c-Jun binds the N terminus of human TAF(II)250 to derepress RNA polymerase II transcription in vitro. J. Biol. Chem. 276:25582-25588. [DOI] [PubMed] [Google Scholar]
- 30.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 31.Mekler, V., E. Kortkhonjia, J. Mukhopadhyay, J. Knight, A. Revyakin, A. N. Kapanidis, W. Niu, Y. W. Ebright, R. Levy, and R. H. Ebright. 2002. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell 108:599-614. [DOI] [PubMed] [Google Scholar]
- 32.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 33.Murakami, K. S., S. Masuda, and S. A. Darst. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296:1280-1284. [DOI] [PubMed] [Google Scholar]
- 34.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 35.Oelgeschlager, T., Y. Tao, Y. K. Kang, and R. G. Roeder. 1998. Transcription activation via enhanced preinitiation complex assembly in a human cell-free system lacking TAFIIs. Mol. Cell 1:925-931. [DOI] [PubMed] [Google Scholar]
- 36.Ptashne, M. 1986. Gene regulation by proteins acting nearby and at a distance. Nature 322:697-701. [DOI] [PubMed] [Google Scholar]
- 37.Ranallo, R. T., K. Struhl, and L. A. Stargell. 1999. A TATA-binding protein mutant defective for TFIID complex formation in vivo. Mol. Cell. Biol. 19:3951-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roeder, R. G. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp. Quant. Biol. 63:201-218. [DOI] [PubMed] [Google Scholar]
- 39.Sanders, S. L., K. A. Garbett, and P. A. Weil. 2002. Molecular characterization of Saccharomyces cerevisiae TFIID. Mol. Cell. Biol. 22:6000-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 42.Tsukihashi, Y., T. Miyake, M. Kawaichi, and T. Kokubo. 2000. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 20:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu, S. Y., M. C. Thomas, S. Y. Hou, V. Likhite, and C. M. Chiang. 1999. Isolation of mouse TFIID and functional characterization of TBP and TFIID in mediating estrogen receptor and chromatin transcription. J. Biol. Chem. 274:23480-23490. [DOI] [PubMed] [Google Scholar]