Abstract
The Sid2p-Mob1p kinase complex is an important component of the septation initiation network (SIN) in the fission yeast Schizosaccharomyces pombe. However, regulation of this complex is still elusive. Here we show that Mob1p is required not only for the subcellular localization of Sid2p but also for its kinase activity. We identified a region at the amino terminus of Sid2p that is required for Mob1p binding and spindle pole body (SPB) localization. Deletion of this region abolishes Mob1p binding and diminishes SPB localization, whereas this region alone is sufficient to associate with Mob1p and SPBs. We further show that a similar region of the N terminus of the Sid2p-related protein kinase Orb6p binds to the Mob1p-related protein Mob2p, suggesting that this may be a conserved mode of interaction for this family of kinases. Phosphorylation of Ser402 and especially Thr578 is important for Sid2p function. Sid2p with a mutation of Thr578 to Ala (T578A) can no longer rescue sid2-250 mutant cells, and this results in reduction of Mob1p binding. Sid2p mutants mimicking phosphorylation at this site (T578D and T578E) can rescue sid2-250 cells, enhance Sid2p kinase activity, and partially rescue growth defects of upstream sin mutants. Interestingly, Sid2p, but not Mob1p, is self-associated. Our experiments suggest that self-associated Sid2p is inactive. This self-association is mediated by a region that overlaps with Mob1p and SPB binding sites. Overexpression of Mob1p is able to disrupt the self-association of Sid2p. Taken together, our results suggest that Sid2p kinase may utilize multiple modes of regulation including self-association, Mob1p binding, and phosphorylation to achieve its full activity at an appropriate time and place in the cell.
Cytokinesis is a fundamental step of cell proliferation by which daughter cells acquire equal amounts of genetic materials and cellular components (13, 16). To maintain genomic stability, cytokinesis does not occur until chromosome segregation is complete. Failed or precocious cell division causes aneuploidy, which is often associated with cancer (20). Therefore, it is crucial that cytokinesis be precisely executed at the correct time and place.
In order to coordinate cytokinesis with mitosis, signaling networks have evolved in eukaryotic organisms to faithfully control late cell cycle progression by triggering cytokinesis once mitotic events have been successfully accomplished (35). In the fission yeast Schizosaccharomyces pombe, this conserved signaling network is referred to as the septation initiation network (SIN), which triggers actomyosin ring constriction and septum formation after chromosome segregation (3, 16, 30). Inactivation of the SIN results in cytokinesis failure, thereby leading to elongated and multinucleated cells. In contrast, hyperactivation of the SIN causes the formation of multiple septa without cell separation. The SIN is a spindle pole body (SPB)-associated, GTPase-regulated protein kinase cascade comprised of the Spg1p GTPase, downstream protein kinases Cdc7p, Sid1p, and Sid2p, and the associated factors Cdc14p (in complex with Sid1p) and Mob1p (in complex with Sid2p) (10, 15, 18, 40, 43, 44). Sid4p and Cdc11p function together as a platform to assemble the SIN components at the SPBs (5, 25, 46). The key output of the SIN seems to be the Sid2p-Mob1p kinase complex. Sid2p kinase activity depends on all other SIN components and peaks at the time of cytokinesis (44). The Sid2p-Mob1p kinase complex presumably transmits the division signal from the SPB to the actomyosin ring, since it accumulates at the division site immediately prior to cytokinesis (18, 40, 44).
Sid2p is a conserved serine/threonine protein kinase that belongs to the Ndr subfamily of the AGC group of kinases (45). Phylogenetic analysis shows that most species have two distinct subclasses of this family. One subclass, characterized by mammalian Ndr1 and Ndr2 (31, 45), Drosophila melanogaster Trc (11), Caenorhabditis elegans Sax-1 (56), Saccharomyces cerevisiae Cbk1p (4, 6, 39, 52), and S. pombe Orb6p (50), seems to play a role in cellular morphogenesis. The second subclass is characterized by the tumor suppressor Lats/Warts in metazoans (22, 36, 49, 53-55), S. cerevisiae Dbf2p and Dbf20p (47, 51), and S. pombe Sid2p (44), which are involved in cell cycle control and cell division. Work with yeasts suggests that one feature of this family of kinases is binding to a Mob1p-family protein (6, 18, 19, 24, 40, 52). Yeasts have two Mob1p-family proteins, Mob1p and Mob2p (18, 27, 40), with Mob1p associating with Sid2p/Dbf2p and Mob2p associating with Orb6p/Cbk1p (6, 18, 19, 24, 40, 52). Budding yeast Mob1p is important for phosphorylation and activation of Dbf2p (28), and fission yeast Mob1p is essential for localization of Sid2p (18, 40). Recent studies with human Ndr1 and budding yeast Dbf2p have shown that phosphorylation plays a critical role in their activation (28, 32). These kinases are phosphorylated at two conserved phosphorylation sites, one in the activation loop and the second in a hydrophobic motif within the C terminus. It was proposed that Mob1p binding with Dbf2p results in a conformational change, enabling its upstream kinase Cdc15p to phosphorylate both phosphorylation sites, thereby stimulating Dbf2p activity (28). Taken together, these results suggest that stimulatory phosphorylation and Mob1 binding may be important for this family of kinases. However, the mechanistic relationship between Mob1p binding and phosphorylation as well as activation of the kinase partner remains to be elucidated.
The Sid2p-Mob1p kinase complex is a crucial component of the SIN, by which upstream SIN signaling is relayed to the division site, thereby triggering cytokinesis (18, 40, 44). In this study, we have investigated regulation of the Sid2p-Mob1p kinase complex. Our results show that Mob1p binding, phosphorylation, and self-association of Sid2p play important roles in its regulation, and we also identified a region containing amino acids 101 to 207 of Sid2p, which is essential for Mob1p binding and self-association. Our results suggest that Sid2p may utilize multiple modes of regulation in order to precisely achieve its full activity at the right time and place.
MATERIALS AND METHODS
Yeast methods and strains.
The S. pombe strains used in this study are listed in Table 1. All strains are isogenic to wild-type strain 972 (26). Fission yeast media, growth conditions, and manipulations were carried out as described previously (33). Except where noted, cells were grown in yeast extract (YE) medium. All experiments involving temperature-sensitive strains were done at the permissive temperature of 25°C and the restrictive temperature of 36°C unless otherwise indicated. Standard genetic and recombinant DNA methods were used as described previously (33, 41). S. pombe transformations were carried out by using either a lithium acetate method (23) or electroporation (38). DNA was prepared from bacteria and isolated from agarose gels by using Qiagen (Valencia, Calif.) kits and from yeast cells as described previously (17). DNA sequencing was performed at the University of Massachusetts Medical School's Nucleic Acid facility. Oligonucleotide primers were purchased from Integrated DNA Technologies (Coralville, Iowa).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YDM105 | ade6-M210 leu1-32 ura4-D18h− | Our stock |
| YDM116 | sid4-A1 ade6 leu1-32 ura4-D18 h− | Our stock |
| YDM275 | cdc11-123 ade6-M210 leu1-32 ura4-D18 h− | Our stock |
| YDM429 | sid2-250 ade6 leu1-32 ura4-D18 h+ | Our stock |
| YDM430 | spg1-106 ade6-M210 leu1-32 ura4-D18 h+ | Our stock |
| YDM435 | sid1-125 ade6-M210 leu1-32 ura4-D18 h+ | Our stock |
| YDM436 | cdc14-118 ade6-M210 leu1-32 ura4-D18 h+ | Our stock |
| YDM514 | sid2-13myc::kan, ade6 leu1-32 ura4-D-18 h+ | Our stock |
| YDM670 | mob1-1 ade6 leu1-32 ura4-D18 | Our stock |
| YDM708 | 13myc-mob1 ade6 leu1-32 ura4-D18 his3-D1 h+ | Our stock |
| YDM748 | GFP-mob1 sid2-13myc ade6 leu1-32 ura4-D18 | Our stock |
| YDM815 | mob1-N1 leu1-32 h− | Simanis lab |
| YDM816 | mob1-M17 leu1 h− | Simanis lab |
| YDM819 | mob1-3HA::kan leu1-32 h− | Simanis lab |
| YDM820 | mob1-J2 h− | Simanis lab |
| YDM862 | sid2-13myc::kan mob1-M17 ade6 | This study |
| YDM863 | sid2-13myc::kan mob1-N1 ade6 leu1-32 ura4-D18 | This study |
| YDM864 | sid2-13myc::kan mob1-J2 ade6 | This study |
| YDM1029 | mob1-3HA::kan sid2-13Myc::kan cdc25-22 ade6 leu1-32 ura4-D18 | This study |
| YDM1827 | GFP-mob1 sid2-13myc cdc7-24 ade6 leu1-32 ura4-D18 | This study |
| YDM1828 | GFP-mob1 sid2-13myc cdc11-123 ade6 leu1-32 ura4-D18 | This study |
| YDM1829 | GFP-mob1 sid2-13myc cdc14-118 ade6 leu1-32 ura4-D18 | This study |
| YDM1830 | GFP-mob1 sid2-13myc cdc15-140 ade6 leu1-32 ura4-D18 | This study |
| YDM1831 | GFP-mob1 sid2-13myc sid1-239 ade6 leu1-32 ura4-D18 | This study |
| YDM1832 | GFP-mob1 sid2-13mycspg1-106 ade6 leu1-32 ura4-D18 | This study |
| YDM1833 | GFP-mob1 sid2-13myc sid4-A1 ade6 leu1-32 ura4-D18 | This study |
| YDM1350 | sid2-13myc::kan ade6 leu1-32 ura4-D18 h− | Our stock |
| YDM1351 | cdc7-24 ade6-M210 leu1-32 ura4-D18 | Our stock |
For the yeast two-hybrid interaction assays, S. cerevisiae strain PJ69-4A was used as described previously (21). A variety of sid2 deletions were amplified by PCR with the full-length sid2 gene used as a template (pRHA41-sid2) and were subcloned into two-hybrid vectors, the Gal4p DNA binding domain-containing vector pGBT9 or the Gal4p activation domain-containing pGAD424 by standard cloning techniques. Other genes to be tested, including mob1, mob2, and orb6, were also subcloned into two-hybrid vectors in a similar way. Strain PJ69-4A was transformed by using the LiAc/PEG procedure (12). Leu+ and Trp+ transformants were selected and scored for positive interaction by being streaked onto synthetic dextrose plates lacking adenine and histidine. Three different transformants were scored for each sample.
Construction of sid2 mutant plasmids.
A variety of sid2 deletions were amplified by PCR using pRHA41-sid2 as a template and then subcloned into pRHA41 and pREP42-GFP vectors, whose expressions are driven by the moderate-strength nmt1 promoter (7, 29). pREP1-mob1 is driven by the strong nmt1 promoter, while pUR19-sid2-GFP is controlled by the native sid2 promoter.
All sid2 phosphorylation site mutants were created by using a QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, Tex.) using pRHA41-sid2 as a template. The following oligonucleotides were used to create individual phosphorylation site mutants at serine 402 or threonine 578: S402A, 5′-GATCCTGTATACGCTCATGCAGTAGTTGGCTCTCCTGAT-3′ and 5′-ATCAGGAGAGCCAACTACTGCATGAGCGTATACAGGATC-3′; T578A, 5′-AATGCTTTTATCGGCTTCGCTTTTAGGCATCAAAAAAAT-3′ and 5′-ATTTTTTTGATGCCTAAAAGCGAAGCCGATAAAAGCATT-3′; T578D, 5′-AATGCTTTTATCGGCTTCGATTTTAGGCATCAAAAAAAT-3′ and ATTTTTTTGATGCCTAAAATCGAAGCCGATAAAAGCATT; T578E, 5′-AATGCTTTTATCGGCTTCGAATTTAGGCATCAAAAAAAT-3′ and 5′-ATTTTTTTGATGCCTAAATTCGAAGCCGATAAAAGCATT-3′. The double mutant S402A;T578A was obtained by sequential mutagenesis. Changes in nucleotides were confirmed by DNA sequencing.
Microscopy.
For fluorescence microscopy, cells were prepared by methanol fixation as described previously (2). DNA was visualized with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma, St. Louis, Mo.) at a concentration of 1 μg/ml. Images were captured by using a Nikon Eclipse E 600 microscope with a Hamamatsu Orca-ER cooled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan) and IPlab Spectrum software (Scanalytics, Fairfax, VA).
Immunoprecipitation, immunoblot analysis, and in vitro kinase assay.
Cell pellets were obtained from 4 × 108 to 5 × 108 (20 to 25 optical density [OD] units at 595 nm) exponentially growing cells, which were collected by centrifugation and were frozen at −80°C if not used immediately. All subsequent manipulations were carried out at 4°C or on ice. Cells were lysed in NP-40 buffer {1% NP-40, 150 mM NaCl, 2 mM EDTA, 6 mM Na2HPO4, 4 mM NaH2PO4, 100 μg of phenylmethylsulfonyl fluoride/ml, 80 mg of benzamidine/ml, 1 mg of pepstatin/ml, 10 mg of leupeptin/ml, and 120 mg of AEBSF, HCl [4-(2-aminoethyl)benzenesulfonyl fluoride/ml, HCl]} by vortexing vigorously with acid-washed glass beads (Sigma) for 1 min. Protein lysates were normalized by utilizing a BCA protein assay kit (Pierce, Rockford, Ill.). Immunoprecipitations were performed by adding either 1 μl of anti-Myc mouse monoclonal immunoglobulin G (IgG) (9E10, 5.5 mg/ml; a gift from K. Gould, Vanderbilt University, Nashville, Tenn.), 1 μl of anti-HA mouse monoclonal IgG (12CA5, 5 mg/ml; a gift from K. Gould), or 1 μl of anti-green fluorescent protein (GFP) rabbit monoclonal IgG (Molecular Probes, Eugene, Oreg.) to the NP-40 cell lysates, followed by incubation on ice for 1 h. Immune complexes were purified by adding 40 μl of a 1:1 slurry of protein G-Sepharose beads (Sigma), followed by incubation on a rocker for 1 h at 4°C and centrifugation in a microcentrifuge for 1 min. The beads were washed three times with 1 ml of NP-40 buffer. The immune complex beads were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), which was then transferred onto an Immobilon P nylon membrane (Millipore, Bedford, Mass.) by use of a tank transfer system (Bio-Rad, Hercules, Calif.). The blotted membranes were probed with the anti-Myc IgG (1:2,000 dilution), anti-HA IgG (1:1,000 dilution), or anti-GFP IgG (1:1,000 dilution) and developed by using an alkaline phosphatase-enhanced chemiluminescent system (Bio-Rad).
For detection of the protein kinase activity, the immune complex beads were washed three times with 100 μl of kinase assay buffer (10 mM Tris [pH 7.4], 10 mM MgCl2 and protease inhibitor cocktail containing 100 μg of phenylmethylsulfonyl fluoride/ml, 80 mg of benzamidine/ml, 1 mg of pepstatin/ml, 10 mg of leupeptin/ml, and 120 mg of AEBSF/ml) and then incubated at 30°C for 30 min in 20 μl of kinase assay buffer with 10 μl of reaction cocktail containing 9.45 μl of 1-mg/ml myelin basic protein (Sigma), 0.05 μl of cold ATP at a concentration of 10 mM and 0.5 μl of [γ-32P]ATP (3,000 Ci/mmol at 10 mCi/ml; Amersham Pharmacia Biotech, Arlington Heights, Ill.). Myelin basic protein is used as an artificial substrate for the kinase activity analysis. Reactions were stopped by addition of 20 μl of 2× SDS-PAGE sample buffer, and the samples were resolved on SDS-15% polyacrylamide gels that were then imaged and quantified on a PhosphorImager (Molecular Dynamics). Relative kinase activity was determined by subtracting the background kinase activity associated with beads incubated with lysates not expressing the tagged Sid2p kinase, followed by normalization for the relative levels of Sid2p protein as determined by immunoblotting.
RESULTS
Mob1p is essential for Sid2p kinase activity.
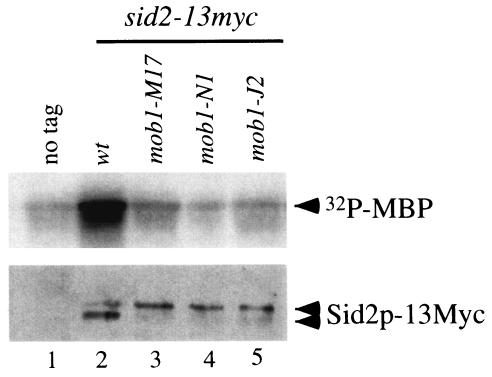
Previous results showed that Sid2p localization requires Mob1p (18, 40). Thus, Mob1p may be important for the regulation of Sid2p kinase. To test whether Mob1p is required for Sid2p kinase activity, we examined Sid2p kinase activity in different mob1 temperature-sensitive mutants, expressing chromosomally tagged Sid2p-13Myc, by using an in vitro kinase assay. At both the permissive and restrictive temperatures, the Sid2p-13Myc in vitro kinase activity in mob1-J2, mob1-M17, and mob1-N1 mutants was completely abolished, in contrast to what was observed with the wild type (Fig. 1 and data not shown). Interestingly, we found that the Sid2p-13Myc kinase activity was unaffected in both mob1-R4 and mob1-R17 mutants (data not shown), consistent with previous results showing that 13Myc-tagged Sid2p was able to complement these two alleles of mob1 (40).
FIG. 1.
Mob1p is required for Sid2p kinase activity. Cells were grown in YE medium overnight at 25°C to mid-log phase. Cultures were diluted to the same density and then shifted to 36°C for 3 h. Sid2p-13Myc kinase activity was analyzed by immunoprecipitation with anti-Myc antibody followed by an in vitro kinase assay. The protein levels were determined by immunoblot analysis following immunoprecipitation with anti-Myc antibody. The following strains were used: lane 1, YDM105; lane 2, YDM514; lane 3, YDM862; lane 4, YDM863; lane 5, YDM864.
Sid2p and Mob1p remain associated throughout the cell cycle.
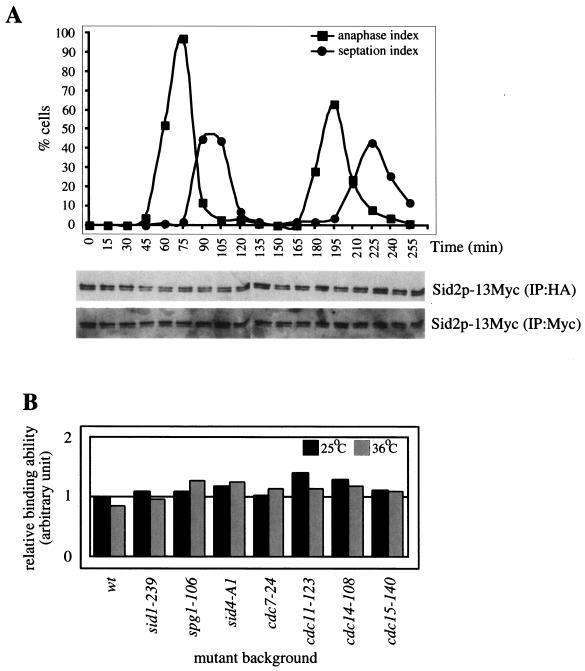
Sid2p kinase activity is regulated in a cell cycle-dependent manner (44); however, the mechanism behind this is still unclear. Since Mob1p interacts with and is required for the Sid2p kinase activity (Fig. 1) (18, 40), we investigated whether the interaction between Sid2p and Mob1p is cell cycle regulated. For this purpose, cells containing Sid2p-13Myc and Mob1p-3HA were synchronized by a block-and-release process utilizing the cdc25-22 mutant. Cell cycle progression was monitored by scoring the percentages of anaphase and septated cells over time points (Fig. 2A). The levels of Sid2p-13Myc that coimmunoprecipitated with Mob1p-3HA were determined through the cell cycle. This showed that at a gross level the Sid2p-Mob1p interaction is not cell cycle regulated (Fig. 2A). The levels of Sid2p and Mob1p were not significantly altered throughout the cell cycle (Fig. 2A and data not shown).
FIG. 2.
The interaction between Sid2p and Mob1p is not sufficient for activation of Sid2p kinase. (A) Sid2p and Mob1p remain associated throughout the cell cycle. sid2-13myc mob1-3HA cdc25-22 cells (YDM1029) were grown in YE medium overnight at 25°C to mid-log phase, shifted to 36°C for 4 h and then returned to 25°C. Samples were collected every 15 min. Cell cycle progression was monitored by scoring anaphase and septating cells for each time point. The interaction between Sid2p-13Myc and Mob1p-3HA was analyzed by immunoprecipitation with anti-HA antibody followed by immunoblot analysis with anti-Myc antibody. The Sid2p-13Myc protein level was also determined by immunoprecipitation with anti-Myc antibody followed by immunoblot analysis. (B) Sid2p and Mob1p remain bound in the absence of functional SIN components. Wild-type and various sin mutant cells containing sid2-13myc GFP-mob1 (YDM748, YDM1827 to -1829, and YDM1831 to -1833) were grown in YE medium overnight at 25°C to mid-log phase and split into two portions, which were then grown at 25 and 36°C, respectively, for 3 h, and then cell pellets were collected. The interaction between Sid2p-13Myc and GFP-Mob1p was analyzed by immunoprecipitation with anti-GFP antibody followed by immunoblot analysis with anti-Myc or anti-GFP antibodies. The ability of GFP-Mob1p to associate with Sid2p-13Myc in different mutant backgrounds relative to wild-type cells at 25°C is shown. This was calculated by measuring the ratio of Sid2p-13Myc immunoprecipitated with GFP-Mob1p to the level of GFP-Mob1p-immunoprecipitated anti-GFP antibody. These ratios were normalized to that of wild-type cells at 25°C, which were arbitrarily set at 1.0. cdc15-140 mutants (YDM1830) were used as a positive control for the Sid2p-Mob1p interaction.
Furthermore, since Sid2p requires upstream SIN components for its kinase activity (44), we examined whether the Sid2p-Mob1p interaction is dependent on these components. For this purpose, the interaction was examined in a variety of SIN mutant cells (cdc7-24, cdc11-123, cdc14-118, sid1-239, spg1-106, and sid4-A1) containing chromosomally tagged Sid2p-13Myc and GFP-Mob1p. At both the permissive and restrictive temperatures, the Sid2p-Mob1p interaction was not significantly affected in upstream SIN mutants (Fig. 2B and data not shown). This result suggests that the Sid2p-Mob1p interaction is not necessarily regulated by upstream SIN components. Therefore, although Mob1p is important for the Sid2p kinase activity, the association between these two proteins does not seem to be regulated. However, it is possible that the relative conformation of binding is regulated or that the Sid2p-Mob1p interaction is regulated in a localized substoichiometric pool of proteins.
Two conserved phosphorylation sites of Sid2p are important for its function.
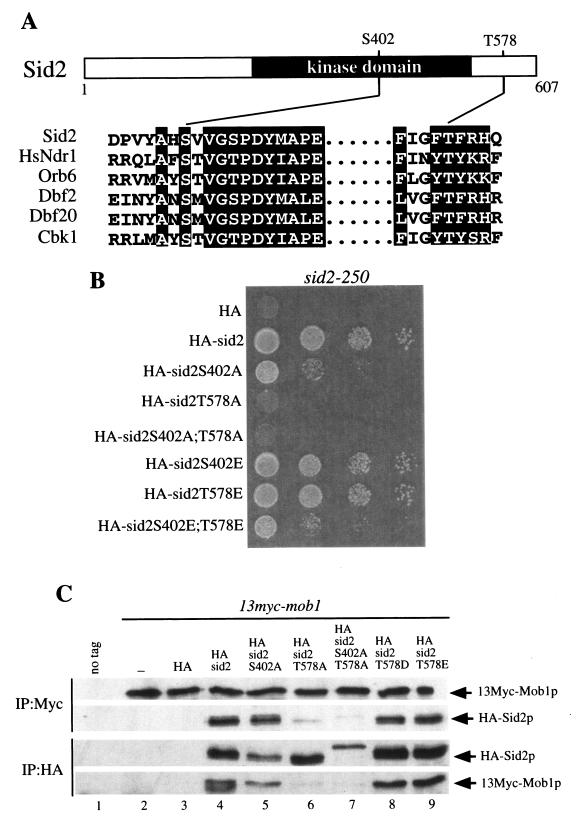
Phosphorylation of human Ndr1 protein kinase at two sites is required for its activation, and these two sites are highly conserved in the relatives of Ndr1, including Sid2p (Fig. 3A) (45). Sid2p is phosphorylated in vivo (data not shown), suggesting that phosphorylation may also be important for its regulation. Thus, to examine whether both corresponding residues of Sid2p (Ser402 and Thr578) may be involved in activation of Sid2p kinase, we mutated these residues to alanine to create single (S402A and T578A) and double (S402A;T578A) phosphorylation site mutants that were then tested for their ability to rescue the growth defect of the sid2 temperature-sensitive mutant, sid2-250. Sid2p-T578A and Sid2p-S402A;T578A did not rescue the sid2-250 growth defect at the restrictive temperature, while Sid2p-S402A only partially rescued the sid2-250 mutant (Fig. 3B). Furthermore, we also mutated both residues to aspartic acid or glutamic acid to create single and double mutants to mimic phosphorylation. These mutants were also tested for their ability to rescue the sid2-250 growth defect. Sid2p-S402E and Sid2p-T578E completely rescued the sid2-250 growth defect, whereas Sid2p-S402E;T578E only partially restored its growth ability (Fig. 3B). The incomplete rescue of the sid2-250 mutant by Sid2p-S402E;T578E may be due to the fact that glutamic acid does not perfectly mimic phosphorylation. These results indicate that Ser402 and Thr578 play an important role in Sid2p function.
FIG. 3.
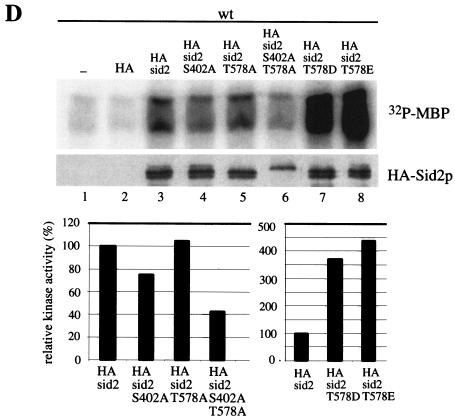
Two conserved putative phosphorylation sites of Sid2p, Ser402 and Thr578, are important for Sid2p function. (A) Alignment of the sequences around two potential phosphorylation sites on Sid2p in the activation loop (S402 in Sid2p) and the hydrophobic motif (T578 in Sid2p) with human Ndr1, budding yeast Cbk1p, Dbf2p, and Dbf20p, and fission yeast Orb6p. (B) sid2-250 cells (YDM429) carrying the pRHA41 vector alone (HA) or with the different phosphorylation site mutants shown were grown in selective medium with thiamine (5 μg/ml) overnight at 25°C to mid-log phase. Cultures were diluted to 0.1 OD unit at 595 nm, and 2 × 104 cells at a 1:10 serial dilution were spotted onto selective plates in the absence of thiamine at 25°C for 22 to 24 h and then shifted to 36°C for 3 or 4 days. (C) Wild-type (YDM105) (no tag) or 13myc-mob1 cells (YDM708) carrying no vector (−), or pRHA41 with no insert (HA) or the different Sid2p variants shown were grown in selective medium with thiamine overnight at 30°C to mid-log phase. Cells were washed three times in minimal medium, diluted to the same cell density, and induced for 22 h in the absence of thiamine. Coimmunoprecipitation was performed with anti-Myc or anti-HA antibody followed by Western blot analysis with anti-Myc or anti-HA antibody. (D) Wild-type cells (YDM105) carrying the same plasmids as in panel C were grown under the same conditions as those described for panel C. The kinase activities of immune complexes from immunoprecipitation with anti-HA antibody were analyzed (top panel), and all 3HA-Sid2p mutant protein levels were determined by immunoblot analysis following immunoprecipitation with anti-HA antibody (middle panel). The relative kinase activity shown in the bottom panel was calculated as described in Materials and Methods.
To further understand how phosphorylation controls Sid2p function, we went on to examine whether Mob1p binding and/or the kinase activity of these Sid2p phosphorylation site mutant proteins was affected. The interaction between Mob1p and the various Sid2p mutant proteins was examined by coimmunoprecipitation with episomally expressed 3HA-Sid2p mutant proteins and endogenous 13Myc-Mob1p. 13Myc-Mob1p immunoprecipitated 3HA-Sid2p-S402A, 3HA-Sid2p-T578D, 3HA-Sid2p-T578E, and wild-type Sid2p, all of which were equally abundant in 13Myc-Mob1p immunoprecipitates and vice versa (Fig. 3C). In contrast, the Mob1p binding of 3HA-Sid2p-T578A and 3HA-Sid2p-S402A;T578A was almost abolished (Fig. 3C). Similar results were observed when the two-hybrid interaction assay was used (data not shown), suggesting that phosphorylation of Sid2p on Thr578 is important for Mob1p binding. We are not sure why the migration of Sid2p-S402A;T578A double mutant is different from the migrations of the wild type and the other mutants. It is possible that this represents an instance where the unphosphorylated form of a protein runs more slowly.
The in vitro kinase activity of these Sid2p mutant proteins was also analyzed. The activity of the double mutant 3HA-Sid2p-S402A;T578A protein was reduced to ∼40% of that of wild-type 3HA-Sid2p. Surprisingly, the activities of the single mutants 3HA-Sid2p-S402A and 3HA-Sid2p-T578A were not significantly changed compared with 3HA-Sid2p kinase activity (Fig. 3D). In contrast, 3HA-Sid2p-T578D and 3HA-Sid2p-T578E kinase activities were dramatically enhanced, three- to fivefold over that of wild-type 3HA-Sid2p (Fig. 3D). The activities of 3HA-Sid2p-S402D and 3HA-Sid2p-S402E were comparable to that of the wild type (data not shown). On the basis of these results, both Ser402 and Thr578 are important for Sid2 function and phosphorylation of Thr578 plays a major role.
Since Sid2p is the most downstream component of the SIN so far, it is likely that phosphorylation of Ser402 and Thr578 is mediated through the upstream SIN components. If this is the case, then mimicking phosphorylation might be able to bypass the requirement of the upstream SIN signaling. To test this possibility, we investigated the abilities of 3HA-Sid2p-T578E and 3HA-Sid2p-S402E to rescue the growth defect of upstream SIN mutants. While 3HA-Sid2p-S402E was unable to rescue the growth defects of upstream SIN mutants (data not shown), 3HA-Sid2p-T578E could weakly restore the growth abilities of sid1-125, spg1-106, cdc14-118, and cdc11-123 but not that of sid4-A1 or cdc7-24 (Table 2). These results suggest that phosphorylation of Thr578 is an important function of the upstream SIN components. Consistent with the direct physical interaction between Sid2p and Mob1p, both 3HA-Sid2p and 3HA-Sid2p-T578E were also able to rescue the growth defect of the mob1-1 mutant (Table 2).
TABLE 2.
Effects of the Thr578 phosphomimetic mutant of Sid2p on growth abilities of sin mutantsa
| sin mutant | Growth ability of mutant carrying indicated plasmidb
|
|||||
|---|---|---|---|---|---|---|
| Vector
|
Sid2
|
Sid2-T578E
|
||||
| +T | −T | +T | −T | +T | −T | |
| Wild type | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| cdc7-24 | − | − | − | − | − | − |
| cdc11-123 | − | − | − | − | − | +++ |
| cdc14-118 | − | − | − | − | − | + |
| sid1-125 | − | − | − | − | ++ | +++ |
| sid2-250 | − | − | ++++ | ++++ | ++++ | ++++ |
| spg1-106 | − | − | − | − | − | ++ |
| sid4-A1 | − | − | − | − | − | − |
| mob1-1 | − | − | +++ | ++++ | +++ | ++++ |
Cells (YDM105, YDM116, YDM275, YDM249, YDM430, YDM435, YDM436, YDM670, and YDM1351) were grown on selective plates at 25°C for 22 to 24 h and then shifted to 36°C for 3 or 4 days.
Vector, pRHA41; Sid2, pRHA41-sid2; Sid2-T578E, pRHA41-sid2-T578E; +T, with thiamine; −T, without thiamine; −, no growth observed; + to ++++, relative growth rates ranging from very poor growth to wild-type growth rates.
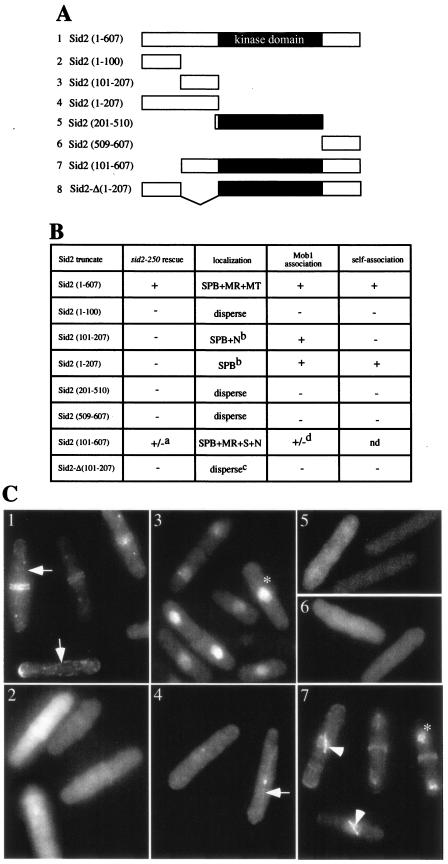
The amino acids 101 to 207 of Sid2p, immediately preceding the kinase domain, are sufficient for Mob1p binding and the SPB localization.
To identify functional domains within Sid2p, we constructed a variety of Sid2p deletions that were 3HA tagged at the N terminus and were expressed from the moderate-strength nmt1 promoter (Fig. 4A). The abilities of these 3HA-tagged Sid2p mutants to rescue the growth defect of sid2-250 cells were investigated at the restrictive temperature in the absence of thiamine. We found that only full-length 3HA-Sid2p could completely restore the growth abilities of mutants while 3HA-Sid2p (amino acids 101 to 607) [hereafter referred to as Sid2p (101-607)] partially rescued the growth defect of mutant cells (Fig. 4B). Other Sid2p mutants were unable to rescue the growth defect of the mutant cells (Fig. 4B). These results indicate that the kinase domain alone is not sufficient for Sid2 function and the regions outside the kinase domain are important for Sid2p function.
FIG. 4.
Amino acids 101 to 207 of Sid2p are sufficient for Mob1p binding and SPB localization. (A) Sid2p deletion constructs used in this study. The kinase domain (amino acids 208 to 508) is shown in black. (B) Summary of complementation, Sid2p localization, Mob1p binding, andself-association in cells exogenously expressing the various Sid2p deletions described for panel A. To test for rescue of the growth defect of the sid2-250 mutant, cells (YDM429) carrying pRHA41 vector with no insert (HA) or the different Sid2p deletions shown were grown on selective plates without thiamine at 25°C for 22 to 24 h and then shifted to 36°C. The growth ability was judged by colony formation at 36°C. For Sid2p localization studies, all of these deletion mutants were tagged with GFP and expressed from the moderate-strength nmt1 promoter of pREP42 vector. Cells (YDM105) containing these GFP-Sid2p fusions were grown in selective medium with thiamine overnight at 30°C to mid-log phase and induced for 18 h in the absence of thiamine. GFP-Sid2p localization was examined in fixed cells. For Mob1p binding and self-association of Sid2p, the two-hybrid interaction assay was performed with cells expressing various Sid2p deletion constructs from pGBT9 and Mob1p or Sid2p from pGAD424 as described in Materials and Methods. Abbreviations: SPB, spindle pole body; MR, medial ring; MT, cytoplasmic microtubules; S, mitotic spindle; N, nucleus; a, partial rescue; b, occasionally found on mitotic spindle; c, see Fig. 5B; d, a weak interaction compared to that observed with full-length Sid2p; nd, not determined. (C) Localization of various Sid2p deletion constructs expressed as described in panel B. The number to the left of each construct in panel A refers to the corresponding image in panel C. Arrows indicate cells with cytoplasmic microtubule-like signals; arrowheads indicate cells with mitotic spindle-like signals; asterisks indicate cells with nuclear signals. Each panel exhibits a montage of representative cells.
To further define the region of Sid2p required for Mob1p binding and the SPB localization, we analyzed the different Sid2p constructs by using the two-hybrid interaction assay and fusion to GFP. For the two-hybrid interaction assay, we tested the abilities of the different mutant proteins to interact with Mob1p. We found that the N terminus of Sid2p, Sid2p (1-207), was sufficient to bind to Mob1p (Fig. 4B). Furthermore, we dissected the N terminus and found that Sid2p (101-207) was sufficient to associate with Mob1p (Fig. 4B). In addition, we found that Sid2p (101-607) exhibited a weaker interaction with Mob1p than did Sid2p (101-207), suggesting that the kinase domain and/or C terminus might interfere with Mob1p binding (Fig. 4B). Taken together, we conclude that amino acids 101 to 207 of Sid2p, a region preceding the kinase domain, are sufficient to interact with Mob1p.
To examine the localization of the above Sid2p truncations, we tagged each of them with GFP at the N terminus and expressed them in S. pombe cells. GFP-Sid2p (101-207) (Fig. 4B and C, panel 3), GFP-Sid2p (1-207) (Fig. 4B and C, panel 4), GFP-Sid2p (101-607) (Fig. 4B and C, panel 7), and full-length GFP-Sid2p (Fig. 4B and C, panel 1) localized to the SPBs, consistent with the requirement of Mob1p for SPB localization of Sid2p, and all of them associate with Mob1p (Fig. 4B). However, we did not find GFP-Sid2p (101-207) or GFP-Sid2p (1-207) at the medial ring, unlike GFP-Sid2p (101-607) and GFP-Sid2p, indicating that other regions in Sid2p might be important for its medial ring localization (Fig. 4B and C). In contrast, GFP-Sid2p (1-100) (Fig. 4B and C, panel 2), GFP-Sid2p (201-510) (Fig. 4B and C, panel 5), and GFP-Sid2p (509-607) (Fig. 4B and C, panel 6) were dispersed throughout the cell. Interestingly, we noticed that GFP-Sid2p (101-207) and GFP-Sid2p (101-607), both devoid of the first 100 amino acids at the N terminus, were also localized in the nucleus, suggesting that the first 100 amino acids might contain a nuclear export sequence (Fig. 4C, panels 3 and 7, respectively). However, the functional significance of Sid2p in the nucleus remains to be elucidated. In addition, we found that full-length GFP-Sid2p and GFP-Sid2p (1-207) showed a pattern like that of interphase microtubule arrays but were not observed on the mitotic spindle (Fig. 4C, panels 1 and 4, respectively). GFP-Sid2p (101-207) and GFP-Sid2p (101-607) showed a spindle-like signal (Fig. 4C, panel 7; also data not shown). These results presumably reflect affinity of Sid2p for microtubules, and because full-length Sid2p is excluded from the nucleus, it is present only on cytoplasmic microtubules, whereas Sid2p (101-207) and Sid2p (101-607) are stuck in the nucleus and thus only localize to the spindle due to the lack of nuclear envelope breakdown in mitosis. The ability to see Sid2p on microtubules is likely due to moderate overexpression, since endogenous Sid2p-GFP only occasionally exhibits a weak microtubule-like signal (M.-C. Hou and D. McCollum, unpublished observations).
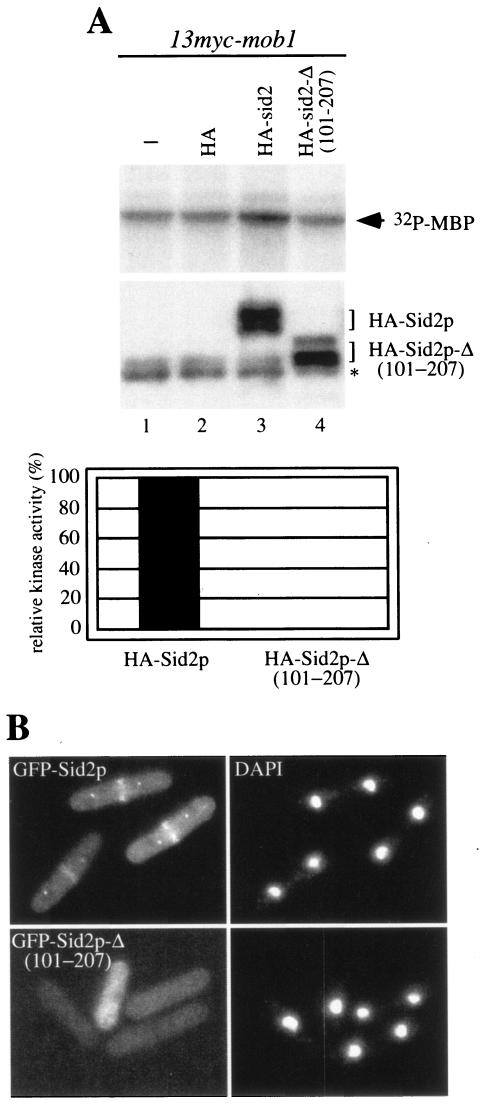
Since Sid2p (101-207) is sufficient for Mob1p binding and SPB localization, we deleted amino acids 101 to 207 from full-length Sid2p to create Sid2p-Δ (101-207) to determine whether this region was essential. We found that Sid2p-Δ (101-207) was no longer capable of binding Mob1p both in the two-hybrid interaction assay and by coimmunoprecipitation from S. pombe lysates (Fig. 4B and data not shown) and its kinase activity was completely abolished (Fig. 5A). We also examined the localization of Sid2p-Δ (101-207) by fusing it to GFP. Localization of Sid2p-Δ (101-207) to the SPBs and division site was largely lost, with only 20% of anaphase cells (30 of 150) showing very faint SPB and division site localization compared to 81% of anaphase cells (122 of 151) containing GFP-Sid2p (Fig. 5B). In addition, Sid2p-Δ (101-207) was not functional in vivo because 3HA-Sid2p-Δ (101-207) was unable to rescue the growth defect of sid2-250 cells (Fig. 4B). Taken together, our results demonstrate that amino acids 101 to 207 are important for Mob1p binding and essential for Sid2p function.
FIG. 5.
Deletion of amino acids 101 to 207 of Sid2p results in a loss of Sid2p function. (A) In vitro kinase activity of 3HA-Sid2p constructs was examined from immune complexes by immunoprecipitation with anti-HA antibody from 13myc-mob1 cells (YDM708) carrying no vector (−), or pRHA41 vector with no insert (HA), pRHA41-sid2 (HA-sid2) or pRHA41-sid2-Δ (101-207) (HA-sid2-Δ (101-207). The protein levels of 3HA-Sid2p and 3HA-Sid2p-Δ (101-207) were determined by Western blot analysis following immunoprecipitation with anti-HA antibody (top panel). The relative kinase activity was calculated as described in Materials and Methods. The asterisk indicates the IgG heavy chain. (B) Wild-type cells (YDM105) expressing episomal GFP-Sid2p and GFP-Sid2p-Δ (101-207) were grown in selective medium with thiamine overnight at 30°C to mid-log phase, washed three times in minimal medium, and induced for 12 h in the absence of thiamine.The localization of GFP-Sid2p and GFP-Sid2p-Δ (101-207) was examined in fixed cells. The left and right columns represent GFP fluorescence and DAPI staining, respectively. Each panel exhibits a montage of representative cells.
The N terminus of Sid2p family kinases is important for binding to Mob1p family proteins.
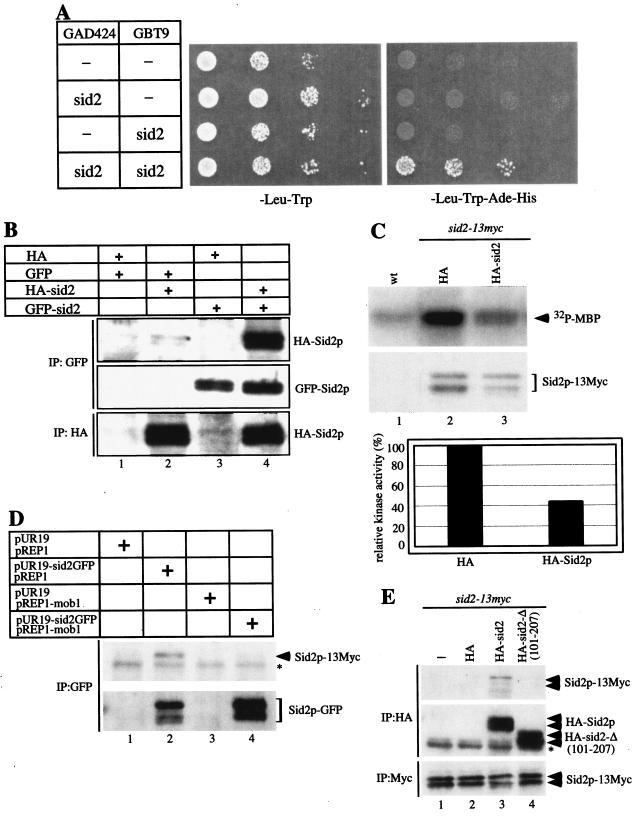
On the basis of the above results, the N terminus of Sid2p is sufficient for Mob1p binding and the SPB localization. Thus, we wondered whether the N terminus of the Sid2p family kinases is the binding region for Mob1p family proteins in general. To test this idea, we employed the two-hybrid interaction assay to examine the interaction between the N terminus of the Sid2p family kinase Orb6p and Mob1p family member Mob2p. We found that the N terminus of Orb6p kinase, Orb6p (1-92), was sufficient to bind to Mob2p (Table 3). We conclude that the N terminus of Sid2p family kinases is sufficient for binding to Mob1p family proteins and furthermore may play a regulatory role in the function of the Sid2p family kinases in general. In addition, we noticed that the N terminus of Orb6p also had a weak interaction with Mob1p, detected by the two-hybrid interaction assay (Table 3), which may not be relevant in vivo since the interaction between Orb6p and Mob1p was not detected in vivo by coimmunoprecipitation (our unpublished observations).
TABLE 3.
Interaction between the N terminus of the Sid2p family of kinases and the Mob1p family of proteins in fission yeasta
| Plasmid expressing Sid2p kinase | Interaction with indicated plasmid expressing Mob1p protein
|
||
|---|---|---|---|
| GAD424 | GAD424-mob1 | GAD424-mob2 | |
| GBT9 | − | − | − |
| GBT9-sid2 (1-207) | − | ++ | − |
| GBT9-orb6 (1-92) | − | + | ++ |
−, no interaction; +, weak interaction; ++, strong interaction.
Sid2p is self-associated in vivo.
During the course of these experiments, we noticed that Sid2p interacted with itself in the two-hybrid interaction assay (Fig. 6A). To determine whether Sid2p could associate with itself in S. pombe cells, coimmunoprecipitation was carried out with 3HA-Sid2p and GFP-Sid2p both expressed episomally from the moderate-strength nmt1 promoter. GFP-Sid2p immunoprecipitated 3HA-Sid2p and vice versa (Fig. 6B). Furthermore, endogenous Sid2p-13Myc also immunoprecipitated Sid2p-GFP expressed episomally from its own promoter (Fig. 6E), indicating that this association between overexpressed GFP-Sid2p and 3HA-Sid2p is not likely to be an artifact of overexpression.
FIG. 6.
Role of Sid2p self-association in regulating Sid2p function. (A) Strain PJ69-4A carrying the two-hybrid vectors expressing Sid2p (pGAD424-sid2 or pGBT9-sid2) was tested for Sid2p self-association as indicated (see Materials and Methods). (B) Cells (YDM105) carrying control vectors pRHA41 and pREP42-GFP (HA and GFP) or pRHA41-Sid2 (HA-Sid2) or pREP42-GFP-Sid2 (GFP-Sid2) as indicated were grown in selective medium with thiamine overnight at 30°C to mid-log phase. Cultures were washed three times in minimal medium, diluted tothe same cell density, and induced for 22 h in the absence of thiamine. Sid2p self-association was detected by immunoprecipitation with anti-GFP or anti-HA antibodies followed by immunoblot analysis with anti-GFP or anti-HA antibody. (C) Wild-type (wt; YDM105) or sid2-13myc (YDM1350) cells carrying pRHA41 vector with no insert (HA) or Sid2p (HA-sid2) were grown in selective medium with thiamine at 30°C overnight to mid-log phase. Cultures were washed three times in minimal medium, diluted to the same cell density, and induced for 20 h in the absence of thiamine. In vitro kinase assays were carried out following immunoprecipitation with anti-Myc antibody. The protein level was determined by immunoblot analysis following immunoprecipitation with anti-Myc antibody. The relative kinase activity is shown in the bottom panel and was calculated as described in Materials and Methods. (D) sid2-13myc cells (YDM1350) carrying the indicated plasmids were grown under the same conditions as those described for panel B. After 16 h of induction in the absence of thiamine, immunoprecipitation was performed with anti-GFP antibody followed by immunoblot analysis with anti-Myc or anti-GFP antibody. The asterisk indicates a cross-reacting protein band. (E) sid2-13myc cells carrying no vector (−), pRHA41 vector with no insert (HA), Sid2p (HA-Sid2), or Sid2p-Δ (101-207) [HA-sid2-Δ (101-207)] were grown under the same conditions as those described for panel C. After 22 h of induction in the absence of thiamine, immunoprecipitation was performed with anti-HA or anti-Myc antibody followed by immunoblot analysis with anti-HA or anti-Myc antibody. The asterisk indicates the IgG heavy chain.
We also investigated whether Mob1p was able to associate with itself. The results from coimmunoprecipitation and the two-hybrid interaction experiments showed that Mob1p is not associated with itself (data not shown). This suggests that Mob1p is not present in Sid2p multimers or else that one molecule of Mob1p binds to multiple molecules of Sid2p, which seems less likely (see below).
If Mob1p is not present in the multimeric Sid2p complex, then at least two forms of Sid2p are present in the cell, the Mob1p-bound and self-associated forms. Because Mob1p is required for Sid2p activation, self-associated Sid2p would presumably be inactive. To test this, we sought to examine the effect of increasing Sid2p self-association on its kinase activity. We hypothesized that overexpression of exogenous Sid2p should increase the fraction of endogenous Sid2p-13Myc in Sid2p multimers. Thus, we assayed in vitro kinase activity of endogenous Sid2p-13Myc following overexpression of exogenous 3HA-Sid2p. We found that the Sid2p-13Myc kinase activity was reduced in the presence of overexpressed 3HA-Sid2p, suggesting that Sid2p self-association may have an inhibitory effect on its function (Fig. 6C). However, an alternative interpretation might be that overexpressed 3HA-Sid2p could have titrated out endogenous Mob1p and prevented endogenous Sid2p-13Myc from binding to Mob1p, thereby eliminating its kinase activity. One model for Mob1p function could be that Mob1p inhibits the formation of inactive Sid2p multimers as one way to promote Sid2p kinase activity. To test this possibility, we analyzed the interaction between endogenous Sid2p-13Myc and Sid2p-GFP episomally expressed from its own promoter in the absence or presence of Mob1p overexpression. In the absence of overexpressed Mob1p, Sid2p-13Myc associated with Sid2p-GFP normally but Sid2p self-association was abolished when Mob1p was overexpressed (Fig. 6D), indicating that Mob1p overexpression disrupted Sid2p self-association, and further supporting the idea that Mob1p is not present in Sid2p multimers.
Since Mob1p binding is affected by phosphorylation of Sid2p on Thr578, we tested whether Thr578 phosphorylation had an effect on Sid2p self-association. We performed coimmunoprecipitation assays with 3HA-Sid2p and GFP-Sid2p phosphorylation site mutant proteins expressed episomally from the moderate-strength nmt1 promoter. The result showed that Sid2p self-association still occurred normally between different combinations of Thr578 phosphorylation site mutant proteins (data not shown).
To determine the region of Sid2p that mediates its self-association, we employed the two-hybrid interaction assay. A variety of Sid2p mutant proteins were tested for association with full-length Sid2p. We found that the N terminus of Sid2p, Sid2p (1-207), but not Sid2p (1-100) or Sid2p (101-207), associated with full-length Sid2p (Fig. 4B). The interaction became very weak or nonexistent when Sid2p (1-207) was tested for interaction with different Sid2p deletions, and thus we were unable to map the exact nature of the intermolecular interaction. We also examined the ability of Sid2p-Δ (101-207) to self-associate with Sid2p by using the two-hybrid interaction assay and coimmunoprecipitation. Both experiments showed that Sid2p-Δ (101-207) was unable to associate with full-length Sid2p, further supporting the importance of the N terminus of Sid2p for its self-association (Fig. 4B and 6E). Taken together, we conclude that the N terminus is important for Sid2p self-association.
DISCUSSION
Role of Mob1p in Sid2p kinase function.
The Mob1p-binding region of Sid2p was mapped to residues 101 to 207 in the N terminus immediately before the kinase domain. Consistent with this, deletion of this region caused loss of Mob1p binding and Sid2p kinase activity. Somewhat surprisingly, this Sid2p mutant could still localize, albeit poorly, to the SPBs, even though previous studies had shown that Mob1p is required for Sid2p localization (18, 40). One explanation for this could be the finding that a region of the N terminus of Sid2p that overlaps partially with residues 101 to 207 binds directly to the SPB scaffold protein Cdc11p (K. Gould, personal communication). Because the Cdc11p and Mob1p binding domains overlap, it is possible that in the context of the regular Sid2p protein, Mob1p binding may help make the Cdc11p binding domain of Sid2p accessible, thereby facilitating Sid2p SPB localization.
Sid2p kinase activity is tightly regulated in a cell cycle-dependent manner (44). Our results show that Mob1p is required not only for proper Sid2p localization but also for its kinase activity, just as cyclin proteins are essential for the kinase activity of Cdk family kinases. However, unlike Cdks and cyclins, Mob1p levels do not vary during the cell cycle and the levels of Mob1p bound to Sid2p also do not vary dramatically during the cell cycle. This could mean that the Sid2p-Mob1p interaction alone is necessary but not sufficient to activate Sid2p kinase and that additional mechanisms regulate Sid2p activation. Alternatively, binding between Sid2p and Mob1p could be regulated only for a subpopulation of each protein perhaps localized to the SPBs, which would be missed by our studies using whole-cell lysates. A third possibility, which we favor, is that the Sid2p-Mob1p interaction could be altered, perhaps by phosphorylation, in a cell cycle-dependent manner to activate the Sid2p kinase activity (see below).
Relationship between Mob1p binding and Sid2p phosphorylation.
We have also found that there seems to be a complex interplay between Mob1p binding and Sid2p phosphorylation. Based on work performed with the Sid2p-related kinase Ndr1 as well as the budding yeast homolog Dbf2p (28, 32), we have found that Sid2p is likely phosphorylated on two conserved residues, Ser402 in the activation loop and Thr578 in a hydrophobic motif within the C terminus. Phosphorylation at both of these residues is important for Sid2p kinase activity, since the Sid2p-S402A;T578A mutant has severely impaired kinase activity. However, the single mutants (especially T578A) did not have kinase activity significantly different from that of the wild type, despite being unable to rescue sid2-250 mutants. Although this result is surprising, we can imagine several possible explanations. First, the T578A mutation may not perfectly mimic T578 in the unphosphorylated state, and this could both have an effect on Mob1p binding and cause the kinase to be activated in the absence of Mob1p. Alternatively, it is possible that these single mutants are more impaired for kinase activity in the cell or that phosphorylation at each of these sites has additional functions besides affecting kinase activity. Further studies will be required in order to distinguish between these possibilities. Phosphorylation of residue Thr578 seems to be the most important of these two sites, since the T578A mutant displayed a more severe phenotype than did the S402A mutant, and given the fact that unlike the S402E phosphomimetic mutation, the T578E phosphomimetic mutation results in elevated Sid2p kinase activity and can rescue the growth defect of upstream sin mutants. Interestingly, we found that the Sid2p-T578A mutant showed a reduced ability to bind Mob1p even though the Mob1p binding domain is on the other end of the protein. Because Mob1p is able to bind Sid2p in the absence of the C terminus containing Thr578 (data not shown), this suggests that the C terminus interferes with Mob1p binding and that this interference is relieved by phosphorylation at Thr578. However, if Thr578 phosphorylation is cell cycle regulated, then we should have observed cell cycle-regulated changes in the Sid2p-Mob1p complex levels, which we did not. One explanation for this could be that the T578A mutation does not perfectly mimic Thr578 in the unphosphorylated state and may have a more severe effect on Mob1p binding. For wild-type Sid2p, phosphorylation may govern how, but not whether, Mob1p binds Sid2p. Alternatively, it is possible that Thr578 is not phosphorylated in a cell cycle-dependent manner. Reciprocally, it has been reported in studies with budding yeast that the Sid2p homolog Dbf2p requires Mob1p for it to be phosphorylated (28). This is quite reminiscent of the interaction between Cdk1 and cyclin, where phosphorylation of Cdk1 at Thr161 depends on cyclin but at the same time cyclin binding is enhanced by phosphorylation at Thr161 (9, 14). Interestingly, both the phosphorylation site analogous to Thr578 in Sid2p and the Mob1p binding region of Sid2p are conserved among all Sid2p-related kinases. We also observe that the Sid2p-related kinase Orb6p binds the Mob1p-family protein Mob2p through an analogous domain at its N terminus, suggesting that this mode of interaction will be conserved among other members of this family of proteins including the mammalian kinases Ndr1/2, Lats/Warts (45), and the various mammalian Mob1p-family members (18, 27).
Sid2p phosphorylation has been shown to be important for its function; however, its upstream kinase is unknown. For budding yeast, Cdc15p has been implicated as the upstream kinase that phosphorylates Dbf2p on Ser374 and Thr544 (28). S. pombe Cdc7p, a homolog of Cdc15p, acts upstream of Sid2p (44), implying that Cdc7p might function in Sid2p phosphorylation. However, our results showed that mimicking Sid2p phosphorylation on Ser402 and/or Thr578 could not bypass the requirement of Cdc7p and hence, a kinase (or kinases) other than Cdc7p might be responsible for Sid2p phosphorylation on both residues. Alternatively, the lack of rescue could be explained if Cdc7p is required for additional aspects of the SIN function. One possible function could be phosphorylation of Mob1p, which has been shown to be also a phosphoprotein (M.-C. Hou and D. McCollum, unpublished observations). Sid1p kinase, in complex with Cdc14p, has also been shown to be upstream of Sid2p (15, 44), and our results showed that mimicking Sid2p phosphorylation on Thr578 is able to partially rescue the growth defect of sid1 and cdc14 mutants, suggesting the involvement of Sid1p kinase in Sid2p phosphorylation at least on Thr578. Further work will be needed to investigate whether Cdc7p, Sid1p, or some other kinase phosphorylates Sid2p.
Self-association and Sid2p regulation.
Accumulating studies have highlighted the importance of homodimerization in the regulation of both tyrosine and serine/threonine protein kinases (1, 37, 42). Our results show that Sid2p is able to interact with itself. It is not known whether Sid2p self-association is direct or mediated by other factors. Our results show that Mob1p does not dimerize and is not present in multimers in vivo, suggesting that Mob1p is not involved in Sid2p self-association. These data could be explained if either Sid2p formed multimers lacking Mob1p or each Sid2p multimer contained a single Mob1p protein. However, we support the former model, since Sid2p self-association is disrupted when Mob1p is overexpressed, suggesting not only that Mob1p is not in Sid2p multimers but also that it antagonizes their formation. Furthermore, amino acids 101 to 207 of Sid2p are important both for its self-association and for Mob1p binding, suggesting that there is a competition between self-association and Mob1p binding. Also consistent with this model, overexpression of exogenous Sid2p, which we would expect to increase multimer formation, results in a decrease in the kinase activity of the endogenous protein, suggesting that Sid2p multimers may be inactive.
A model for Sid2p-Mob1p interaction.
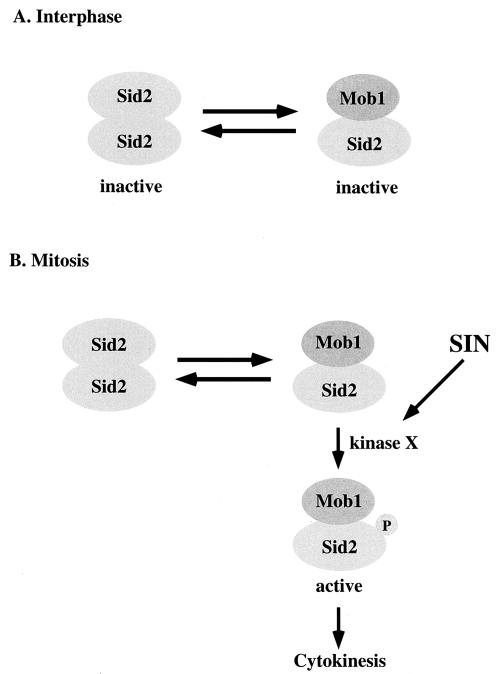
We propose a model for the role of Mob1p binding, phosphorylation, and self-association in the regulation of Sid2p kinase (Fig. 7). Mob1p-associated Sid2p is in equilibrium with self-associated Sid2p, both of which are in an inactive state during interphase (Fig. 7A). Upon entry into mitosis, Mob1p-associated Sid2p localizes strongly to the SPBs, where its activators are, in a manner that depends on the SIN scaffold components Cdc11p and Sid4p. This may allow Sid2p to be activated by phosphorylation by an upstream SIN kinase or kinases, which in turn could help stabilize or change the mode of Mob1p binding to activate the kinase (Fig. 7B). Because Mob1p is also phosphorylated (M.-C. Hou and D. McCollum, unpublished observations), it is likely that the model will prove to be more complex when the functions of Mob1p phosphorylation and the upstream kinase(s) are determined. Sid2p self-association may help prevent leaky Sid2p activity prior to activation of the SIN, while the Sid2p-Mob1p interaction allows for reversal of this inactive state, SPB localization, and timely activation by the SIN. Furthermore, phosphorylation provides another level of regulation to ensure that Mob1p-associated Sid2p is not improperly activated until the SIN is activated. Thus, Sid2p activation is tightly regulated by Mob1p binding, phosphorylation, and self-association so that cytokinesis does not occur prior to the completion of all mitotic events.
FIG. 7.
A model for regulation of Sid2p-Mob1p complex during the cell cycle (described in the text). Kinase X is a yet-to-be-identified upstream kinase of Sid2p.
Localization to the SPB, cell division site, and microtubules is mediated by the N terminus of Sid2p.
As part of our studies mapping the Mob1p binding domain of Sid2p, we also characterized regions of Sid2p important for its localization. We found that episomally expressed full-length GFP-Sid2p localized to the SPB weakly in interphase and strongly during mitosis. It also localized to the cell division site and to structures that resembled cytoplasmic microtubules. This contrasts somewhat with our previous results with chromosomally tagged Sid2p-GFP, which localized constitutively to the SPBs and was not commonly observed on microtubules. However, the SPB localization pattern was similar to chromosomally tagged GFP-Mob1p, which localized faintly to interphase SPB and strongly to mitotic SPBs (18). These differences could be due to differences in which end of Sid2p has the GFP tag and in levels of expression. GFP-Sid2p (101-207), GFP-Sid2p (1-207), GFP-Sid2p (101-607), and GFP-Sid2p localized to the SPBs, but GFP-Sid2p (101-207) and GFP-Sid2p (1-207) were unable to localize to the division site, suggesting that other regions in Sid2p are required for localization to the division site. The C-terminal region of Sid2p seems to be necessary but not sufficient for its localization to the division site because GFP-Sid2p (101-510) is capable of localizing to the SPBs but unlike GFP-Sid2p (101-607) does not localize to the cell division site (M.-C. Hou and D. McCollum, unpublished observations). However, GFP-Sid2p (509-607) alone does not localize to the cell division site. The significance of microtubule association in Sid2p function is not known. Intriguingly, previous results showed that Sid2p localized poorly to the division site in the absence of microtubules (44), suggesting the possibility that Sid2p could be delivered to the division site and/or SPB along microtubules.
Interestingly, all Sid2p mutants lacking amino acids 1 to 100 accumulated in the nucleus. This suggests that this region has a nuclear export or cytoplasmic retention signal and that full-length Sid2p shuttles between the nucleus and cytoplasm. Because GFP-Sid2p (201-510) and GFP-Sid2p (509-607) do not localize to the nucleus but GFP-Sid2p (101-207) does, the Sid2p nuclear localization signal probably is in residues 101 to 207. Since deletion of amino acids 1 to 100 resulted in poor rescue of the sid2-250 temperature-sensitive mutants, nuclear shuttling may be important for Sid2p function. The SIN has been implicated in regulating the Cdc14p-like phosphatase Clp1p/Flp1p, which is in the nucleolus for much of the cell cycle (8, 48). Thus, regulation of Clp1p/Flp1p is one possible function for nuclear Sid2p. However, precise mutations in putative nuclear import and export sequences will be required to settle this issue.
Sid2p family kinases may utilize conserved mechanisms to regulate their activity.
Our results show that Sid2p utilizes multiple modes of regulation to control its activity. We suspect that many of the regulatory mechanisms that govern Sid2p function may be conserved in other members of this family of kinases. We and others have shown that the N terminus of the Sid2p family kinases in both budding (Cbk1p) and fission (Sid2p and Orb6p) yeasts is required for association with Mob1p-family proteins (Mob1p and Mob2p) (Table 3) (34). This family of kinases is regulated by phosphorylation at conserved sites in the activation loop and the hydrophobic motif. It will be interesting to see if there is an interrelationship between phosphorylation and Mob1p binding, as work from our lab and others suggests. Additionally, Sid2p is the first member of this family of kinases reported to be able to self-associate. Whether self-association plays a role in regulation of other kinases in this family remains to be investigated.
Acknowledgments
We are grateful to Kathy Gould and Jennifer Morrell for strains, advice, and sharing results prior to publication and to Viesturs Simanis for strains.
This work was supported by National Institutes of Health (NIH) grant GM58406 to D.M.
REFERENCES
- 1.Bakkenist, C. J., and M. B. Kastan. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421:499-506. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, M. K., D. McCollum, and K. L. Gould. 1997. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283:494-506. [DOI] [PubMed] [Google Scholar]
- 3.Bardin, A. J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2:815-826. [DOI] [PubMed] [Google Scholar]
- 4.Bidlingmaier, S., E. L. Weiss, C. Seidel, D. G. Drubin, and M. Snyder. 2001. The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:2449-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, L., and K. L. Gould. 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA 97:5249-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colman-Lerner, A., T. E. Chin, and R. Brent. 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107:739-750. [DOI] [PubMed] [Google Scholar]
- 7.Craven, R. A., D. J. Griffiths, K. S. Sheldrick, R. E. Randall, I. M. Hagan, and A. M. Carr. 1998. Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene 221:59-68. [DOI] [PubMed] [Google Scholar]
- 8.Cueille, N., E. Salimova, V. Esteban, M. Blanco, S. Moreno, A. Bueno, and V. Simanis. 2001. Flp1, a fission yeast orthologue of the S. cerevisiae CDC14 gene, is not required for cyclin degradation or rum1p stabilisation at the end of mitosis. J. Cell Sci. 114:2649-2664. [DOI] [PubMed] [Google Scholar]
- 9.Ducommun, B., P. Brambilla, M. A. Felix, B. R. Franza, Jr., E. Karsenti, and G. Draetta. 1991. Cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 10:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fankhauser, C., and V. Simanis. 1994. The Cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 13:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geng, W., B. He, M. Wang, and P. N. Adler. 2000. The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 13.Glotzer, M. 2001. Animal cell cytokinesis. Annu. Rev. Cell Dev. Biol. 17:351-386. [DOI] [PubMed] [Google Scholar]
- 14.Gould, K. L., S. Moreno, D. J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guertin, D. A., L. Chang, F. Irshad, K. L. Gould, and D. McCollum. 2000. The role of the Sid1p kinase and Cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 19:1803-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guertin, D. A., S. Trautmann, and D. McCollum. 2002. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 66:155-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 18.Hou, M. C., J. Salek, and D. McCollum. 2000. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10:619-622. [DOI] [PubMed] [Google Scholar]
- 19.Hou, M. C., D. J. Wiley, F. Verde, and D. McCollum. 2003. Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116:125-135. [DOI] [PubMed] [Google Scholar]
- 20.Jallepalli, P. V., and C. Lengauer. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer 1:109-117. [DOI] [PubMed] [Google Scholar]
- 21.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Justice, R. W., O. Zilian, D. F. Woods, M. Noll, and P. J. Bryant. 1995. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9:534-546. [DOI] [PubMed] [Google Scholar]
- 23.Keeney, J. B., and J. D. Boeke. 1994. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komarnitsky, S. I., Y. C. Chiang, F. C. Luca, J. Chen, J. H. Toyn, M. Winey, L. H. Johnston, and C. L. Denis. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol. Cell. Biol. 18:2100-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krapp, A., S. Schmidt, E. Cano, and V. Simanis. 2001. S. pombe Cdc11p, together with Sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11:1559-1568. [DOI] [PubMed] [Google Scholar]
- 26.Leupold, U. 1970. Genetical methods for Schizosaccharomyces pombe. Methods Cell Physiol. 4:169-177. [Google Scholar]
- 27.Luca, F. C., and M. Winey. 1998. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9:29-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mah, A. S., J. Jang, and R. J. Deshaies. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98:7325-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maundrell, K. 1993. Thiamine-repressible vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 30.McCollum, D., and K. L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11:89-95. [DOI] [PubMed] [Google Scholar]
- 31.Millward, T., P. Cron, and B. A. Hemmings. 1995. Molecular cloning and characterization of a conserved nuclear serine(threonine) protein kinase. Proc. Natl. Acad. Sci. USA 92:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Millward, T. A., D. Hess, and B. A. Hemmings. 1999. Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274:33847-33850. [DOI] [PubMed] [Google Scholar]
- 33.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast, Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 34.Nelson, B., C. Kurischko, J. Horecka, M. Mody, P. Nair, N. Pratt, A. Zougman, L. D. B. McBroom, T. R. Hughes, C. Boone, and F. C. Luca. 2003. RAM: a conserved signaling that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell 14:3782-3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigg, E. A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21-32. [DOI] [PubMed] [Google Scholar]
- 36.Nishiyama, Y., T. Hirota, T. Morisaki, T. Hara, T. Marumoto, S. Iida, K. Makino, H. Yamamoto, T. Hiraoka, N. Kitamura, and H. Saya. 1999. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 459:159-165. [DOI] [PubMed] [Google Scholar]
- 37.Parrini, M. C., M. Lei, S. C. Harrison, and B. J. Mayer. 2002. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol. Cell 9:73-83. [DOI] [PubMed] [Google Scholar]
- 38.Prentice, H. L. 1992. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 20:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Racki, W. J., A. M. Becam, F. Nasr, and C. J. Herbert. 2000. Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19:4524-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salimova, E., M. Sohrmann, N. Fournier, and V. Simanis. 2000. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 113:1695-1704. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schlessinger, J. 2002. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110:669-672. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, S., M. Sohrmann, K. Hofmann, A. Woollard, and V. Simanis. 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 11:1519-1534. [DOI] [PubMed] [Google Scholar]
- 44.Sparks, C. A., M. Morphew, and D. McCollum. 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146:777-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamaskovic, R., S. J. Bichsel, and B. A. Hemmings. 2003. NDR family of AGC kinases—essential regulators of the cell cycle and morphogenesis. FEBS Lett. 546:73-80. [DOI] [PubMed] [Google Scholar]
- 46.Tomlin, G. C., J. L. Morrell, and K. L. Gould. 2002. The spindle pole body protein Cdc11p links Sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13:1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toyn, J. H., and L. H. Johnston. 1994. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 13:1103-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trautmann, S., B. A. Wolfe, P. Jorgensen, M. Tyers, K. L. Gould, and D. McCollum. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931-940. [DOI] [PubMed] [Google Scholar]
- 49.Turenchalk, G. S., M. A. St John, W. Tao, and T. Xu. 1999. The role of lats in cell cycle regulation and tumorigenesis. Biochim. Biophys. Acta 1424:M9-M16. [DOI] [PubMed] [Google Scholar]
- 50.Verde, F., D. J. Wiley, and P. Nurse. 1998. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95:7526-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Visintin, R., and A. Amon. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2. Mol. Biol. Cell 12:2961-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss, E. L., C. Kurischko, C. Zhang, K. Shokat, D. G. Drubin, and F. C. Luca. 2002. The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158:885-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia, H., H. Qi, Y. Li, J. Pei, J. Barton, M. Blackstad, T. Xu, and W. Tao. 2002. LATS1 tumor suppressor regulates G2/M transition and apoptosis. Oncogene 21:1233-1241. [DOI] [PubMed] [Google Scholar]
- 54.Xu, T., W. Wang, S. Zhang, R. A. Stewart, and W. Yu. 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121:1053-1063. [DOI] [PubMed] [Google Scholar]
- 55.Yang, X., D. M. Li, W. Chen, and T. Xu. 2001. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene 20:6516-6523. [DOI] [PubMed] [Google Scholar]
- 56.Zallen, J. A., E. L. Peckol, D. M. Tobin, and C. I. Bargmann. 2000. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell 11:3177-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]