Significance
The ability to control and direct differentiation of stem cells for research or therapeutic applications relies on the availability of ligands that control specific signaling pathways. Natural ligands exhibit promiscuous interactions and have limited availability because of their poor expression/stability profiles. In contrast, antibodies exhibit exquisite specificity and have optimal expression properties. Antibodies that block or activate receptor signaling have great potential for controlling differentiation, but their identification is laborious. Herein we describe an innovative system for identifying functional antibodies by introducing antibody gene populations into ES cells. Antibody-expressing ES clones with altered differentiation outcomes can be readily identified using lineage-specific gene-expression markers. The antibody gene can then be recovered for antibody production and use.
Keywords: phage display, phenotypic selection, functional screen
Abstract
Antibodies that modulate receptor function have great untapped potential in the control of stem cell differentiation. In contrast to many natural ligands, antibodies are stable, exquisitely specific, and are unaffected by the regulatory mechanisms that act on natural ligands. Here we describe an innovative system for identifying such antibodies by introducing and expressing antibody gene populations in ES cells. Following induced antibody expression and secretion, changes in differentiation outcomes of individual antibody-expressing ES clones are monitored using lineage-specific gene expression to identify clones that encode and express signal-modifying antibodies. This in-cell expression and reporting system was exemplified by generating blocking antibodies to FGF4 and its receptor FGFR1β, identified through delayed onset of ES cell differentiation. Functionality of the selected antibodies was confirmed by addition of exogenous antibodies to three different ES reporter cell lines, where retained expression of pluripotency markers Oct4, Nanog, and Rex1 was observed. This work demonstrates the potential for discovery and utility of functional antibodies in stem cell differentiation. This work is also unique in constituting an example of ES cells carrying an inducible antibody that causes a functional protein “knock-down” and allows temporal control of stable signaling components at the protein level.
Cellular communication mediated through surface receptors plays a critical role in development and in maintaining homeostasis in adulthood. Antibodies, such as Humira and Avastin, which activate or block receptor function, have proven value in therapeutic applications (1, 2). Functional antibodies also have great potential in stem cell biology, but the realization of this has been limited because only a fraction of antibodies affect receptor function and their identification is laborious. Functional antibodies are required to recognize the native conformation of the target receptor or ligand and to bind an appropriate epitope with high affinity. Identifying such antibodies therefore requires ELISA screening of large numbers of candidates to identify binding clones, followed by expression, purification, and assessment of individual antibodies using target-specific reporter cell assays. The required antibody diversity can be accessed by phage or yeast display, which can generate hundreds of antibodies to a single target (3, 4). The ready availability of the antibody gene from display technologies permits reformatting and production in mammalian cells to generate antibody products for cell-based screening. Importantly, access to the antibody gene also creates the potential for direct expression within mammalian reporter cells, thereby permitting antibody production and functional screening in one cell. Alterations in the characteristics of the antibody-expressing reporter cell could then identify clones encoding functional antibodies. This potential has recently been exemplified through the lentiviral infection of a TF1 reporter cell line with an antibody population, leading to the identification of secreted antibodies, which activate the erythropoietin receptor (5). In an alternative approach, antibodies were retained at the cell surface of BaF3 reporter cells, leading to the identification of an agonistic antibody to the granulocyte colony-stimulating receptor (6).
Pluripotent embryonic stem cells (ES cells) represent an ideal reporter cell system for identifying functional antibodies because they are poised to differentiate into many different cell types in vitro (7). ES cell fate decisions are influenced by a wide range of cell-surface receptors (7–9), creating the potential to target many different classes of ligands and receptors (e.g., receptor tyrosine kinases, G protein-coupled receptors, ion channels, integrins, and cadherins) with antibodies. Irrespective of the signaling pathway involved, the differentiation status of antibody-producing ES clones can be conveniently monitored using lineage-specific promoters driving fluorescent reporter genes or immunostaining with appropriate antibodies. Thus, measuring perturbations in ES cell differentiation represents a sensitive and flexible approach to identify antibodies modifying receptor function.
Results
Establishing an In-Cell Expression and Reporting System Allowing Inducible Antibody Expression and Secretion in ES Cells.
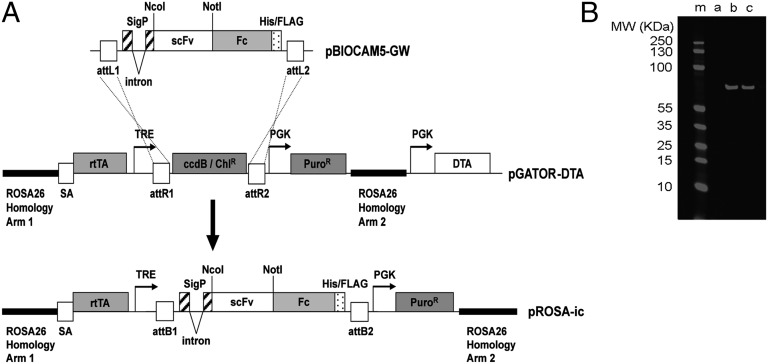
An in-cell expression and reporting system (ICER system) was developed using ES cells to efficiently identify functional antibodies within a population of target-specific binders. Mouse ES cells are readily modified through gene targeting, allowing insertion of individual antibody genes into a single locus in each cell. This process normalizes expression within the cell population and facilitates the identification of functional antibody genes. In the work presented here, homologous recombination was used to target antibody gene populations into the ubiquitously expressed reverse orientation splice acceptor betageo line 26 (Rosa-26) locus. An antibody expression cassette was introduced within a 10-kb region of the Rosa-26 gene, which has previously been used for targeting the mouse genome (10). Within the resulting targeting vector (pROSA-ic) (Fig. 1A), expression of the antibody gene was controlled by a doxycyline-responsive promoter allowing control of antibody expression.
Fig. 1.
Construction of the pROSA-ic vector for targeting antibody expression cassettes into the ubiquitously expressed Rosa-26 locus of mouse ES cells. (A) Antibody genes (in the form of single-chain Fvs) were subcloned into the NcoI and NotI sites of the pBIOCAM5-GW entry vector to create a fusion with a human IgG1 Fc gene. LR Gateway recombination was used to facilitate the introduction of this scFv-Fc fusion gene into the large (14 kb) pGATOR-DTA targeting vector. The resultant pROSA-ic construct encompasses an antibody gene expression cassette within a 10-kb region of the Rosa-26 gene. This region is used to drive homologous recombination of the antibody expression cassette into the ubiquitously expressed Rosa-26 locus in mouse ES cells. The diphtheria toxin gene (DTA) is used to reduce frequency of clones with random integration of the cassette in the genome. In pROSA-ic antibody, expression is controlled by a doxycycline-responsive “Tet-On” promoter system. The final construct encodes a signal peptide directing secretion of a human scFv fused to a human IgG1 Fc domain (scFv-Fc fusion). The Fc domain drives dimerization of the resulting antibody. Abbreviations include: attL1, attL2, attR1, attR2, attB1, attB2, gateway recombination sites; sigP, signal peptide; His/FLAG, hexa-histidine Tri-FLAG peptide tag; scFv, single-chain Fv; Fc, human IgG1 Fc with C-terminal His-FLAG tag; SA, splice acceptor; rtTA, Tet-on transactivator; TRE, Tet response element; PuroR, puromycin resistance gene; DTA, diptheria toxin gene fragment A. (B) Doxycycline-inducible antibody expression in E14 ES cells. An anti-Notch antibody N1_E7 (11) was subcloned into pROSA-ic and the resultant targeting vector was used to transfect E14 ES cells. Correctly targeted puromycin-resistant colonies were selected and grown under self-renewal conditions with 0, 1, or 2 μg/mL doxycycline (lanes a–c, respectively). After 4 d expressed antibody was affinity-purified using anti-FLAG agarose. Samples were run on SDS/PAGE, blotted, and the expressed scFv-Fc was detected with an anti-FLAG mouse antibody and a secondary anti-mouse IR680-labeled antibody (Licor).
The capacity of the system to achieve gene targeting and controlled antibody expression was initially tested using control anti-Notch antibodies (11). Following transfection, stable puromycin-resistant clones were selected, genomic DNA prepared, and correct integration of the targeting vectors was confirmed by PCR. Western blot analysis (Fig. 1B) confirmed that doxycycline-dependant antibody secretion occurred within the resultant clones. ELISA was used to measure antibody concentration (1–2 μg/mL after 4-d induction) and to confirm binding to antigen.
FGF4 Signaling as a Target for Antibody-Mediated Control of Differentiation.
The potential of the ICER system for selection of functional antibodies was exemplified in ES cells (ES-ICER) by the generation of novel antagonists of FGF4 (fibroblast growth factor 4) signaling. In mouse ES cells, autocrine FGF4 activates FGF receptors (FGFRs), providing a permissive signal via extracellular-signal-regulated kinases (ERKs) that allows cells to respond to differentiation cues (12–14). This FGF-mediated differentiation capacity can be blocked by selective small-molecule inhibitors of either FGFR tyrosine kinases or ERK kinase (12). In mouse ES cells, FGF4 is the major activator of ERK. It would therefore be anticipated that ES cells expressing blocking antibodies to FGF4 activity would be shielded from the FGF4-mediated response to differentiation cues, resulting in retained expression of “pluripotency genes,” such as Oct4 (15), Rex1 (16), and Nanog (17), even under differentiation culture conditions.
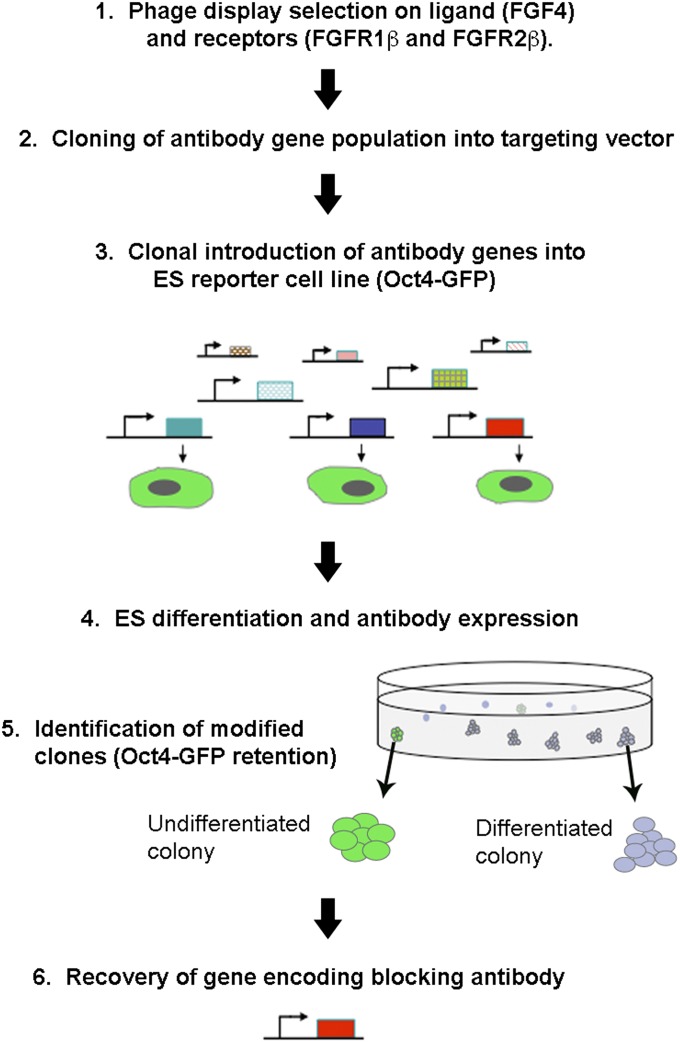
Fig. 2 summarizes the experimental strategy used. Antibody populations were derived from phage display by selection on target antigen (although the ICER system could equally be applied to antibody gene populations derived from other display methods or from immunized animals). Phage display selections were carried out on FGF4, FGFR1β, and FGFR2β [the IIIc splice variants of FGFR (18)] and generation of binding antibodies was confirmed by ELISA (Fig. S1). Selected antibody populations were cloned into pROSA-ic before targeting into the Rosa-26 locus of mouse Oct4-ΔPE-GFP ES reporter cells (19). In these cells, green fluorescent protein (GFP) is expressed under the control of the distal enhancer of the Oct4 gene, which specifically reports the ES cell state. Following selection of puromycin-resistant colonies, culture medium was changed from self-renewal medium [serum/Leukemia inhibitory factor (LIF)] to medium inducing differentiation (ES-Cult/N227). Expression and secretion of the introduced antibodies was induced by addition of doxycycline. To enable screening of the population en masse, it was necessary to retain the antibodies in the vicinity of the producer cell to prevent “cross-talk” between clones, and this was achieved through growth in semisolid medium. This process has the added benefit of allowing accumulation of higher antibody concentrations in the vicinity of the producer colony compared with liquid culture.
Fig. 2.
Overview of the experimental strategy (numbered 1–6) to identify antibody-expressing ES colonies that resist differentiation through blockade of FGF signaling. Antibody populations binding to FGF4, FGFR1β, and FGFR2β were selected from a phage display library and were cloned into the targeting vector pROSA-ic. Homologous recombination was used to direct integration into the ubiquitously expressed Rosa-26 genomic locus of individual Oct4–ΔPE-GFP ES cells. Puromycin-resistant colonies were selected, antibody expression was induced, and ES colonies were subjected to differentiation in semisolid medium for 2.5–4.5 d. Clones that resisted differentiation (judged by retention of round morphology and Oct4-ΔPE-GFP expression) were picked and the antibody genes retrieved by PCR for further use.
ES Clones Expressing FGF4-Blocking Antibodies Resist Differentiation.
The potential for antibody-induced retention of Oct4-GFP expression was initially assessed using flow cytometry of Oct4−ΔPE-GFP ES transfected with the anti-FGF4 antibody population (Fig. S2). After 3–4 d differentiation in the absence of antibody induction, only 76.7% of transfected Oct4−ΔPE-GFP cells retained GFP expression. In contrast, addition of the ERK kinase inhibitor PD0325901 (PD03), which blocks FGFR-mediated signaling (14), inhibited differentiation and caused retention of GFP expression in 98.8% of cells. Induction of antibody expression by addition of doxycycline also maintained GFP expression in 95.2% of cells, suggesting the existence of a proportion of blocking antibodies within the population. Maintenance of the undifferentiated state in the resultant population was independently confirmed through quantification of alkaline phosphatase-positive colony formation following a return of the cells to self-renewal conditions (Fig. S3). This result showed increased clonogenicity of the cells grown in the presence of doxycycline during earlier exposure to differentiation conditions.
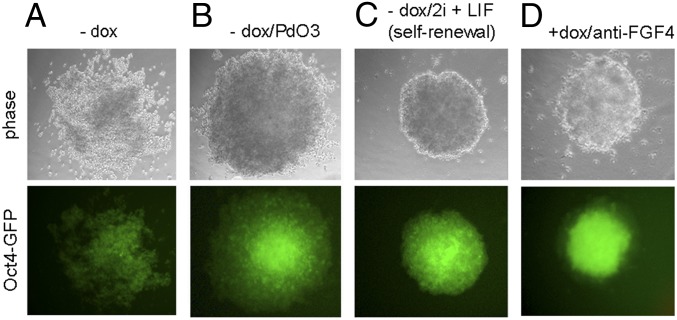
Direct observation by fluorescent microscopy also demonstrated antibody-directed inhibition of differentiation within individual colonies. In control cultures differentiated for 3 d in the absence of doxycycline, the expected reduction in GFP expression was observed along with a loss of the compact, round colony shape seen in undifferentiated cultures (Fig. 3A). PD03 helped retain the compact morphology and fluorescence (Fig. 3B), whereas all ES colonies grown in self-renewal conditions (−dox/2i) retained their compact shape and high levels of GFP expression (Fig. 3C). In the presence of doxycycline, ES colonies grown under differentiation coditions showed a range of phenotypes (Fig. S4). Among these colonies, ∼5% retained the compact shape and GFP fluorescence of the undifferentiated colonies observed when grown under self-renewal conditions (Fig. 3D and Fig. S4). This approach therefore allowed the in situ identification of the few select ES colonies maintaining an undifferentiated phenotype. Four colonies (of 217) from the anti-FGF4 population were picked and propagated under self-renewal conditions, where they demonstrated their retained undifferentiated state, despite earlier exposure to differentiation conditions. Furthermore, upon secondary testing in liquid differentiation culture (i.e., in the absence of methylcellulose) for 7 d, a doxycycline-dependent resistance to differentiation was demonstrated in two clones, FGF4_A and FGF4_C (Fig. 4 and Fig. S5).
Fig. 3.
Colonies obtained following transfection of Oct4–ΔPE-GFP ES cells with an anti-FGF4 antibody population, after 3 d of growth in differentiation medium ES-Cult/N227. (A–C) typical colonies obtained in the absence of doxycycline under the indicated conditions. (D) Example of colony retaining an undifferentiated phenotype, obtained in the presence of doxycycline (1 μg/mL) (magnification 20×).
Fig. 4.
Selected anti-FGF4 clones maintain Oct4-ΔPE-GFP expression in liquid differentiation in the presence of doxycycline. (A) Cells from the αFGF4_A and αFGF4_C clones were analyzed by flow cytometry after 7-d growth in differentiation conditions in the absence or presence of doxycycline (1 μg/mL) or in the presence of 2 μM control Erk kinase inhibitor PD0325901 (PD03). (B) Phase and fluorescent images of Oct4–ΔPE-GFP cells expressing the α-FGF4A antibody after 7 d of differentiation in N227 in the presence or absence of doxycycline. The differentiation experiment was performed two times with similar results (magnification 20×).
Exogenous Anti-FGF4 Antibodies Cause Retained Expression of Pluripotency Markers During Growth in Differentiation Conditions.
The antibody genes from clones FGF4_A and FGF4_C were recovered by PCR and subcloned into a mammalian expression vector (pBIOCAM5) (11). The encoded antibodies were expressed in HEK293 cells, purified, and added exogenously to unmodified Oct4−ΔPE-GFP ES cells. The ability of exogenously purified antibody to reduce receptor function was confirmed by the significant inhibition of differentiation observed after adding antibody (αFGF4_A or αFGF4_C) to unmodified Oct4−ΔPE-GFP ES cells (see Fig. 5A for the αFGF4_A result). Antibody-directed control of stem cell fate was independently confirmed using Nanog and Rex1 ES reporter cells [Nanog-GFP (TNGA) and Rex1-GFPd2], in which GFP is inserted in the Nanog and Rex1 loci, respectively (17, 20) (Fig. 5 B and C, and Fig. S6). Nanog and Rex1 are more sensitive markers of the pluripotent state of ES cells than Oct4, and in the respective reporter cell lines a decline in GFP expression was apparent after only 2–3 d in differentiation culture conditions. Rex1-GFPd2 ES cells were found to be the most sensitive reporter of differentiation status. The addition of either αFGF4_A or αFGF4_C antibodies was shown to inhibit differentiation as judged by retained Rex1-GFP expression (Fig. 5D). Thus, using three different markers of pluripotency, the ES-ICER system was shown to permit the identification of functional antibodies that delayed the onset of ES cell differentiation. Furthermore the selected antibodies were specific to FGF4 as judged by their lack of binding to acidic and basic FGF (FGF1 and FGF2) by ELISA (Fig. S7A, antibody sequences shown in Fig. S7C).
Fig. 5.
Exogenous anti-FGF4 antibodies cause retained expression of pluripotency markers when added to the three independent ES cell lines. (A) Oct4–ΔPE-GFP ES cells grown in differentiation medium for 7 d in the presence or absence of purified αFGF4_A antibody (20 μg/mL) and compared with PDO3 control (magnification 20×). (B) Flow cytometry of Rex1-GFPd2 ES cells after 3 d of differentiation in the presence of αFGF4_A and compared with PDO3 control. (C) Flow cytometry of Nanog-GFP ES cells after 3 d of differentiation in the presence of αFGF4_A and compared with PDO3 control. (D) Percentages of cells expressing Rex1-GFP after 3 d of differentiation in the presence or absence of the indicated purified anti-FGF4 antibodies (20 μg/mL), negative (α-desmin and Fc only) and positive (PDO3) controls. The bars represent the average of triplicates of one flow cytometry experiment ± SD. (E) The histogram shows the fraction of Rex1-GFP+ cells (normalized to PDO3 response and using “minus antibody” as 0) from three independent experiments as described in D. The bars represent the average of triplicates of three separate flow cytometry experiments ± SD The asterisk denotes statistical significance. P = 0.0017 for αFGF4_A vs. antidesmin. In addition, P = 0.004 for αFGF4_C vs. α-desmin.
Selection of Anti-FGFR Antibodies, Which Inhibit ES Cell Differentiation.
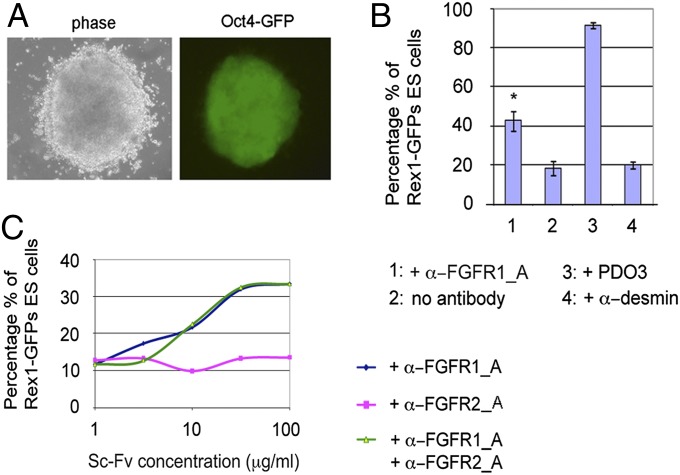
Using the strategy outlined above, Oct4−ΔPE-GFP ES cells were transfected with antibody populations selected by phage display on FGFR1β and FGFR2β, (Fig. 6, and Figs. S8 and S9). For FGFR1β and FGFR2β, 3 of 462 and 4 of 132 colonies, respectively, were found to retain a clear undifferentiated phenotype in semisolid medium (Fig. 6A and Fig. S9B) and could be subsequently propagated under self-renewal conditions. Upon addition of exogenous FGFR1β antibodies, only one clone retained Rex-1 GFP expression when the respective purified antibody (α-FGFR1_A) was added exogenously to Rex1-GFPd2 ES cells grown in differentiation conditions (Fig. 6 B and C). To investigate this further, an in vitro biochemical assay was established, which recapitulated the interaction of FGF receptors with immobilized FGF4/heparin. In this assay all three antibodies inhibited the ligand:receptor interaction, although α-FGFR1_A demonstrated the greatest potency (Fig. S8F). In the case of FGFR2β, addition of exogenous antibodies failed to inhibit differentiation of Rex1-GFPd2 cells (Fig. 6C and Fig. S9C), although all showed blocking activity in the biochemical assay (Fig. S9D). All antibodies were shown by ELISA to be specific for the appropriate FGF receptor (Fig. S7B). These results suggest that the primary differentiation assay was sufficiently sensitive to identify specific functional antibodies covering a range of potencies (probably aided by the high local antibody concentration in the semisolid medium). Only those antibodies with the greatest potency (demonstrated for FGF4 and FGFR1β) inhibited differentiation when exogenously added to the cells. For less-potent clones, affinity maturation (21) could be used to improve the affinity/potency following their identification from the initial screen.
Fig. 6.
Identification of antibodies blocking FGFR1β using the ICER system. (A) Example of a colony maintaining Oct4-GFP fluorescence and ES-like morphology after 4 d of differentiation in ES-Cult/N227 following transfection with the anti-FGFR1 antibody library (magnification 20×). (B) Percentages of cells expressing Rex1-GFP after 3 d of differentiation in the presence or absence of the purified anti FGFR1β antibodies (100 μg/mL), negative (antidesmin), and positive (PDO3) controls. The bars represent the average of duplicates of a single flow cytometry experiment ± SD. (C) α-FGFR1_A and α-FGFR2_A antibodies were added alone or in combination at a range of concentrations to Rex-GFP ES cells. The percentage of Rex1-GFP+ cells from a single experiment was quantified with flow cytometry.
Discussion
In this article we introduce a system for selecting functional antibodies within populations through the expression of secreted antibodies causing perturbation of receptor function in embryonic stem cells (ES cells). Zhang et al. (5) have recently described a similar approach, using secreted antibodies retained in semisolid medium, to identify agonists of the erythropoietin receptor (using TF1 erythroleukemic reporter cells transfected with the erythropoietin receptor). As an alternative, agonistic antibodies recognizing G-CSF receptor (G-CSFR) were also identified by anchoring the expressed antibodies to the membrane of a G-CSFR–transfected BaF3 cell line (6). Importantly the isolated anti–G-CSFR antibody was shown on subsequent addition to CD34 hematopoetic stem cells to cause transdifferentiation, unlike the natural ligand. In the ES-ICER system described here, antagonistic antibodies were selected. These antibodies were identified using pluripotent ES cells and were targeted to either autocrine ligand (FGF4) or endogenous FGF receptor.
The key elements of the ES-ICER system involve targeting of single antibody genes from preselected populations into the Rosa-26 locus of mouse ES cells to create a combined expression/reporting cell system, allowing autoexpression and detection of an altered phenotype. The approach could be extended to identify functional peptides or alternative binding scaffolds. This and the previously described systems (5, 6) make no assumptions about the mode of receptor inhibition/activation and can work even if the other partners or cofactors are unknown or unavailable for biochemical assay. Although an important output of this approach are antibodies that can be added exogenously to affect cell differentiation, the resultant ES clones also have potential utility in the exploration of receptor function both in vitro and in vivo. These clones, encoding an inducible functional antibody, will permit exquisite temporal control of signaling components at the protein level, enabling protein knockdown in diverse developmental contexts.
Although the approach was exemplified by blockade of autocrine FGF4-mediated ES cell differentiation, the basic strategy could be adapted to generate antibodies, both agonistic and antagonistic, to the many ligands/receptors that are expressed during ES differentiation. Our growing knowledge of the role of these molecules in cell fate determination, combined with the development of more efficient protocols for directed stem cell differentiation, can be exploited to extend the versatility of this approach. In this study, Oct4 gene expression was used to monitor differentiation status but phenotypic changes could be detected by other lineage-specific reporters, by immunostaining or through changes in morphology.
The principle use of functional antibodies to date has been as disease-modifying drugs in oncology and immunology, and the presented ICER system will facilitate future drug discovery efforts. With their optimal stability and expression properties, functional antibodies also have great, untapped potential in the control of stem cell fate. In particular, the exquisite specificity of antibodies confers advantages over natural receptor agonists, which often exhibit promiscuous interactions and are subject to regulatory feedback mechanisms. Functional antibodies could therefore transform methods for the derivation and control of human ES/iPS cells lines, creating human cell lines for research and valuable models of embryogenesis. Ultimately, the ability to control differentiation of genetically unmodified human stem cells through the administration of functional antibodies or peptides, could unleash new ex vivo or in vivo approaches to stem cell-based therapeutics.
Methods
Targeting Antibody Genes to the ROSA-26 Locus.
The Rosa-26 targeting vector pGATOR was modified to insert the Diptheria toxin fragment A (DTA) within the vector backbone. Homologous recombination will result in the loss of the DTA gene, whereas random plasmid integration will result in DTA expression, which will cause cell death, thereby reducing the background from random integration. The DTA gene was PCR-amplified from pL3L4_DTA (EUCOMMtools) and cloned into the SfiI site of the pGATOR vector to create pGATOR-DTA (SI Methods). pBIOCAM5-GW was created by PCR amplification from the pBIOCAM5 plasmid to give a PCR product encoding a single-chain Fv (scFv)-Fc fusion flanked by the attB1 and attB2 GATEWAY recombination sites (22), which was then BP recombined with pDONR221 (Invitrogen) to give pBIOCAM5-GW. Selected antibody gene populations (or the previously created anti-Notch clones) (11) were amplified by PCR, as described previously (4), restriction digested, and ligated into the NcoI/NotI sites of pBIOCAM5-GW. Ligated DNA was electroporated into Escherichia coli DH5α cells to produce an average library size of 2 × 105 clones. DNA was prepared from this pBIOCAM5-GW library entry plasmid and was LR recombined with pGATOR-DTA. The resultant plasmid was introduced into chemically competent E. coli DH10B cells, where an average library size of 3,300 clones was created for the anti-FGF4, anti-FGFR1β, and anti-FGFR2β populations. Plasmid DNA was prepared, linearized with SfiI, extracted with phenol/chloroform, ethanol-precipitated, and dissolved in TE (pH 8.0) to give a concentration of 1–2 µg/µL of linearized targeting vector.
ES Cell Lines and Culture.
E14Tg2a ES cells were described previously (23). Oct4–ΔPE-GFP ES cells were from gain of function (GOF) mice, in which GFP is specifically expressed in the preimplantation embryo and primordial germ cells but not the postimplantation epiblast (19). In culture, GFP is expressed in the ES cells but not the epiblast stem cell (EpiSCs). Both E14Tg2a and Oct4–ΔPE-GFP ES cells were maintained on tissue-culture dishes coated with gelatin (0.1% swine skin), in GMEM (Sigma) supplemented with: recombinant LIF (100 U/mL), 15% (vol/vol) FCS (Sigma selected batch), 1 mM sodium pyruvate (Invitrogen), 2 mM l-Glutamine (Invitrogen), 1× MEM nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma), 100 U/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen). For experiments involving doxycycline-induced antibody secretion in the medium, 1.5 x 103 cells/cm2 were plated and cultured in ES medium in the presence or absence of doxycycline (1 µg/mL). For colony-forming assays, 0.5 x 103 cells/cm2 were plated. Nanog-GFP and Rex-GFPd2 ES cells were grown in NDiff-N227 (previously NDiff-N2B27) (StemCells) containing 1 μM of CHIR99021 (a GSK-3 inhibitor) and 2 μM PD0325901 (an ERK kinase inhibitor; Stratech) plus LIF (2i/LIF) (14).
Differentiation of ES Colonies in Semisolid, Serum-Free Medium (ES-Cult/N227).
After 4 d of puromycin selection and 36 h before differentiation, doxycycline was added, where appropriate, to initiate antibody expression before the autocrine production of FGF4. At the fifth day of selection, the ES medium was replaced with a semisolid differentiation medium, consisting of 60% (vol/vol) N227 and 40% (vol/vol) ES-Cult M3120 (StemCell Technologies) plus puromycin (1 µg/mL) and doxycycline (1 µg/mL), where appropriate. Differentiation progress was monitored daily by observing the morphology and the fluorescence of the colonies. The maximum time the colonies could be sustained in semisolid medium without detaching from the dish was around 4 d. This timing was sufficient to allow for discrimination between colonies retaining and those down-regulating Oct4-GFP fluorescence.
Recovery of Antibody Genes from ES Cell Clones.
PCR reactions were carried out using 100 ng of genomic DNA from selected clones as template in 20 μL of KOD buffer with 100 μM dNTPs, 1.5 mM MgSO4, 5% (vol/vol) DMSO, 0.4 units KOD Hot Start Polymerase (EMD Millipore) using 0.25 μM primers: 2544 5′-CTTTCTCTCCACAGGCGCCATGG-3′ and 2545 5′- GTGTGGGTCTTGTCTGCGGC-3′.
Cycling conditions were 95 °C for 2 min followed by 40 cycles of 95 °C, 20 s; 57 °C, 10 s; 70 °C, 40 s. PCR products were purified with GeneJET PCR purification kit (Thermo Scientific), digested with NcoI and NotI and purified by electophoresis on a 1% agarose-TBE gel, followed by excision of the band running at ∼800 bp and extraction of the DNA with the GeneJET gel extraction kit (Thermo Scientific).
Development and Use of in Vitro Biochemical Assay Replicating Interaction of FGFR1 or FGFR2 Binding to FGF4.
scFv gene inserts from stem cell genomic DNA of selected clones were subcloned into the NcoI/NotI-digested expression vector pBIOCAM5 and scFv-Fc expressed by transient transfection of HEK293F suspension cells (11). Affinity purification by Ni-NTA Sepharose was carried out as described previously (11). Black Maxisorp 96-well plates (437111; Thermo Scientific Nunc) were coated with heparin sulfate proteoglycan (3 μg/mL in PBS; Sigma) overnight at 4 °C and blocked with PBS-M [2% (wt/vol) milk powder in PBS] for 1 h at room temperature. Dilutions of scFv-Fcs were preincubated with FGFR1β-Fc or FGFR2β-Fc in TBS-BSA (TBS, 0.2 mg/mL BSA) for 1 h at room temperature in a total volume of 112.5 μL in a 96-well polypropylene plate. FGF4 (37.5 μL in TBS-BSA) was then added and two separate 50-μL aliquots (for duplicate assays), transferred to separate wells of the heparin sulfate proteoglycan-coated plate, and washed with TBS. Final concentrations of the FGFR1β/FGF4 and FGFR2β/FGF4 receptor/ligand pairs were 158 ng/mL and 2 μg/mL, respectively. After 1-h incubation, plates were washed three times each with TBS-T and TBS. Anti-Fc-biotin (0.5 μg/mL in TBS-BSA, 109–065-006; Jackson ImmunoResearch) was added, plates incubated for 1 h, washed as above and DELFIA Eu-N1 Streptavidin (50 μL, 1:100 dilution in TBS-BSA; Perkin-Elmer) added, incubated for 30 min, washed as above and DELFIA Dissociation-enhancement (50 μL) added before time-resolved fluorescence measurement (λexc = 320 nm λem = 615 nm) with a Fusion plate reader (Perkin-Elmer).
Supplementary Material
Acknowledgments
We thank Austin Smith’s and Jenny Nichols’ laboratories, especially Gillian Morrison, Melanie Rittirsch, Tüzer Kalkan, Elena Tzouanacou, and Kenneth Jones at the Cambridge Stem Cell Institute for the E14Tg2a, Nanog-GFP (TNGA), and Rex1-GFPd2 mouse embryonic stem cells, advice on ES cell differentiation and reagents; the Azim Surani laboratory for the Oct4–ΔPE-GFP mouse embryonic stem cells; Marko Hyvonen for FGF4; Nigel Miller for help with flow cytometry; Kothai Pathiban and Tony Pope for technical assistance; Peter Slavny and Aneesh Karatt Velatt for useful discussions; and Barry Rosen for helpful comments on the manuscript. This work was supported by the Isaac Newton Trust, the Wellcome Trust, and IONTAS Ltd.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 17608.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312062110/-/DCSupplemental.
References
- 1.Lloyd C, et al. Modelling the human immune response: Performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng Des Sel. 2009;22(3):159–168. doi: 10.1093/protein/gzn058. [DOI] [PubMed] [Google Scholar]
- 2.Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9(10):767–774. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 3.Edwards BM, et al. The remarkable flexibility of the human antibody repertoire; Isolation of over one thousand different antibodies to a single protein, BLyS. J Mol Biol. 2003;334(1):103–118. doi: 10.1016/j.jmb.2003.09.054. [DOI] [PubMed] [Google Scholar]
- 4.Schofield DJ, et al. Application of phage display to high throughput antibody generation and characterization. Genome Biol. 2007;8(11):R254. doi: 10.1186/gb-2007-8-11-r254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Wilson IA, Lerner RA. Selection of antibodies that regulate phenotype from intracellular combinatorial antibody libraries. Proc Natl Acad Sci USA. 2012;109(39):15728–15733. doi: 10.1073/pnas.1214275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie J, Zhang H, Yea K, Lerner RA. Autocrine signaling based selection of combinatorial antibodies that transdifferentiate human stem cells. Proc Natl Acad Sci USA. 2013;110(20):8099–8104. doi: 10.1073/pnas.1306263110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132(4):661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton DA, Benvenisty N. Effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc Natl Acad Sci USA. 2000;97(21):11307–11312. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X, Browning VL, Odorico JS. Activin, BMP and FGF pathways cooperate to promote endoderm and pancreatic lineage cell differentiation from human embryonic stem cells. Mech Dev. 2011;128(7-10):412–427. doi: 10.1016/j.mod.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2(4):292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falk R, et al. Generation of anti-Notch antibodies and their application in blocking Notch signalling in neural stem cells. Methods. 2012;58(1):69–78. doi: 10.1016/j.ymeth.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunath T, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007;134(16):2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 13.Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134(16):2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 14.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niwa H, Miyazaki J-i, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 16.Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 17.Chambers I, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450(7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 18.Bellosta P, et al. Identification of receptor and heparin binding sites in fibroblast growth factor 4 by structure-based mutagenesis. Mol Cell Biol. 2001;21(17):5946–5957. doi: 10.1128/MCB.21.17.5946-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6(5):468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray J, et al. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol. 2011;13(7):838–845. doi: 10.1038/ncb2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyson MR, et al. Mapping protein interactions by combining antibody affinity maturation and mass spectrometry. Anal Biochem. 2011;417(1):25–35. doi: 10.1016/j.ab.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10(11):1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326(6110):292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.