Abstract
9G4+ IgG Abs expand in systemic lupus erythematosus (SLE) in a disease-specific fashion and react with different lupus Ags including B cell Ags and apoptotic cells. Their shared use of VH4-34 represents a unique system to understand the molecular basis of lupus autoreactivity. In this study, a large panel of recombinant 9G4+ mAbs from single naive and memory cells was generated and tested against B cells, apoptotic cells, and other Ags. Mutagenesis eliminated the framework-1 hydrophobic patch (HP) responsible for the 9G4 idiotype. The expression of the HP in unselected VH4-34 cells was assessed by deep sequencing. We found that 9G4 Abs recognize several Ags following two distinct structural patterns. B cell binding is dependent on the HP, whereas anti-nuclear Abs, apoptotic cells, and dsDNA binding are HP independent and correlate with positively charged H chain third CDR. The majority of mutated VH4-34 memory cells retain the HP, thereby suggesting selection by Ags that require this germline structure. Our findings show that the germline-encoded HP is compulsory for the anti–B cell reactivity largely associated with 9G4 Abs in SLE but is not required for reactivity against apoptotic cells, dsDNA, chromatin, anti-nuclear Abs, or cardiolipin. Given that the lupus memory compartment contains a majority of HP+ VH4-34 cells but decreased B cell reactivity, additional HP-dependent Ags must participate in the selection of this compartment. This study represents the first analysis, to our knowledge, of VH-restricted autoreactive B cells specifically expanded in SLE and provides the foundation to understand the antigenic forces at play in this disease.
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease in which faulty B cell tolerance promotes multiple autoantibodies including some like anti-ds DNA, anti-Smith, and anti-nucleosome Abs with high disease specificity (1, 2). Elucidating the molecular basis of SLE-specific autoantibodies in the naive and memory compartments is important to understand fundamental aspects of the disease pathogenesis including the relative role of different Ags in the initial breakdown of tolerance and the subsequent expansion of pathogenic B cells. Yet, probing studies of these questions are hampered by challenges in the identification of disease-specific autoreactive B cells. Consequently, extant studies have pursued only analyses of unselected B cells and have been largely restricted to assessing general anti-nuclear Abs (ANA) and dsDNA binding (3–5). To circumvent these limitations, we have resorted to the study of autoantibodies that express the 9G4 idiotype (9G4 Abs), for which sensitivity (45–70%) and specificity (>95%) for SLE is similar to that of anti-dsDNA Abs (6). The relevance of 9G4 Abs is further illustrated by their correlation with overall disease activity and several clinical manifestations including lupus nephritis (6–12). These features, the shared expression of a single VH gene (VH4-34), and the ability to purify 9G4+ B cells with a highly specific anti-idiotype Ab render the study of 9G4 Abs a powerful experimental system for the study of SLE-specific autoreactivity. The 9G4 system is also highly suitable owing to the high degree of intrinsic autoreactivity engrained in the germline sequence of the VH4-34 H chain expressed by 9G4 Abs and the inability of SLE patients to appropriately censor autoreactive 9G4 cells (12, 13). Indeed, most unmutated IgM 9G4 Abs studied recognize N-acetyl-lactosamine (LN) glycans expressed by Ii blood group Ags. Strikingly, anti-Ii 9G4 Abs comprise virtually 100% of the powerful IgM cold-agglutinin responses triggered by acute Mycoplasma pneumoniae and EBV infections. Anti-i reactivity also underlies the ability of IgM 9G4 to bind B cells (14–20). The canonical anti-i B cell binding (BCB) of 9G4 Abs is further documented by presence of this reactivity IgM 9G4 Abs derived from fetal spleen B cells representing the innate repertoire without previous antigenic selection. Of great relevance for our work, both the expression of the 9G4 idiotype and the 9G4 canonical LN autoreactivity are determined by a framework 1 (FR1) hydrophobic patch (HP) formed by residues Q6W7 and A23V24Y25of the VH4-34 germline sequence (13, 21, 22).
In healthy subjects, effective tolerance ensures that 9G4 responses are restricted to acute responses and do not persist in the long-lived IgG memory and plasma cell compartments. In contrast, we have shown that in SLE, 9G4 B cells are substantially expanded in the IgG memory compartment, and 9G4 Abs contribute disproportionally to circulating IgG levels owing to defective germinal center (GC) censoring that is unique to SLE among the autoimmune diseases (12). Yet, the Ags responsible for the selection of 9G4 IgG Abs in SLE GC remain poorly understood. Nonetheless, important clues can be gleaned from extensive serological analyses performed by our group. Thus, serum 9G4 IgG Abs bind B cells both in vitro and in vivo by reacting with LN chains on a B220 glycoform preferentially expressed on naive B cells (9). In vivo, 9G4 BCB is prominent in active SLE and correlates with naive B cell lymphopenia possibly due to a direct lytic activity of these Abs (9, 23). Yet, 9G4 Abs may also react with ssDNA, dsDNA (11, 24–26), as well as apoptotic cells as shown own by our studies of SLE sera (27–29) and by chronic lymphocytic leukemia studies (30). The latter reactivity is of great interest because apoptotic cells accumulate in SLE GC (31) and may induce anti-apoptotic cell Abs (APCA) with pathogenic functions (32–40).
Of course, serological studies cannot discriminate whether BCB and APCA binding is due to cross-reactivity or polyreactivity against both cell types or is instead mediated by different types of 9G4 Abs. To clarify these questions and to further characterize the contribution of 9G4 Abs to lupus autoreactivity, we analyzed a large panel of recombinant 9G4 mAbs. Our results demonstrate high autoreactivity of 9G4 Abs against several lupus Ags of pathogenic significance that can be segregated into HP-dependent and HP-independent categories. Combined with deep sequencing analysis of the relative frequency of VH4-34 B cells that express the HP, the aggregate of our findings indicate that both types of autoantigens participate in the selection of these autoreactive B cells into the SLE memory compartment with apoptotic cells contributing a major source of HP-independent Ags. Contrary to previous studies of the global human B cell repertoire (41), L chains play a major modulatory role in VH4-34-determined autoreactivity but rarely eliminate it. This study represents the first systematic analysis, to our knowledge, of a VH-restricted autoreactive B cell population specifically expanded in SLE and provides the basis for a detailed understanding of the antigenic forces at play in this disease.
Materials and Methods
Patient selection
Two SLE patients who fulfilled the American College of Rheumatology revised criteria for the classification of this disease were systematically studied. One patient displayed very high titers of serum 9G4 Abs as well as ex vivo and in vitro binding of serum 9G4 Abs to B cells and apoptotic cells. In contrast, the other patient did express low levels of serum 9G4 Abs and the corresponding B cell and apoptotic cell reactivities. Isolated 9G4 mAbs derived from additional lupus patients were also analyzed as indicated. Two sex- and age-matched healthy control subjects (HC) were also included in the study of 9G4 mAbs. Five additional SLE patients were recruited for deep sequencing analysis of the VH4-34 repertoire. All patients were recruited from the Lupus Clinic of the University of Rochester Medical Center.
Sample procurement and cell isolation
Detailed written informed consent was obtained from all patients and healthy donors in accordance with protocols approved by the Human Subjects Institutional Review Board of the University of Rochester Medical Center. PBMCs from HC and from two patients with SLE who fulfilled at least 4 out of 11 American College of Rheumatology criteria for the classification of SLE were isolated by VACUTAINER CPT tubes (BD Biosciences, Bedford, MA) according to the manufacturer’s instructions. Tonsil samples were obtained from individuals undergoing routine tonsillectomy. Mononuclear cells were isolated from tonsil using Ficoll-Hypaque density gradient centrifugation (GE Healthcare, Piscataway, NJ). Naive B cells were isolated from tonsil mononuclear cells by negative magnetic bead purification using the Human Naive B cell Isolation Kit (Miltenyi Biotec, Auburn, CA).
Flow cytometry
The following anti-human Abs were used in this study using a three-laser LSR2 instrument: anti-CD19 allophycocyanin-Cy7, anti-CD24 FITC, anti-IgD FITC, anti-IgD PE, anti-IgM FITC, anti-IgG PE (BD Biosciences), anti-CD27 APC, anti-CD38 PE-Cy7 (eBioscience, San Diego, CA), and anti-IgA PE (Southern Biotechnology Associates, Birmingham, AL). Anti-rat idiotypic 9G4 (kindly provided by Prof. F. Stevenson, Tenovus Laboratories, Southampton, U.K.) was fluorescently labeled using the Pacific Blue Protein Labeling Kit (Molecular Probes, Carlsbad, CA). AnnexinV FITC (Southern Biotechnology Associates) and 7-aminoactinomycin D (7-AAD) were used to stain apoptotic and dead cells, respectively. Purified mononuclear cells were stained at 4°C in PBS. Flow cytometric analysis was performed with FlowJo software from TreeStar.
Single-cell sorting
Single B cells were sorted on an FACSAria (BD Biosciences) into 96-well PCR plates (Bio-Rad, Hercules, CA) containing 4 μl/well 0.5× PBS containing 10 mM DTT (Invitrogen, Carlsbad, CA), 8 U RNAsin (Promega, Madison, WI), and 0.4 U 5′-3′ Prime RNAse Inhibitor (Eppendorf). Plates were sealed with Microseal F Film (Bio-Rad) and immediately frozen on dry ice before storage at −80°C until used for RT-PCR. The following populations were sorted: naive cells (CD19+IgD+CD27−CD24+CD38+) and isotype-switched memory (CD19+IgD−CD27+).
Single-cell RT-PCR
cDNA was synthesized in a total volume of 20 μl/well in the original 96-well sorting plate using the iScript cDNA Synthesis Kit (Bio-Rad). IgH, Igλ, and Igκ V gene transcripts were amplified independently by nested PCR using 4 μl cDNA or unpurified PCR1 product as template, as appropriate. All PCR reactions were performed in 96-well plates in a total volume of 40 μl/well containing 50 nM each primer, 250 μM each dNTP (Fermentas, Glen Burnie, MD), and 0.25 U HotStar Taq DNA polymerase (Qiagen, Valencia, CA). Primers and cycling conditions for PCR amplification of Ig L chains have been described elsewhere (42). Primer sequences for PCR amplification of Ig H chains were as follows: H chain forward: VH1 exterior, 5′-ATGGACTGCACCTGGAGGAT-3′; VH1 interior, 5′-CCTCTTCTTGGTGGCAGCAG′-3′; VH2 exterior, 5′-CATACTTTGTTCCACGCTCC-3′; VH2 interior, 5′-TCCACGCTCCTGCTGCTGAC-3′; VH3 exterior, 5′-ATGGAGTTTGGGCTGAGCTG; VH3 interior, 5′-GGGGCTGAGCTGGGTTTTCC-3′; VH4 exterior, 5′-GAAACACCTGTGGTTCTTCC′-3′; VH4 interior, 5′-CTGGTGGCAGCTCCCAGATG-3′; VH5 exterior, 5′-ATGGGGTCAACCGCCATCCT-3′; VH5 interior, 5′-CCCTCCTCCTGGCTGTTCTC-3′; VH6 exterior, 5′-CTCCTTCCTCATCTTCCTGC-3′; VH6 interior, 5′-CCTCATCTTCCTGCCCGTGC; H chain reverse: IgA exterior, 5′-GTGTAGTGCTTCACGTGGCA-3′; IgA interior, 5′-GGCATGTCACGGACTTGCCG-3′; IgG exterior, 5′-GAGTCCTGAGGACTGTAGGA-3′; IgG interior, 5′-GCGCCTGAGTTCCACGACAC-3′; IgM exterior, 5′-GTGATGGAGTCGGGAAGGAA-3′; IgM interior, 5′-CGACGGGGAATTCTCACAGG-3′; and sequencing reverse: SeqCK, 5′-GTCCTTGCTGTCCTGCT-3′; and SeqCL, 5′-GGYGGGAACAGAGTG-3′.
PCR products were visualized on a 1% agarose gel containing Sybersafe (Invitrogen). Bands were excised and purified using the Qiaquick Gel Extraction Kit (Qiagen). All samples were stored at −20°C.
Ig gene sequencing and analysis
Purified PCR2 products were sequenced at Genewiz Sequences were analyzed by IgBLAST comparison with GenBank (www.ncbi.nlm.nih.gov/igblast) to identify germline V(D)J gene segments with highest identity. Sequences analyzed by IgBLAST were aligned with the published VH4-34 germline sequence using Megalign program from Seqman (Laser Gene Software). Ig CDR3 length and charge was determined by Joinsolver (http://joinsolver.niams.nih.gov).
Expression vector cloning and Ab production
Expression vector cloning, transfection of human embryonic kidney 293T cells (American Type Culture Collection, Manassas, VA), and mAb expression and purification were performed as described elsewhere (42). Expression vectors were provided by Dr. Eric Meffre (Yale University).
ELISA
Quantitative IgG and 9G4 ELISAs were performed on Polysorp 96-well plates (Nunc, Rochester, NY) as previously described (21, 41).
ANA assays
QUANTALite ANA and dsDNA ELISAs (INOVA Diagnostics, San Diego, CA) were performed according to the manufacturer’s directions. Positive binding was defined according to instructions for the low positive control as an OD of more than twice the negative control. Abs were only considered reactive if positive reactivity was confirmed in at least two independent experiments. ANA was also measured using Hep-2 substrates by immunofluorescence. Hep-2–coated slides (Nova lite HEp-2, catalog number 708101; INOVA Diagnostics) were incubated in a moist chamber at ambient temperature with 20 μl purified Abs at 30 μg/ml for 30 min, washed in PBS, and incubated for 30 min with FITC-labeled goat anti-human IgG (20 μl). ANA-negative and ANA-positive control sera were included in all experiments. Samples were examined on a Fluorescence microscope (Fluorescence illumination system, Nikon Eclipse 501 Camera, Diagnostic Instruments X-cite 120; Nikon) using IPlab 4.0 software. Positive staining was determined by comparison with the controls at equal exposure times.
Antichromatin ELISA
QUANTALite Chromatin ELISA (INOVA Diagnostics) was performed according to the manufacturer’s directions. Positive binding was defined according to instructions for the low positive control as an OD of more than twice the negative control. Abs were only considered reactive if positive reactivity was confirmed in at least two independent experiments.
Anticardiolipin ELISA
Anticardiolipin (ACL) ELISA (ALPCO Diagnostics) was performed according to the manufacturer’s directions. Any valve >11 U/ml was defined as strong positive (+++); 11 U/ml 1:4 (++); and 11 U/ml 1:16 (+). Abs were only considered reactive if positive reactivity was confirmed in at least two independent experiments.
BCB assay
Naive B cells were isolated from human tonsils. A total of 100,000 cells was resuspended in 50 μl mAb at 100 μg/ml and incubated at 4°C for 30 min. Cells were washed three times in cold PBS and subsequently stained with Abs to CD19, CD27, IgD, and IgG, as well as with 7-AAD. Flow cytometric analysis was performed on an LSRII (BD Biosciences) as previously described (21). Dead cells and doublets were excluded. The median fluorescence intensity (MFI) of mAb (IgG) was determined on the naive B cell population.
Apoptotic cell binding assay
The CD45-negative Jurkat cell line (J45.1; American Type Culture Collection) was cultured at 1 × 106 cells/ml with 20 μg/ml camptothecin for 18 h to induce apoptosis. A total of 100,000 cells was resuspended in 50 μl mAb at 100 μg/ml and incubated at 4°C for 30 min. Cells were washed three times in binding buffer (0.01 M HEPES [pH 7.4], 0.14 M NaCl, and 2.5 mM CaCl2) and subsequently stained with anti-IgG, anti–Annexin V, and 7-AAD. Flow cytometric analysis was performed on an LSRII (BD Biosciences). The MFI of mAb (IgG) was measured on J45.1 cells.
Autoantigen microarrays
The three mAbs at 10 μg/ml were incubated with autoantigen array (Microarray Core, University of Texas Southwestern Medical Center, Dallas, TX) and the autoantibodies binding to the Ags on the array detected with laser wavelengths 532 nm (Cy3-labeled anti-IgG) to generate .tif images. Genepix Pro 6.0 software was used to analyze the image and generate the .GPR file (GenePix Results format). Net fluorescence intensities were defined as the spot minus background fluorescence intensity; data obtained from duplicate spots were averaged. Signal-to-noise ratio (SNR) was used as a quantitative measure of the ability to resolve true signal from background noise. A higher SNR indicates higher signal over background noise. An SNR ≥3 was considered true signal from background noise. A heat map was generated using Cluster and Treeview software (http://rana.lbl.gov/EisenSoftware.htm).
Glycan microarray
We used the Consortium for Functional Glycomics (CFG) mammalian glycan microarrays version 5.1 for the analysis of samples at the Protein-Glycan Interaction Core (Emory University School of Medicine, Atlanta, GA) following standard CFG procedures (http://www.functionalglycomics.org). Briefly, the 75G12 Ab was incubated on the glycan microarray slide at 150 μg/ml in binding buffer for 1 h at room temperature. Then 9G4 at 20 μg/ml was added to the microarray, followed by detection with Alexa 546–labeled anti-rat IgG (Invitrogen). Image acquisition was performed with ScanArrayExpress (PerkinElmer) and data analysis was performed with Imagene software (BioDiscovery).
Deep sequencing of VH4-34 H chains
Five thousand cells were sorted from CD19+IgD−CD27+ cells, and total cellular RNA was isolated using the RNeasy Mini Kit (Qiagen) by following the manufacturer's protocol. After DNase I treatment (Life Technologies, Carlsbad, CA), 1 μg RNA was subjected to reverse transcription using the iScript RT Kit (Bio-Rad). Aliquots of the resulting single-stranded cDNA products were included with 50 nM VH4-specific primers and 50 nM Cg-specific primers in a 20-μl nested PCR reaction using Platinum PCR Supermix (Life Technologies) and amplified using a Bio-Rad C1000 Thermal Cycler (Bio-Rad) with the following conditions: PCR1: after an initial step of 95°C for 5 min, 20 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, ending with a final extension step of 72°C for 5 min; PCR2: after an initial step of 95°C for 5 min, 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, ending with a final extension step of 72°C for 5 min. Specific primer sequences used are as follows: PCR1, VH4: 5′-CACCTGTGGTTCTTCCTCCT-3′; Cg: 5′-GAGTCCTGAGGACTGTAGGA-3′; PCR2, VH4: 5′-CTGGTGGCAGCTCCCAGATG-3′; and Cg: 5′-GCGCCTGAGTTCCACGACAC-3′. Lib-L 454 fusion tags (forward: 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′ and reverse: 5′-CCTATCCCCTGTGTGCCTTGGCAGTCTCAG-3′) were included in PCR2 primer sequences along with one of four MID (Multiplex Identifier) tags for bar-coding sequences (MID1: TCTCTATGCG; MID2: ACGAGTGCGT; MID3: CGTGTCTCTA; and MID4: TAGTATCAGC).
After amplification, PCR products were purified with Agentcourt Ampure XP beads (Beckman Coulter, Indianapolis, IN) according to the manufacturer’s protocol, quantified, and pooled for multiplex sequencing. Emulsion-based clonal amplification, breaking, and 454 sequencing were carried out according to manufacturer’s protocol using a 454 GS Junior System (Roche/454 Life Sciences, Branford, CT).
Resulting sequences were filtered for size by excluding sequences <300 and >600 bp. Sequences were then aligned with iHMMune-align software and IMGT.org/HighVquest. Aligned sequences were compared, and unmatched, poor, and nonfunctional results were filtered. The integrity of the 9G4 HP was assessed based on the conservation of the VH4-34 FR1 germline sequence previously identified as responsible for this structure (Fig. 3).
FIGURE 3.
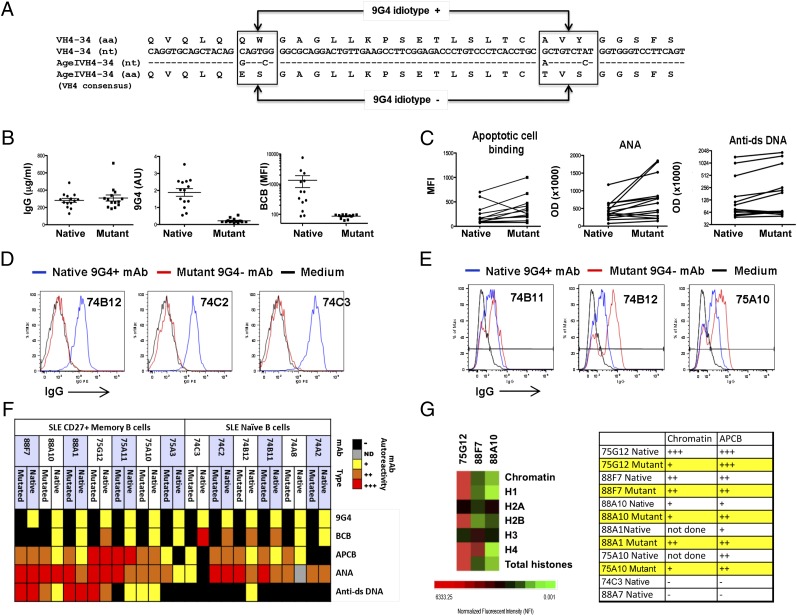
HP-dependent and -independent 9G4 autoreactivity. (A) All functional VH4 family members are represented. Differences between VH4-34 and the VH4 consensus sequence accounting for the structure of the 9G4 HP are boxed. The AgeIVH4-34M primer mutates four bases in the 5′ end of VH4-34, leading to amino acid changes at positions 6, 7, 23, and 25 that generate a consensus VH4 sequence devoid of the 9G4 HP. (B) A total of 14 native and 14 mutant mAbs quantified by IgG ELISA and tested for binding to 9G4. Binding to 9G4 was normalized to IgG concentration and is expressed in arbitrary units (AU). Mutants lost all reactivity to 9G4 (p < 0.0001, paired t test). BCB was lost on all mutant mAbs tested (p = 0.0005, paired t test). (C) Average APCB increased with mutation (p < 0.02, paired t test). APCB increased in 11 out of 14 mutants tested, 1 decreased, and 2 negative binders remained negative. Average reactivity to nuclear Ags increased (p < 0.02, paired t test), with 12 out of 14 mutants increasing in reactivity and 2 decreasing. Reactivity to dsDNA increased with mutation (p < 0.05, paired t test). All eight negative binders remained negative, whereas three weak and three strong binders increased in reactivity. (D) BCB of three representative mAbs and mutants is shown. (E) Late APCB of three representative mAbs and mutants. (F) Heat map of Ag binding by 14 9G4+ mAbs and corresponding mutants. −, black; +, weak, yellow; ++, orange, moderate; and +++, red, strong. (G) Ag microarray showing normalized fluorescent intensity of three mAbs to chromatin and histones. The scale is shown at the bottom. Reactivity of APCB corresponds to chromatin binding by ELISA as shown in the table in the right panel.
Statistical analysis
Statistical analyses were performed using one of the following: paired t test, unpaired t test, Pearson correlation, Spearman correlation, Fisher exact test, χ2 test, or one-way ANOVA with post hoc Tukey test. Differences were considered significant at p < 0.05. Error bars denote SEM.
Results
Autoreactivity of SLE 9G4 Abs
In addition to the canonical VH4-34–encoded anti-i reactivity that determines binding to B cells (43), SLE serum 9G4 Abs can also react with other lupus autoantigens including dsDNA (6) and apoptotic cells (anti-apoptotic cell binding [APCB]), which represent a rich source of pathogenic lupus autoantigens (38, 43). Our initial recognition of 9G4 APCB in SLE (9, 27, 28) has been recently consolidated by extensive serological analysis in our laboratory showing that such autoreactivity is present in 60% of unselected SLE patients, is present in the vast majority of patients with elevated serum 9G4 Ab titers (80% of 9G4+), and correlates with disease activity and active lupus nephritis (29). A summary of these data are provided in Supplemental Fig. 1. To better understand the structural underpinnings of the different types of 9G4 autoreactivity, we generated a panel of rIgG mAbs from SLE 9G4+ isotype-switched memory cells expressing either surface IgG or IgA (44). For comparison, single 9G4+ cells were also sorted from naive cells of SLE patients and HC. Finally, 9G4+ memory cells, scarcely represented in HC (12), were similarly analyzed. In all, we generated 98 unique 9G4 mAbs that were tested for autoreactivity against viable B cells (BCB) and apoptotic cells (APCB) and multiple other lupus Ags. In accord with other studies of human mAb autoreactivity, individual Abs were classified as polyreactive if they bound two or more of the antigenic targets tested (3). Both BCB and APCB binding assays are illustrated in Fig. 1, and the global reactivity of the full mAb panel is presented in detail in Fig. 2 and summarized in Table I, which also describes their genetic and molecular features. Ab sequences are provided in Supplemental Figs. 2 and 3 (GenBank accession numbers KF495432–KF495529; http://www.ncbi.nlm.nih.gov/Genbank).
FIGURE 1.
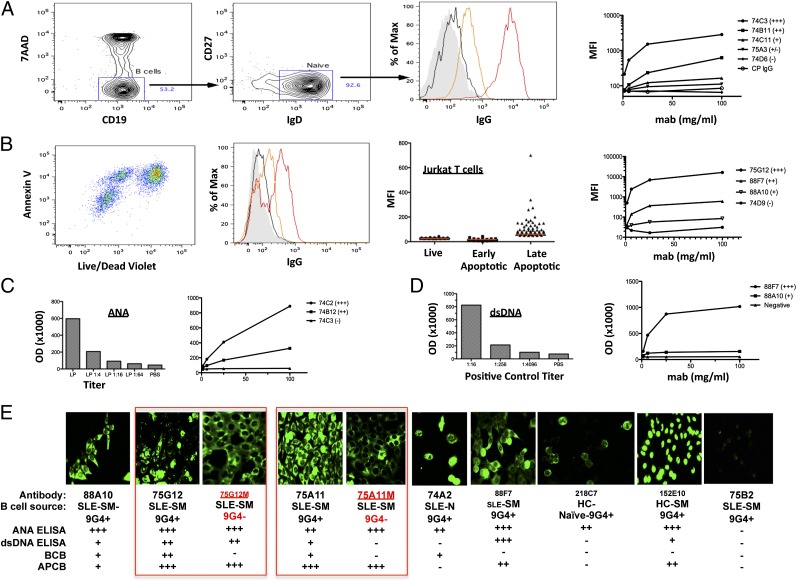
Autoreactivity assays. (A) BCB assay: 100,000 naive tonsil B cells incubated with mAb (100 μg/ml), followed by anti-IgG PE. Shown is the gating strategy for BCB by flow cytometry. MFI was quantified on 7-AAD− naive B cells. Examples are shown of negative (black), weak (orange), and strong (red) BCB compared with background staining (filled histogram). BCB titration curves are shown of five representative Abs and CP IgG (polyclonal purified human serum IgG). (B) APCB assay: camptothecin treatment of J45.1 cells provides live (AnnexinV−LD−), early apoptotic (AnnexinV+LDlow), and late apoptotic cells (AnnexinV+LDhi). Cells were stained with mAb (100 μg/ml) followed by anti-IgG PE, and different staining intensities are depicted as for BCB. In all cases, binding was mediated by reactivity with late apoptotic cells (middle panel). Titration binding curves are shown for representative Abs. (C) ANA ELISA: binding range was defined using a 4-fold limiting dilution of the supplied low positive control (+++, >LP; ++, >LP 1:4; +, >LP 1:16) as OD. Binding curves of three representative mAbs are shown. (D) dsDNA positive control (+++, >1:16; ++, >1:256; +, >1:4096). Binding curves of two representative mAbs and the supplied negative control are shown. (E) HEp-2 cell immunofluorescence assay showing diverse staining patterns. Cell source and autoreactivity results are indicated. Mutant Abs with disrupted HP (75G12M and 75A11M) are also shown side by side with the parental Abs 75G12 and 75A11. Original magnification ×40.
FIGURE 2.
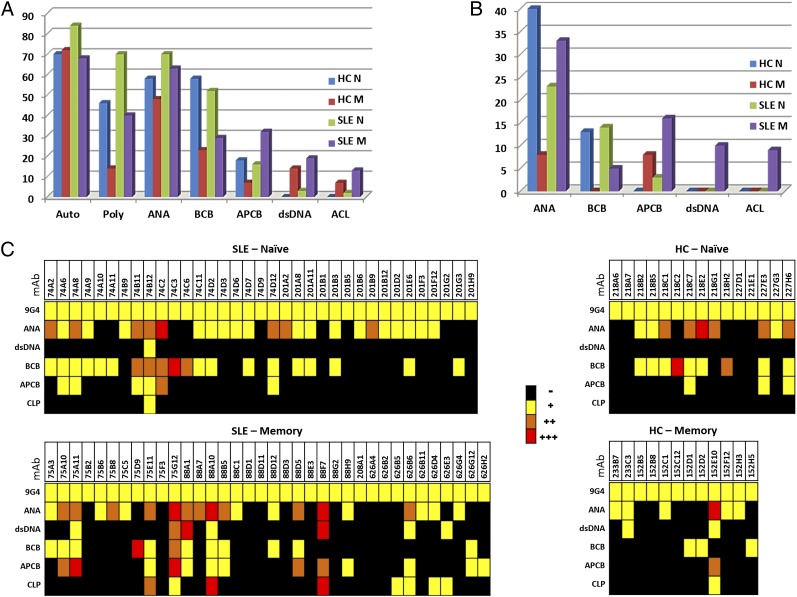
Autoreactivity of 9G4+ mAbs. 98 9G4+ mAbs were tested for Ag reactivity as illustrated in Fig. 1 and analyzed based on disease state (HC and SLE) and cell origin (naive [N] and isotype-switched memory [M]). (A) Frequency of individual autoreactivities (ANA, BCB, APCB, dsDNA, and ACL). Total autoreactivity (Auto) and polyautoreactivity (Poly) defined by binding to two or more Ags. (B) Percentage of strong binders (++/+++). (C) Heat map summary of Ag binding: −, black; +, weak, yellow; ++, orange, moderate; and +++, red, strong.
Table I. Autoreactivity and molecular characteristics of 98 9G4+ mAbs.
| 9G4 ELISA | ANA ELISA | dsDNA ELISA | B Cell Binding | Apoptotic Cardiolipin |
VH | JH | CDR3H Length | CDR3H Charge | VL | JL | CDR3L Length | CDR3L Charge | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Binding | ELISA | CH | |||||||||||||
| mAb | |||||||||||||||
| HC-Naive | |||||||||||||||
| 218A6 | + | − | − | − | − | − | M | VH4-34 | J5 | 10 | −1 | K3-20 | J2 | 10 | 0 |
| 218A7 | + | − | − | − | − | − | M | VH4-34 | J4 | 15 | 0 | K3-11 | J1 | 5 | 1 |
| 218B2 | + | + | − | + | − | − | M | VH4-34 | J6 | 18 | 1 | K3-20 | J5 | 11 | 0 |
| 218B5 | + | + | − | + | − | − | M | VH4-34 | J6 | 20 | −2 | K3-20 | J1 | 9 | 1 |
| 218C1 | + | ++ | − | + | − | − | M | VH4-34 | J4 | 20 | 0 | K3-15 | J1 | 8 | 0 |
| 218C2 | + | − | − | +++ | − | − | M | VH4-34 | J4 | 25 | −4 | L2-11 | J3 | 9 | 1 |
| 218C7 | + | ++ | − | + | + | − | M | VH4-34 | J4 | 23 | 0 | K1-5 | J2 | 11 | 0 |
| 218E2 | + | +++ | − | + | − | − | M | VH4-34 | J4 | 17 | 0 | L3-21 | J3 | 11 | −1 |
| 218G1 | + | ++ | − | − | − | − | M | VH4-34 | J4 | 14 | 2 | K1-5 | J2 | 11 | 1 |
| 218H2 | + | − | − | ++ | − | − | M | VH4-34 | J4 | 21 | 0 | L1-40 | J3 | 12 | −1 |
| 227D1 | + | − | − | − | − | − | M | VH4-34 | J4 | 14 | −1 | K3-20 | J1 | 9 | 0 |
| 227E1 | + | − | − | − | − | − | M | VH4-34 | J5 | 17 | −1 | L2-11 | J1 | 9 | 2 |
| 227E3 | + | ++ | − | + | + | − | M | VH4-34 | J3 | 15 | 1 | K3-11 | J2 | 8 | 1 |
| 227G3 | + | + | − | − | − | − | M | VH4-34 | J6 | 15 | 1 | K3-15 | J2 | 10 | 0 |
| 227H6 | + | ++ | − | + | + | − | M | VH4-34 | J6 | 15 | 0 | L2-14 | J3 | 11 | 0 |
| HC-SM | |||||||||||||||
| 233B7 | + | + | − | − | − | − | G | VH4-34 | J6 | 24 | −1 | K3-11 | J3 | 10 | 2 |
| 233C3 | + | + | + | − | − | − | G | VH4-34 | J6 | 16 | 2 | L2-23 | J3 | 10 | 2 |
| 152B5 | + | − | − | − | − | − | A1 | VH4-34 | J5 | 19 | 1 | K1-39 | J2 | 9 | 1 |
| 152B8 | + | − | − | − | − | − | G2 | VH4-34 | J5 | 16 | 0 | K4-1 | J1 | 9 | 0 |
| 152C1 | + | + | − | − | − | − | G1 | VH4-34 | J6 | 25 | 3 | K2-28 | J5 | 9 | 0 |
| 152C12 | + | − | − | − | − | − | G4 | VH4-34 | J3 | 15 | 2 | K3-20 | J1 | 9 | 1 |
| 152D1 | + | − | − | + | − | − | G1 | VH4-34 | J5 | 14 | 0 | K1-5 | J1 | 9 | 1 |
| 152D2 | + | − | − | + | − | − | A1 | VH4-34 | J5 | 10 | 2 | K1-39 | J2 | 9 | 0 |
| 152E10 | + | +++ | + | − | ++ | + | G1 | VH4-34 | J4 | 19 | 3 | K1-39 | J1 | 9 | 1 |
| 152F12 | + | + | − | − | − | − | G3 | VH4-34 | J5 | 14 | 2 | K1-17 | J1 | 9 | 0 |
| 152H3 | + | + | − | − | − | − | G1 | VH4-34 | J6 | 24 | 0 | K1-39 | J2 | 9 | 1 |
| 152H5 | + | − | − | + | − | − | G1 | VH4-34 | J5 | 14 | 0 | K1-5 | J1 | 9 | 1 |
| SLE-Naive | |||||||||||||||
| 74A2 | + | ++ | − | + | − | − | M | VH4-34 | J3 | 21 | 2 | K1D33 | J4 | 9 | −1 |
| 74A6 | + | + | − | + | + | − | M | VH4-34 | J4 | 11 | 1 | K3-15 | J1 | 9 | 1 |
| 74A8 | + | ++ | − | ND | + | − | M | VH4-34 | J6 | 30 | 3 | L1-40 | J3 | 12 | 0 |
| 74A9 | + | + | − | + | − | − | M | VH4-34 | J5 | 21 | 1 | K1-5 | J2 | 10 | 0 |
| 74A10 | + | − | − | + | − | − | M | VH4-34 | J5 | 17 | 0 | L2-23 | J3 | 14 | 0 |
| 74A11 | + | − | − | + | − | − | M | VH4-34 | J4 | 13 | 0 | K3-20 | J1 | 9 | 1 |
| 74B9 | + | + | − | − | − | − | M | VH4-34 | J4 | 15 | 2 | K3-20 | J1 | 9 | 0 |
| 74B11 | + | ++ | − | ++ | + | − | M | VH4-34 | J5 | 18 | 2 | K3-15 | J2 | 10 | 0 |
| 74B12 | + | ++ | + | ++ | + | + | M | VH4-34 | J4 | 24 | 3 | K3-11 | J4 | 10 | 1 |
| 74C2 | + | +++ | − | ++ | ++ | − | M | VH4-34 | J4 | 21 | 3 | L2-23 | J3 | 11 | 0 |
| 74C3 | + | − | − | +++ | − | − | M | VH4-34 | J4 | 13 | 0 | L1-40 | J1 | 12 | −1 |
| 74C6 | + | − | − | ++ | − | − | M | VH4-34 | J5 | 16 | 1 | L1-40 | J3 | 12 | −1 |
| 74C11 | + | + | − | + | − | − | M | VH4-34 | J3 | 21 | 2 | L1-40 | J3 | 11 | −1 |
| 74D2 | + | + | − | + | − | − | M | VH4-34 | J4 | 20 | 3 | K1-5 | J1 | 9 | 0 |
| 74D3 | + | + | − | − | − | − | M | VH4-34 | J5 | 18 | 0 | K4-1 | J5 | 9 | 0 |
| 74D6 | + | + | − | − | − | − | M | VH4-34 | J4 | 11 | 1 | K3-20 | J1 | 9 | 0 |
| 74D7 | + | + | − | + | − | − | M | VH4-34 | J2 | 15 | 0 | K1-5 | J2 | 10 | 0 |
| 74D9 | + | − | − | − | − | − | M | VH4-34 | J5 | 14 | −1 | K1-5 | J2 | 10 | 0 |
| 74D12 | + | ++ | − | + | + | − | M | VH4-34 | J4 | 14 | 2 | K1-39 | J2 | 9 | 1 |
| 201A2 | + | ++ | − | − | − | − | M | VH4-34 | J4 | 15 | 1 | K4-1 | J1 | 9 | 1 |
| 201A8 | + | + | − | + | − | − | M | VH4-34 | J5 | 15 | 1 | K1-39 | J2 | 9 | 0 |
| 201A11 | + | + | − | + | − | − | M | VH4-34 | J3 | 17 | −1 | K1-39 | J1 | 9 | 1 |
| 201B1 | + | − | − | − | − | − | M | VH4-34 | J3 | 14 | 2 | K1-39 | J1 | 8 | 1 |
| 201B3 | + | + | − | + | − | − | M | VH4-34 | J4 | 12 | 0 | K2-28 | J4 | 8 | 0 |
| 201B5 | + | − | − | − | − | − | M | VH4-34 | J6 | 16 | −3 | K4-1 | J2 | 9 | 1 |
| 201B6 | + | + | − | − | − | − | M | VH4-34 | J5 | 9 | 0 | K1-12 | J4 | 9 | 0 |
| 201B9 | + | ++ | − | − | − | − | M | VH4-34 | J6 | 11 | 0 | K1-33 | J2 | 9 | −1 |
| 201B12 | + | + | − | − | − | − | M | VH4-34 | J3 | 11 | −1 | K4-1 | J2 | 9 | 0 |
| 201D2 | + | + | − | − | − | − | M | VH4-34 | J3 | 13 | −1 | K1-39 | J2 | 9 | 1 |
| 201E6 | + | + | − | + | − | − | M | VH4-34 | J6 | 17 | 0 | K1-39 | J2 | 9 | 0 |
| 201F3 | + | + | − | − | − | − | M | VH4-34 | J1 | 18 | 4 | K1-15 | J2 | 10 | 0 |
| 201F12 | + | + | − | − | − | − | M | VH4-34 | J2 | 18 | 1 | K1-39 | J2 | 9 | 1 |
| 201G2 | + | − | − | − | − | − | M | VH4-34 | J5 | 14 | −1 | K1-39 | J2 | 9 | 0 |
| 201G3 | + | − | − | + | − | − | M | VH4-34 | J6 | 19 | 0 | K1-39 | J4 | 9 | 0 |
| 201H9 | + | − | − | − | − | − | M | VH4-34 | J6 | 21 | −2 | K3-11 | J5 | 5 | 1 |
| SLE-SM | |||||||||||||||
| 75A3 | + | + | − | + | − | − | A1 | VH4-34 | J4 | 22 | 2 | K3-15 | J2 | 10 | 0 |
| 75A10 | + | ++ | − | + | ++ | − | G1 | VH4-34 | J4 | 14 | 3 | K1-8 | J3 | 9 | 0 |
| 75A11 | + | ++ | + | + | +++ | − | G3 | VH4-34 | J4 | 17 | 2 | K3-20 | J1 | 9 | 2 |
| 75B2 | + | − | − | − | − | − | A1 | VH4-34 | J4 | 15 | 0 | L2-11 | J1 | 10 | 1 |
| 75B6 | + | + | − | − | − | − | A1 | VH4-34 | J4 | 12 | 4 | K1-33 | J2 | 9 | −1 |
| 75B8 | + | ++ | − | − | − | − | A1 | VH4-34 | J5 | 7 | 1 | K2-30 | J4 | 9 | 1 |
| 75C5 | + | + | − | − | − | − | A1 | VH4-34 | J4 | 13 | 3 | K4-1 | J3 | 9 | 0 |
| 75D9 | + | − | − | +++ | − | − | A1 | VH4-34 | J6 | 14 | 0 | K4-1 | J2 | 9 | 2 |
| 75E11 | + | ++ | − | + | + | ++ | A1 | VH4-34 | J6 | 21 | 2 | L1-51 | J1 | 11 | −1 |
| 75F3 | + | − | − | − | − | − | A1 | VH4-34 | J5 | 14 | 3 | L1-40 | J3 | 11 | −1 |
| 75G12 | + | +++ | ++ | ++ | +++ | + | G3 | VH4-34 | J4 | 17 | 2 | K3-20 | J1 | 9 | 2 |
| 88A1 | + | ++ | +++ | + | + | − | G1 | VH4-34 | J4 | 16 | 4 | L1-51 | J3 | 11 | −1 |
| 88A7 | + | ++ | − | − | − | − | A1 | VH4-34 | J3 | 25 | 3 | K3-15 | J1 | 10 | −1 |
| 88A10 | + | +++ | + | + | + | +++ | A1 | VH4-34 | J5 | 17 | 3 | K1-12 | J4 | 9 | 0 |
| 88B5 | + | ++ | − | + | + | − | A1 | VH4-34 | J4 | 17 | 2 | K3-20 | J1 | 9 | 0 |
| 88C1 | + | + | − | − | − | − | G1 | VH4-34 | J5 | 22 | 3 | K3-20 | J2 | 9 | −1 |
| 88D1 | + | − | − | − | − | − | G1 | VH4-34 | J6 | 20 | −2 | L1-40 | J3 | 9 | 1 |
| 88D11 | + | − | − | − | − | − | G1 | VH4-34 | J6 | 20 | −2 | L1-40 | J3 | 9 | 1 |
| 88D12 | + | + | − | + | − | − | A1 | VH4-34 | J6 | 17 | 1 | K3-20 | J4 | 9 | 1 |
| 88D3 | + | − | − | − | − | − | G1 | VH4-34 | J4 | 11 | 0 | K2-29 | J2 | 9 | 3 |
| 88D5 | + | ++ | − | + | ++ | − | A1 | VH4-34 | J1 | 16 | 4 | K1-39 | J2 | 9 | 0 |
| 88E3 | + | − | − | − | − | − | G1 | VH4-34 | J3 | 14 | −4 | L2-11 | J3 | 9 | 0 |
| 88F7 | + | +++ | +++ | − | ++ | +++ | G1 | VH4-34 | J5 | 19 | 4 | K3-20 | J2 | 10 | 0 |
| 88G2 | + | − | − | − | − | − | A1 | VH4-34 | J6 | 24 | 1 | K3-20 | J1 | 10 | −1 |
| 88H9 | + | + | − | − | + | − | G1 | VH4-34 | J3 | 9 | −1 | K3D-15 | J3 | 9 | 0 |
| 208A1 | + | − | − | − | − | − | G | VH4-34 | J5 | 21 | 0 | K3-11 | J1 | 9 | 1 |
| 626A4 | + | − | − | − | − | − | G1 | VH4-34 | J6 | 17 | 1 | K1-9 | J2 | 9 | −1 |
| 626B2 | + | − | − | − | − | − | G1 | VH4-34 | J6 | 20 | 1 | K1-39 | J4 | 9 | 1 |
| 626B5 | + | − | − | − | − | + | G1 | VH4-34 | J4 | 12 | 0 | K1-5 | J1 | 9 | 1 |
| 626B6 | + | ++ | + | − | + | + | G1 | VH4-34 | J4 | 17 | 3 | K1-39 | J1 | 9 | 0 |
| 626B11 | + | + | − | − | − | − | G1 | VH4-34 | J4 | 20 | −2 | L1-51 | J3 | 11 | −1 |
| 626D4 | + | + | − | − | − | + | G1 | VH4-34 | J4 | 13 | 0 | K2-28 | J5 | 9 | 0 |
| 626E3 | + | − | + | − | − | + | G1 | VH4-34 | J6 | 16 | −1 | K1-39 | J2 | 9 | 0 |
| 626G4 | + | + | − | − | − | − | G1 | VH4-34 | J6 | 14 | 0 | K3-20 | J4 | 8 | 0 |
| 626G12 | + | − | − | + | + | − | G1 | VH4-34 | J6 | 14 | −1 | K1-16 | J5 | 9 | 0 |
| 626H2 | + | − | − | − | + | − | G1 | VH4-34 | J6 | 20 | 2 | K3-20 | J2 | 9 | 0 |
9G4 single-cell mAbs are listed according to their cellular origin (SM, switched memory cells). The isotype expressed by the corresponding cells is noted. The germline H chain D and J segments as well as the L chain V and J segments used by each Ab are listed. The length and charge of the third hypervariable region of H and L chain (CDR3H and CDR3L) are shown. The intensity of autoreactivity to different Ags is also shown.
As shown in Fig. 2A, the large majority of 9G4 Abs (67–82%) from different cellular sources were autoreactive against at least one Ag, and 40–70% were positive in two or more autoreactivity assays (poly-autoreactive Abs). The notable exception was the lower level of strong autoreactivity and poly-autoreactivity (12%) of the scarce 9G4 memory cells present in healthy subjects. Overall, ANA autoreactivity was most common, as it was present in up to 70% of all SLE Abs and 57 and 69% of 9G4 Abs derived from healthy and SLE naive cells, respectively. Of note, this level of 9G4 ANA autoreactivity is substantially higher than previously detected in previous studies of total naive cells with similar numbers of mAbs (∼20% for healthy naive cells and 40% for SLE naive cells) by different groups and our own laboratory (3, 4, 44). Therefore, these results conclusively establish the high degree of autoreactivity against SLE-related Ags of Abs using VH4-34 H chains. Of significant interest, similar degrees of autoreactivity were found in memory cells from both patients despite differences in disease activity and levels of serum 9G4 Abs. This finding suggests that, as it has been shown for ANA and anti-DNA B cells, autoreactive memory 9G4 B cells can be at least temporarily silenced by late tolerance mechanisms (31, 45, 46).
Immunofluorescence analysis (Fig. 1E) revealed several ANA nuclear binding patterns commonly observed with lupus serum including homogeneous, speckled, and perinuclear. Cytoplasmic staining was frequently observed in addition to nuclear reactivity. In contrast, nucleolar reactivity was detected only in a subset of mutated Abs derived from SLE memory cells. Although more typically found in scleroderma, anti-nucleolar Abs have been reported in SLE associated with cytoplasmic reactivity (47) and allow stratification of this disease in African-American families (48).
Only 15% of Abs were autoreactive in the absence of ANA reactivity. Except for one SLE memory Ab with both BCB and APCB, in every case, their autoreactivity was directed only against B cells, and virtually all of these anti–B cell–restricted Abs (80%) originated from naive B cells, whether from HC or SLE. Overall, BCB was found in up to 60% of all 9G4 naive B cells in HC and SLE but in only 27% of SLE memory cells. APCB was present in all compartments, but its frequency was highest in SLE memory cells (31%) and lowest in healthy memory B cells. As is the case for murine and human polyclonal SLE Abs (48), 9G4 APCB was predominantly directed against Ags expressed in late apoptotic cells (Fig. 1B). When only strong autoreactivity (++/+++) was considered, APCB, dsDNA, and ACL binding was preferentially if not exclusively found in 9G4 SLE memory cells, which conversely, had decreased strong BCB (Fig. 2B, 2C). In contrast, 9G4 healthy memory cells expressed the lowest level of strong autoreactivity for ANA, BCB, and APCB and absent dsDNA and ACL (Fig. 2B).
Given that both B cells and apoptotic cells could express multiple Ags, we tested selected mAbs to further understand these reactivities. We had previously demonstrated through immunoprecipitation and enzymatic modification studies that LN-bearing B220 was the B cell target recognized by serum 9G4 IgG Abs. This specificity was directly confirmed by testing mAb 75G12 against a high-density glycan microarray containing 610 glycans (Supplemental Fig. 4).
The glycan array data has been deposited at: https://www.functionalglycomics.org/glycomics/publicdata/primaryscreen.jsp; the number that links to samples is: cfg_request_2712 Ignacio Sanz, and the specific primary screen numbers for each sample is: primscreen_5949-primscreen_5964.
Regarding APCB reactivity selected mAbs with strong APCB also recognized chromatin and individual histones as documented by glomerular proteome microarrays previously used by some of us (C.M. and Q.-Z.L.) with lupus serum Abs (49) (Fig. 3G). The autoantigen protein array data has been deposited at http://www.ncbi.nlm.nih.gov/geo/info/linking.html; accession numbers for each sample are GSM1224646, GSM1224647, and GSM1224648.
Interestingly, our unbiased single-cell analysis revealed similar frequencies of IgG and IgA cells in the SLE switched memory compartment (46 and 54%, respectively). No major differences in ANA or BCB were observed between the two isotypes. In contrast, strong APCB was clearly overrepresented in IgG Abs, which also contained all of the strong dsDNA binders, with only one IgA Ab displaying modest dsDNA binding (ANA/dsDNA ratio: 1.8 for IgG and 9.0 for IgA; BCB/APCB ratio: IgG 0.75; IgA 2.0). IgG3 Abs were also represented in our panel. Of significant interest, both IgG3 Abs found in SLE memory cells had extended autoreactivity against multiple Ags, including strong APCB and dsDNA, a finding consistent with the reported contribution of IgG3 Abs to lupus nephritis through the recognition of nucleosomal Ags expressed by apoptotic glomerular cells (50) and with the binding of 75G12 to chromatin and histones previously discussed (Fig. 3G). Remarkably, both IgG3 Abs were clonally related with a single amino acid difference in H chain second CDR and identical H chain third CDR (HCDR3) (75G12 and 75A11; Supplemental Fig. 2). The finding of two clonally related Abs out of 35 sorted memory cells (6%) suggests the presence of sizable clonal expansions in the SLE memory compartment.
As expected, all of the Abs generated from 9G4+ B cells also reacted positively with the rat anti-human mAb that defines the 9G4 idiotype. Interestingly however, different degrees of 9G4 reactivity were present with a strong correlation noted (p < 0.05), between the degree of 9G4 expression and the strength of BCB but not with autoreactivity against any other Ags tested (not shown). Among different autoreactivities, the strongest correlation was found between APCB binding and antinuclear reactivity (p < 0.0001) as all 20 apoptotic cell binders displayed antinuclear reactivity, 18 of them with strong binding. Correlations between other autoreactivities were significantly weaker or not present (not shown).
The VH4-34 FR1 HP plays a differential role in separate 9G4 autoreactivities
The VH4-34 HP is required for I/i binding of 9G4 IgM Abs but may be dispensable for reactivity against DNA (51–55). Our previous analysis of SLE serum polyclonal 9G4+ IgG predicted that the HP would be required for BCB but not for binding to the other lupus-associated self-Ags. To test this hypothesis, 14 autoreactive SLE 9G4+ mAbs (6 from naive cells and 8 from memory cells; Fig. 3) were modified to eliminate the HP and generate an FR1 sequence concordant with the consensus VH4 sequence present in all other VH4 members. The sequence determinants of the VH4-34 HP as compared with other VH4 genes have been described elsewhere in detail (56) and are shown in Fig. 3A together with our mutagenesis strategy. 9G4 mutations did not prevent proper pairing of H and L chains nor did they inhibit Ab expression (Fig. 3B). The 9G4 mutation, however, universally resulted in a complete loss of BCB, thereby establishing that, irrespective of the HCDR3, mutations outside the HP, and associated L chain, BCB is HP dependent. Consistently, BCB was also absent in Abs that had accumulated in vivo mutations in FR1 residues responsible for the HP structure. Thus, all four Abs (74D6, 201B9, 152B8, and 88C1) with spontaneous mutations in either A23 or Y25 were devoid of any BCB despite retaining some expression of the 9G4 idiotype. This preservation of 9G4 expression in the presence of partial mutation of the FR1 HP is consistent with previous mutagenesis studies (56) and indicates an absolute requirement of full integrity of the HP for BCB. Strikingly, nucleolar reactivity but not other types of ANA binding was also completely abolished by the disruption of the 9G4 idiotype through in vitro mutagenesis (Fig. 1E). A notable difference between the two HP-dependent autoreactivites, however, is that nucleolar binding was only observed in SLE memory cells. In contrast to the stringent requirement for BCB, disruption of the HP resulted in increased average binding to late apoptotic cells (p < 0.02, paired t test; Fig. 3C, 3F), as 11 out of 14 mutants showed increase in APCB. Enhancement was also observed for ANA reactivity (p < 0.02, paired t test) in 12 out of 14 mutants (Fig. 3C, 3F). Average dsDNA reactivity also increased with HP mutation (p < 0.05, paired t test). All eight negative binders remained negative, whereas three weak and three strong binders increased in reactivity (Fig. 3C, 3F). Binding to chromatin was preserved, albeit moderately decreased, by the 9G4 HP mutation (Fig. 3G, right panel).
The VH4-34 FR1 HP is preserved in the majority of VH4-34 SLE memory cells
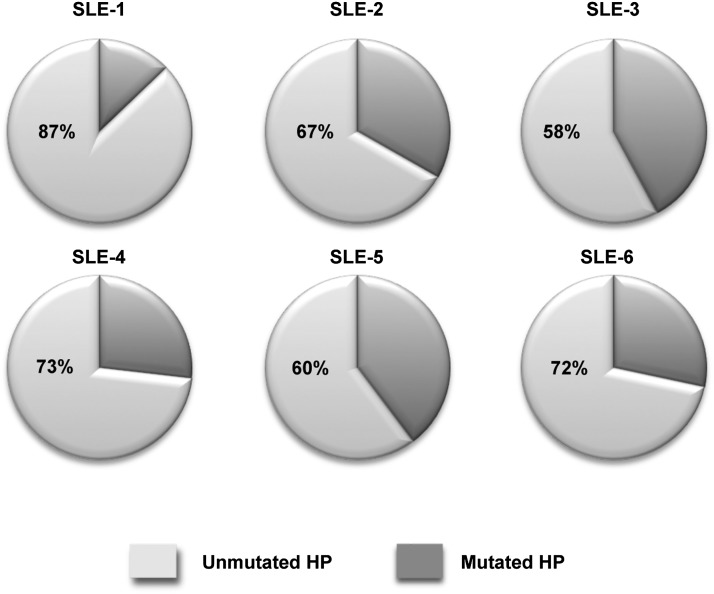
Given the critical role of the 9G4 HP in BCB and its modulating impact on other SLE autoreactivities, we wanted to ascertain the degree to which this determinant was preserved in the SLE memory B cell repertoire, as this would provide important information regarding the antigenic pressures that shape autoimmune memory. Thus, we performed deep sequencing analysis of global SLE memory B cells to determine the relative frequency of cells that had lost expression of the HP secondary to somatic hypermutation. In all, ∼30,000 IgG SLE sequences were analyzed from 6 SLE patients. VH4-34 contributed ∼10% of all sequences, a frequency in keeping with the abundance of 9G4+ memory cells we previously described in this disease (12). Interestingly, the HP was retained in most SLE memory cells with a frequency of ∼90% in some patients (average 75%; Fig. 4).
FIGURE 4.
Mutation analysis of VH4-34 sequences by 454 amplicon sequencing. VH4-specific primers were used to amplify rearranged H chains from total cellular RNA of sorted CD19+IgD−CD27+ cells from SLE patients. The 454 amplicon sequencing for six SLE subjects was performed, and resulting sequences were aligned to germline sequences using IMGT/High Vquest and iHMMune-align software. 9G4 idiotype regions were analyzed for mutations in aa 23 or 25 in the FR1 region. Each subject is represented by an individual pie chart. Dark slices represent the percent of VH4-34 sequences predicted to have lost expression of the HP due to somatic hypermutation of the A23 and/or Y25 residues.
The HCDR3 charge correlates with APCB, ANA, and dsDNA binding but not with BCB
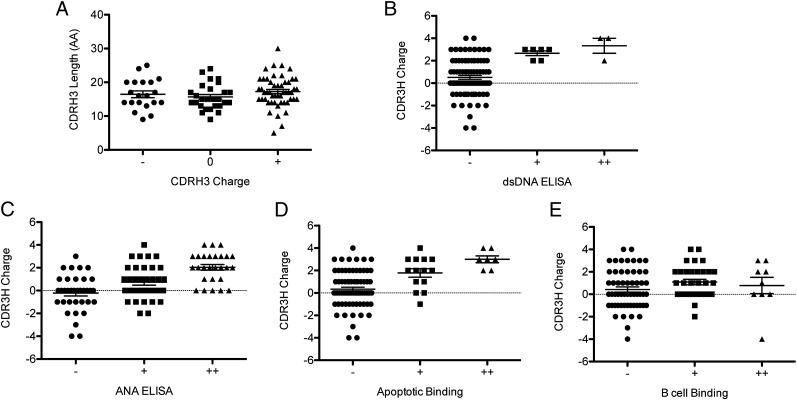
HCDR3 generates the largest degree of diversity in the human Ab repertoire and has a critical contribution to the conformation and antigenic specificity of the Ag binding pocket (57). However, the influence of the HCDR3s is less understood in 9G4 Abs because the FR1 HP may be dominant in binding to the canonical I/i Ags, and, yet, at least for IgM cold-agglutinins, the HCDR3 can determine binding to other self-Ags while modulating anti-I reactivity (26, 52, 56). Accordingly, we hypothesized that the nature of the HCDR3 would preferentially influence HP-independent 9G4 autoreactivities (APCB, ANA, and dsDNA). No correlation between charge and amino acid length of the HCDR3 was found for the 98 9G4 mAbs (p = 0.2841, Pearson; Fig. 5A). The length of the HCDR3 did not correlate with BCB (p = 0.4170, Pearson), APCB (p = 0.3658), ANA (p = 0.1495), or dsDNA binding (p = 0.7655). Interestingly, negatively charged HCDR3s correlated with lack of significant autoreactivity including BCB (Fig. 5). In contrast, in keeping with their well-established contribution to dsDNA binding (58), a strong correlation was observed between positively charged HCDR3 and dsDNA reactivity (p < 0.005) (Fig. 5B). A positive correlation also existed between positively charged HCDR3 and ANA autoreactivity (p < 0.005, Pearson) collectively and when the Abs were grouped according to their strength of ANA reactivity (negative/−; weak/+; and strong/++; Fig. 5C). After removal of dsDNA binders from the analysis, strong and weak antinuclear binders each had a statistically higher average charge than negative binders (p < 0.05, ANOVA). Hence, positively charged HCDR3 contribute to ANA reactivity irrespective of their anti-dsDNA reactivity. Positively charged HCDR3 also showed a positive correlation with APCB (p < 0.005, Pearson). This correlation remained after removal of the eight dsDNA binders from the analysis (p < 0.005). When apoptotic cell binders were divided into three groups as before, there still was a statistical difference in the average charge of the CDRH3 among each of the three groups (Fig. 5D). In contrast to the other types of autoreactivity, the charge of the HCDR3 had no influence in the ability of 9G4 Abs to bind B cells (p = 0.3372, Pearson; Fig. 5E).
FIGURE 5.
CDR3 charge, but not length, influences 9G4 autoreactivity. Charge and amino acid (AA) length of the H chain CDR3 were calculated from the sequence of all 98 mAbs. Differences in groups were determined by ANOVA. (A) The mAbs were divided into three groups based on the charge of the CDRH3. There were no differences in length of the CDR3H (p = 0.4568, ANOVA). (B–E) The mAbs were divided into three groups based on the strength of reactivity (−, negative; +, weak; and ++, strong) to self-Ags. (B) dsDNA binding (p = 0.0017, ANOVA). (C) ANA ELISA (p < 0.0001, ANOVA). (D) APCB (p = 0.0001, ANOVA). (E) BCB (p = 0.3372, ANOVA).
Influence of associated L chains on the autoreactivity of 9G4 Abs
As illustrated by anti-ds DNA Abs, L chains have the ability to modify Ag binding, and their secondary rearrangement plays a major role in editing mouse and human B cell autoreactivity (41, 59). Although L chain genetic analysis can reveal informative associations with different autoreactivities (60), the contribution of L chains to 9G4 Abs autoreactivity has not been systematically studied. Overall, 9G4 Abs had significant underrepresentation of λ chains (κ/λ ratio = 3.6), which may be more efficient at editing autoreactivity in humans (41). Moreover, we found a strong preference for Vκ-proximal upstream Jκ segments with use of the more distal Jκ4-5 in only 14 out of 98 Abs (14%) with all Jκ4-5 Abs found in SLE B cells. Combined, these features argue against active secondary L chain recombination in 9G4 B cells.
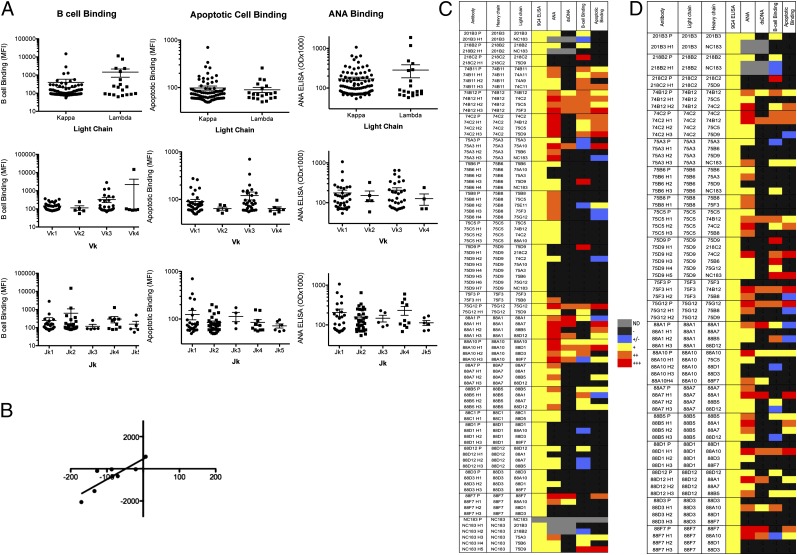
No significant skewing in terms of L chain isotype, Vκ, or Jκ gene use was found for BCB Abs (Fig. 6A). Similarly, no differences were found between positive and negative binders when B cell reactivity was analyzed as a dichotomous variable. No significant correlations between the charge and amino acid length of the L chain CDR3 and the degree of BCB were observed (Table I). A similar analysis was performed for APCB and ANA binding. There was no statistical difference in the average MFI of apoptotic cell or ANA binding between Abs expressing κ or λ L chains (Fig. 6B, 6C). The vast majority of binders used either Vκ1 or Vκ3 L chains. The three strongest apoptotic binders used Jκ1, and no significant binding was found in Jκ5 Abs. A statistical analysis of dsDNA reactivity was not performed due to the small number of dsDNA binders in this study. However, both κ and λ L chains were used (five and one Abs, respectively) in a ratio similar to the total Ab sample.
FIGURE 6.
L chain contribution to Ag-binding by 9G4+ Abs. A total of 97 (BCB) and 98 (APCB and ANA) 9G4+ mAbs were analyzed. (A) mAbs were divided by L chain type, Vκ family, or Jκ gene segment and analyzed for differences in the MFI of BCB and APCB and nuclear Ag reactivity (OD). No significant differences were found in any of the analyses. (B) 9G4 expression levels and BCB activity values are plotted for hybrid Abs for which the level of 9G4 expression decreased due to L chain replacements. The trend line shows a strong correlation between loss of 9G4 expression and attenuated BCB (p < 0.0075). (C) Heat map showing the degree of autoreactivity of parental Abs compared with hybrids expressing the same H chain in combination with different L chains. (D) A similar display of hybrid Abs grouped on the basis of their expression of a shared L chain recombined with different H chains.
The impact of L chains was directly assessed by the generation of 55 hybrid Abs formed by recombination of several H chains and assorted L chains. Of significant interest, L chain swapping had a significant quantitative impact on the expression of the 9G4 idiotype, which declined in 8 out of 12 hybrid Abs as compared with the native Ab expressing the same H chain, whereas 4 Abs experienced increased 9G4 expression. Consistent with the mutagenesis experiments and the role of the HP, decline in 9G4 expression through L chain replacement correlated with decreased BCB (Fig. 6B). In terms of autoreactivity, the overall trend observed for all autoreactivities tested was one of preservation of the binding properties of the parental Abs contributing the H chain (Fig. 6C). In isolated instances, however, the swapped L chain did significantly alter autoreactivity in either direction (decrease or increase). Interestingly, most of the swapped L chains that abolished binding were permissive of autoreactivity in other Abs (whether native or hybrids; Fig. 6D). Similarly, autoreactivity was observed in some hybrid Abs using L chains derived from parental Abs devoid of the same type of autoreactivity.
Spontaneous somatic mutation is present in a significant fraction of SLE 9G4 naive B cells
Our genetic analysis of 9G4 Abs uncovered an unexpected rate of spontaneous VH4-34 mutation rates (3–10% amino acid mutation) in a significant fraction of SLE naive B cells (25%; Supplemental Fig. 2). This rate is well above the expected error rate introduced by the PCR amplification and was not observed in healthy naive cells nor in our previous studies of healthy naive B cells (45). These results indicate the presence of early mutations in SLE naive B cells presumably secondary to Ag-driven activation. Of note, this phenomenon has been well documented in different murine lupus models (46).
Discussion
The structural basis of human autoantibodies has been the subject of intensive investigation for more than four decades (41, 61). Initial studies of rheumatoid factor paraproteins provided critical insights into the structural basis of idiotypes and Ab diversity while also hinting at a role for somatic hypermutation first recognized by seminal studies of mouse L chains (62). Taking advantage of rDNA technology, we provided original evidence for the ability of unmutated VH genes to encode human IgM autoantibodies including natural autoantibodies and SLE-specific anti-Sm Abs (63, 64). Yet, to this day, the structural basis and repertoire use of mature IgG autoantibodies in human autoimmune diseases remains poorly understood despite isolated studies of limited numbers of Abs in some cases using phage-display technology that precludes identification of the original H and L chain pairs (65, 66). These limitations have been largely resolved by the generation of mAbs from single cells (3, 4) and more recently by the emergent use of large scale next-generation sequencing (67, 68). However, although single-cell studies have identified multiple tolerance checkpoints, their open-ended analysis of relatively small numbers of unselected B cells have limited their ability to detect SLE-associated autoreactivity beyond the global ANA reactivity present in a significant fraction of all human B cells (3, 4). Moreover, molecular analysis has been limited to global reversion of mutated sequences from diverse VH genes to their germline counterpart, a process generally leading to loss of autoreactivity. Consequently, these studies could only confirm a general role for somatic hypermutation in lupus autoreactivity (5).
In this manuscript, we sought to circumvent these limitations by taking advantage of the properties of 9G4 Abs and applying both single-cell analysis and next-generation sequencing. Given their autoreactivity against different lupus Ags and the shared expression of VH4-34, the study of 9G4 cells represents a highly informative experimental model to understand the breakdown of B cell tolerance in SLE (12, 13). A central advantage of this system stems from the strong autoreactivity engrained in the VH4-34 germline sequence, which in many cases is largely independent of the HCDR3 and L chain. In this respect, the 9G4 system also represents an important departure from other human (auto)Ab systems.
To the best of our knowledge, this study represents the largest of its kind involving a prespecified SLE-specific autoimmune response in humans. It is important to emphasize that in addition to studying a number of mAbs that compare favorably with other studies, the focus on a single VH gene considerably enhances the power to understand the structural features that underlie the participation of 9G4 Abs in several SLE-related autoreactivities. It should be noted that although our mAbs were derived from only two patients, their autoreactivity mirrors the 9G4 serum autoreactivity of large numbers of SLE patients tested in our laboratory and is therefore representative of this disease population at large (9, 28, 29). Accordingly, we slanted our experimental design toward the analysis of large numbers of original Abs, mutants, and L chain hybrids rather than increasing the number of donors, as this design is better suited to the strength of any structural correlates. The choice of patients is also of major relevance to our study design. Thus, as it is the case for other SLE autoreactivities such as anti-ds DNA and ACL Abs, some SLE patients do not express high levels of 9G4 Abs despite the presence of significant number of 9G4 memory cells (9, 12), presumably reflecting late tolerance checkpoints that prevent their differentiation into Ab-producing cells (5, 31, 45, 46). We therefore intentionally selected two patients representing opposite ends of the spectrum of serum 9G4 Ab levels and the associated serum BCB and AACA reactivities. The concordance of results obtained with both patients provides further validation for our structural conclusions and indicate that differences in terminal differentiation of 9G4 cells into the plasma cell compartment in individual SLE patients may be dependent on effective functional censoring rather than the inability to generate autoreactive memory cells of pathogenic potential.
Our study provides conclusive evidence for the central postulate of the 9G4 model, namely the almost universal autoreactivity of naive cells expressing VH4-34 (upwards of 70%), with predominance of anti-i BCB and ANA binding, which substantially surpasses the level of autoreactivity (20%) previously detected by our group and others in non-9G4 naive cells in HC and SLE patients (3, 4, 44, 69). Although it is possible that a component of 9G4-associated autoreactivity may be eliminated during B cell maturation in the bone marrow, these results indicate the importance of peripheral tolerance mechanisms and are consistent with our earlier studies pointing to the GC as the main site of censoring for these cells in healthy subjects (12, 13). The efficiency of this mechanism is known to result in very low levels of 9G4 memory cells in healthy subjects that prevented the study of larger number of these cells. Nevertheless, of 12 Abs analyzed, only 1 displayed strong autoreactivity, and several showed binding to ANA or B cells despite the absence of serological autoreactivity in the corresponding healthy donors. As in SLE patients, these findings are consistent with the existence of late censoring checkpoints that prevent the differentiation of autoreactive 9G4 memory B cells into autoantibody-secreting cells (31, 45, 46).
A central finding of our studies is the delineation of SLE 9G4 autoreactivity into two distinct structural patterns: HP-dependent (BCB and ANA nucleolar binding) and HP-independent (global ANA, dsDNA, and APCB), as elimination of the HP universally extinguished both BCB and nucleolar ANA binding while frequently enhancing global ANA, APCB, and dsDNA reactivity (Fig. 3). Limited testing of the impact of the HP in antichromatin binding indicates that this region may influence but is not required for this autoreactivity. However, although necessary, expression of the HP is not sufficient for either BCB or nucleolar binding. Moreover, although BCB was strong in a large fraction of naive B cells, nucleolar reactivity was only found in SLE memory cells. Combined with our L chain–swapping experiments, these observations indicate that, in a significant departure from other SLE studies, the intrinsic BCB potential of VH4-34 H chains is modulated by the HCDR3 and/or the associated L chain but does not require somatic hypermutation. In contrast, antinucleolar reactivity appears to require somatic hypermutation. Formal validation of this interpretation will require in vitro reversal to the VH4-34 germline sequence of mutated anti-nucleolar 9G4 Abs.
The strong correlation found between HCDR3 charge and APCB, dsDNA, and ANA reactivity indicates a critical role for this central component of the conventional Ag binding pocket in HP-independent autoreactivity. This interpretation is consistent with studies of IgM 9G4 Abs showing that, conformationally, the HCDR3 lies in close proximity to the HP, may promote anti-dsDNA binding if positively charged, and may interfere with canonical 9G4 Ab I/i binding and 9G4-induced RBC agglutination (26, 52, 56). However, ours is the first demonstration, to our knowledge, of the reverse situation, in which the presence of the HP attenuates or even blocks binding to ANA, dsDNA, and APCB in Abs that otherwise possess the structural requirements for binding to this group of SLE Ags.
Given the inherent autoreactivity of the VH4-34 H chain, 9G4 cells also represent a useful tool to understand the role of L chains in SLE autoreactivity and the participation of receptor editing in the modulation of human B cell autoreactivity (41). Our results conclusively demonstrate that, despite the L chain independence reported for IgM cold-agglutinins, L chains have a strong modifying potential of SLE 9G4 autoantibodies (70). However, contrary to previous studies indicating the ability of most L chains to edit the autoreactivity present in non-9G4 human B cells (41), most L chains tested failed to eliminate VH4-34-determined autoreactivity. On that basis, one might have expected 9G4 B cells to undergo enhanced secondary L chain recombination in an attempt to edit autoreactivity. However, the Jκ and λ L chain use patterns were not consistent with this prediction in HC or SLE. Moreover, the use of Vκ-distal Jκ genes or λ L chains, more likely to be found in edited autoreactive B cells (56, 60), did not correlate with lack of 9G4 autoreactivity. Instead, our findings are reminiscent of the traditional 3H9 mouse H chain, which imparts dominant anti-dsDNA autoreactivity that can be edited by very few L chains (59) and also with recent studies of the global B cell repertoire in SLE indicating lack of consistent positive or negative selection of individual L chains (70). Larger repertoire studies as well as more extensive testing of selected L chains expressed by 9G4 cells devoid of autoreactivity, currently underway in our laboratory, will be required to conclusively answer these important questions.
The intrinsic predisposition to autoreactivity of VH4-34 H chains notwithstanding, a small but significant fraction of naive Abs (30% in HC and 15% in SLE) did not demonstrate autoreactivity against B cells or other Ags tested. Whether they react with the i Ag or related Ags is currently under investigation using the glycan microarrays shown in Supplemental Fig. 4. Autoreactivity against other self-Ags is also being explored using several approaches including Ag microarray screening using different platforms, which have been useful to disclose unexpected autoreactivity of anti-HIV Abs (71).
Interestingly, both HC and SLE naive cells showed similar patterns of BCB and ANA autoreactivity despite the remarkable rate of somatic hypermutation we found in naive SLE B cells (3–10% in 25% of cells; Supplemental Fig. 2). These findings indicate that SLE naive 9G4 B cells may experience substantial activation-induced somatic hypermutation largely induced by binding to i and/or ANA Ags and suggest that the expansion of other SLE-related autoreactivites requires additional somatic hypermutation and selection presumably in the GC upon interaction, at least in part, with apoptotic cells. This conclusion is supported by the increased reactivity against apoptotic cells, dsDNA, and cardiolipin found in SLE memory cells further discussed below.
Understanding the participation of apoptotic cells, a rich source of self-Ags including chromatin, in the diversification and selection of autoreactive memory B cells is particularly important in SLE, in which these cells accumulate in the GC (31, 34–37) and may activate pathogenic autoreactive B cells (31). The ensuing APCA may play a powerful effector pathogenic role not only through conventional type III hypersensitivity reactions but also through their ability to induce type I IFN production by dendritic cells (40). Again, 9G4 cells represent a highly informative model as they are specifically expanded in the GC of SLE patients but not in other autoimmune diseases (12), and 9G4 APCB binding is strongly present in SLE serum and correlates with disease activity population at large (28, 29). Indeed, we observed enhanced APCB in a significant fraction (31%) of SLE 9G4 memory cells in the context of substantially lower levels of BCB relative to SLE and HC naive cells. Although the actual Ags mediating APCB remain to be formally elucidating, our initial studies indicate that binding to histone/chromatin may mediate such autoreactivity in at least a fraction of these Abs.
SLE memory cells were also enriched for binding to other lupus Ags that could be enriched on apoptotic cells including dsDNA and cardiolipin (39), as illustrated by the following naive/memory ratios for these reactivities respectively: BCB: 1.8; APCB: 0.5; dsDNA: 0.15; ACL: 0.2 (total reactivity); BCB: 2.0; APCB: 0.15; dsDNA: 0; and ACL: 0 (strong reactivity). Even more informative, in contrast to their consistently positive ANA reactivity, multiple anti-APCB Abs showed little to none BCB activity (Figs. 2C, 3B). Combined, our observations have important implications for our understanding of the structural correlates of 9G4 autoreactivities and provide the basis for testable hypotheses regarding the selection of autoimmune memory responses in SLE, although a definitive answer to the latter question will require the study of larger numbers of patients. Thus, although the intrinsic HP-dependent anti-i/ANA autoreactivity might provide the initial trigger for the activation of 9G4 in SLE, HP-independent Ags, in part contributed by apoptotic cells, appear to play an important role in subsequent memory cell selection. Of note, apoptotic cells could also provide HP-dependent Ags including nucleolar Ags (this work) and glycosphingolipids (39, 72–74), and might select cells with dual binding through the HP and the HCDR3, respectively. This model is consistent with the behavior illustrated by memory Abs 75G12 and 75A11 in which disruption of the HP eliminated nucleolar binding while preserving strong APCB (Fig. 1E). A similar role could be postulated for chromatin given the attenuated binding observed with 9G4 mutants that retained APCB. Nonetheless, preferential positive selection dominated by HP-independent Ags should result in a VH4-34 memory repertoire dominated by HP-negative Abs. This prediction, however, runs contrary to the retention of the HP we observed through deep sequencing analysis in a major fraction (up to 90%) of VH4-34 lupus memory B cells. Thus, an integrated explanation of our results must invoke the participation not only of apoptotic cells but also of hitherto unrecognized HP-dependent Ags distinct from B cell and ANA Ags.
A more conclusive understanding of the Ags driving the expansion of this important population of autoreactive memory cells in SLE will require additional studies of naturally occurring 9G4-negative VH4-34 Abs and of naturally mutated 9G4+ Abs after in vitro reversion to the VH4-34 germline sequence. In addition, both currently available and future Ab panels will require extensive testing against diverse arrays of proteins, carbohydrates, and glycolipids. Nonetheless, the results present in this study firmly establish VH4-34–encoded 9G4 Abs as a major source of SLE-associated autoreactivity and provide a critical first step toward our understanding of the natural autoreactivity of a major population of human B cells and of the processes leading to its pathogenic participation in SLE.
Acknowledgments
We thank Prof. Freda. Stevenson (Tenovus Laboratories, Southampton, U.K.) for the 9G4 hybridoma and Dr. Eric Meffre (Yale University) for expression vectors and Dr. Steve Gill and Ann Gill (University of Rochester Medical Center) for invaluable help with 454 sequencing. We also thank the blood donors and research coordinators, the CFG for services by the Glycan Array Synthesis Core (The Scripps Research Institute, La Jolla, CA) that produced the mammalian glycan microarray and the Protein-Glycan Interaction Core (Emory University School of Medicine, Atlanta, GA) that performed the analysis of samples on the array.
This work was supported in part by the following grants from the National Institutes of Health (NIH): NIH-National Institute of Allergy and Infectious Diseases (NIAID) R37AI049660 (to I.S.), NIH-NIAID ACE U19 AI56390 (Autoimmunity Centers of Excellence), and NIH-NIAID P01 AI078907. A.S.C. was also supported by Training Grants NIH-T32 DE 007202 and NIH-T90 DE 21985. The Consortium for Functional Glycomics was funded by NIH-National Institute of General Medical Sciences Grants GM62116 and GM98791.
The sequences presented in this article have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank) under accession numbers KF495432–KF495529 and to the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/info/linking.html) under accession numbers GSM1224646, GSM1224647, and GSM1224648 for autoantigen protein array data.
The online version of this article contains supplemental material.
- 7-AAD
- 7-aminoactinomycin D
- ACL
- anticardiolipin
- ANA
- anti-nuclear Ab
- APCA
- anti-apoptotic cell Ab
- APCB
- anti-apoptotic cell binding
- BCB
- B cell binding
- CFG
- Consortium for Functional Glycomics
- FR1
- framework 1
- GC
- germinal center
- HC
- healthy control subject
- HCDR3
- H chain third CDR
- HP
- hydrophobic patch
- LN
- N-acetyl-lactosamine
- MFI
- median fluorescence intensity
- SLE
- systemic lupus erythematosus
- SNR
- signal-to-noise ratio.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Hargraves M. M. 1949. Production in vitro of the L.E. cell phenomenon; use of normal bone marrow elements and blood plasma from patients with acute disseminated lupus erythematosus. Proc. Staff Meet. Mayo Clin. 24: 234–237 [PubMed] [Google Scholar]
- 2.Tan E. M., Cohen A. S., Fries J. F., Masi A. T., McShane D. J., Rothfield N. F., Schaller J. G., Talal N., Winchester R. J. 1982. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25: 1271–1277 [DOI] [PubMed] [Google Scholar]
- 3.Wardemann H., Yurasov S., Schaefer A., Young J. W., Meffre E., Nussenzweig M. C. 2003. Predominant autoantibody production by early human B cell precursors. Science 301: 1374–1377 [DOI] [PubMed] [Google Scholar]
- 4.Yurasov S., Wardemann H., Hammersen J., Tsuiji M., Meffre E., Pascual V., Nussenzweig M. C. 2005. Defective B cell tolerance checkpoints in systemic lupus erythematosus. J. Exp. Med. 201: 703–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiller T., Tsuiji M., Yurasov S., Velinzon K., Nussenzweig M. C., Wardemann H. 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26: 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Isenberg D., Spellerberg M., Williams W., Griffiths M., Stevenson F. 1993. Identification of the 9G4 idiotope in systemic lupus erythematosus. Br. J. Rheumatol. 32: 876–882 [DOI] [PubMed] [Google Scholar]
- 7.Isenberg D. A., McClure C., Farewell V., Spellerberg M., Williams W., Cambridge G., Stevenson F. 1998. Correlation of 9G4 idiotope with disease activity in patients with systemic lupus erythematosus. Ann. Rheum. Dis. 57: 566–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Vollenhoven R. F., Bieber M. M., Powell M. J., Gupta P. K., Bhat N. M., Richards K. L., Albano S. A., Teng N. N. 1999. VH4-34 encoded antibodies in systemic lupus erythematosus: a specific diagnostic marker that correlates with clinical disease characteristics. J. Rheumatol. 26: 1727–1733 [PubMed] [Google Scholar]
- 9.Cappione A. J., Pugh-Bernard A. E., Anolik J. H., Sanz I. 2004. Lupus IgG VH4.34 antibodies bind to a 220-kDa glycoform of CD45/B220 on the surface of human B lymphocytes. J. Immunol. 172: 4298–4307 [DOI] [PubMed] [Google Scholar]
- 10.Bhat N. M., Lee L. M., van Vollenhoven R. F., Teng N. N., Bieber M. M. 2002. VH4-34 encoded antibody in systemic lupus erythematosus: effect of isotype. J. Rheumatol. 29: 2114–2121 [PubMed] [Google Scholar]
- 11.Isenberg D. A., Collins C. 1985. Detection of cross-reactive anti-DNA antibody idiotypes on renal tissue-bound immunoglobulins from lupus patients. J. Clin. Invest. 76: 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappione A., III, Anolik J. H., Pugh-Bernard A., Barnard J., Dutcher P., Silverman G., Sanz I. 2005. Germinal center exclusion of autoreactive B cells is defective in human systemic lupus erythematosus. J. Clin. Invest. 115: 3205–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugh-Bernard A. E., Silverman G. J., Cappione A. J., Villano M. E., Ryan D. H., Insel R. A., Sanz I. 2001. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J. Clin. Invest. 108: 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feizi T. 1967. Monotypic cold agglutinins in infection by mycoplasma pneumoniae. Nature 215: 540–542 [DOI] [PubMed] [Google Scholar]
- 15.Feizi T. 1968. Cold agglutinin titres, cold agglutinin structure and serum immunoglobulin levels in a variety of syndromes including mycoplasma pneumoniae infection. Bibl. Haematol. 29: 322–326 [DOI] [PubMed] [Google Scholar]
- 16.Feizi T. 1975. Blood group antigens. Ii antigens. Proc. R. Soc. Med. 68: 799–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecomte J., Feizi T. 1975. A common idiotype on human macroglobulins with anti-I and anti-i specificity. Clin. Exp. Immunol. 20: 287–302 [PMC free article] [PubMed] [Google Scholar]
- 18.Loomes L. M., Uemura K., Childs R. A., Paulson J. C., Rogers G. N., Scudder P. R., Michalski J. C., Hounsell E. F., Taylor-Robinson D., Feizi T. 1984. Erythrocyte receptors for Mycoplasma pneumoniae are sialylated oligosaccharides of Ii antigen type. Nature 307: 560–563 [DOI] [PubMed] [Google Scholar]
- 19.Silberstein L. E., Jefferies L. C., Goldman J., Friedman D., Moore J. S., Nowell P. C., Roelcke D., Pruzanski W., Roudier J., Silverman G. J. 1991. Variable region gene analysis of pathologic human autoantibodies to the related i and I red blood cell antigens. Blood 78: 2372–2386 [PubMed] [Google Scholar]
- 20.Grillot-Courvalin C., Brouet J. C., Piller F., Rassenti L. Z., Labaume S., Silverman G. J., Silberstein L., Kipps T. J. 1992. An anti-B cell autoantibody from Wiskott-Aldrich syndrome which recognizes i blood group specificity on normal human B cells. Eur. J. Immunol. 22: 1781–1788 [DOI] [PubMed] [Google Scholar]
- 21.Potter K. N., Li Y. C., Capra J. D. 1994. The cross-reactive idiotopes recognized by the monoclonal antibodies 9G4 and LC1 are located in framework region 1 of two non-overlapping subsets of human VH4 family encoded antibodies. Scand. J. Immunol. 40: 43–49 [DOI] [PubMed] [Google Scholar]
- 22.Manjarrez-Orduño N., Quách T. D., Sanz I. 2009. B cells and immunological tolerance. J. Invest. Dermatol. 129: 278–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhat N. M., Bieber M. M., Hsu F. J., Chapman C. J., Spellerberg M., Stevenson F. K., Teng N. N. 1997. Rapid cytotoxicity of human B lymphocytes induced by VH4-34 (VH4.21) gene-encoded monoclonal antibodies, II. Clin. Exp. Immunol. 108: 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat N. M., Bieber M. M., Spellerberg M. B., Stevenson F. K., Teng N. N. 2000. Recognition of auto- and exoantigens by V4-34 gene encoded antibodies. Scand. J. Immunol. 51: 134–140 [DOI] [PubMed] [Google Scholar]
- 25.Stevenson F. K., Longhurst C., Chapman C. J., Ehrenstein M., Spellerberg M. B., Hamblin T. J., Ravirajan C. T., Latchman D., Isenberg D. 1993. Utilization of the VH4-21 gene segment by anti-DNA antibodies from patients with systemic lupus erythematosus. J. Autoimmun. 6: 809–825 [DOI] [PubMed] [Google Scholar]
- 26.Thomas M. D., Clough K., Melamed M. D., Stevenson F. K., Chapman C. J., Spellerberg M. B., Potter K. N., Newton-Bishop J. A., Gregson N., Dawson K. 1999. A human monoclonal antibody encoded by the V4-34 gene segment recognises melanoma-associated ganglioside via CDR3 and FWR1. Hum. Antibodies 9: 95–106 [PubMed] [Google Scholar]
- 27.Milner E. C., Anolik J., Cappione A., Sanz I. 2005. Human innate B cells: a link between host defense and autoimmunity? Springer Semin. Immunopathol. 26: 433–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenks S., Palmer E., Marin E., Sanz I. 2010. 9G4 Autoantibodies Dominate the Anti-Apoptotic Cell Autoimmune Response in SLE. Arthritis Rheum. 62: S486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jenks S., Palmer E., Marin E., Hartson L., Chida A. S., Richardson C., Sanz I. 9G4+ auto-antibodies are an important source of apoptotic cell reactivity associated with high disease activity in systemic lupus erythematosus. Arthritis Rheum. 2013 doi: 10.1002/art.38138. DOI: 10.1002/art.38138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu C. C., Catera R., Zhang L., Didier S., Agagnina B. M., Damle R. N., Kaufman M. S., Kolitz J. E., Allen S. L., Rai K. R., Chiorazzi N. 2010. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood 115: 3907–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheid J. F., Mouquet H., Kofer J., Yurasov S., Nussenzweig M. C., Wardemann H. 2011. Differential regulation of self-reactivity discriminates between IgG+ human circulating memory B cells and bone marrow plasma cells. Proc. Natl. Acad. Sci. USA 108: 18044–18048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGaha T. L., Karlsson M. C. I., Ravetch J. V. 2008. FcgammaRIIB deficiency leads to autoimmunity and a defective response to apoptosis in Mrl-MpJ mice. J. Immunol. 180: 5670–5679 [DOI] [PubMed] [Google Scholar]
- 33.Hutcheson J., Scatizzi J. C., Siddiqui A. M., Haines G. K., III, Wu T., Li Q. Z., Davis L. S., Mohan C., Perlman H. 2008. Combined deficiency of proapoptotic regulators Bim and Fas results in the early onset of systemic autoimmunity. Immunity 28: 206–217 [DOI] [PubMed] [Google Scholar]
- 34.Gaipl U. S., Kuhn A., Sheriff A., Munoz L. E., Franz S., Voll R. E., Kalden J. R., Herrmann M. 2006. Clearance of apoptotic cells in human SLE. Curr. Dir. Autoimmun. 9: 173–187 [DOI] [PubMed] [Google Scholar]
- 35.Bijl M., Reefman E., Horst G., Limburg P. C., Kallenberg C. G. M. 2006. Reduced uptake of apoptotic cells by macrophages in systemic lupus erythematosus: correlates with decreased serum levels of complement. Ann. Rheum. Dis. 65: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reefman E., Horst G., Limburg P. C., Kallenberg C. G. M., Bijl M. 2007. Opsonization of apoptotic cells by systemic lupus erythematosus autoantibodies inhibit their uptake by macrophages. Arthritis Rheum. 26:3399–3411 [DOI] [PubMed] [Google Scholar]
- 37.Denny M. F., Chandaroy P., Killen P. D., Caricchio R., Lewis E. E., Richardson B. C., Lee K. D., Gavalchin J., Kaplan M. J. 2006. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J. Immunol. 176: 2095–2104 [DOI] [PubMed] [Google Scholar]
- 38.Muñoz L. E., Lauber K., Schiller M., Manfredi A. A., Herrmann M. 2010. The role of defective clearance of apoptotic cells in systemic autoimmunity. Nat Rev Rheumatol 6: 280–289 [DOI] [PubMed] [Google Scholar]
- 39.Hall J. C., Casciola-Rosen L., Rosen A. 2004. Altered structure of autoantigens during apoptosis. Rheum. Dis. Clin. North Am. 30: 455–471, vii [DOI] [PubMed] [Google Scholar]
- 40.Båve U., Magnusson M., Eloranta M. L., Perers A., Alm G. V., Rönnblom L. 2003. Fc γ RIIa is expressed on natural IFN-α-producing cells (plasmacytoid dendritic cells) and is required for the IFN-α production induced by apoptotic cells combined with lupus IgG. J. Immunol. 171: 3296–3302 [DOI] [PubMed] [Google Scholar]
- 41.Wardemann H., Hammersen J., Nussenzweig M. C. 2004. Human autoantibody silencing by immunoglobulin light chains. J. Exp. Med. 200: 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbonaviciute V., Fürnrohr B. G., Meister S., Munoz L., Heyder P., De Marchis F., Bianchi M. E., Kirschning C., Wagner H., Manfredi A. A., et al. 2008. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 205: 3007–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M. C., Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329: 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quách T. D., Manjarrez-Orduño N., Adlowitz D. G., Silver L., Yang H., Wei C., Milner E. C. B., Sanz I. 2011. Anergic responses characterize a large fraction of human autoreactive naive B cells expressing low levels of surface IgM. J. Immunol. 186: 4640–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Culton D. A., O’Conner B. P., Conway K. L., Diz R., Rutan J., Vilen B. J., Clarke S. H. 2006. Early preplasma cells define a tolerance checkpoint for autoreactive B cells. J. Immunol. 176: 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlomchik M. J. 2008. Sites and stages of autoreactive B cell activation and regulation. Immunity 28: 18–28 [DOI] [PubMed] [Google Scholar]
- 47.Watanabe N., Fisher H. M., Epstein W. V. 1969. Specificity and reactivity of cytoplasmic and nucleolar antibody in SLE sera. Arthritis Rheum. 12: 173–180 [DOI] [PubMed] [Google Scholar]
- 48.Sawalha A. H., Namjou B., Nath S. K., Kilpatrick J., Germundson A., Kelly J. A., Hutchings D., James J., Harley J. 2002. Genetic linkage of systemic lupus erythematosus with chromosome 11q14 (SLEH1) in African-American families stratified by a nucleolar antinuclear antibody pattern. Genes Immun. 3(Suppl 1): S31–S34 [DOI] [PubMed] [Google Scholar]
- 49.Li Q. Z., Xie C., Wu T., Mackay M., Aranow C., Putterman C., Mohan C. 2005. Identification of autoantibody clusters that best predict lupus disease activity using glomerular proteome arrays. J. Clin. Invest. 115: 3428–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalaaji M., Mortensen E., Jørgensen L., Olsen R., Rekvig O. P. 2006. Nephritogenic lupus antibodies recognize glomerular basement membrane-associated chromatin fragments released from apoptotic intraglomerular cells. Am. J. Pathol. 168: 1779–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mockridge C. I., Chapman C. J., Spellerberg M. B., Isenberg D. A., Stevenson F. K. 1996. Use of phage surface expression to analyze regions of human V4-34(VH4-21)-encoded IgG autoantibody required for recognition of DNA: no involvement of the 9G4 idiotope. J. Immunol. 157: 2449–2454 [PubMed] [Google Scholar]
- 52.Li Y., Spellerberg M. B., Stevenson F. K., Capra J. D., Potter K. N. 1996. The I binding specificity of human VH 4-34 (VH 4-21) encoded antibodies is determined by both VH framework region 1 and complementarity determining region 3. J. Mol. Biol. 256: 577–589 [DOI] [PubMed] [Google Scholar]
- 53.Chapman C. J., Spellerberg M. B., Smith G. A., Carter S. J., Hamblin T. J., Stevenson F. K. 1993. Autoanti-red cell antibodies synthesized by patients with infectious mononucleosis utilize the VH4-21 gene segment. J. Immunol. 151: 1051–1061 [PubMed] [Google Scholar]
- 54.Potter K. N., Li Y., Pascual V., Williams R. C., Jr., Byres L. C., Spellerberg M., Stevenson F. K., Capra J. D. 1993. Molecular characterization of a cross-reactive idiotope on human immunoglobulins utilizing the VH4-21 gene segment. J. Exp. Med. 178: 1419–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spellerberg M. B., Chapman C. J., Mockridge C. I., Isenberg D. A., Stevenson F. K. 1995. Dual recognition of lipid A and DNA by human antibodies encoded by the VH4-21 gene: a possible link between infection and lupus. Hum. Antibodies Hybridomas 6: 52–56 [PubMed] [Google Scholar]
- 56.Isnardi I., Ng Y. S., Srdanovic I., Motaghedi R., Rudchenko S., von Bernuth H., Zhang S. Y., Puel A., Jouanguy E., Picard C., et al. 2008. IRAK-4- and MyD88-dependent pathways are essential for the removal of developing autoreactive B cells in humans. Immunity 29: 746–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanz I. 1991. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J. Immunol. 147: 1720–1729 [PubMed] [Google Scholar]
- 58.Radic M. Z., Weigert M. 1994. Genetic and structural evidence for antigen selection of anti-DNA antibodies. Annu. Rev. Immunol. 12: 487–520 [DOI] [PubMed] [Google Scholar]
- 59.Li H., Jiang Y., Prak E. L., Radic M., Weigert M. 2001. Editors and editing of anti-DNA receptors. Immunity 15: 947–957 [DOI] [PubMed] [Google Scholar]
- 60.Samuels J., Ng Y. S., Coupillaud C., Paget D., Meffre E. 2005. Impaired early B cell tolerance in patients with rheumatoid arthritis. J. Exp. Med. 201: 1659–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Capra J. D., Kehoe J. M. 1974. Structure of antibodies with shared idiotypy: the complete sequence of the heavy chain variable regions of two immunoglobulin M anti-gamma globulins. Proc. Natl. Acad. Sci. USA 71: 4032–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weigert M. G., Cesari I. M., Yonkovich S. J., Cohn M. 1970. Variability in the lambda light chain sequences of mouse antibody. Nature 228: 1045–1047 [DOI] [PubMed] [Google Scholar]
- 63.Sanz I., Casali P., Thomas J. W., Notkins A. L., Capra J. D. 1989. Nucleotide sequences of eight human natural autoantibody VH regions reveals apparent restricted use of VH families. J. Immunol. 142: 4054–4061 [PubMed] [Google Scholar]
- 64.Sanz I., Dang H., Takei M., Talal N., Capra J. D. 1989. VH sequence of a human anti-Sm autoantibody. Evidence that autoantibodies can be unmutated copies of germline genes. J. Immunol. 142: 883–887 [PubMed] [Google Scholar]
- 65.Rey E., Zeidel M., Rhine C., Tami J., Krolick K., Fischbach M., Sanz I. 2000. Characterization of human anti-acetylcholine receptor monoclonal autoantibodies from the peripheral blood of a myasthenia gravis patient using combinatorial libraries. Clin. Immunol. 96: 269–279 [DOI] [PubMed] [Google Scholar]
- 66.del Rincon I., Zeidel M., Rey E., Harley J. B., James J. A., Fischbach M., Sanz I. 2000. Delineation of the human systemic lupus erythematosus anti-Smith antibody response using phage-display combinatorial libraries. J. Immunol. 165: 7011–7016 [DOI] [PubMed] [Google Scholar]
- 67.Zuckerman N. S., Hazanov H., Barak M., Edelman H., Hess S., Shcolnik H., Dunn-Walters D., Mehr R. 2010. Somatic hypermutation and antigen-driven selection of B cells are altered in autoimmune diseases. J. Autoimmun. 35: 325–335 [DOI] [PubMed] [Google Scholar]
- 68.Logan A. C., Gao H., Wang C., Sahaf B., Jones C. D., Marshall E. L., Buño I., Armstrong R., Fire A. Z., Weinberg K. I., et al. 2011. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc. Natl. Acad. Sci. USA 108: 21194–21199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duty J. A., Szodoray P., Zheng N.-Y., Koelsch K. A., Zhang Q., Swiatkowski M., Mathias M., Garman L., Helms C., Nakken B., et al. 2009. Functional anergy in a subpopulation of naive B cells from healthy humans that express autoreactive immunoglobulin receptors. J. Exp. Med. 206: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schoettler N., Ni D., Weigert M. 2012. B cell receptor light chain repertoires show signs of selection with differences between groups of healthy individuals and SLE patients. Mol. Immunol. 51: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang G., Holl T. M., Liu Y., Li Y., Lu X., Nicely N. I., Kepler T. B., Alam S. M., Liao H. X., Cain D. W., et al. 2013. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J. Exp. Med. 210: 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eda S., Yamanaka M., Beppu M. 2004. Carbohydrate-mediated phagocytic recognition of early apoptotic cells undergoing transient capping of CD43 glycoprotein. J. Biol. Chem. 279: 5967–5974 [DOI] [PubMed] [Google Scholar]
- 73.Suzuki T., Kiyokawa N., Taguchi T., Sekino T., Katagiri Y. U., Fujimoto J. 2001. CD24 induces apoptosis in human B cells via the glycolipid-enriched membrane domains/rafts-mediated signaling system. J. Immunol. 166: 5567–5577 [DOI] [PubMed] [Google Scholar]
- 74.Cocca B. A., Seal S. N., D’Agnillo P., Mueller Y. M., Katsikis P. D., Rauch J., Weigert M., Radic M. Z. 2001. Structural basis for autoantibody recognition of phosphatidylserine-β 2 glycoprotein I and apoptotic cells. Proc. Natl. Acad. Sci. USA 98: 13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]