Abstract
Recent studies have suggested that Spt6 participates in the regulation of transcription by RNA polymerase II (RNAPII). However, its underlying mechanism remains largely unknown. One possibility, which is supported by genetic and biochemical studies of Saccharomyces cerevisiae, is that Spt6 affects chromatin structure. Alternatively, Spt6 directly controls transcription by binding to the transcription machinery. In this study, we establish that human Spt6 (hSpt6) is a classic transcription elongation factor that enhances the rate of RNAPII elongation. hSpt6 is capable of stimulating transcription elongation both individually and in concert with DRB sensitivity-inducing factor (DSIF), comprising human Spt5 and human Spt4. We also provide evidence showing that hSpt6 interacts with RNAPII and DSIF in human cells. Thus, in vivo, hSpt6 may regulate multiple steps of mRNA synthesis through its interaction with histones, elongating RNAPII, and possibly other components of the transcription machinery.
Accumulating evidence suggests that Spt6 is an evolutionarily conserved, positive regulator of transcription by RNA polymerase II (RNAPII). Circumstantial evidence points to the possibility that Spt6 may play a role similar to that of Spt5. For example, in the yeast Saccharomyces cerevisiae, Spt6 is essential for growth and important for transcription of a subset of genes (28). Yeast Spt6 interacts genetically with Spt5, whereas their physical interaction is weak or absent (8, 15, 16). In Drosophila spp., Spt6 is associated with several transcriptionally active chromosomal loci, including hsp70 loci, after heat shock induction (1, 11, 21). Immunofluorescence revealed that localization of Drosophila Spt6 and Spt5 significantly overlaps the chromosomes (1, 11, 21). In zebra fish, homozygous mutations within the Spt5 or Spt6 gene cause similar pleiotropic developmental defects (13). In addition, both mutant embryos are defective in heat shock response (13).
Despite the wealth of information available, the molecular mechanism by which Spt6 regulates RNAPII transcription remains largely unknown. One established supposition is that it acts as a regulator of chromatin structure. In S. cerevisiae, Spt6 genetically interacts with histones H2A and H2B and with components of the Swi/Snf protein complex (3, 5, 24). In addition, biochemical analysis has shown that yeast Spt6 associates physically with histones H3 and H4 and directs nucleosome assembly from a plasmid DNA and core histones in vitro (3). Recently, it has been reported that Spt6 plays a critical role in maintaining normal chromatin structure (12). A second possibility is that Spt6 may regulate mRNA synthesis through its association with the exosome protein complex, which is probably involved in mRNA surveillance (2). Although the interaction between Drosophila Spt6 and components of the exosome has been clearly demonstrated (2), the functional importance of this interaction remains to be elucidated. A third possibility, which we have specifically addressed, is that Spt6 may directly regulate transcription elongation by RNAPII. This inference is engendered in the striking similarities between Spt6 and Spt5 in several aspects, as noted above. Human Spt5 (hSpt5) and human Spt4 (hSpt4) form a biochemically defined transcription elongation factor complex termed DSIF. DSIF associates with RNAPII and is able to both repress and activate transcription elongation in vitro (4, 18, 25, 30, 31). An additional protein complex, termed negative elongation factor (NELF), is required for DSIF-dependent transcription inhibition (19, 32); both the stimulatory and repressive activities of DSIF are regulated by positive transcription elongation factor (P-TEFb) (10, 25, 26). The idea that Spt6 may directly regulate transcription elongation is also supported by genetic analyses of S. cerevisiae. Yeast Spt6 interacts genetically with the transcription elongation factor TFIIS. Furthermore, a mutation in the SPT6 gene causes a 6-azauracil-sensitive phenotype, indicating a defect in transcription elongation (8).
This study investigated the possible roles of human Spt6 (hSpt6) in transcription elongation by using naked DNA templates in vitro. We demonstrate that hSpt6 enhances the rate of transcription elongation, probably through its interaction with RNAPII elongation complexes. We also provide evidence that hSpt6 interacts with RNAPII and DSIF in human cells. Thus, in vivo, Spt6 may regulate multiple steps of mRNA synthesis through its interaction with histones, RNAPII elongation complexes, and the exosome complex.
MATERIALS AND METHODS
Preparation of hSpt6-specific antibodies.
Peptide 1 (acetylated LNKKPHVVTVAGENRDAQMLIED), corresponding to amino acids 841 to 863 of hSpt6, and peptide 2 (CNVTGIAHRRPQGESYDQAIRNDE), corresponding to amino acids 1169 to 1192 of hSpt6, were synthesized chemically. Mice were immunized with these peptides, and then monoclonal antibodies were prepared according to the protocol described by Harlow and Lane (7).
Immunoblot analysis.
Immunoblot assays were carried out as described previously (25). The blot filter was developed with the ECL system. Reprobing was then carried out according to the manufacturer's protocol (Amersham Biosciences Corp.).
Cloning of hSpt6 cDNA.
HeLa cell cDNA synthesized from total RNA by using random primers and SuperScript II (Gibco Invitrogen Co.) was used as a template for PCR amplification of hSpt6 cDNA. Information on the hSpt6 cDNA found in GenBank was used to prepare the following six primers: hSpt6-1 (GCGTGTCAAACATATGTCAGATGACGAGGA), hSpt6-2 (TCCTTTCTACCTTTCTTGGTGGGGGT), hSpt6-3 (CCTACTCCAGAAGCTGTGCTAGAAG), hSpt6-4 (TGAAGATCTCCTCTGTGTTGGGAGAG), hSpt6-5 (AGCTGTCGATATAAGGACCTCCGGA), and hSpt6-6 (GACCACCAAAGAGAACGTCTCACTTCA).
PCR with KOD-plus polymerase (Toyobo Co.) was carried out with HeLa cell cDNA and the following primer combinations: hSpt6-1 and hSpt6-2, hSpt6-3 and hSpt6-4, and hSpt6-5 and hSpt6-6. The amplified DNA fragments were phosphorylated with T4 polynucleotide kinase (Toyobo Co.) and cloned into the EcoRV site within the polylinker region of pBluescriptSK(+) (Stratagene). The DNA fragments amplified by the first, second, and third sets of primers encoded the N-terminal, central, and C-terminal regions of hSpt6, respectively. In turn, these fragments yielded the plasmids pBS-hSpt6Nterm, pBS-hSpt6center, and pBS-hSpt6Cterm, respectively. Subsequently, pBS-hSpt6Nterm was digested with NdeI-HaeII, pBS-hSpt6center was digested with HaeII-MroI, and pBS-hSpt6Cterm was digested with MroI and partially digested with BamHI. Each fragment was cloned in the correct order into the NdeI-BamHI sites of pET-14b (Novagen Inc.) to generate the pET-hSpt6 (mutant 1) expression vector.
Construction of expression plasmids for wild-type and mutant hSpt6.
Baculovirus expression vectors for recombinant hSpt6 proteins were constructed from hSpt6 cDNA in the pET-hSpt6 (mutant 1) plasmid by using a PCR method. The sequences of oligonucleotides used in PCRs are as follows: mut1-5′ (ATGGTCGACGACTACAAGGACGACGATGACAAGCATATGTCAGATGACGAGG ACGA), mut1-3′ (CTAGCGGCCGCGTAACCAGAGTCCGAGGAGC), mut2-3′ (CTAGCGGCCGCCTAGGATGCCATGGGCTGGACAT), and mut3-5′ (ATGGTCGACGACTACAAGGACGACGATGACAAGCATATGAGAGTCTGGCAGTGGGATGA).
All 5′ primers were designed to include the FLAG tag sequence. PCR products were inserted into the pFastbacHTc plasmid (Invitrogen Corp.) between the SalI and NotI sites, which were created in the 5′ primers and 3′ primers, respectively. Mutant 1 cDNA amplified by primers mut1-5′ and mut1-3′ was inserted into pFastbacHTc to generate pFastbacHTc-hSpt6mutant1. Mutant 2 cDNA amplified by primers mut1-5′ and mut2-3′ was inserted into pFastbacHTc to generate pFastbacHTc-hSpt6mutant2. Mutant 3 cDNA amplified by primers mut3-5′ and mut1-3′ was inserted into pFastbacHTc to generate pFastbacHTc-hSpt6mutant3. To generate pFastbacHTc-hSpt6 for expression of wild-type hSpt6 protein, an NdeI- and NheI-digested DNA fragment (1,083 bp) of pFastbacHTc-hSpt6mutant1 was exchanged with an NdeI-NheI-digested DNA fragment (1,452 bp) encoding the N-terminal region of wild-type hSpt6. The DNA fragment (1,452 bp) was generated using primer hSpt6-0 (5′-GGGAATTCCATATGTCTGATTTTGTGGAAAGCGA-3′) and primer hSpt6-7 (5′-CAAACTGCTCGGGAGTAAGC-3′) from HeLa cell cDNA. Plasmids containing amplified cDNA fragments were analyzed using a capillary sequencer (ABI3100). We found three different amino acids at positions 491 (Leu), 1024 (Glu), and 1122 (Ala) instead of Arg, Gly, and Thr, respectively, in the GenBank database (accession number NP_003161). The differences were not caused by PCR error (unpublished data), but we have not otherwise attempted to elucidate the difference. Then, plasmids containing the insert DNA transposed to bacmid via the attachment sites for the Tn7 transposon on pFastbacHTc. Transfection of extracted bacmids into Sf9 cells was carried out with Lipofectin reagent (Invitrogen Corp.).
We carried out two independent immunoblot assays to confirm whether the wild-type hSpt6 cDNA that we isolated actually encodes the full length of the hSpt6 protein. A FLAG-hemagglutinin (HA)-tagged version of hSpt6 protein was expressed in HeLa cells, and we found that it migrated slightly more slowly than native hSpt6 on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels as a result of the two tags (data not shown). In vitro transcription and translation analysis via a rabbit reticulocyte lysate showed that the cloned cDNA of hSpt6 in the pET-14b vector produced a protein with a mobility on SDS-PAGE gels that was similar to that of native hSpt6 (data not shown). Details concerning expression vectors for the FLAG-HA-tagged version of hSpt6 protein in HeLa cells are available from the authors upon request.
Preparation of recombinant Spt proteins.
A baculovirus expression system (Invitrogen Corp.) was used to produce recombinant baculoviruses, which were used to infect Sf9 cells at a multiplicity of infection of 10 or higher. Infected cells were lysed by sonication in HGKEN (20 mM HEPES [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.1% NP-40) containing 0.1 M KCl. Cell lysates were incubated with anti-FLAG gel (Sigma-Aldrich Corp.) for 2 h at 4°C. After being washed twice with HGKEN containing 0.1 M KCl and HGKEN containing 0.5 M KCl and then once with HGKEN containing 0.1 M KCl, bound proteins were eluted four times with HGKEN containing 0.1 M KCl and 150 ng of FLAG peptide (Sigma-Aldrich Corp.)/μl. The eluates were supplemented with NaCl and imidazole at final concentrations of 500 and 10 mM, respectively, and incubated with Ni-NTA (Qiagen Inc.) columns for 2 h. The columns were washed with buffer A (20 mM NaH2PO4, 30 mM Na2HPO4, 5% glycerol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 20 mM imidazole; then, the bound proteins were eluted with buffer A containing 0.25 M imidazole. The eluates were dialyzed against buffer B (20 mM HEPES-NaOH [pH 7.9], 20% [vol/vol] glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol [DTT], 0.5 mM PMSF) containing 100 mM KCl.
Recombinant hSpt4 and hSpt5 proteins were prepared as described previously (31).
Immunodepletion of hSpt6.
Immunodepletion of hSpt6 from nuclear extracts (NE) was carried out as described previously (25). NE (200 μl, 1.2 mg of protein) were incubated four times with 135 μg of anti-hSpt6 antibody coupled to protein G-Sepharose.
In vitro transcription assays.
In vitro transcription reactions using 125 ng of supercoiled DNA template (pTF3-6C2AT) were carried out as described previously (25). Briefly, 12.5 μl of reaction mixtures that contained pTF3-6C2AT and NE, hSpt6-depleted NE (dNE), or the concentrated P1.0 fraction (see below) with or without Spt proteins or Spt protein fractions in TRX buffer (25 mM Tris-HCl [pH 7.9], 10% [vol/vol] glycerol, 50 mM KCl, 0.5 mM DTT, 0.5 mM EDTA) was incubated for 45 min at 30°C. Nucleoside triphosphates (NTPs), 50 U of RNase T1 (Ambion Inc.), and 80 μM 3′-O-methylguanosine 5′-triphosphate (3′-OMe-GTP) in TRX buffer were then added, and the mixtures were incubated for an additional 10 min. When three different templates were used, reactions were terminated by the addition of α-amanitin (1 ng/μl), followed by incubation for 20 min at 37°C with RNase T1 (500 U) to isolate G-less transcripts. Transcripts were deproteinized, precipitated with ethanol, and analyzed on 8% acrylamide denaturing gels as described previously (25).
We used two primers to construct DNA templates that produce 100- and 50-nucleotide (nt) G-less transcripts: C2AT-100 (CCAAGCTTAGATTTGGGAAATATAA) and C2AT-50 (CCAAGCTTAATAATGAGGAAAGGAGA). We performed PCR using a universal M13 primer and either C2AT-100 or C2AT-50, and pTF3-6C2AT was used as a template. Amplified DNA fragments were cloned into EcoRI-HindIII sites within the polylinker region of pBluescriptSK(+) to generate pTF3-6C2AT-100 and pTF3-6C2AT-50. pTF3-6C2AT, pTF3-6C2AT-100, and pTF3-6C2AT-50 each have eight ATF binding sites and the adenovirus early 4 promoter region.
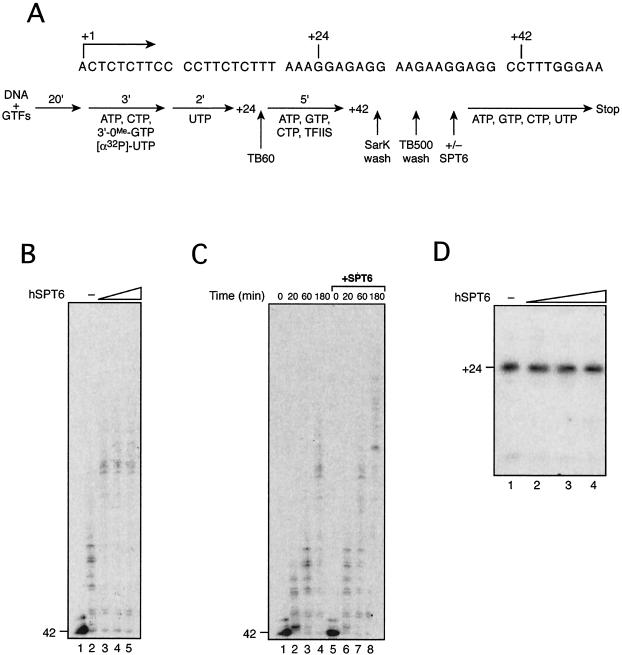
Reconstituted transcription assays using immobilized templates were carried out as described previously (17) with slight modifications. The EcoRI-ScaI fragment of plasmid pML20-47 (20) was biotinylated at the EcoRI end and coupled to streptavidin beads. Preinitiation complexes (PICs) were formed by incubating the beads carrying 100 ng of DNA with general transcription factors (GTFs) and RNAPII for 20 min at 30°C in TB60 [13 mM HEPES, 5.2 mM MgCl2, 2% polyethylene glycol, 45 μg of bovine serum albumin, 10 mM DTT, 10 mM (NH4)2SO4, 0.12 mM PMSF, 0.06 mM EDTA, 60 mM KCl]. The PICs were then incubated with RNasin (16 U) and a mixture of NTPs (0.5 mM 3′-OMe-GTP, 50 μM ATP, 5 μM CTP, 0.3 μM [α-32P]UTP) for 3 min at 30°C and chased with 1 μM UTP for 2 min to produce the +24 complex (see Fig. 7A). The stalled +24 complex was washed with TB60 buffer, equilibrated with TB60 buffer, and then incubated with 5 μM ATP, GTP, and CTP in the presence of TFIIS for 5 min to produce the +42 complex (see Fig. 7A). The +42 complex was washed once with TB60 containing 0.05% Sarkosyl and twice with TB500 containing 500 mM KCl and equilibrated with TB60. The +42 complex was used as starting material for transcription elongation assays. Mutant 1 was used for the reactions shown in Fig. 7 because it has an activity similar to that of wild type.
FIG. 7.
hSpt6 enhances the rate of transcription elongation. (A) An outline of transcription assays using an immobilized DNA template is presented with a partial nucleotide sequence of the template. (B) The +42 complex was isolated and incubated with buffer (lane 2) or 16, 32, or 160 ng (lanes 3 to 5) of r-hSpt6 mutant 1 for 10 min at 30°C; the reactions were chased for 60 s by addition of 30 μM NTPs. (C) The +42 complex was incubated with (lanes 5 to 8) or without (lanes 1 to 4) purified r-hSpt6 mutant 1 for 10 min at 30°C, and the reactions were chased for 20, 60, or 180 s, or not, as indicated. (D) PICs were formed by incubating the DNA template on magnetic beads, GTFs, and RNAPII in the absence (lane 1) or presence of 8, 16, and 160 ng (lanes 2 to 4) of r-hSpt6 mutant 1 for 20 min. Transcription was allowed to proceed to position +24 by incubating the reaction mixtures with NTPs lacking GTP for 8 min at 30°C.
Preparation of the P1.0 fraction.
HeLa NE were prepared as described previously (6), and ca. 180 mg of protein was fractionated on a P11 phosphocellulose column (Whatman plc.) as described previously (25). After being washed with 0.05 and 0.3 M KCl, proteins were eluted from the column with 1.0 M KCl (0.3 to 1.0 M KCl fraction). This 1.0 M fraction was used as the P1.0 fraction. The P1.0 fractions were then concentrated about sixfold by (NH4)2SO4 precipitation as described previously (25).
Formaldehyde cross-linking and immunoprecipitation.
Approximately 1.0 × 107 cells expressing either FLAG- and histidine-tagged (FH)-RNAPII or FLAG-DSIF were incubated at room temperature for 10 min in the presence of 1% formaldehyde in 50 mM HEPES (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA. Stop solution (1.25 M glycine-10 mM Tris-HCl [pH 8.0]) was added to stop cross-linking. Cells were washed with cold phosphate-buffered saline and then scraped into 1 ml of ice-cold radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 8.0], 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl) containing protease inhibitors (5 μg of aprotinin/ml, 5 μg of antipain/ml, 5 μg of leupeptin/ml, 5 μg of pepstatin/ml, 5 μg of chymostatin/ml, and 1 mM PMSF). After 30 min on ice, cell lysates were transferred to microcentrifuge tubes and sonicated with a 3-mm-diameter probe for 20 min. The supernatants were recovered by centrifugation at 20,000 × g for 20 min and used as starting material (input) for immunoprecipitation.
An anti-FLAG M2 agarose affinity gel (20 μl; Sigma-Aldrich Corp.) was incubated with 1 mg of the input material on a rotator for 2 h at 4°C. The gel was separated from unbound proteins by centrifugation at 2,300 × g for 1 min and washed three times with 200 μl of RIPA buffer containing protease inhibitors and three times with 200 μl of RIPA buffer containing protease inhibitors and 500 mM NaCl. Bound proteins were eluted by the addition of 20 μl of elution buffer containing 0.2 mg of FLAG peptide/ml. The elution step was repeated three additional times. Cross-linking was reversed by incubating the samples at 65°C overnight in the presence of SDS sample buffer (25) prior to loading them onto an SDS polyacrylamide gel. We also used anti-Myc agarose affinity gel (20 μl; Sigma-Aldrich Corp.) and anti-HA agarose affinity gel (20 μl; Sigma-Aldrich Corp.) as irrelevant antibodies.
RESULTS
Preparation of hSpt6-specific monoclonal antibodies.
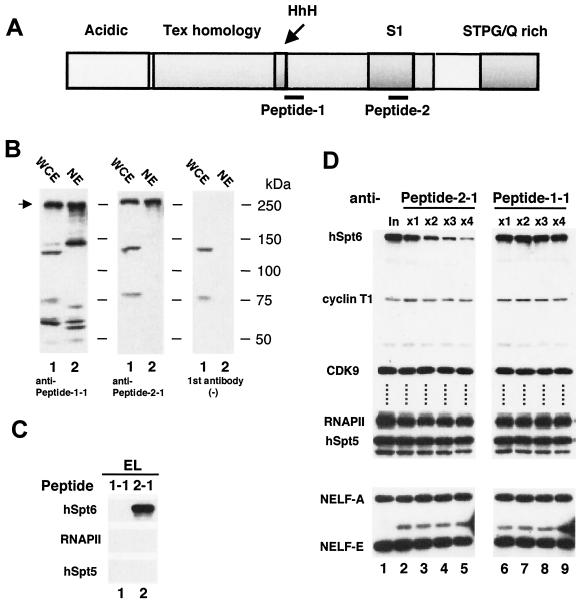
Mouse monoclonal antibodies were raised against two 24-amino-acid segments of hSpt6, designated peptide 1 (amino acids 841 to 863) and peptide 2 (amino acids 1169 to 1192) (Fig. 1A). These antibodies were tested by immunoblotting of whole-cell extracts (WCE) and NE obtained from HeLa cells. As shown in Fig. 1B, the monoclonal anti-peptide 1-1 antibody reacted with several bands in both extracts; however, the monoclonal anti-peptide 2-1 antibody detected three bands in WCE and a single band in NE. Two of those detected in WCE are thought to be nonspecific because they were observed without the first antibody (Fig. 1B, right). We infer that the band with a molecular mass of ca. 250 kDa, a size that roughly agrees with the expected size of the hSpt6 protein, corresponds to a native Spt6 protein. These results suggest that the anti-peptide 2-1 antibody recognizes hSpt6 specifically and that anti-peptide 1-1 reacts with the native Spt6 protein, but it can also react with other proteins.
FIG. 1.
Immunological analysis of hSpt6. (A) Schematic representation of hSpt6. The acidic, Tex homology, HhH, S1, and STPG/Q-rich domains are indicated. Thin lines show locations of peptides 1 and 2. (B) Immunoblot analysis of HeLa cell extracts using monoclonal antibodies against hSpt6. WCE (7.5 μl; lanes 1) and NE (3 μl; lanes 2) were subjected to immunoblot analysis. Shown at right is the immunoblot analysis performed without the first antibody. (C) Immunoblot analysis of anti-peptide 1-1 and anti-peptide 2-1 immunocomplexes. Aliquots (2.0 μl) of the eluates from the anti-peptide 1-1 and anti-peptide 2-1 immunocomplexes were subjected to immunoblot analysis using anti-peptide 1-1 (hSpt6), anti-CTD (RNAPII; Covance Inc.), and anti-DSIF p160 (hSpt5) (22) antibodies. (D) Immunodepletion of hSpt6 from HeLa NE. NE (200 μl) were treated four times with anti-peptide 2-1 (lanes 2 to 5) or anti-peptide 1-1 (lanes 6 to 9) antibodies coupled to protein G-Sepharose; aliquots (2.0 μl) of the flowthrough fractions were subjected to immunoblot analysis using anti-peptide 1-1 (hSpt6), anti-cyclin T1 (cyclin T1; kindly provided by David Price), anti-CDK9 (CDK9; Santa Cruz Biotechnology, Inc.), anti-CTD (RNAPII), anti-DSIF p160 (hSpt5), anti-NELF-A (NELF-A) (33), and anti-RD (NELF-E) (32) antibodies.
Subsequently, we used these antibodies to affinity-purify hSpt6 from HeLa NE. We incubated HeLa NE with the antibodies coupled to protein G-Sepharose; after extensive washing of the beads, the immunoprecipitated proteins were eluted using a high concentration of antigen peptides. Immunoblot analysis of the eluate materials by using anti-peptide 1-1 antibody showed that anti-peptide 2-1, but not anti-peptide 1-1, can be used to immunoprecipitate the 250-kDa hSpt6 protein (Fig. 1C). The largest subunits of RNAPII and hSpt5 were not detected in the eluate materials (Fig. 1C), suggesting that these proteins are not closely associated with hSpt6.
hSpt6 is important for efficient transcription in vitro.
We immunodepleted hSpt6 from HeLa NE by using anti-peptide 2-1 antibody to examine possible roles of hSpt6 in transcription. After immunodepletion was repeated four times, more than 80% of hSpt6 was removed from the NE, whereas no changes in the levels of hSpt5, RNAPII, P-TEFb, and NELF were detected (Fig. 1D, lanes 1 to 5). The same procedure carried out using anti-peptide 1-1 antibody did not decrease the levels of hSpt6 and the other proteins examined (Fig. 1D, lanes 6 to 9), demonstrating that immunodepletion with anti-peptide 2-1 antibody is specific and efficient.
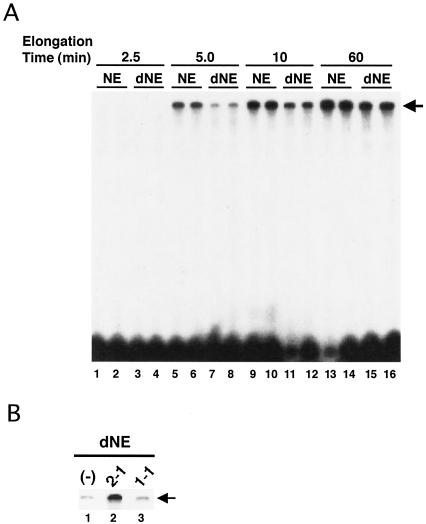
Next we performed in vitro transcription assays using mock-depleted NE and dNE. These extracts were preincubated with a supercoiled plasmid, pTF3-6C2AT; after addition of NTPs, transcription reactions were allowed to proceed for various times. Each reaction was carried out in duplicate, and each experiment was repeated four times. Figure 2A shows a representative result. We found that the synthesis of the 380-nt full-length G-free transcripts proceeded with markedly slower kinetics when the dNE were used than when NE were used. The transcriptional defect was complemented by addition of the anti-peptide 2-1 immunoprecipitates but not by addition of the anti-peptide 1-1 immunoprecipitates, which lack hSpt6 (Fig. 2B). These results suggest that hSpt6 is critical at some point during transcription on a naked DNA template.
FIG. 2.
hSpt6 is important for transcription on a naked DNA template in vitro. (A) Immunodepletion of hSpt6 causes a transcriptional defect. Transcription assays were carried out using the NE that were immunodepleted using either anti-peptide 1-1 (NE) or anti-peptide 2-1 (dNE) as shown in Fig. 1. The NE (2 μl) were incubated with 125 ng of the supercoiled DNA template pTF3-6C2AT for 45 min at 30°C. NTPs (final concentrations of 80 μM 3′-OMe-GTP, 60 μM ATP, 600 μM CTP, and 1 μM UTP containing 5 μCi of [α-32P]UTP) were added to the reaction mixtures, which were then incubated for the times indicated. An arrow indicates the position of 380-nt, full-length transcripts. (B) Add-back of the immunoprecipitates. Reactions were carried out as described for panel A, except that aliquots (2.0 μl) of the eluates from the anti-peptide 2-1 and anti-peptide 1-1 immunoprecipitates were added to the reactions carried out using dNE. The immunoprecipitates and dNE were incubated for 45 min at 30°C with 125 ng of the supercoiled DNA template pTF3-6C2AT before addition of NTPs. An arrow indicates the position of the full-length transcripts.
hSpt6 is important for efficient transcription elongation in vitro.
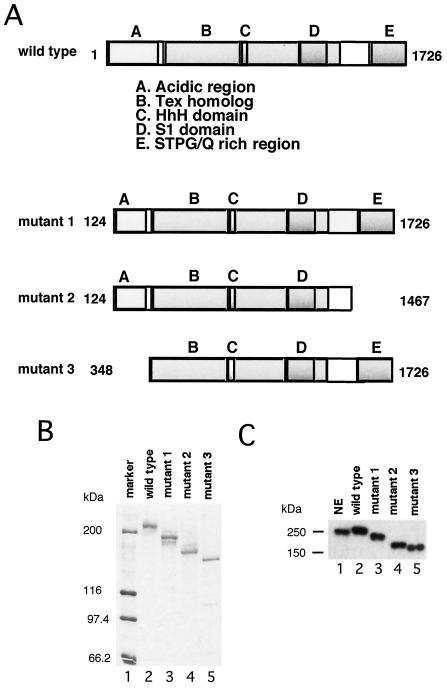
We cloned the hSpt6 cDNA (see Materials and Methods) and overexpressed it as a histidine-tagged protein (r-hSpt6) in insect cells. The protein, purified by metal-chelate affinity chromatography, migrated to a relative molecular mass of ca. 250 kDa on an SDS gel (Fig. 3B and C, lane 2), which is similar in size to the native hSpt6 protein. We also produced three deletion mutants of hSpt6 (Fig. 3A and lanes 3 to 5 of Fig. 3B and C). Mutants 1 and 3 lack either part or all of the N-terminal acidic region, respectively. Mutant 2 lacks part of the acidic region and the C-terminal STPG/Q-rich region.
FIG. 3.
Preparation of recombinant hSpt6 and its mutants. (A) Schematic representation of wild-type and mutant Spt6 proteins used in this study. Letters A to E above the boxes stand for the conserved domains of hSpt6. Numbers indicate amino acid positions at the N and C termini of each mutant. (B) Silver staining. Purified recombinant hSpt6 proteins (ca. 10 ng) were subjected to SDS-PAGE and visualized by silver staining. (C) Immunoblot analysis using anti-peptide 2-1 antibody. HeLa NE (12 μg; lane 1) and purified recombinant hSpt6 proteins (ca. 10 ng; lanes 2 to 5) were analyzed by immunoblotting.
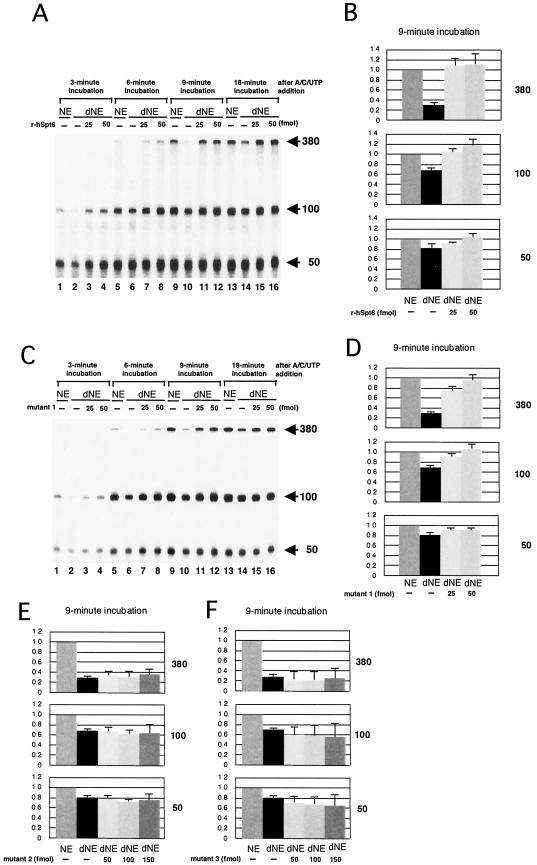
To determine the step at which hSpt6 affects transcription, we carried out transcription assays using three different DNA templates containing the same promoter and a G-less cassette of 380, 100, or 50 nt. When normal NE were used, 100- and 50-nt transcripts were detected as early as 3 min after NTP addition whereas 380-nt transcripts were detected as early as 6 min (Fig. 4A). The amounts of these transcripts reached maximal levels around 9 min, suggesting that reinitiation does not occur efficiently under the conditions employed. When the dNE were used instead, the synthesis of 380-nt transcripts, first detected at the 9-min time point (Fig. 4A), proceeded with greatly slower kinetics. On the other hand, synthesis of 100- and 50-nt transcripts was only modestly affected by immunodepletion, which was clearly seen at the 3-min time point but not at later points. For that reason, the transcriptional defect caused by immunodepletion was prominent in the synthesis of longer transcripts (Fig. 4B). To determine whether depletion of the hSpt6 polypeptide is solely responsible for the transcriptional defect, we added r-hSpt6 to the dNE and found that transcription was restored to a level similar to that achieved with normal NE (Fig. 4A). These results strongly suggest that hSpt6 is essential for efficient transcription elongation.
FIG.4.
hSpt6 is important for transcription elongation in vitro. Transcription assays were carried out as described for Fig. 2B except that three different templates, pTF3-6C2AT (12.5 ng), pTF3-6C2AT-100 (22.5 ng), and pTF3-6C2AT-50 (90 ng) were used; they yielded 380-, 100-, and 50-nt transcripts, respectively. Purified wild-type r-hSpt6 (A and B), mutant 1 (C and D), mutant 2 (E), or mutant 3 (F) was included in the reactions where indicated. Transcription was allowed to proceed for the indicated times; then the reactions were processed as described in Materials and Methods. Arrows indicate the positions of the 380-, 100-, and 50-nt transcripts. In panels B, D, E, and F, the counts of each transcript at the 9-min time point were quantified and normalized against the counts obtained from a reaction carried out using mock-depleted NE (mean ± standard deviation [SD], P = 0.05, n = 4).
We then compared the abilities of the mutant hSpt6 proteins to restore transcriptional activities of dNE. Mutant 1 had a restoration activity similar to that of wild-type r-hSpt6 (Fig. 4C and D). In contrast, mutants 2 and 3 were unable to restore the transcriptional activities of dNE (Fig. 4E and F). Therefore, we infer that the acidic region and the STPG/Q-rich region are essential for hSpt6 activity in vitro.
Possible interactions between hSpt6 and DSIF.
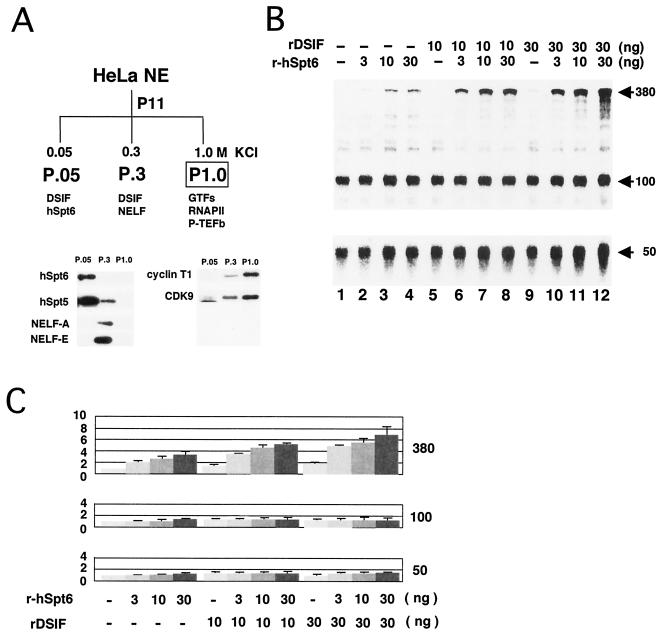
Previously, it has been shown that, after passing a HeLa NE through a phosphocellulose column, the combination of 0.5 and 0.85 M KCl fractions, which lack DSIF, supports transcription in vitro (25). In this study, we used a 0.3 to 1.0 M KCl fraction (P1.0 fraction) to investigate possible interactions between hSpt6 and DSIF. The distributions of DSIF, hSpt6, and other transcription factors in the phosphocellulose P11 column fractions were analyzed by immunoblotting. hSpt5 was detected mainly in the 0.05 M KCl fraction P.05 and to a lesser extent in the 0.3 M KCl fraction P.3 (Fig. 5A); hSpt6 and NELF were found in P.05 and P.3, respectively (Fig. 5A). CDK9, cyclin T1, RNAPII, and GTFs, including TATA-binding protein, TFIIB, TFIIE, TFIIF, and TFIIH, were mainly detected in the P1.0 fraction (Fig. 5A and data not shown). Thus, P1.0 lacks DSIF, hSpt6, and NELF but contains P-TEFb, RNAPII, and GTFs.
FIG. 5.
Possible interaction between hSpt6 and DSIF. (A) Distribution of various transcription factors in phosphocellulose P11 column fractions was analyzed by immunoblotting. The results are summarized schematically at top. (B) Transcription assays were carried out using the concentrated P1.0 fraction as described for Fig. 4. Purified recombinant hSpt6 and DSIF were added where indicated. NTPs (final concentrations of 80 μM 3′-OMe-GTP, 60 μM ATP, 600 μM CTP, and 5 μM UTP containing 10 μCi [α-32P]UTP) were added to the reactions, and the reactants were then incubated for 9 min. Arrows indicate the positions of the 380-, 100-, and 50-nt transcripts. (C) The counts of each transcript at the 9-min time point were quantified and normalized against counts obtained from a reaction without recombinant proteins (mean ± SD, P = 0.05, n = 3).
We carried out in vitro transcription assays using P1.0, as shown in Fig. 4. As reported previously (25), transcription elongation was strongly compromised when P1.0 was used (Fig. 5B). Individual addition of r-hSpt6 and recombinant DSIF to the reaction modestly increased the synthesis of 380-nt transcripts in a dose-dependent manner, whereas simultaneous addition of these proteins remarkably increased the synthesis of the transcripts (Fig. 5B and C). hSpt6 and DSIF appear to have an additive effect on transcription elongation. Taken together, these results suggest that hSpt6 and DSIF can stimulate transcription elongation both individually and in concert.
hSpt6 directly affects RNAPII elongation complexes.
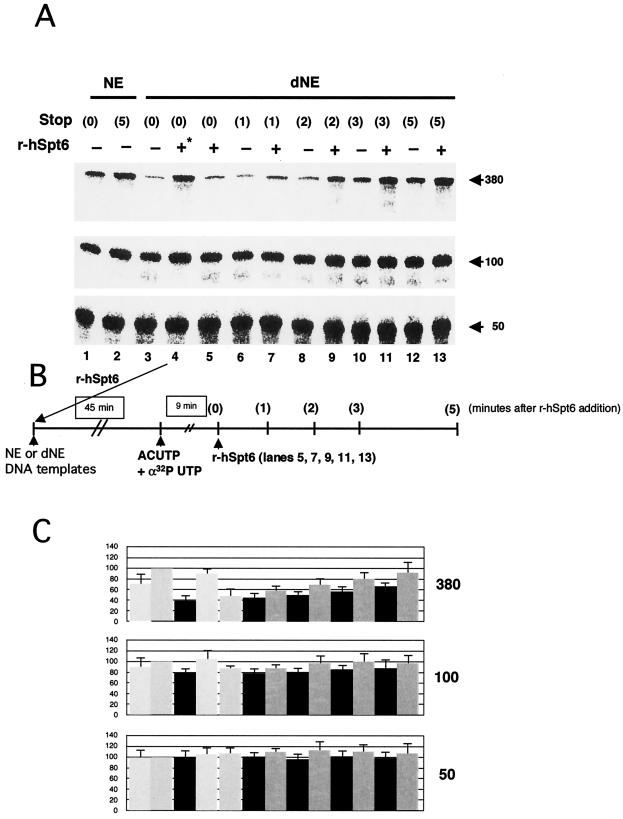
Next, using two strategies, we investigated whether hSpt6 directly affects RNAPII elongation complexes. In the first set of experiments (Fig. 6), r-hSpt6 was added to the dNE transcription system 9 min after addition of NTPs; then, reaction mixtures were incubated for 1 to 5 min more. During the additional incubation period, de novo synthesis of 380-nt transcripts probably did not occur because such an event would require at least 6 min (Fig. 4). Thus, the stimulatory effect of hSpt6, evident as early as 1 min after its addition (Fig. 6A, lanes 6 and 7), is attributable to its effect on already initiated transcription complexes. A highly purified transcription system and an immobilized DNA template were used in the second set of experiments (Fig. 7). RNAPII elongation complexes at position +42 were isolated as described previously (17); then, transcription elongation was allowed to proceed by addition of the four NTPs. Addition of r-hSpt6 (mutant 1) to the chase reaction strongly increased the rate of transcription elongation (Fig. 7B and C). In contrast, addition of r-hSpt6 (mutant 1) to the labeling reaction had no detectable effect on production of 24-nt transcripts (Fig. 7D), demonstrating that hSpt6 does not affect transcription initiation. These data strongly suggest that hSpt6 enhances the rate of transcription elongation through its interaction with the RNAPII elongation complex.
FIG. 6.
Evidence that hSpt6 affects already initiated transcription complexes. (A) Transcription assays were carried out as described for Fig. 4. Purified r-hSpt6 (50 fmol) was added during preincubation (lane 4) or 9 min after NTP addition (lanes 5, 7, 9, 11, and 13). The reactions were terminated at various time points as indicated. Arrows indicate the positions of the 380-, 100-, and 50-nt transcripts. (B) The scheme for this experiment. (C) Counts of each transcript at the 9-min time point were quantified and normalized against counts obtained in lane 2 of panel A (mean ± SD, P = 0.05, n = 3).
hSpt6 interacts with hSpt5 and RNAPII in HeLa cells.
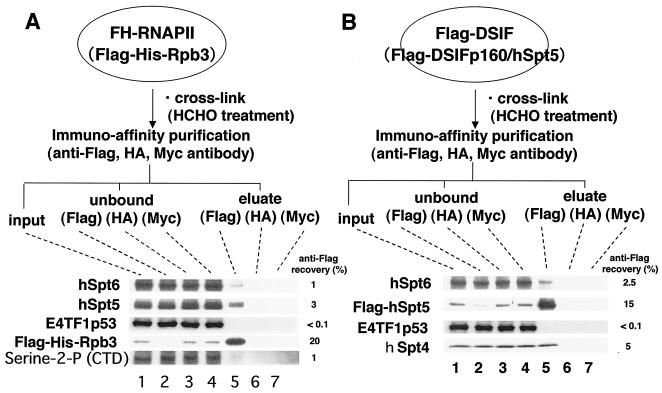
The data presented above suggest that hSpt6 may interact physically with RNAPII and DSIF. However, no such association was detected by immunoprecipitation using anti-hSpt6 antibody (Fig. 1). Thus, interactions of hSpt6 with hSpt5 and RNAPII may be very weak or may occur only during transcription elongation. We used two cell lines derived from HeLa cells to examine this issue: one expressing FLAG-tagged hSpt5 (30) and the other expressing FLAG- and histidine-tagged Rpb3, a small subunit of RNAPII (9). Immunoprecipitation with anti-FLAG antibody from lysates of these cells allows isolation of DSIF and RNAPII complexes, both of which are functionally active in in vitro transcription assays (data not shown). The cells were treated with formaldehyde before lysis to identify weak protein-protein interactions; also, the lysates were subjected to immunoprecipitation with anti-FLAG antibody. Immunoblotting revealed that hSpt6 was coimmunoprecipitated with FLAG-Rpb3, FLAG-hSpt5, and serine 2-phosphorylated Rpb1, the largest subunit of the RNAPII C-terminal domain (CTD), whereas an irrelevant protein, E4TF1p53, was not (Fig. 8). E4TF1p53, which is a subunit of the transcription activator E4TF1, contains a transcription activation domain and does not bind to DNA (22). In addition, hSpt6 was not immunoprecipitated by control antibodies and anti-HA and anti-Myc antibodies. Therefore, these results suggest that hSpt6 interacts with hSpt5 and elongating RNAPII within the cells. We note that similar results were obtained for DNase treatment of immunoprecipitates.
FIG. 8.
Interactions of hSpt6 with DSIF and RNAPII. Clonal cell lines expressing FH-RNAPII (9) (A) and FLAG-DSIF (31) (B) were treated with 1% formaldehyde for 10 min at room temperature. Cell extracts were prepared and incubated with anti-FLAG M2-agarose gel, anti-Myc-agarose gel, and anti-HA-agarose gel, and immunoprecipitated materials were recovered as described in Materials and Methods. Input (1%), unbound (1%), wash (10%), and eluate (20%) materials were subjected to immunoblot analysis using anti-peptide 1-1 (hSpt6), anti-hSpt5, anti-E4TF1-p53 (22), anti-hSpt4 (14), and anti-phosphoserine-2 of the RNAPII CTD (Covance Inc.). We quantified band density and calculated the recovery rate of each protein.
DISCUSSION
Despite accumulated evidence that suggests a role for Spt6 in transcription, its exact function remains largely unknown. Here we present, for the first time, biochemical evidence showing that hSpt6 directly regulates transcription at the level of elongation. We infer that this is a specific function of hSpt6 because the elongation-stimulatory activity of hSpt6 depends critically on its structural integrity. Denaturation of wild-type r-hSpt6 with guanidine hydrochloride and subsequent removal of the denaturant by dialysis removed this activity (unpublished data). Moreover, small deletions at either end or internal deletions of the polypeptide abolished this activity for all of the mutants examined in this study except mutant 1 (Fig. 3 and unpublished data). For these reasons, several evolutionarily conserved structural domains of the large Spt6 protein seem to be required for stimulation of transcription elongation. We are currently performing crystal structure analysis to address this question. On the other hand, Winkler et al. have reported that the carboxyl terminus of hSpt6 binds to the human cytomegalovirus-encoded transcription activator pUL69 as well as histone H3. They speculate that the pUL69 function of stimulating gene expression in vivo may be mediated by its behavior as an antirepressor by competing for binding of histones to hSpt6 (27). Mutant 2 lacks the CTD and lost its activity in transcription from naked DNA templates, suggesting that the CTD may be a surface for Spt6 interactors.
The latest report from John Lis' group, using a cross-linked chromatin immunoprecipitation assay, showed that Spt6 is recruited to the promoter proximal region of activated hsp70 gene and travels with the RNAPII elongation complex through chromatin of Drosophila polytene chromosomes (21). In addition, elegant work by Fred Winston's group also provided strong evidence that yeast Spt6 regulates transcription by direct interaction with histones (12). Thus, it was postulated that Spt6 acts as a histone chaperone during transcription elongation while moving on activated genes from the 5′ to 3′ region together with RNAPII by mobilizing histones during the passage of RNAPII through chromatin structures (23).
On the other hand, it has also been reported that Spt6 physically associates with RNAPII and Spt5 in yeast (15, 16), fly (1, 21), and human (this study); it is likely that these interactions are important for efficient transcription elongation by RNAPII as well. For that reason, we employed simplified in vitro transcription systems using DNA templates that were free of nucleosomes. We showed that Spt6 also affects transcription without histones. Data obtained using a highly purified transcription assay (Fig. 7) showed that the single hSpt6 polypeptide is capable of affecting an RNAPII elongation complex that comprises immobilized template DNA, nascent RNA, and RNAPII.
Nevertheless, these observations do not exclude the possibility that other polypeptides may play regulatory roles in transcriptional activation by hSpt6. For instance, the interaction between Spt6 and histones, as mentioned earlier, appears to be essential for efficient transcription elongation (12). In addition, DSIF is an important factor that we found. Although DSIF is not essential for transcriptional activation by hSpt6 because hSpt6 functioned in reactions devoid of DSIF (Fig. 5 and 7), data presented in Fig. 5 showed that hSpt6 and DSIF have an additive effect on transcription elongation. We carried out formaldehyde cross-linking experiments (Fig. 8) because hSpt5 and RNAPII were not detected in the anti-hSpt6 immunoprecipitates from HeLa NE (Fig. 1). Those experiments demonstrated that hSpt6 indeed interacts with DSIF and RNAPII in human cells, although these interactions may be relatively weak or may occur only during transcription elongation. We also speculate that some features specific to the RNAPII elongation complex, such as a hyperphosphorylated form of the RNAPII CTD and nascent RNA, might be important for stable association with hSpt6. In this regard, it is noteworthy that Spt6 has an evolutionarily conserved RNA-binding motif called the S1 domain.
Interaction between Spt6 and DSIF may also explain how Spt6 is recruited to the promoter proximal region, as observed by Lis' group (21). In contrast to the behavior of Spt6, Spt5 has been shown to be present in abundance in the promoter proximal region of the Drosophila hsp70 gene even when the gene is not activated; also, Spt5 is known to act in concert with NELF to suppress RNAPII elongation in the early phases of elongation. However, this inhibition was quickly reversed by heat shock conditions, after which RNAPII was allowed to transcribe the hsp70 gene (21, 29). These results suggest that Spt6 can associate with RNAPII that is stalled by Spt5 and NELF at the promoter proximal region. Subsequently, Spt6 moves to the 3′ region with the RNAPII elongation complex in response to heat stimuli (21, 29). Thereby, Spt5 may recruit Spt6 to transcription elongation complexes. Once at its site of action, Spt6 activates the stalled RNAPII and affects chromatin structure such that processive transcription by the RNAPII elongation complex can take place.
In conclusion, recent data from our team and others collectively suggest that Spt6 is a multifunctional transcription factor. Using naked DNA templates, we have established that hSpt6 can function as a classic transcription elongation factor that enhances the rate of transcription elongation by RNAPII. However, the interactions between Spt6 and other essential players in transcription, such as DSIF and histones, also appear to be important. As such, they merit further investigation to elucidate their complex mechanisms that underlie transcription.
Acknowledgments
We are indebted to Grant Hartzog for many helpful discussions. We also thank Sachiko Okabe, Makoto Hasegawa, and Tetsu Yung for technical support and manuscript preparation and members of the Handa Lab for helpful suggestions. We thank David Price for providing anti-cyclin T1 antibodies.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (to T.W. and H.H.), a research grant from CREST of the JST Corporation, and a grant from NEDO (to H.H). D.R. received support from the NIH (GM37120) and the Howard Hughes Medical Institute. This study was also supported in part by the Grant of the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrulis, E. D., J. Werner, A. Nazarian, H. Erdjument-Bromage, P. Tempst, and J. T. Lis. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420:837-841. [DOI] [PubMed] [Google Scholar]
- 3.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 4.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Compagnone-Post, P. A., and M. A. Osley. 1996. Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics 143:1543-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harlow E. D., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa, M., T. Endou, T. Narita, Y. Yamada, T. Yamaguchi, T. Wada, and H. Handa. 2003. A rapid purification method for human RNA polymerase II by two-step affinity chromatography. J. Biochem. 133:133-138. [DOI] [PubMed] [Google Scholar]
- 10.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 13.Keegan, B. R., J. L. Feldman, D. H. Lee, D. S. Koos, R. K. Ho, D. Y. Stainier, and D. Yelon. 2002. The elongation factors Pandora/Spt6 and Foggy/Spt5 promote transcription in the zebra fish embryo. Development 129:1623-1632. [DOI] [PubMed] [Google Scholar]
- 14.Kim, D. K., N. Inukai, T. Yamada, A. Furuya, H. Sato, Y. Yamaguchi, T. Wada, and H. Handa. 2003. Requirement for human Spt4 in RNA polymerase II elongation control in vitro. Genes Cells 8:371-378. [DOI] [PubMed] [Google Scholar]
- 15.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal, S. S., H. Cho, S. Kim, K. Cabane, and D. Reinberg. 2002. FCP1, a phosphatase specific for the heptapeptide repeat of the largest subunit of RNA polymerase II, stimulates transcription elongation. Mol. Cell. Biol. 22:7543-7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parada, C. A., and R. G. Roeder. 1999. A novel RNA polymerase II-containing complex potentiates Tat-enhanced HIV-1 transcription. EMBO J. 18:3688-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 20.Samkurashvili, I., and D. S. Luse. 1998. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol. Cell. Biol. 18:5343-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saunders, A., J. Werner, E. D. Andrulis, T. Nakayama, S. Hirose, D. Reinberg, and J. Lis. 2003. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301:1094-1096. [DOI] [PubMed] [Google Scholar]
- 22.Sawada, J., M. Goto, C. Sawa, H. Watanabe, and H. Handa. 1994. Transcriptional activation through the tetrameric complex formation of E4TF1 subunits. EMBO J. 13:1396-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svejstrup, J. Q. 2003. Transcription. Histones face the FACT. Science 301:1053-1055. [DOI] [PubMed] [Google Scholar]
- 24.Swanson, M. S., M. Carlson, and F. Winston. 1990. SPT6, an essential gene that affects transcription in Saccharomyces cerevisiae, encodes a nuclear protein with an extremely acidic amino terminus. Mol. Cell. Biol. 10:4935-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler, M., T. aus Dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winston, F. 1992. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast, p. 1271-1293. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Wu, C. H., Y. Yamaguchi, L. R. Benjamin, M. Horvat-Gordon, J. Washinsky, E. Enerly, J. Larsson, A. Lambertsson, H. Handa, and D. Gilmour. 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17:1402-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 27:179-197. [DOI] [PubMed] [Google Scholar]
- 31.Yamaguchi, Y., T. Wada, D. Watanabe, T. Takagi, J. Hasegawa, and H. Handa. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085-8092. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi, Y., N. Inukai, T. Narita, T. Wada, and H. Handa. 2002. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 22:2918-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]