Abstract
Leukoencephalopathy with vanishing white matter (VWM) is a severe inherited human neurodegenerative disorder that is caused by mutations in the genes for the subunits of eukaryotic initiation factor 2B (eIF2B), a heteropentameric guanine nucleotide exchange factor that regulates both global and mRNA-specific translation. Marked variability is evident in the clinical severity and time course of VWM in patients. Here we have studied the effects of VWM mutations on the function of human eIF2B. All the mutations tested cause partial loss of activity. Frameshift mutations in genes for eIF2Bɛ or eIF2Bβ lead to truncated polypeptides that fail to form complexes with the other subunits and are effectively null mutations. Certain point mutations also impair the ability of eIF2Bβ or -ɛ to form eIF2B holocomplexes and also diminish the intrinsic nucleotide exchange activity of eIF2B. A point mutation in the catalytic domain of eIF2Bɛ impairs its ability to bind the substrate, while two mutations in eIF2Bβ actually enhance eIF2 binding. We provide evidence that expression of VWM mutant eIF2B may enhance the translation of specific mRNAs. The variability of the clinical phenotype in VWM may reflect the multiple ways in which VWM mutations affect eIF2B function.
Leukoencephalopathy with vanishing white matter (VWM) is a severe inherited neurodegenerative disease (35). This condition is also known as childhood ataxia with diffuse central nervous system hypomyelination (34). Clinically, this condition is characterized by progressive ataxia, spasticity, and variable optic atrophy, as well as seizures. Mental decline, if seen, is usually mild. More rapid deterioration is associated with minor head injury or fever, and the latter often results in coma. Death occurs after a variable time period, from several years up to decades. In magnetic resonance imaging, a diagnostic pattern with diffuse abnormality of cerebral white matter is observed (35, 36). Over time, the abnormal white matter vanishes to be replaced by cerebrospinal fluid or tissue water. There is considerable phenotypic variation. For example, the so-called “Cree” leukoencephalopathy seen in North American Indians is especially severe, leading to very early onset of neurological deterioration (3 to 9 months) and death before 21 months (8). Recent work has revealed additional serious forms of VWM that involve very early onset and early death (7, 8). In some cases, the ovaries are also affected (35).
VWM shows autosomal recessive inheritance. Linkage analysis initially assigned a VWM locus to a 5-centimorgan interval on chromosome 3q27 (21). Subsequent work showed that that mutations in the gene for the ɛ subunit of the translation factor eukaryotic initiation factor 2B (eIF2B) were associated with VWM. These included several point or frameshift mutations within this gene. In some patients, linkage analysis did not point to a role for the eIF2Bɛ (or EIF2B5) gene. In several of them, VWM was found to be associated with mutations in the β subunit of eIF2B, a heteropentamer (22). Subsequent work showed that mutations in any eIF2B gene can cause VWM (37). All VWM patients have now been found to have mutations in one or other of the genes for the subunits of eIF2B. Affected individuals carry two mutated copies of a given eIF2B gene; i.e., inheritance shows an autosomal recessive pattern. The particularly severe Cree form of this disease is linked to a point mutation (R195H) in eIF2Bɛ (8), and a second severe form is linked to a different point mutation in the same gene (7).
eIF2B is a guanine nucleotide exchange factor that plays a key role in the initiation of mRNA translation and in its control (28). It promotes GDP/GTP exchange on initiation factor eIF2. In its GTP-bound state, eIF2 recruits the initiator Met-tRNA to the 40S subunit during translation initiation. Since it is the anticodon of this tRNA that recognizes the start codon in the mRNA, active eIF2 is required for all normal translation initiation events. eIF2B activity can be regulated in a number of ways. The best documented of these is the phosphorylation of the α subunit of eIF2 at Ser51 (4), which converts it from a substrate of eIF2B into a potent competitive inhibitor (30). Phosphorylation of eIF2α is increased under a wide range of cellular stress conditions and can be catalyzed by several different protein kinases (3). Since active eIF2.GTP is required for all translation initiation events, inhibition of eIF2B activity causes a decrease in general protein synthesis. However, reduced eIF2B activity actually enhances the translation of certain mRNAs by virtue of the presence of upstream open reading frames (uORFs) in their 5′ untranslated regions (UTRs). Examples include the mRNAs for the transcription regulators GCN4 in (16) and ATF4 in mammals (13). Since these proteins modulate transcription of multiple genes, decreased eIF2B activity may also affect the expression of many genes.
eIF2B is composed of five nonidentical subunits, α through ɛ (16). Of these, eIF2Bɛ displays catalytic activity when expressed alone, and the minimal catalytic region of the yeast ortholog was recently delineated (9). The corresponding region of the mammalian protein contains the site that is phosphorylated by glycogen synthase kinase 3 and that inactivates eIF2B (39). The other subunits do not possess a region homologous to this catalytic segment and appear not to be involved in catalysis. However, eIF2Bγ shows similarity to the N-terminal part of eIF2Bɛ and forms a binary complex with eIF2Bɛ in yeast, which is referred to as the catalytic subcomplex (25). The other three subunits, α, β and δ, also show mutual sequence similarity and form a trimeric subcomplex in yeast. Evidence from careful genetic studies with yeast indicates that these subunits play a key role in sensitizing the eIF2B holocomplex to inhibition by eIF2[αP] (20, 24, 40). Indeed, recombinant eIF2α can bind to the αβδ subcomplex, which has been termed the regulatory subcomplex (25). Although eIF2Bɛ itself can mediate guanine nucleotide exchange, its activity is greatly enhanced by its association with the other subunits (6, 10).
VWM is the first example of an inherited human disease caused by mutations in a component of the basal translational machinery. A clear priority is now to establish how mutations in eIF2B lead to VWM and the other lesions associated with this condition. It will also be important to understand why they give rise primarily to a neurological disease and why its severity and age of onset are so variable. Here, as an essential first step, we have studied how they affect the integrity and function of the eIF2B complex. Our data show that all the VWM mutations tested cause partial loss of activity of human eIF2B. However, the magnitude of the effect differs between mutations. Most interestingly, the basis of the decreased activity differs between mutations, prompting the possibility that the variable phenotype of the condition may reflect the distinct ways in which different mutations affect the eIF2B complex.
MATERIALS AND METHODS
Materials.
Chemicals and biochemicals were obtained as described previously (40). Antibodies to eIF2B subunits were obtained from Santa Cruz, apart from the anti-eIF2Bɛ antibody which was described in reference 38. Anti-myc was from Sigma. Antihemagglutinin (anti-HA) was from Boehringer Mannheim, and anti-eIF4A has been described previously (23). Antibodies to phosphorylated eIF2α were from Cell Signaling Technology, and anti-eIF2α is a monoclonal kindly provided by the late E. C. Henshaw.
Construction of expression vectors.
Full-length eIF2B cDNAs were obtained from the IMAGE consortium or were generated by reverse transcriptase PCR (RT-PCR) (Table 1). VWM mutations were made by PCR-based technology (for further information, see Table 2). These mutant cDNAs or the wild-type (WT) cDNA were cloned into an expression vector with N-terminal hexahistidine (His6) and myc tags. As the C terminus of eIF2Bɛ contains the catalytic domain (9) and the extreme C-terminal tip interacts with eIF2 (1), we reasoned that N-terminal tags were less unlikely to interfere with the function of the eIF2B complex than were C-terminal ones. All DNAs were confirmed by sequencing. A mutant lacking 47 residues within the catalytic domain (9) (ΔCAT) was created as a negative control for activity studies. Mouse ATF4 uORF was amplified from pATF4luc (a kind gift from David Ron, New York, N.Y.) and was inserted into the NheI-BamHI site of pEGFP-N1 (Clontech). Sequences encoding an HA tag were introduced into the PCR primer to facilitate detection by immunoblotting.
TABLE 1.
Sources of cDNAs and WT human eIF2B constructs
| Gene | Source(s)a |
|---|---|
| eIF2B1 | RT-PCR |
| CAGGATCCATGGACGACAAGGAGTTAATTG | |
| GAAAGCTTACAGATAGAGCTTGATGAG | |
| eIF2B2 | IMAGE clone 3534548 |
| eIF2B3 | IMAGE clone 4555992, RT-PCR for C terminus |
| TTGCTAGCATGGAATTTCAAGCAGTAGTG | |
| AAGTCGACTCAGATCTCCATGAGCTGGTC | |
| eIF2B4 | IMAGE clone 3534267, RT-PCR for C terminus |
| TCAAGCTTATGGCTGCTGTGGCCGTG | |
| ACGTCGACTCACTGGTCACTGCTCTTG | |
| eIF2B5 | IMAGE clone 3876105 |
IMAGE clones were bought from MRC gene service (London, United Kingdom) or Invitrogen (Paisley, United Kingdom). RT-PCR was carried out with human HEK 293 cells RNA to generate full-length cDNA for eIF2B1, eIF2B3, and eIF2B4 cDNAs. All products were confirmed by DNA sequencing. WT eIF2B2 and eIF2B5 were cloned into pHM with His and myc tags, at the N terminus. myc-tag-only constructs of five individual subunits were made by cloning into pCMV-tag3 (Stratagene).
TABLE 2.
Primers used for mutagenesis
| Mutation | Primer(s) (5′-3′)a |
|---|---|
| eIF2B2 | |
| E213G | GTCCAAAGCAGGTATTGGGACAACTGTCATGACTGATGCTG |
| CAGCATCAGTCATGACAGTTGTCCCAATACCTGCTTTGGAC | |
| V316D | GCCCTGTGTTTGACTACGATCCCCCAGAGCTCATTAC |
| GTAATGAGCTCTGGGGGATCGTAGTCAAACACAGGGC | |
| ΔC terminusb | Deletion from ApaI site |
| eIF2B5 | |
| ΔC terminusc | Deletion from XhoI site |
| AAGGATCCATGGCGGCCCTGTAGTG | |
| T91A | CCACAGGTGTACAGGAAGCATTTGTCTTTTGTTGCTGG |
| AAGGATCCATGGCGGCCCTGTAGTG | |
| R113Hd | |
| R195H | TACGACATTGTCTTCGTGGCAATGAGTTGGGTGGCTGG |
| GAAGACAATGTCGTAGTGGCTGTGG | |
| R315H | CTGTGCTGACGTCATCGGCCGATGGGTCTACCCTC |
| GAGGGTAGACCCATCGGCCGATGACGTCAGCACAG | |
| R339P | GAGCTGCACTCATTCCCCGCACAACATCTACCGAG |
| CTCGGTAGATGTTGTGCGGGGAATGAGTGCAGCTC | |
| W628R | CCTCTGCTAAAGGCCCGGAGCCCTGTTTTTAGGAAC |
| GTTCCTAAAAACAGGGCTCCGGGCCTTTAGCAGAGG | |
| ΔCATe | GTGGATGTCATCCATCTGAGG |
| CTGTGCTGACGTCATCGGCCGATGGGTCTACCCTC |
Mutations were introduced either by QuikChange or conventional PCR.
This mutant is similar to the VWM frameshift mutation in eIF2B2 (F264fs).
This mutant is similar to the VWM frameshift mutation in eIF2B5 (M203fs).
An EcoRI-SacI (140-bp) fragment was amplified from a patient DNA sample.
The PCR fragment was cut with ApaI and was ligated to an ApaI-PmII-digested eIF2B5 expression construct, resulting in deletion of 47 amino acids within the catalytic domain.
Analysis of eIF2B complexes and activity.
For analysis of eIF2B expression, lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting, as described earlier (27). To isolate complexes containing the recombinant His-myc-tagged eIF2B subunits, lysates were purified on Ni-nitrilotriacetic acid (NTA)-agarose (Qiagen). Typically 300 μg of protein was used for this type of analysis. The bound material was analyzed by SDS-PAGE-Western blotting as described above. To measure eIF2B activity, complexes were isolated in the same way and GDP/GTP exchange activity was determined by using our standard assay with complexes containing purified human eIF2 and [3H]GDP as substrate. Since the levels of expression of WT or mutant eIF2Bβ/ɛ could vary significantly within a given experiment, we first assessed the amount of the purified complexes by SDS-PAGE-immunoblotting for myc and then adjusted the amounts of material for the assay accordingly to ensure that similar amounts of recombinant eIF2B were used in all assays.
Cell culture, transfection, treatment, and lysis.
Human embryonic kidney 293 (HEK 293) cells were grown and lysed exactly as described earlier (40). Cells were transfected by using the calcium phosphate precipitation method as described in reference 11. Unless otherwise stated, cells were lysed about 48 h after transfection. For studies on the ATF-linked report, HEK 293 cells (in 10-cm-diameter dishes) were transfected with pEGFP or pATF4-EGFP plus the eIF2Bβ construct under study (or empty vector as a control) as indicated. Twenty-four hours after transfection, the media were changed to Dulbecco's modified Eagle's medium without methionine. Four-and-a-half microliters of [35S]methionine was then added. Sixteen hours later, cells were lysed and lysates were used for immunopreciptation (IP) with anti-GFP antibody (Roche). One quarter of the sample was loaded in each lane. For Northern blot analyses, 48 h after transfection, one dish of cells for each combination of vectors was collected for RNA isolation with Trizon (Invitrogen). One quarter of the total RNA isolated was loaded per lane in a 1.4% formaldehyde-agarose gel. Northern blot hybridization with probes for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and GFP were done as described by Sambrook et al. (31). Films were developed after 6 h (GFP) or 2 days (GAPDH) exposure at −80°C.
For thermal treatment of cells, dishes were transferred from the incubator (37°C) to a water bath at the desired temperature (usually 41°C) for 2 h. A water bath was used to ensure that cells quickly reached the new temperature.
RESULTS
Choice of VWM mutants for investigation in this study.
Our aim was to gain insight into the effects of VWM mutations on the function of human eIF2B complexes. We therefore overexpressed WT or mutant human eIF2B subunits in HEK 293 cells, as His6- and/or myc-tagged proteins. We chose HEK 293 cells because they are highly transfectable human cells and because it has already been shown that eIF2Bɛ expressed in them is incorporated into functional eIF2B complexes (39). Many VWM mutations occur in eIF2Bɛ, and we chose several for analysis. These were the frameshift mutant 792delTinsACA, which truncates eIF2Bɛ at residue F264, and the point mutations T91A, R113H, R315G, R339P, W628R (22), and R195H (8) (Table 3). They were chosen as they give rise to VWM when present in a simple homozygous fashion (T91A, R113H, R195H, and R315G) and/or involve a substantial amino acid change and may thus also have marked effects on eIF2B itself (R339P and W628R). We also wished to study the effects of mutations in another subunit of eIF2B. eIF2Bβ is a component of the regulatory, rather than the catalytic, subcomplex. We therefore chose a frameshift mutant in eIF2Bβ, which is predicted to yield a severely truncated polypeptide, and the point mutants E213G (homozygous) and V316D (major change) (reference 22 and Table 3).
TABLE 3.
Mutations of eIF2B studied in this reporta
| Subunit | DNA mutation | Effect on protein sequence | Comment on mutation | Other remark(s) |
|---|---|---|---|---|
| eIF2Bɛ | 792delinsACA | F264fs | Compound heterozygous | Gives truncated protein |
| eIF2Bɛ | A271G | T91A | Homozygous | T conserved in mammals |
| eIF2Bɛ | G338A | R113H | Homozygous | R in rabbit; H in rodents; varies in rest |
| eIF2Bɛ | G984A | R195H | Homozygous | R in mammals |
| eIF2Bɛ | G943G | R315G | Homozygous | Varies |
| eIF2Bɛ | G1016C | R339P | Compound heterozygous with T91A | R in mammals |
| eIF2Bɛ | T1882C | W628R | Compound heterozygous with T91A | W in mammals; W, Y, or L in rest |
| eIF2Bβ | 607_612delinsTG | M203fs | Compound heterozygous | Gives truncated protein |
| eIF2Bβ | A638G | E213G | Homozygous | E in all spp. except A. thaliana |
| eIF2Bβ | T947A | V316D | Compound heterozygous with E213G | V in all known sequences |
Conservation refers to the known sequences: mammalian sequences are from rat, rabbit, and mouse; others are from Caenorhabditis elegans, A. thaliana, D. melanogaster, and S. cerevisiae.
Requirements for the formation of mammalian eIF2B holocomplexes.
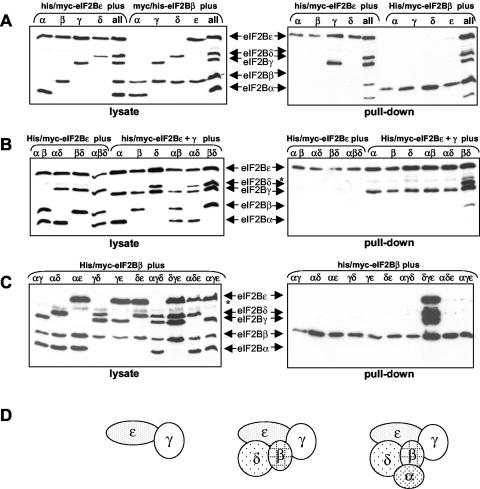
Studies with eIF2B are complicated because it is a heteropentamer. To assess and interpret possible effects of VWM mutations on formation of eIF2B complexes, it was first important to understand the assembly of WT human eIF2B. To do this, we expressed His-myc-tagged WT eIF2Bɛ or eIF2Bβ together with one or more of the other myc-tagged subunits. The His-myc-tagged polypeptide was then isolated on Ni-NTA-agarose, and the bound material was analyzed for other myc-tagged eIF2B subunits. When His-myc-tagged eIF2Bɛ was expressed with each of the other subunits singly, only eIF2Bγ bound to eIF2Bɛ, suggesting that, as in yeast (25), the γ and ɛ subunits form a binary subcomplex (Fig. 1A). eIF2Bα, -β, or -δ could not form ternary complexes with eIF2Bγɛ (Fig. 1B). Without eIF2Bγ, eIF2Bɛ was unable to bind even the combination of eIF2Bα, -β, and -δ (Fig. 1B). Indeed, the only four-subunit combination that forms stable complexes is that of β, γ, δ, and ɛ (Fig. 1B), consistent with the findings that eIF2B lacking eIF2Bα can be generated in insect cells (6) or isolated from mammalian cells (5).
FIG. 1.
Formation of eIF2B complexes in HEK 293 cells. HEK 293 cells were transfected with constructs encoding His-myc-tagged human eIF2Bɛ or -β as indicated, together with vectors encoding myc-tagged versions of the other subunits, as indicated. Samples of cell lysate were either analyzed directly by SDS-PAGE and immunoblotting with anti-myc (lysate) or were first subjected to chromatography on Ni-NTA-agarose prior to this type of analysis (pull-down). Positions of the tagged eIF2B subunits are shown. *, a band that cross-reacts nonspecifically with the anti-myc antibody; all, cells were cotransfected with cDNAs for all four of the other eIF2B subunits. The weak signals in, e.g., the lanes for αδɛ with His-myc-eIF2Bβ in panel C may reflect some “bridging” of the recombinant subunits by the endogenous eIF2B subunits. Panel D presents a cartoon summarizing the data, showing which complexes between eIF2B subunits can form in mammalian cells, i.e., eIF2Bγɛ, eIF2Bβγδɛ, and eIF2Bαβγδɛ.
His-myc-eIF2Bβ was unable to associate with any of the other individual subunits (Fig. 1A). We did not detect an eIF2Bαβδ complex, suggesting that association with eIF2Bγɛ may be needed for stable interaction between the mammalian α, β, and δ subunits (in contrast with the situation for yeast eIF2B (reference 40 and Fig. 1C). These data are the first detailed information on the assembly of mammalian eIF2B. Figure 1D is a summary cartoon showing which complexes can form.
Certain VWM mutations affect the ability of eIF2B subunits to form holocomplexes.
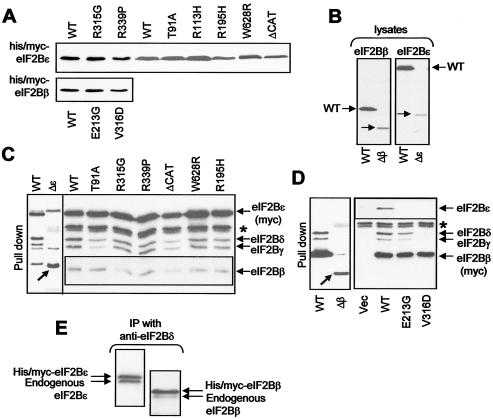
To study whether VWM mutations affected assembly of eIF2B complexes, HEK 293 cells were transfected with vectors for VWM/WT eIF2B subunits, either with the cDNA for a single subunit (Fig. 2) or with the cDNAs for all five (Fig. 3). The missense mutants expressed as polypeptides of sizes similar to that of the WT protein (Fig. 2A). As expected, the constructs mimicking the frameshift mutants gave small polypeptides (Fig. 2B).
FIG. 2.
Formation of complexes containing a VWM mutant version of eIF2Bβ or ɛ. HEK 293 cells were transfected with vectors for WT or selected VWM mutants of the β or ɛ subunits of eIF2B, as fusions with His and myc tags, or with the empty vector (Vec). (A and B) Samples of cell lysate were analyzed by SDS-PAGE and Western blotting by using anti-myc. (C and D) Samples of cell lysate were subjected to chromatography on Ni-NTA-agarose prior to immunoblot analysis of the bound material with anti-myc, -eIF2Bγ, and -eIF2Bδ as well as anti-eIF2Bβ (C) or -eIF2Bɛ (D). Δɛ and Δβ in panels B and D indicate the frameshift mutants, the positions of the truncated polypeptides that they encode being indicated by diagonal arrows. Positions of the other, endogenous eIF2B subunits are also shown. Weak signals for the anti-eIF2Bβ and ɛ antisera in panels C and D, respectively, mean that their signals are shown for longer exposures as insert panels. The asterisk indicates a nonspecific detection by anti-myc of a protein that binds nonspecifically to Ni-NTA-agarose. (E) Cells were transfected with vectors for myc/His-eIF2Bɛ or -β. Lysates were subjected to IP with anti-eIF2Bδ, and precipitates were analyzed by SDS-PAGE and immunoblotting with anti-eIF2Bɛ (left side) or anti-eIF2Bβ (right side).
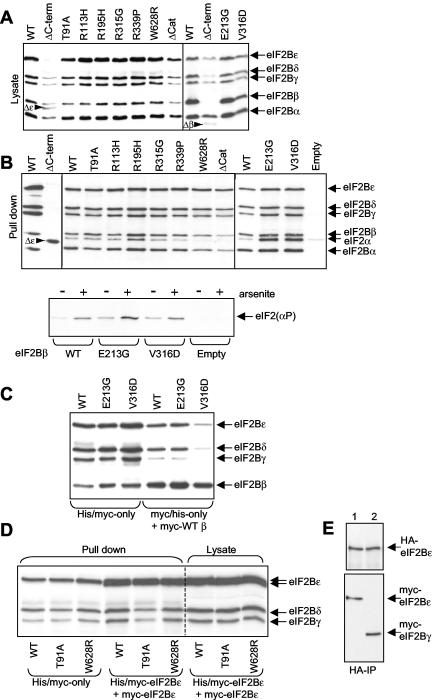
FIG. 3.
Formation of complexes containing VWM variants of eIF2Bβ or -ɛ. HEK 293 cells were transfected with vectors for WT or selected VWM mutants of the β or ɛ subunits of eIF2B, or the ΔCAT mutant of eIF2Bɛ, as fusions with His and myc tags, and with vectors encoding myc-tagged version of the other four subunits. As a negative control, cells were transfected with empty vectors (indicated). Samples of cell lysate were analyzed directly by SDS-PAGE and Western blotting (A) or first subjected to chromatography on Ni-NTA-agarose prior to analysis by SDS-PAGE-immunoblotting (B). Blots were developed with anti-myc (A and B) and anti-eIF2α (B). Positions of the myc-tagged eIF2B subunits and of eIF2α are shown. Δɛ and Δβ indicate the positions of the truncated polypeptides arising from the frameshift mutations. In the lower part of panel B, only the vectors for WT or mutant eIF2Bβ were used (or empty vector as negative control). Samples of cell lysate were subjected to chromatography on Ni-NTA-agarose prior to analysis by SDS-PAGE and Western blotting with anti-eIF2(Ser51[P]). Equal expression of eIF2B subunits was confirmed by probing with anti-myc (data not shown). Where indicated, cells were pretreated with sodium arsenite (0.5 mM, 30 min) prior to lysis. Cells were also transfected with the empty vector, and similar pull-downs were performed to confirm that very little, if any, eIF2 was retained nonspecifically on the resin (B, upper and lower sections). (C and D) As done for panel B, vectors for the indicated His-myc-tagged eIF2B subunits were used together with myc-tagged versions of the other four WT subunits. Where indicated, cells also received DNA for the myc-tagged versions of eIF2Bɛ or β. Data are immunoblots of material retained on Ni-NTA-agarose (except where indicated, “lysate”). In panel D, the two arrows for eIF2Bɛ indicate that the broad band seen for this subunit actually consists of a poorly resolved doublet of species, the lower one being the myc-tagged eIF2Bɛ and the upper one being the myc/His-tagged subunit. (E) HEK 293 cells were transfected with HA- and myc-tagged versions of eIF2Bɛ; those shown in lane 2 also received the vector for myc-eIF2Bγ. Samples of lysates were subjected to IP with anti-HA followed by SDS-PAGE and immunoblotting, development being with anti-HA (upper section) or anti-myc (lower section).
Initially, we expressed the VWM mutant His-myc-tagged eIF2B subunits alone and used Ni-NTA-agarose to isolate them and study their ability to interact with the other, endogenous, subunits. A limitation here is that we are unable to detect eIF2Bα by using the available antisera. However, as shown above, eIF2Bα is not required for complex formation between the other four subunits. The β, γ, and δ subunits were clearly detected in pull-downs from cells expressing WT His-myc-eIF2Bɛ and for each of the five point mutants of eIF2Bɛ tested (Fig. 2C). In contrast, the other subunits were not observed in pull-downs from cells expressing the eIF2Bɛ frameshift mutant, presumably because the truncated polypeptide lacks regions required for interaction with them. It also lacks the catalytic domain (9) and is thus functionally null. Lastly, the pull-downs for the T91A and ΔCAT mutants consistently showed less of the other four subunits than were seen in pull-downs from WT eIF2Bɛ, suggesting that they have reduced ability to form eIF2B holocomplexes (Fig. 2C).
eIF2Bβ(E213G) formed complexes with the other eIF2B subunits to almost the same extent as WT eIF2Bβ (Fig. 2D). In contrast, only very weak signals were observed for the other subunits following affinity isolation of eIF2Bβ(V316D) on NTA-agarose (Fig. 2D), indicating that it cannot efficiently form holocomplexes. The polypeptide that mimics the frameshift M203fs failed to interact with the other subunits, suggesting that the C terminus of eIF2Bβ is needed for this. Thus, certain VWM mutations affect eIF2B function by impairing holocomplex formation. As complex formation greatly enhances eIF2Bɛ activity (10), this is likely to reduce markedly eIF2B activity in patients' cells.
To assess what proportion of total eIF2B complexes in the transiently transfected cells contain the recombinant subunit, we immunoprecipitated complexes with anti eIF2Bδ and analyzed the precipitates through SDS-PAGE-immunoblotting by using anti-eIF2Bɛ or -β. As seen in Fig. 2E, >50% and almost 80% of the complexes contain the recombinant eIF2Bɛ and -β proteins, respectively.
The potencies of antisera for eIF2Bβ, -γ, -δ, and -ɛ differ considerably, and there is no suitable antibody for eIF2Bα. We therefore also used an additional approach in which we coexpressed the eIF2B subunit under study with the other four polypeptides, each with a myc tag. All five subunits were fairly evenly expressed as assessed by immunoblotting of lysates with anti-myc (Fig. 3A). However, eIF2Bɛ(ΔCAT) and the truncation mutants of eIF2Bɛ and -β were expressed at lower levels, perhaps because they are less able to form complexes. When His-myc-tagged eIF2Bɛ missense mutant polypeptides were isolated on Ni-NTA-agarose, the other four myc-tagged subunits were readily detected in every case (Fig. 3B, top), showing that eIF2Bα is incorporated into these complexes.
Similar data were obtained for WT and VWM mutant eIF2Bβ (Fig. 3B). The data for the eIF2Bɛ(T91A) and eIF2Bβ(V316D) mutants obtained here differ from those of Fig. 2, in which only single subunits were expressed and where both showed decreased ability to form eIF2B complexes. In the earlier experiments, the overexpressed subunit must compete with the corresponding endogenous polypeptide for binding to a small pool of the other four eIF2B polypeptides. It thus appears that eIF2Bɛ(T91A) and eIF2Bβ(V316D) can indeed form complexes but fail to compete efficiently with the endogenous WT eIF2Bβ.
To test this, we examined whether the mutant eIF2B proteins could compete with WT myc-tagged eIF2Bβ and -ɛ to form holocomplexes. As expected, coexpression of WT myc-eIF2Bβ with His-myc WT eIF2Bβ or the E213G mutant reduced somewhat the amounts of the other subunits that bound to the His-myc-tagged polypeptide (Fig. 3C). However, coexpression of WT eIF2Bβ almost completely blocked the association of the eIF2Bβ(V316D) mutant with the other subunits.
In the case of eIF2Bɛ(T91A), we saw only a modest effect from expressing WT-myc eIF2Bɛ alongside the T91A-His-myc protein (Fig. 3D). Surprisingly, we also saw that, along with the His-myc-eIF2Bɛ polypeptide, the slightly smaller myc-tagged eIF2Bɛ polypeptide was also present in the pull-down, suggesting that eIF2Bɛ might dimerize, which could account for the small decrease in recovery of the T91A mutant. To confirm this, we coexpressed HA-tagged eIF2Bɛ with His-myc-eIF2Bɛ. After IP with anti-HA, His-myc-eIF2Bɛ was indeed clearly seen (Fig. 3E, lower part, lane 1). As described above, eIF2Bɛ interacts with eIF2Bγ in eIF2B holocomplexes (2, 25). Parts of their sequences are homologous (2) and could be involved in the γ-ɛ interaction. Thus, one explanation is that, when eIF2Bɛ is expressed at high levels, it self dimerizes via the region resembling eIF2Bγ. To test this, we overexpressed myc-eIF2Bγ with both HA- and myc-tagged eIF2Bɛ: strikingly, only myc-eIF2Bγ was now seen in the anti-HA IP (Fig. 3E, lower part, lane 2). Dimerization of eIF2Bɛ is thus likely an artifact of overexpression.
VWM mutations decrease the activity of eIF2.
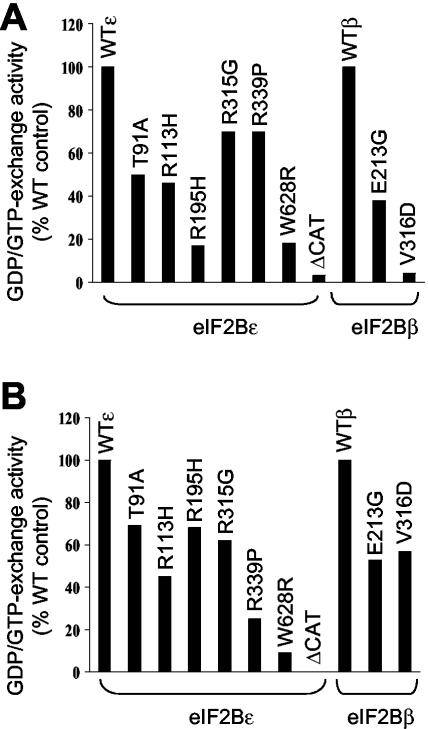
The major aim of this study was to assess whether, and how, VWM mutations affect the function of eIF2B. The above data show that most of the mutated subunits can efficiently form eIF2B complexes, at least when overexpressed with the four other subunits. We therefore assayed the activity of complexes containing recombinant WT or VWM mutant subunits after isolation from extracts of transfected cells by using NTA-resin and our standard assay with preformed eIF2-[3H]GDP complexes as substrate. Initially, we expressed a single WT and/or mutant eIF2B subunit and isolated complexes containing this polypeptide and endogenous subunits. Although eIF2Bɛ displays some activity on its own (6, 10), this is greatly enhanced by association with the other subunits (10), as verified by earlier studies with HEK 293 cells (39). Thus, although the pull-down material contains both free His-myc-eIF2Bɛ and complexes containing this polypeptide, the activity detected overwhelmingly reflects that of complexes. In all cases, complexes containing VWM mutants of eIF2Bɛ showed lower activity than WT complexes (Fig. 4A). The defect varied from mild for, e.g., eIF2Bɛ(R315G) to severe for the W628R mutation. No exchange activity was detected for NTA-pull-downs of the eIF2Bβ or eIF2Bɛ truncation mutants, consistent with their inability to form eIF2B holocomplexes and with the absence from the eIF2Bɛ frameshift mutant of the C-terminal catalytic domain (9).
FIG. 4.
Effects of VWM mutations on the activity of eIF2B. HEK 293 cells were transfected with vectors for WT or selected VWM mutant versions of the β or ɛ subunits of eIF2B, or the ΔCAT mutant of eIF2Bɛ, as fusions with His and myc tags (A and B), and with vectors encoding myc-tagged version of the other four subunits (B). Samples of cell lysate were subjected to chromatography on Ni-NTA-agarose prior to analysis. Levels of expression were carefully assessed by SDS-PAGE and Western blot analysis (using anti-myc) of the bound material to normalize the amounts used in each assay. Appropriate amounts of this bound material were then assayed for GDP/GTP exchange activity by using eIF2-[3H]GDP complexes as the substrate. Data are shown as percentage of activity with corresponding WT subunit and are typical of between 4 and 10 experiments performed.
By using the above approach, we could not obtain accurate data for the eIF2Bɛ(T91A) and eIF2Bβ(V316D) mutants as they do not efficiently form eIF2B holocomplexes. Thus, we tested the VWM mutant under study expressed together with the other four WT subunits, since they can form holocomplexes under this set of conditions (Fig. 3B, upper part). The eIF2Bɛ(T91A) and eIF2Bβ(V316D) mutants still displayed low intrinsic GDP/GTP exchange activity, in addition to their defective abilities to form complexes (Fig. 4B). Since Western analysis of these pull-downs shows similar amounts of all five recombinant subunits (Fig. 3B, upper part), these data also accurately reflect the activities of other VWM mutant eIF2B complexes. The data in Fig. 1 rule out the occurrence of diverse complexes of differing subunit compositions.
Some VWM mutations affect the binding of eIF2B to eIF2.
Since neither of the truncation mutants forms complexes and both are thus effectively null mutations, we focused our studies on the point mutations, which all reduce eIF2B activity. One way in which they might do this is by altering the ability of eIF2B to bind eIF2. This involves multiple interactions. A lysine block in eIF2β interacts with the extreme C terminus of eIF2Bɛ (1, 18, 19). The eIF2Bαβδ subcomplex interacts with eIF2α, especially when eIF2α is phosphorylated (20, 25). To study binding of eIF2B to eIF2, we analyzed Ni-NTA-agarose pull-downs from cells transfected with vectors for the His-myc-tagged WT-VWM mutant eIF2Bɛ or β- and myc-tagged versions of the other four subunits for the presence of eIF2α. In most cases, complexes containing VWM mutants showed binding to eIF2 similar to that shown by the WT complexes (Fig. 3B, upper part). However, in three cases marked differences were seen. Firstly, complexes containing eIF2Bɛ(W628R) bound markedly less eIF2. Secondly, both the eIF2Bβ point mutations tested here actually increase the binding of eIF2. In other experiments, NTA-agarose pull-down experiments were performed and the bound material was analyzed for eIF2(αS51[P]). The amount of eIF2α(P) bound to complexes containing the VWM mutants of eIF2Bβ was greater than for the WT complexes (consistent with the increased binding of total eIF2α) (Fig. 3B). Treatment of cells with sodium arsenite, which enhances eIF2α phosphorylation, increased this binding to similar extents for the complexes containing WT and mutant eIF2B (Fig. 3B, lower part). It thus seems that the VWM point mutations do not affect association of eIF2B with eIF2α(P), and it is thus unlikely that they abolish regulation of eIF2B by eIF2α(P). This is consistent with the data for the effects of VWM mutations on the properties of yeast eIF2B (29).
The VWM mutations do not confer a temperature-sensitive phenotype.
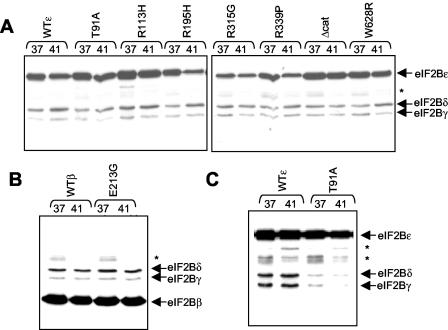
Clinically, the onset or deterioration of VWM symptoms in children is associated with febrile infection (12, 34-36). It has therefore been suggested that VWM mutant eIF2B complexes might show a temperature-sensitive phenotype. We used a range of approaches to test this. WT or VWM eIF2B polypeptides were expressed in HEK 293 cells, as detailed above. In some experiments, we subjected cells to thermal stress by incubating them at an elevated temperature, usually 41°C. Cells were lysed and samples were processed for analysis of formation of eIF2B complexes and an activity assay. Elevated temperatures did not generally affect the integrity of eIF2B complexes containing WT or VWM mutant eIF2Bɛ or -β (Fig. 5A and B): amounts of other subunits copurifying with the myc-His-tagged eIF2B subunit under study were the same from lysates of cells kept at 37°C or were transferred for 2 h to a higher temperature. In some cases, the total amount of the tagged subunit fell at the higher temperature but not the amount of the other subunits copurifying with it. Effects were similar for the WT and VWM mutant polypeptides and likely reflect greater instability of the excess single subunits than of those that incorporated into complexes. In some experiments, eIF2B complexes containing eIF2Bɛ(T91A) did appear less stable than those containing WT eIF2Bɛ (Fig. 5C). It was also possible that the intrinsic activity of VWM eIF2B is thermosensitive. However, in multiple experiments, we saw no thermosensitivity of WT or VWM eIF2B at temperatures that ranged up to suprapathological values (44°C). This was true whether the elevated temperature was applied to the cells prior to isolation of eIF2B complexes, following their isolation but prior to assay or during the assay.
FIG. 5.
Analysis of potential thermosensitivity of eIF2B complexes containing WT or VWM mutant eIF2B subunits. HEK 293 cells were transfected with vectors for WT or selected VWM mutant versions of the ɛ (A and C) or β (B) subunits of eIF2B, as fusions with His and myc tags. In some cases, cells were held at 41° for 2 h prior to lysis instead of at the normal temperature of 37°C. Samples of lysate were subjected to chromatography on Ni-NTA-agarose prior to analysis by SDS-PAGE and Western blotting with anti-myc for His-myc-eIF2Bɛ (A and C) and for His-myc-eIF2Bβ (B) and anti- eIF2Bγ and eIF2Bδ. *, nonspecific detection by anti-myc of proteins that bind nonspecifically to Ni-NTA-agarose.
Functional consequences of VWM mutations for specific protein synthesis in vivo.
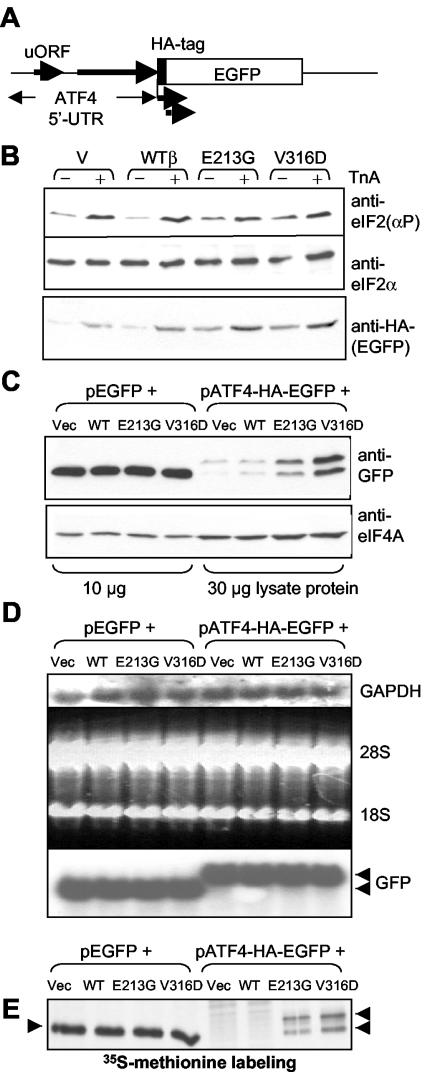
It was important to assess the effects of VWM mutations on the function of eIF2B in vivo. However, even though free eIF2Bɛ has only low activity, overall eIF2B activity in cells expressing it will increase, obscuring any effects due to impaired activity due to VWM mutations. We therefore focused on eIF2Bβ as it lacks intrinsic activity and 80% of eIF2B complexes in cells overexpressing eIF2Bβ contain the recombinant subunit (Fig. 2E). The translation of certain mRNAs whose 5′ UTRs contain multiple uORFs provides a sensitive readout for small decreases in eIF2B activity; e.g., the ATF4 mRNA is regulated in this way (13). We therefore used a vector encoding enhanced green fluorescent protein (EGFP) downstream of the ATF4 5′ UTR to assess alterations in eIF2B activity in vivo. The EGFP was cloned behind the ATF4 5′ UTR with an HA tag to facilitate detection (Fig. 6A). The control vector pEGFP yields an mRNA with a very short 5′ UTR devoid of uORFs.
FIG. 6.
VWM mutants of eIF2Bβ enhance expression of EGFP from a reporter construct containing the ATF4 5′ UTR. (A) Diagram showing the reporter construct based on the 5′ UTR of the ATF4 mRNA. The broad arrows indicate that the reporter vector gives rise to two translation products, one of which lacks the HA tag (lower, broken arrow) (see panels B, C, and E). (B to E) HEK 293 cells were transfected with vectors for WT eIF2Bβ or the indicated VWM mutants or the empty vector (Vec), along with the reporter construct containing the 5′ UTR of mouse ATF4 ahead of the coding region for HA-tagged EGFP. (C) The control reporter (pEGFP) was used in place of the ATF4-based vector where indicated. In panels B and C, samples were analyzed by SDS-PAGE and Western blotting by using the indicated antisera. (Anti-eIF4A was used as a loading control.) For panel B, where indicated, cells were treated with TnA (2.5 μg/ml) for 16 h prior to lysis. The two bands seen with anti-GFP for the ATF4-based vector in panel C reflect the fact that a second in-frame AUG within the HA-EGFP cistron gives rise to a product lacking the HA tag (only the upper band is recognized by anti-HA). This downstream start codon is likely the start codon for the EGFP itself. This phenomenon probably reflects continued ribosome movement beyond the ATF4 5′ UTR leading up to acquisition by the ribosome of active eIF2, allowing use of the downstream start codon. (D) Samples of lysate were processed for Northern blot analysis. Shown are the autoradiography data obtained with probes for GAPDH (top) and GFP (bottom) and the stained gel showing the positions of the 18S and 28S rRNAs (middle: the top and middle panels serve as loading controls). The ATF4-EGFP mRNA is, as expected, larger than the EGFP one. (E) Cells were labeled with [35S]methionine (see Materials and Methods), and samples of cell lysate were analyzed by IP with anti-GFP, SDS-PAGE, and fluorography to detect the labeled HA-EGFP polypeptides (positions indicated by arrowheads). Again, two bands are seen here, for the reason described above.
As shown in Fig. 6B, treatment of HEK 293 cells with tunicamycin A (TnA) led to increased phosphorylation of eIF2α (due to activation of the eIF2α kinase PERK) (14). In cells transfected with empty vector or with vector for WT eIF2Bβ, the level of expression of HA-tagged EGFP was low under control conditions and increased after treatment with TnA (Fig. 6B). This is as expected, since the TnA-induced increase in eIF2α phosphorylation inhibits eIF2B, promoting translation of the ATF4-HA-EGFP mRNA. WT eIF2Bβ had little effect on the expression of HA-EGFP. In cells expressing either of the eIF2Bβ mutants, increased basal expression of HA-EGFP was observed even without TnA treatment (Fig. 6B and C), consistent with the data showing that both mutations impair the activity of eIF2B. These findings also imply that, in VWM patients, these mutations may increase translation of mRNAs such as that for ATF4, potentially leading (as ATF4 is a transcription factor) to changes in the expression of multiple genes. No change in expression of EGFP from the control vector was seen (Fig. 6C), implying that there are no gross changes in protein synthesis (Fig. 2E). This is consistent with data from direct measurements of protein synthesis (our unpublished findings) and the fact that no growth defects have been reported for VWM patients with these mutations.
When cells expressing VWM mutants of eIF2Bβ were treated with TnA, a further increase in HA-EGFP expression was seen. Given that a high proportion of eIF2B complexes in cells transfected with eIF2Bβ cDNAs contain the recombinant subunit (Fig. 2E), the data suggest that the mutant complexes probably remain sensitive to inhibition by eIF2(αP). Northern blot analysis revealed that mRNA expression was not altered by VWM eIF2B (Fig. 6D) and that increased rates of synthesis of EGFP were directly confirmed by [35S]methionine labeling (Fig. 6E).
Further analysis revealed that expression of the VWM mutant eIF2Bβ subunits, but not WT eIF2Bβ, by itself increased the phosphorylation of eIF2α (Fig. 6B, see also Fig. 3B, lower part)). Thus, the elevated level of expression of EGFP from the ATF4-based reporter may stem both from increased eIF2α phosphorylation and the lower intrinsic activity of the eIF2B complexes containing the mutant eIF2Bβ. The overall point remains the same, that expression of the WVM mutants activates translation of mRNAs containing uORFs. Since eIF2B binds phosphorylated eIF2 more tightly than it binds dephosphorylated eIF2 (30), this finding may explain why more eIF2 is found associated with eIF2B complexes containing the eIF2Bβ mutants (Fig. 3B).
DISCUSSION
The aim of this study was to examine the effects of VWM mutations in eIF2B genes on the properties of human eIF2B. This is the first key step to understanding the molecular and cellular pathology of this devastating condition. The data described here provide the first information on the effects on the human eIF2B complex of mutations that give rise to VWM in humans and reveal that all the VWM mutations tested in either the β or ɛ subunit of eIF2B result in a partial loss of function. This is probably as expected, as VWM shows a recessive pattern of inheritance, and is in agreement with the data in reference 29 on the effects of VWM mutations on yeast eIF2B. A striking feature of our data is the diverse ways in which VWM mutations affect eIF2B.
The frameshift mutants give rise to severely truncated proteins that are expressed poorly in HEK 293 cells. They cannot form eIF2B holocomplexes and thus fail to give rise to productive eIF2B complexes. These mutations essentially give rise to a complete loss of function and therefore are null, but not interfering, mutants. It is therefore not surprising that these mutations occur only in a compound heterozygous manner, since simple homozygotes would lack functional eIF2B (22) (a situation that is almost certainly lethal). Patients with these mutations therefore rely on their second (point-mutated) copy of the relevant gene.
All the missense mutations tested give rise to full-length polypeptides that can form eIF2B holocomplexes, although some show reduced abilities to do so. These are the eIF2Bɛ(T91A) mutant, which shows a modest impairment of complex formation, and the eIF2Bβ(V316D) mutant, which shows a more severe deficit in complex formation. T91 is conserved in the known mammalian eIF2Bɛ sequences but is not found in sequences from other species. V316 may be a more critical residue, as this residue is also found at the corresponding positions in eIF2Bβ sequences from other mammals and from Saccharomyces cerevisiae, Drosophila melanogaster, and Arabidopsis thaliana. This may explain the more drastic effect of a mutation at this position on eIF2B complex assembly. When coexpressed with the other four human polypeptides at high levels, both variants were able to form complexes, showing that they are not completely unable to interact with the other subunits but are merely less able to do so. Measurements of the GDP/GTP exchange activity of such complexes showed that they have lower activity than do complexes containing WT eIF2B. Our data for the eIF2Bβ(V316D) mutant agree closely with those of the authors of reference 29, who found that the corresponding yeast mutation (V341D) also impaired complex formation and activity.
To try to explain the basis of the reduced activity of these VWM eIF2B complexes, we examined their ability to bind to eIF2, the substrate for eIF2B. Three mutations altered the binding of eIF2B to eIF2. The eIF2Bɛ(W628R) mutation markedly decreased the ability of the eIF2B complex to bind eIF2. The fact that this residue lies within the catalytic domain of eIF2Bɛ (9) may explain why it impairs substrate binding. It is important that this tryptophan residue is not one of the conserved tryptophans within the cluster of aromatic and acidic residues that constitute the eIF2β binding site at the extreme C terminus of eIF2Bɛ (1). This decrease in substrate binding may explain the reduced nucleotide exchange activity of eIF2B complexes containing the eIF2Bɛ(W628R) mutant. Both the eIF2Bβ missense mutants tested here showed increased binding to eIF2. This appears surprising since their activities are also lower than for the WT complex. However, their expression increases the level of phosphorylation of eIF2α, and the increased binding may thus reflect the higher affinity of eIF2α(P) for eIF2B than that of the nonphosphorylated factor (30). This would lead to inhibition of eIF2B activity. Thus, the observed effects of these eIF2Bβ mutants on the ATF4-based reporter may reflect both the reduced activity of complexes containing them and the increased phosphorylation of eIF2α (Fig. 6). It is unclear why expression of mutant eIF2Bβ increases basal eIF2α phosphorylation. The accumulation of (unfolded) mutant eIF2Bβ may elicit a stress response leading to increased eIF2α phosphorylation, or the lower activity of the VWM mutant eIF2B complexes may lead to increased translation of stress genes (e.g., ATF4). It will be important to study whether eIF2α phosphorylation and stress protein expression are increased in cells from VWM. Our data provide no information to suggest that the VWM mutations affect the susceptibility of eIF2B to regulation by eIF2α phosphorylation.
Episodes of infection accompanied by fever cause deterioration of VWM patients, and it has been suggested (37) that VWM mutations might confer on the eIF2B complex a thermosensitive phenotype. However, in extensive studies that used a range of complementary approaches, we did not detect any general enhancement of the temperature sensitivity of VWM eIF2B. Like the WT complex, its activity was not affected by temperatures up to 44°C. Our data agree with those of Richardson et al. (29), who also found no evidence for thermosensitivity of yeast eIF2B complexes containing VWM mutant subunits. The link between VWM and pyrexia thus appears more complex than a simple model where eIF2B complexes containing VWM mutant subunits are thermosensitive: perhaps the already decreased eIF2B activity in brain tissue of VWM patients is further reduced due to the increased phosphorylation of eIF2α that occurs during pyrexia (reviewed in reference 32) or after traumatic brain injury (26). This may have marked effects either on total protein synthesis or, more likely, on the translation of specific mRNAs.
It is known from other studies that inhibition of eIF2B due to increased phosphorylation of its substrate eIF2 leads to upregulation of the translation of certain mRNAs, e.g., those for GCN4 in yeast (16) and ATF4 in mammals (13). eIF2α phosphorylation—and thus regulation of eIF2B activity—also appears to be important for the control of other transcription factors (15, 17, 33). As both proteins are transcriptional regulators, this effect could in principle affect the expression of multiple genes. Given that VWM mutations impair the activity of eIF2B, we asked whether expression of VWM mutant eIF2B had a similar effect. Using a reporter construct in which the ATF4 5′ UTR was cloned upstream of the coding region for an HA-tagged version of EGFP, we showed that both the eIF2Bβ point mutants do indeed enhance the expression of the reporter protein. Thus, one anticipates that the translation of mRNAs that are regulated in this way would be upregulated in VWM patients. Examples of such mRNAs are already known in mammals (e.g., ATF4), and others likely await discovery. If such mRNAs were expressed in a tissue-specific manner (e.g., in brain), this could explain why the phenotype of VWM is primarily neurological. Alternatively, it could be that overall protein synthesis in neuronal tissue is especially sensitive to perturbations in eIF2B activity.
Further progress requires cells (in particular, brain cells) that homozygously express VWM eIF2B, e.g., the generation of transgenic knock-in mice bearing VWM mutations. Work towards this goal is already under way.
Acknowledgments
This work was supported by the Wellcome Trust (grants 066330 and 067782).
We are grateful to David Ron for generously providing reagents.
REFERENCES
- 1.Asano, K., T. Krishnamoorthy, L. Phan, G. D. Pavitt, and A. G. Hinnebusch. 1999. Conserved bipartite motifs in yeast eIF5 and eIF2Bɛ, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 18:1673-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushman, J. L., A. I. Asuru, R. L. Matts, and A. G. Hinnebusch. 1993. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:1920-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemens, M. J. 2001. Initiation factor eIF2α phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell. Biol. 27:57-89. [DOI] [PubMed] [Google Scholar]
- 4.Colthurst, D. R., D. G. Campbell, and C. G. Proud. 1987. Structure and regulation of eukaryotic initiation factor eIF-2. Sequence of the site in the alpha subunit phosphorylated by the haem-controlled repressor and by the double-stranded RNA-activated inhibitor. Eur. J. Biochem. 166:357-363. [DOI] [PubMed] [Google Scholar]
- 5.Craddock, B. L., and C. G. Proud. 1996. The α-subunit of mammalian initiation factor eIF-2B is essential for catalytic activity. Biochem. Biophys. Res. Commun. 220:843-847. [DOI] [PubMed] [Google Scholar]
- 6.Fabian, J. R., S. R. Kimball, N. K. Heinzinger, and L. S. Jefferson. 1997. Subunit assembly and guanine nucleotide exchange factor activity of eukaryotic initiation factor eIF2B subunits expressed in Sf9 cells. J. Biol. Chem. 272:12359-12365. [DOI] [PubMed] [Google Scholar]
- 7.Fogli, A., D. Rodriguez, E. Eymard-Pierre, F. Bouhour, P. Labauge, B. F. Meaney, S. Zeesman, C. R. Kaneski, R. Schiffmann, and O. Boespflug-Tanguy. 2003. Ovarian failure related to eukaryotic initiation factor 2B mutations. Am. J. Hum. Genet. 72:1544-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fogli, A., K. Wong, E. Eymard-Pierre, J. Wenger, J. P. Bouffard, E. Goldin, D. N. Black, O. Boespflug-Tanguy, and R. Schiffmann. 2002. Cree leukoencephalopathy and CACH/VWM disease are allelic at the EIF2B5 locus. Ann. Neurol. 52:506-510. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, E., S. S. Mohammad, and G. D. Pavitt. 2002. Characterization of the minimal catalytic domain within eIF2B: the guanine-nucleotide exchange factor for translation initiation. EMBO J. 21:5292-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez, E., and G. D. Pavitt. 2000. Identification of domains and residues within the ε subunit of eukaryotic translation initiation factor 2B (eIF2Bε) required for guanine nucleotide exchange reveals a novel activation function promoted by eIF2B complex formation. Mol. Cell. Biol. 20:3965-3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall-Jackson, C. A., D. A. Cross, N. Morrice, and C. Smythe. 1999. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 18:6707-6713. [DOI] [PubMed] [Google Scholar]
- 12.Hanefeld, F., U. Holzbach, B. Kruse, E. Wilichowski, H. J. Christen, and J. Frahm. 1993. Diffuse white matter disease in three children: an encephalopathy with unique features on magnetic resonance imaging and proton magnetic resonance spectroscopy. Neuropediatrics 24:244-248. [DOI] [PubMed] [Google Scholar]
- 13.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 14.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 15.Harding, H. P., Y. Zhang, H. Zeng, I. Novoa, P. D. Lu, M. Calfon, N. Sadri, C. Yun, B. Popko, R. Paules, D. F. Stojdl, J. C. Bell, T. Hettmann, J. M. Leiden, and D. Ron. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11:619-633. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, A. G. 2000. Mechanism and regulation of methionyl-tRNA binding to ribosomes, p. 185-243. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Scheuner, R. J. Kaufman, D. R. Cavener, and R. C. Wek. 2003. Phosphorylation of the α subunit of eukaryotic initiation factor 2 is required for activation of NF-κB in response to diverse cellular stresses. Mol. Cell. Biol. 23:5651-5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimball, S. R., N. K. Heinzinger, R. L. Horetsky, and L. S. Jefferson. 1998. Identification of interprotein interactions between the subunits of eukaryotic initiation factors eIF2 and eIF2B. J. Biol. Chem. 273:3039-3044. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V. 1995. Multidomain organization of eukaryotic guanine nucleotide exchange factor eIF-2B revealed by analysis of conserved sequence motifs. Protein Sci. 4:1608-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamoorthy, T., G. D. Pavitt, F. Zhang, T. E. Dever, and A. G. Hinnebusch. 2001. Tight binding of the phosphorylated α subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 21:5018-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leegwater, P. A. J., A. A. Konst, B. Kuyt, L. A. Sandkuijl, S. Naidu, C. B. Oudejans, R. B. Schutgens, J. C. Pronk, and M. S. Van der Knaap. 1999. The gene for leukoencephalopathy with vanishing white matter is located on chromosome 3q27. Am. J. Hum. Genet. 65:728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leegwater, P. A. J., G. Vermeulen, A. A. M. Koenst, S. Naidu, J. Mulders, A. Visser, P. Kersbergen, D. Mobach, D. Fonds, C. G. M. van Berkel, R. J. L. F. Lemmers, R. Frants, C. B. M. Oudejans, R. B. H. Schutgens, J. C. Pronk, and M. van der Knaap. 2001. Subunits of the translation initiation factor eIF2B are mutated in leukoencephaly with vanishing white matter. Nat. Genet. 29:383-388. [DOI] [PubMed] [Google Scholar]
- 23.Li, W., G. J. Belsham, and C. G. Proud. 2001. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J. Biol. Chem. 276:29111-29115. [DOI] [PubMed] [Google Scholar]
- 24.Pavitt, G., W. Yang, and A. G. Hinnebusch. 1997. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 17:1298-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pavitt, G. D., K. V. A. Ramaiah, S. R. Kimball, and A. G. Hinnebusch. 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyse and regulate guanine-nucleotide exchange. Genes Dev. 12:514-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrov, T., B. D. Underwood, B. Braun, S. S. Alousi, and J. A. Rafols. 2001. Upregulation of iNOS expression and phosphorylation of eIF-2α are paralleled by suppression of protein synthesis in rat hypothalamus in a closed head trauma model. J. Neurotrauma 18:799-812. [DOI] [PubMed] [Google Scholar]
- 27.Price, N. T., S. F. Nakielny, S. J. Clark, and C. G. Proud. 1989. The two forms of the beta-subunit of initiation factor-2 from reticulocyte lysates arise from proteolytic degradation. Biochim. Biophys. Acta 1008:177-182. [DOI] [PubMed] [Google Scholar]
- 28.Proud, C. G. 2001. Regulation of eukaryotic initiation factor eIF2B. Prog. Mol. Subcell. Biol. 26:95-114. [DOI] [PubMed] [Google Scholar]
- 29.Richardson, J., S. S. Mohammad, and G. D. Pavitt. 2004. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 24:2352-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowlands, A. G., R. Panniers, and E. C. Henshaw. 1988. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 263:5526-5533. [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 7.39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Scheper, G. C., R. van Wijk, and A. A. M. Thomas. 2001. Regulation of the activity of eukaryotic initiation factors in stressed cells. Prog. Mol. Subcell. Biol. 27:39-56. [PubMed] [Google Scholar]
- 33.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonners-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 34.Schiffmann, R., J. R. Moller, B. D. Trapp, H. H. Shih, R. G. Farrer, D. A. Katz, J. R. Alger, C. C. Parker, P. E. Hauer, C. R. Kaneski, et al. 1994. Childhood ataxia with diffuse central nervous system hypomyelination. Ann. Neurol. 35:331-340. [DOI] [PubMed] [Google Scholar]
- 35.Van der Knaap, M. S., P. G. Barth, F. J. Gabreels, E. Franzoni, J. H. Begeer, H. Stroink, J. J. Rotteveel, and J. Valk. 1997. A new leukoencephalopathy with vanishing white matter. Neurology 48:845-855. [DOI] [PubMed] [Google Scholar]
- 36.Van der Knaap, M. S., W. Kamphorst, P. G. Barth, C. L. Kraaijeveld, E. Gut, and J. Valk. 1998. Phenotypic variation in leukoencephalopathy with vanishing white matter. Neurology 51:540-547. [DOI] [PubMed] [Google Scholar]
- 37.Van der Knaap, M. S., P. A. J. Leegwater, A. A. M. Könst, A. Visser, S. Naidu, C. B. M. Oudejans, R. B. J. Schutgens, and J. C. Pronk. 2002. Mutations of each of the five subunits of translation initiation factor eIF2B can cause leukoencephalopathy with vanishing white matter. Ann. Neurol. 51:264-270. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., M. Janmaat, A. Beugnet, F. E. M. Paulin, and C. G. Proud. 2002. Evidence that the dephosphorylation of Ser535 in the ɛ-subunit of eukaryotic initiation factor 2B is insufficient for the activation of eIF2B by insulin. Biochem. J. 367:475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, X., F. E. M. Paulin, L. E. Campbell, E. Gomez, K. O'Brien, N. Morrice, and C. G. Proud. 2001. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon subunit and their roles in vivo. EMBO J. 20:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, W., and A. G. Hinnebusch. 1996. Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol. Cell. Biol. 16:6603-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]