Summary
Chalkbrood and stonebrood are two fungal diseases associated with honey bee brood. Chalkbrood, caused by Ascosphaera apis, is a common and widespread disease that can result in severe reduction of emerging worker bees and thus overall colony productivity. Stonebrood is caused by Aspergillus spp. that are rarely observed, so the impact on colony health is not very well understood. A major concern with the presence of Aspergillus in honey bees is the production of airborne conidia, which can lead to allergic bronchopulmonary aspergillosis, pulmonary aspergilloma, or even invasive aspergillosis in lung tissues upon inhalation by humans. In the current chapter we describe the honey bee disease symptoms of these fungal pathogens. In addition, we provide research methodologies and protocols for isolating and culturing, in vivo and in vitro assays that are commonly used to study these host pathogen interactions. We give guidelines on the preferred methods used in current research and the application of molecular techniques. We have added photographs, drawings and illustrations to assist bee-extension personnel and bee scientists in the control of these two diseases.
Keywords: Ascosphaera, Ascosphaerales, Aspergillus, chalkbrood, diagnostics, methods, recommendations, stonebrood, COLOSS, BEEBOOK, honey bee
Introduction
Insect pathogenic fungi can be found throughout the fungal kingdom (Humber, 2008), all being capable of invading their hosts and overcoming their immune systems. Two fungal genera (Ascosphaera and Aspergillus) are known to infect honey bee brood, causing chalkbrood and stonebrood diseases. Both are ascomycetes within the Eurotiomycetes. The fungus causing chalkbrood in honey bees has a narrow host range and a unique infection route, it relies solely on sexual reproduction and has many host-specific adaptations. Therefore many methods known from common insect pathogenic fungi are not easily adopted to study chalkbrood.
In contrast, the fungi causing stonebrood are facultative pathogens with a broad host range, they produce asexual conidia and their infection biology resemble many well-known insect pathogenic fungi, like Beauveria and Metarhizium; so several standard insect pathological methods can be directly transferred to this system. In the current chapter, we compile, discuss and provide detailed protocols for various methods to assist beekeepers and bee scientists entering this area of research.
In addition to fungal pathogens causing chalkbrood and stonebrood, two species of microsporidia, Nosema apis and Nosema ceranae, are known to infect adult honey bees. Although recently suggested to be fungi at the very base of the fungal tree (James et al., 2006), these intracellular pathogens have a very different biology and are considered in a separate paper in the BEEBOOK (Fries et al., 2013).
1. Chalkbrood
1.1. introduction
Chalkbrood, the most common fungal bee brood disease, is caused primarily by the fungus Ascosphaera apis (Maassen ex Claussen) Olive and Spiltoir (Spiltoir, 1955). It was recognized in the honey bee in the early 20th Century (Maassen, 1913). The field diagnosis of chalkbrood is based on visual detection of diseased, mummified brood, commonly known as “chalkbrood mummies”. Chalkbrood can reduce colony productivity by lowering the number of newly emerged bees, and in some cases may lead to colony losses. The disease is found infecting honey bee brood in most regions of the world, including warm and dry climates. Clinical symptoms of chalkbrood often appear for only a short time, typically under cold and damp weather conditions (Aronstein and Murray, 2010).
The genus Ascosphaera spp. comprises species that are adapted to eusocial and solitary bees and their habitats. Some of the species are saprophytic, growing on nest materials, such as stored food, faecal matter, and nest debris; others have evolved as opportunistic and / or obligate bee pathogens (Bissett, 1988). In honey bees Ascosphaera major and Arrhenosphaera cranae (both belonging to Ascospheraceae) have been reported only a few times from chalkbrood infected colonies, but Koch’s Postulates have never been demonstrated for them, so they might be secondary invaders.
Ascosphaera apis primarily infects honey bee brood by entering the host through the gut lining. In an infected larva, hyphae penetrate the gut wall, and mycelium develops inside the body cavity. After a few days, mycelium breaks out of the posterior end of the larva leaving the head unaffected (Maurizio, 1934). Fruiting bodies with the new ascospores are formed on aerial hyphae outside the dead larvae. The ascospores are a result of sexual reproduction of fungal mycelia with the opposite sex, unlike most other insect pathogenic fungi where asexually conidia are the infective units, as e.g. seen in Aspergillus. Ascospores of A. apis (hereafter referred to as spores) are believed to be adapted to the harsh gut environment of the host. Ascosphaera apis infect the host larvae if ingested, unlike other insect pathogenic fungi which mainly infect through the external cuticle. Spores of A. apis can stay dormant and viable for years, but upon exposure to CO2 the spores becomes activated, resulting in spore swelling and subsequently germ tube formation that extends to form hyphae (Bamford and Heath, 1989). Presumably, CO2 produced by the larval tissues accumulates in the closed hindgut of the larvae aiding in spore germination (Heath and Gaze, 1987). However additional factors that may be involved in spore germination within the host still remain to be discovered. In addition, it has been shown that chilling of the brood below the optimal rearing temperature increases the number of diseased larvae (Vojvodic et al., 2011a; Flores et al., 1996a). All these specific adaptations have to be taken into account when working with A. apis, and specific protocols have been developed.
1.2. Diagnostics and qualitative detection
1.2.1 Morphological description
1.2.1.1. Macroscopic diagnosis
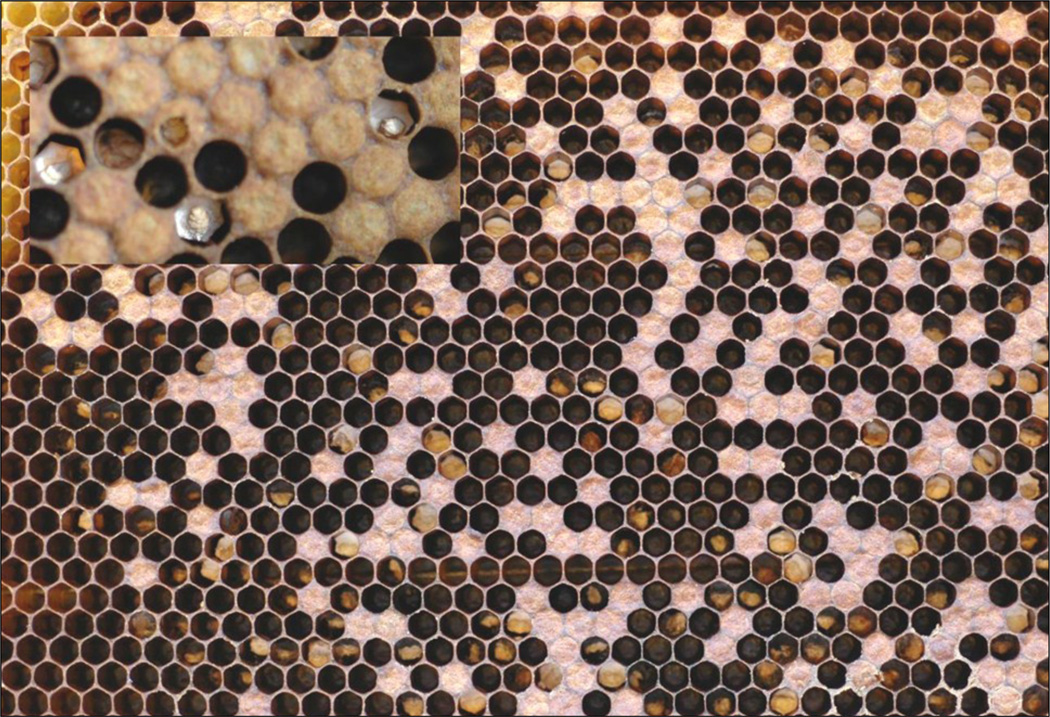
The typical symptoms observed in a colony affected by chalkbrood are irregular wax cappings over the brood and uncapped cells scattered over the brood frames (Fig. 1). The cell capping may also have small holes or appear slightly flattened. Chalkbrood mummies can often be seen in the combs, at the hive entrance or found on the bottom board (Fig. 2). Observation of combs may reveal different stages of the disease; fresh larval cadavers covered with white cotton-like mycelium and desiccated mummies that appear as white, dark or a combination of white and dark solid clumps. White desiccated mummies look like small pieces of chalk giving rise to the name of the disease and dark mummies are coloured by fungal fruiting bodies.
Fig. 1.
A brood frame from a honey bee colony with clinical symptoms of chalkbrood and a close up insert of fresh mummies. In many cells chalkbrood mummies have been partly removed by the worker bees.
Photo: F Padilla, J M Flores and A B Jensen
Fig. 2.
Dark and white chalkbrood mummies. The black mummies contain millions of new infective spores.
Photo: A B Jensen
Diagnosis in the field is generally based on the presences of chalkbrood mummies (as described above). Following field diagnosis, a microscopic examination is usually required to confirm the presence of spore cysts in the samples using the microscope slide smear technique. The spores can be mounted on a microscope slide with a drop of distilled water and observed at 100–400 × magnification.
1.2.1.2. Microscopic diagnosis
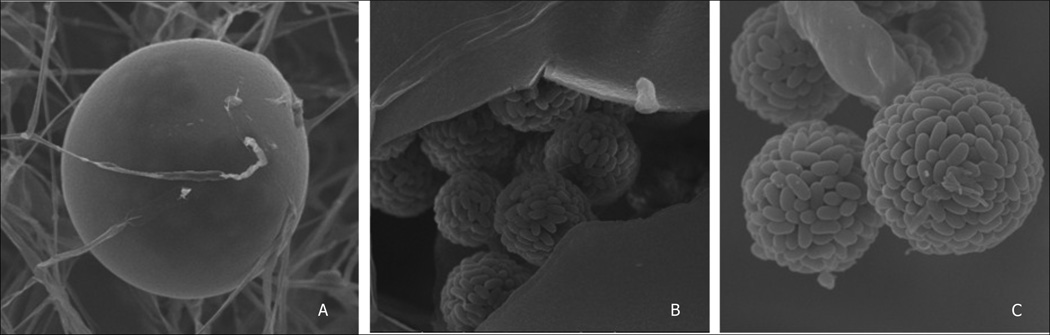
Ascosphaera apis has septate hyphae (2.5 – 8.0 µm in diameter) which show pronounced dichotomous branching (Spiltoir, 1955; Skou, 1988). As mentioned above, this is a heterothallic fungus with two mating types and only when + and − strains are grown in close proximity does the formation of fruiting bodies occur. The fruiting bodies are spherical spore cysts (47 – 140 µm in diameter; Fig. 3a) which contain numerous spore balls (9–19 µm, Fig. 3b) composed of hyaline spores (2.7 – 3.5 × 1.4 – 1.8 µm, Fig. 3c) (Skou, 1972; Bissett, 1988).
Fig. 3.
Scanning electron microscope pictures of Ascosphaera apis fruiting body. A. Spherical fruiting body; B. Cracked fruiting body with sporeballs; C. Sporeballs with multiple ascospores.
Photos: J Bresciani and A B Jensen
1.2.1.3. Biological diagnosis
Identification of the vegetative stages of the fungus can be done by a mating test with two reference strains (AFSEF 7405 and ARSEF 7406). Production of spore cysts with one of the reference strains would prove the identify of A. apis mating. The mating test is described under paragraph 1.4.1.4. Production and quality of inoculums.
1.2.2. Molecular methods
The Polymerase Chain Reaction (PCR) has increasingly been used for detection of microorganisms. The internal transcribed spacer (ITS) region of the nuclear ribosomal repeat unit is the locus most often used for molecular identification of fungal species (Nilsson et al., 2008) and it is now accepted as the general fungal barcode marker (Schoch et al., 2012). Almost no variation between A. apis strain was detected in the ITS region (Anderson et al., 1998; Jensen et al., 2012) and several A. apis species specific primers have been designed targeting the ITS (Table 1). Irrespective of which primers are used, the presence of a band from PCR amplification indicates the presence of A. apis DNA.
Table 1.
List of Ascosphaera apis specific primers
| Primer name | Annealing temp. (°C) |
Primer sequence | Citation |
|---|---|---|---|
| 3-F1* | 62 | TGTCTGTGCGGCTAGGTG | James and Skinner, 2005 |
| 3-F2 | 62 | GGGTTCTCGCGAGCCTG | James and Skinner, 2005 |
| 3-R1* | 62 | CCACTAGAAGTAAATGATGGTTAGA | James and Skinner, 2005 |
| AscoF3 | 64 | GCACTCCCACCCTTGTCTA | Murray et al., 2005 |
| AapisR3 | 64 | CCCACTAGAAGTAAATGATGGTTA | Murray et al., 2005 |
Primer pair recommended to be used as standard
DNA can be extracted with standard kits like DNeasy® Plant Mini Kit (Qiagen), Ultra Clean plant DNA isolation kits (Mo Bio Laboratories) or PrepMan Ultra reagent (Applied Biosystems) using the manufacturers’ protocols (for other protocols of DNA extractions consult the BEEBOOK paper on molecular methods (Evans et al., 2013)
1.2.2.1 PCR for species identification
Genomic DNA can be amplified in a 25 µl reaction containing:
2.5 µl Taq polymerase buffer (100 mM tris-HCl, pH 8.3; 500 mM KCL)
2 U of Taq DNA polymerase
1.5–2.5 mM MgCl2
250 µM of each deoxyribonucleotide triphosphate
0.25 µM of an forward and reverse primer (Table 1)
1 µl of DNA
Reaction conditions are as follows:
initial denaturing for 10 min at 94°C
30 cycles of 45 s denaturing at 94°C
45 s annealing at 62–65°C
1 min extension at 72°C
final extension for 5 min at 72°C.
Results are visualised using Gel Doc (BioRad) or any other DNA imaging systems. We would recommend that the primer pair 3-F1 and 3-R1 (Table 1) (James and Skinner, 2005) be used as a standard for identification of A. apis. Both primers are A. apis specific as opposed to the primer pair in Murray et al. (2005). Furthermore, 3-F1/3-R1 amplifies a longer PCR product that could be very useful for sequencing efforts (James and Skinner, 2005). The PCR protocol described above can be optimised depending on the type of equipment and reagents used in the laboratory. We would recommend sequencing the PCR product as a quality control, the first time the protocol is implemented in a new laboratory (for more information on sequencing consult the BEEBOOK paper on molecular methods (Evans et al., 2013).
1.3. Quantitative detection
1.3.1. Quantify the level of fungal infection in bee colonies
Different methods have been used to assess the level of chalkbrood infection in the honey bee hive, but none of the methods are completely reliable. The most direct method is to remove brood combs from the hives and count the number of mummies inside capped and uncapped cells. These values can be added to the number of mummies found on the bottom board of the hive. It is however important to add an entrance hive trap like a pollen trap (see the BEEBOOK paper on miscellaneous methods (Human et al., 2013) or a screened bottom board (Flores, unpublished data), to hinder the bees from removing mummies from the hive.
Assessing chalkbrood infection by counting mummies has important drawbacks: it does not include the number of mummies inside capped cells, unless all capped cells are opened and inspected; and it does not include the mummies that are removed from the hive in small pieces by worker bees which are thus not collected by the entrance traps. Furthermore, these counting methods are heavily influenced by environmental conditions. High nectar flow induces higher removal of mummies (Thompson, 1964; Momot and Rothenbuhler, 1971) and during a nectar flow more mummies may be recovered in the traps, leading to possible error in the estimation of disease prevalence.
For the above reasons, we propose that the level of colony infection be evaluated by chilling the brood to 25°C (Flores et al., 1996a). The number of mummified larvae after chilling allows for an assessment of the degree of potential infection in bee colonies. This method has also been used to compare the degree of infection among colonies before and after a treatment or as a routine colony inspection (Flores et al., 2001; 2004a). To minimize the effects of brood removal by workers, it is important to place the brood comb in an incubator, as described in paragraph 1.3.1.1.
1.3.1.1. Procedure for quantifying the level of colony infection
Remove one brood comb from the hive containing 5th instar larvae that have not yet been sealed (i.e., the larvae are close to being capped with wax)
Mark on a plastic transparency the area with these unsealed larvae (this step is only important if the frame also contains capped brood)
Return the brood comb to the colony
Remove the brood comb from the hive a maximum of 20 hours later
Cut a piece of comb containing at least 100 recently capped cells (capped within the last 20 hours). These larvae can be identified using the plastic transparency. (It is best to remove unsealed brood from the comb before placed in the incubator)
Place one or more pieces of comb with capped cells in an incubator at 25°C at approximately 65% humidity for 5 days. The chilling will ensure disease development. An open water bottle placed in the incubator will provide sufficient humidity
After 5 days, open all capped cells and record the results
Repeat the experiment three times to obtain individuals from enough patrilines and cover the variation in individual susceptibility to the fungus.
1.3.2. Molecular methods qPCR
In recent years, real-time PCR (RT-PCR) has been used to identify and quantify viral, bacterial and microsporidial pathogens in the honey bee (see the BEEBOOK papers on nosema (Fries et al., 2013), European foulbrood (Forsgren et al., 2013), viruses (de Miranda et al., 2013) and molecular methods (Evans et al., 2013)). There are, however, no methods yet available to quantify A. apis spores or the A. apis hyphal biomass.
1.4. Production and quality of inoculums
1.4.1. Isolation techniques
1.4.1.1. Growth media
In culture, A. apis isolates can grow on many different media such as potato-dextrose agar (PDA), yeast-glucose-starch agar (YGPSA), and Sabouraud dextrose agar) (Bailey, 1981; Heath, 1982; Anderson and Gibson, 1998; Hornitzky, 2001), but they grow very well on media with high sugar content like MY-20 (Udagawa and Horie, 1974).
MY-20:
- Dissolve the following components in approximately 750 ml deionised water:
- 4.0 g peptone
- 3.0 g yeast extract
- 200.0 g glucose
- 20.0 g agar
Adjust the final volume to 1000 ml.
Autoclave at 115°C for 15 minutes.
1.4.1.2. Isolation of A. apis strains
Ascosphaera apis can be isolated from fresh mummies or dry mummies collected directly from brood frames or mummies from the bottom board or hive entrance. Both white and dark mummies can be used, but it is generally easier with white mummies, since the latter will often readily produce spore cysts following incubation when + and - strains come into contact.
Surface sterilize the mummy in 10% sodium hypochlorite for 10 min
Rinse the mummy twice in sterile distilled water for 2 min
Cut the mummy into smaller pieces, place the pieces on an agar plate (SDA, PDA,YGPSA or MY20)
Incubate the plate in dark at 30–34°C
Hyphal growth is visible usually within 2–4 days
Addition of antibacterial compounds (e.g. chloramphenicol, cycloheximide, streptomycin or dodine) in the media can be advantageous to prevent bacterial growth. Ascosphaera apis growth will often be inhibited, but once transferred back to non-antibiotic media it can be quickly recovered.
1.4.1.3. Hyphal tip isolation of A. apis strains
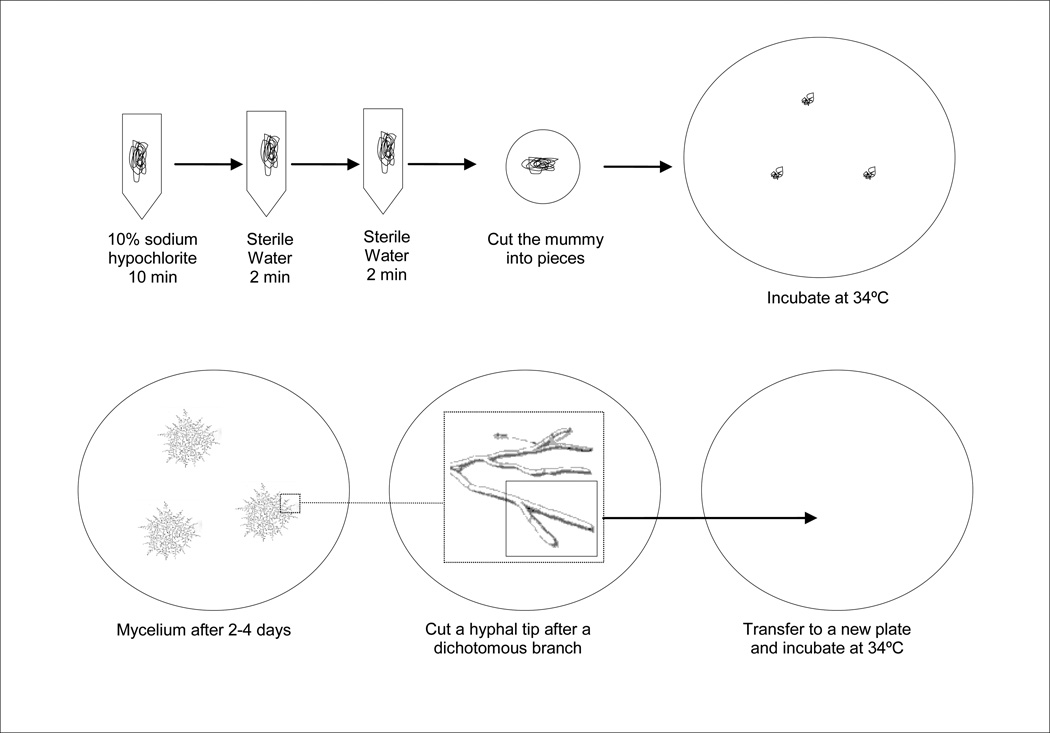
A single honey bee larva can ingest a large number of fungal spores, thus to ensure that the isolate is composed of a single strain, hyphal tip isolation is preferred (Fig. 4).
Place a dissecting microscope in a sterile hood and wipe with 70% ethanol
Use a fresh A. apis isolate (approx. 4 cm in diameter)
Find a single hypha at the edge of the culture under the microscope, and cut a hyphal tip just before the last branching point using a scalpel and a minutien pin
Transfer the tip to a new culture plate
Incubate the plate in darkness at 30–34°C
Fig. 4.
Hyphal tip isolation of Ascosphaera apis
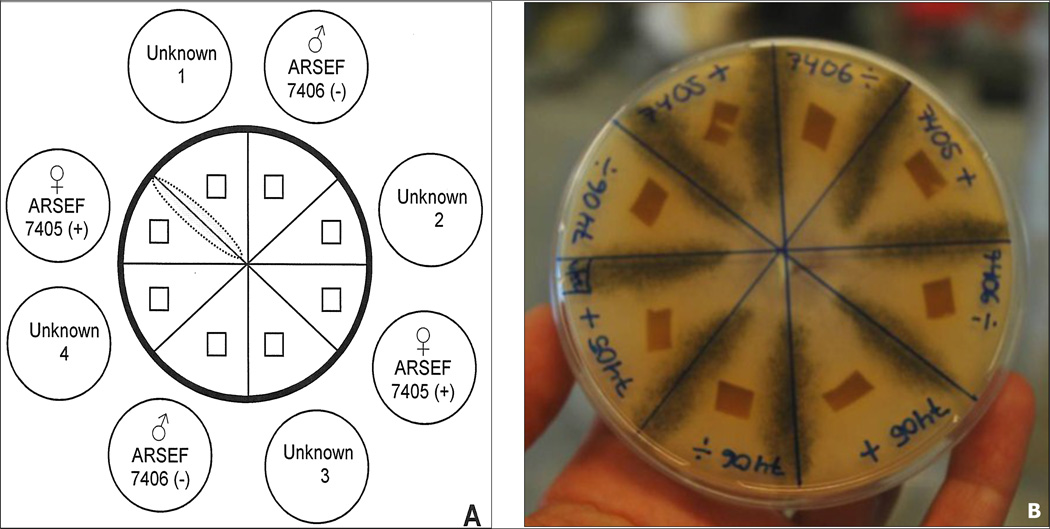
1.4.1.4. Mating test
Once a single isolate is obtained, the mating type can be determined using a sexual compatibility test. We recommend that the two reference strains (AFSEF 7405 and ARSEF 7406) be used in the mating tests.
Cut agar plugs from the border of well growing isolates
Place the plugs of the new isolate between agar plugs of the two reference strains with a distance of approximately 4 cm
Incubate the plate in darkness at 30–34°C
After 5–7 days the presence of dark spore cysts (Fig. 5) normally can be observed as a black zone where hyphae of two strains with opposite mating types meet, indicating successful reproduction
If the test isolates are sexually compatible with the reference + strain, the other is the − mating type, or vice versa On a standard agar plate (diameter 9 cm) 4 new isolates can be tested simultaneously (Fig. 5).
Fig. 5.
Ascosphaera apis mating test and production of inoculums. A. Mating test with four isolates of unknown mating types. Unknown 1 mates with ARSEF 7405 (+) and therefore harbours the opposite mating type (−). B. Production of inoculums between two isolates of opposite mating types. Fruiting bodies with spores inside are formed where the hyphae meet (black zone).
Photo: A B Jensen
1.4.2. PCR for strain differentiation
Genetic variation is crucial for strain differentiation, and the arms race in host-pathogen relationships has been recorded with genetically different A. apis strains (Vojvodic et al., 2011b). The PCR fingerprinting method, using BOX, REP, and ERIC as random primers (Reynaldi et al., 2003), has been used to identify A. apis isolates, and microsatellite markers have been developed (Rehner and Evans, 2009). Recently several sequences of intergenetic regions or introns were screened. Three loci were found to be highly variable and can be used in identifying and differentiating A. apis strains (Jensen et al., 2012) (Table 2). The advantages of DNA sequences of polymorphic loci with specific primers over the PCR fingerprinting and microsatellite markers is that the sequences produced in one study can easily be compared with those in another study.
Table 2.
List of qRT-PCR primers developed for targeting A. apis transcripts. The first three primer sets target genes that are involved in fungal mating and reproduction. Actin can be used as a reference gene
| Primer name | Annealing temp. (°C) |
Primer sequence | Citation |
|---|---|---|---|
| Mat1–2–1 F | 62 | AAAATACCAAGGCCACCGA | Aronstein et al., 2007 |
| Mat1–2–1 R | 62 | GGAGCATATTGGTAATTTGG | Aronstein et al., 2007 |
| Ste11-like F | 62 | GGGAAGATTGCCAGGCC | Aronstein et al., 2007 |
| Ste11-like R | 62 | CAAACTTGTAGTCCGGATG | Aronstein et al., 2007 |
| Htf F | 62 | AAAATCCCAAGGCCTCGTA | Aronstein et al., 2007 |
| Htf R | 62 | CTGGTAGCGGTAGTCAGG | Aronstein et al., 2007 |
| Actin_Aapis F | 58 | CATGATTGGTATGGGTCAG | Aronstein et al., 2007 |
| Actin_Aapis R | 58 | CGTTGAAGGTCTCGAAGAC | Aronstein et al., 2007 |
1.4.2.1. PCR
Genomic DNA can be amplified in a 50 µl reaction containing:
10 µl of 5× HF buffer (1.5 mM MgCL2)
1 U of Phusion® High-Fidelity DNA Polymerase (New England Biolabs, Inc)
200 µM of each deoxyribonucleotide triphosphate
1 µM forward and reverse primer (Table 2)
1 µl of DNA
Reaction conditions:
initial denaturing for 30 s
denaturing at 98°C
10 touchdown cycles: 98°C for 30 s, 70–60°C (decrease of 1° C per cycle) for 30 s, and 72°C for 30 s,
30 cycles of 98°C for 30 s, 60°C for 30 s, and 7 °C for 30 s;
10 min extension at 72°C.
Amplified products can be analysed on a 1.5% (wt/vol) agarose gel stained with ethidium bromide or EZ-vision dye. Alternatively, PCR results could be visualised using DNA imaging. Sharp bands can be sequenced directly (further advice on sequencing and analyses can be found in the BEEBOOK paper on molecular methods (Evans et al., 2013). Note: Alternatively another polymerase can be used and the touch down step can be replaced by adding five normal cycles; however, it will require some optimization.
1.4.3. Preservation of in vitro cultures
An A. apis isolate starts to show signs of aging after 30 days of growth (Ruffinengo et al., 2000) and thus requires monthly transfer. However, frequent culture transfers are expensive, time consuming and increase the risk of contamination. In addition, A. apis isolates, as seen for many other microbes, can lose virulence when kept in culture with repeated transfers, thus it is important to utilize a long term storage method soon after an isolate is obtained. Two different culture preservation methods have been successfully used: cryopreservation (Jensen et al., 2009a) and propagation on Integral Rice Kernels (IRK) (Palacio et al., 2007). Contamination during storage seems to be less risky during cryopreservation at −80°C, however for larger quantities and for laboratories not equipped with cryopreservation capacity, the use of rice kernels is a great alterative.
1.4.3.1. Cryopreservation in glycerol at−80°C
Cut three plugs from the border of a growing agar colony and put the plugs in a cryogenic vial containing 1 ml 10% sterile glycerol
Place the vials in a Cryo 1°C Freezing container, “Mr. Frosty” (Nalgene Co; Rochester, NY). Mr. “Frosty” ensures a 1°C /min cooling rate
Set the “Mr. Frosty” at 5°C for 4 hours and then in −80°C for 24 hours
After 24 hours transfer the vials to standard freezer boxes
Store at −80°C until use
Upon use, thaw the vial in a 34°C water bath and place the hyphal plugs on new agar plates
1.4.3.2. Integral Rice Kernels (IRK) storage
Autoclave (1 atmosphere and 120°C for 15 min) 25 g moistened IRK (2:1 wt/vol) in Petri dishes (9-cm diameter)
Inoculate the IRK with 4-mm diameter plugs from the border of a growing agar colony
Incubate at 30°C for 15 days
Store the IRK at 19°C until use
1.4.4. Spore production
Ascosphaera apis spores are the only infective units of chalkbrood. It is essential to have a high quantity and quality of spores for experiments; e.g. for testing antifungal compounds or for breeding experiments. Spores can be retrieved from black mummies or produced in vitro. From a standardization point of view, the use of in vitro spores from reference strains are strongly recommended, but spores retrieved from black mummies might be chosen in certain situations. Ascosphaera apis will mate on various media but the yield of spores is increased on malt yeast with 20% dextrose agar (MY20) (Ruffinengo et al., 2000). It is recommended that a fresh spore solution be made for every experiment.
1.4.4.1. Production and harvest of in vitro spores
Cut agar plugs from the border of well-growing isolates with different mating types
Place the plugs of the + and − strain a distance of approximately 4 cm on MY20 agar (see paragraph 1.4.1.1.)
Incubate the plate in the dark at 30–34°C at least 3 weeks to ensure production of mature spores
Use a scalpel to scrape off the spore cysts from the black mating stripes
Transfer them to a glass mortar with a ground-glass pestle (glass tissue grinder; Fig. 6a and b)
Add 5–10 µl sterile water and grind the suspension 1 min to release spores from the spore cysts and balls
Add 50 µl sterile water and grind again 1 min
Add 200 µl sterile water to the mortar
Pipette spores from the mortar into an 1.5 ml Eppendorf tube
Add additional 750 µl water to get a medium density of spores
Let large particles, unbroken sporocysts, spore clumps etc. in the suspension settle for 30 min
Pipette, from just below the surface, approximately 500 µl into a new tube, which should contain mostly released individual spores
Prepare a dilution series to 10−2 or 10−3 for counting and calculating the concentration of the undiluted suspension with a haemocytometer (refer to the BEEBOOK paper on miscellaneous methods (Human et al., 2013).
Fig. 6.
Spore germination test: A & B. First grind the sporocysts to release spores from cysts and balls; C & D. incubate the spores in liquid medium and flush with CO2 incubate the spores 32 hours at 34°C; E & F. check for spore enlargement.
Photos: A B Jensen
1.4.4.2. Harvest of in vivo spores
Mix black mummies with 5 ml of sterile distilled water
Shake for 5 min
Homogenize the spore solution with a glass tissue grinder for 5 min
Centrifuge to spin-dry at 3500 rpm for 5 min
Discard the supernatant
Add 5 ml iodine-povidone (50% in distilled sterile water). Iodine-povidone is added to avoid bacterial contamination
Shake for 10 min
Centrifuge to spin-dry at 3500 rpm for 5 min
Discard the supernatant
Wash three times by adding 5 ml of sterile distilled water
Shake for 5 min
Spin-dry at 3500 rpm for 5 min
Add 1 ml water
Prepare a dilution series to 10−2 or 10−3 for counting and calculating the concentration of the undiluted suspension with a haemocytometer (refer to the BEEBOOK paper on miscellaneous methods (Human et al., 2013).
Spores from mummies can also be harvested using the in vitro harvest method described above. To get a better homogenization, scrape the spore cysts from the outside of the mummy before grinding. In vivo spores must be used the same day they are prepared, due to the presence of other microorganisms on the mummies even though the broad-spectrum bactericide iodine-povidone is used to avoid bacterial contamination in the spore solution preparation. Dry dark mummies with viable spores can be preserved for several years (at least 9 years) when stored dry in a closed bottle (Flores, pers. comm.)
1.4.5. Quality test of inoculums
Chalkbrood spores can contaminate all surfaces of the bee hive and accumulate in hive products. They are very resistant to the environment and can remain viable and infective for more than 15 years (Toumanoff, 1951). However, spore viability will decline over time and this will vary from isolate to isolate and from batch to batch. Thus it is important to check spore viability prior to an experiment.
1.4.5.1. Spore viability and germination test (Fig. 6)
Place a sterile Teflon coated slide in a sterile petri dish lined with wet filter paper
Prepare a concentration of 2.0 × 107 spores per ml
Mix 100 µl spore suspension with 500 µl GLEN (see recipe below)
Place 10 µl of the GLEN / spore mix onto the spot of the Teflon coated slides
Place the petri dish in an airtight container and flush with CO2 30 sec to ensure a minimum of 10% CO2
Incubate 32 hours at 34°C
Add a cover slip and count the spores directly on the Teflon slide under a microscope at 400 magnification
Evaluate 100 spores for enlargement or germ tube formation in three different fields of view on the slide
The AnaeroGen system (Oxoid; Basingstoke, UK) has been used with success in cases where the laboratory is not equipped with CO2. A packet is placed together with the petri dish in a sealed jar and will generate 9 to 13% CO2.
1.4.5.2. GLEN medium
- Dissolve the following components in approximately 900 ml deionised water:
- 4.000 g glucose-dextrose
- 7.680 g natrium chloride
- 6.500 g lactalbumin hydrolysate
- 5.000 g yeast extract
- 1.952 g MES-buffer (2-(N-morpholino)ethanesulfonic acid)
Adjust the pH to 7.0 using 1 M NaOH
Readjust the volume to 1000 ml
Sterilize by autoclaving at 121°C for 20 minutes.
1.4.6. Availability and recommended reference isolates
Microbial isolates including fungi are deposited and stored in several culture collections around the world. They are available for both commercial and scientific use. Isolates can be ordered from the culture collection for a fee that is usually lower for non-profit organizations. We would recommend that researchers deposit isolates that have been used in publications in one of these culture collections, so that they can be retrieved for future research.
Different strains of A. apis have shown variable virulence towards honey bees (Vojvodic et al., 2011a) and may also vary in other biological traits. Thus to be able to compare different studies we recommend to always use AFSEF 7405 and ARSEF 7406 as reference isolates. These two isolates have been involved in the A. apis genome sequencing project (Qin et al., 2006); they represent each of the two mating types and both can readily be retrieved from either the USDA-ARSEF or the ATCC culture collections.
Below is the list of the three most important culture collections we recommend for retrieving A. apis isolates:
- USDA-ARSEF; ARS Collection of Entomopathogenic Fungi (7 isolates deposited, March 2012)
- ATCC; American Type Culture Collection (9 isolates deposited, March 2012)
- CBS; Centraalbureau voor Schimmelcultures (7 isolates deposited, March 2012)
1.5. Infection bioassays
A bioassay is a measurement of the effect of a substance on living organisms. Bioassays with A. apis can be conducted using in vitro reared larvae or directly in colonies. An in vitro bioassay has very important advantages over the whole colony bioassay; mainly, it excludes the effects of social immunity and allows for replicable experiments in controlled environment. However, to address questions related to hygienic behaviour, bioassays must be conducted directly within honey bee colonies.
1.5.1. Infection bioassays using in vitro rearing of larvae
Difficulties associated with controlling experimental conditions using honey bee colonies prompted the development of in vivo rearing conditions that allow rearing honey bee larvae in the laboratory and also a way to test the effects of pathogens, toxins and drugs. Refer to the BEEBOOK paper on larval rearing for a detailed protocol (Crailsheim et al., 2013). Exposure bioassays of in vitro reared larvae have been used to test differences in virulence of various A. apis strains (Vojvodic et al., 2011b), to compare the temperature response of chalkbrood and stonebrood (Vojvodic et al., 2011a), to test susceptibility of various honey bee subspecies towards A. apis (Jensen et al., 2009b), to test virulence of A. apis with or without presence of other Ascosphaera species commonly associated with solitary bees (Vojvodic et al., 2012) and to explore the host response to infection of A. apis at the molecular level (Aronstein et al., 2010).
Ascosphaera apis spores have to be ingested by the larvae, hence they must be incorporated in the larval food. There are two ways spores can be administered - either the larvae can be fed a small quantity (5 µl) of spore contaminated diet, which they will ingest quickly, or the larvae can be fed spore-contaminated diet ad libitum for a certain period. The advantage of feeding small quantities is that the exact dose per larvae can be controlled, and if fed with different dosages an exact LD50 can be calculated (Jensen et al., 2009b). Depending on the larval ages, additional feeding has to be done on the same day, approximately 2 hours later, after the first 5 µl is consumed. However if the aim is to produce a high numbers of infected individuals it can be more efficient to use surplus diet, as it is less time consuming.
All larval instars can be infected by A. apis (Jensen, unpublished), but it is recommended to use 2nd–4th instar larvae. When using 5th instar larvae, the A. apis spores have a limited time period in the gut for germination and penetration of the gut wall before the bee defecates, and 1st instar larvae are very fragile.
1.5.2. Infection bioassay of colonies
When it is necessary to provoke chalkbrood infection in bee colonies, it may be sufficient to infect a group of larvae, or it may be necessary to cause widespread disease in the colony. The procedure differs for each case. In the first case, the larvae are directly fed spore-contaminated food. In the second case, the entire colony is exposed to spores either by spraying, feeding sugar/honey with spores or feeding pollen patties with spores (Moffett et al., 1978; Flores et al., 2004a). Flores et al. (2004a) evaluated different ways to inoculate colonies with A. apis spores using pieces of comb containing bee brood that have been subjected to controlled chilling. A mummification of 90.63% of the brood was reached by spraying water containing spores over combs, and colonies fed spore-pollen mixtures reached 86.32% infection. The use of sugar syrup with spores only reached 60.13% mummification and proved to be less effective. We recommend spraying or feeding inoculated pollen since these methods have proven to be most effective.
1.5.2.1. Direct exposure of individual larvae
Prepare a spore solution of 2 × 108 spores/ml in sterile distilled water.
Mix (1:1, v:v) of the solution with honey food (50% honey : 50% distilled sterile water)
Remove sections of comb containing 5th instar larvae from the hive.
Supply each of the 5th instar larvae with 5 µl of contaminated food (equals a dose of 5 × 105 spores per larvae)
Place the food near the mouthpart. Use extreme care to avoid touching the larvae and confirm consumption by direct observation
Immediately after feeding, store the pieces of comb for 2 hours at 25°C
Return the comb to the hive so that the bees will cap the cells containing the inoculated larvae
Remove the comb a maximum of 20 hours later and store it at 25°C and approximately 65% humidity for 5 days. The chilling of the larvae will ensure disease development. An open water bottle placed in the incubator will provide sufficient humidity
After 5 days open all cell cappings and record the results. (See paragraph 3.1. Quantify infection degree in colonies)
1.5.2.2. Infection of colonies with a water spore suspension
Prepare a spore solution of 1.25 × 107 spores/ml in sterile distilled water. (See paragraph 1.4. Production and quality of inoculums)
Spray 10 ml on the each brood comb surface
This quantity has been used successfully on colonies with 8–10 combs (Flores et al., 2004a).
1.5.2.3. Infection of colonies with spores in pollen
Prepare a spore solution of 1.25 × 107 spores/ml in sterile distilled water. (See paragraph 1.4.3. and 1.4.4. Production and quality of inoculums)
Add 100 ml spore solution to 150 g pollen
Distribute the mixture on the top of the combs
An alternative method, if spore concentration is not very important, is to homogenize 15 black mummies and mix with 150 g pollen.
1.6. Expression of fungal genes
Studies investigating host-pathogen interactions have taken a new dimension utilizing mRNA quantification (qRT-PCR). Here we describe qRT-PCR approach used for identification of A. apis mating type idiomorphs and quantification of A. apis transcripts in culture and in host tissue (Aronstein et al., 2007).
Total RNA can be isolated using standard kits, TRIzol (Invitrogen; Carlsbad, California) and RNeasyR Mini Kit (Qiagen; Valencia CA). These reagents have been used successfully to make cDNA from mycelia or honey bee larvae (Aronstein et al., 2007; 2010), for RNA extraction method, see the BEEBOOK paper on molecular methods (Evans et al., 2013)
1.6.1. qRT-PCR for quantification of A. apis transcripts
- cDNA can be amplified in a 20 µl reaction containing,
-
1.1.1.0 U of GoTaq® Flexi DNA polymerase (Promega Co; Madison, WI),
-
1.2.5 × GoTaq® Flexi buffer,
-
1.3.0.25 mM dNTP mix,
-
1.4.2.5 mM MgCl2,
-
1.5.0.3 µM of each primer,
-
1.6.0.75 µl of a 1/1000 stock dilution of SYBR-Green (Invitrogen Corp),
-
1.7.1 µl of cDNA
-
1.1.
- Reaction conditions are as follows:
-
2.1.Initial denaturing at 95° C for 3 min,
-
2.2.40 cycles at 95°C (20 s), 58–62°C (30 s) depending on the gene (Table 3) and 72°C (30 s).Negative controls (all reaction components except the DNA which is replaced with water) must be included in each run.
-
2.1.
For standardization and normalization against housekeeping genes and data analysis, see the BEEBOOK paper on molecular methods (Evans et al., 2013).
Table 3.
List of qRT-PCR primers developed for targeting A. apis transcripts. The first three primer sets target genes that are involved in fungal mating and reproduction. Actin can be used as a reference gene
| Primer name | Annealing temp. (°C) |
Primer sequence | Citation |
|---|---|---|---|
| Mat1–2–1 F | 62 | AAAATACCAAGGCCACCGA | Aronstein et al., 2007 |
| Mat1–2–1 R | 62 | GGAGCATATTGGTAATTTGG | Aronstein et al., 2007 |
| Ste11-like F | 62 | GGGAAGATTGCCAGGCC | Aronstein et al., 2007 |
| Ste11-like R | 62 | CAAACTTGTAGTCCGGATG | Aronstein et al., 2007 |
| Htf F | 62 | AAAATCCCAAGGCCTCGTA | Aronstein et al., 2007 |
| Htf R | 62 | CTGGTAGCGGTAGTCAGG | Aronstein et al., 2007 |
| Actin_Aapis F | 58 | CATGATTGGTATGGGTCAG | Aronstein et al., 2007 |
| Actin_Aapis R | 58 | CGTTGAAGGTCTCGAAGAC | Aronstein et al., 2007 |
1.7. Hygienic behaviour
Hygienic behaviour is defined as the bees’ ability to detect and remove diseased brood from the nest (Rothenbuhler, 1964). Hygienic behaviour was first described in the 1930s when researchers sought to determine the mechanism by which some honey bee colonies were resistant to American foulbrood (reviewed in Spivak and Gilliam, 1993). In the 1980s, it was shown that hygienic behaviour was also the primary mechanism of resistance to chalkbrood (Gilliam et al., 1983), although resistance to this disease involves other factors as well, such as differences in the susceptibility of different colonies or even between patrilines within colonies (Invernizzi et al., 2009; Jensen et al., 2009b).
Hygienic behaviour assays, involving killing brood by freezing or by piercing pupae with a pin (methods described in the BEEBOOK paper on queen rearing and selection (Büchler et al., 2013)) are indirect, and record the proportion of dead brood removed by a colony after a particular amount of time. Most, but not all colonies, show a good correlation between removal of freeze-killed brood and resistance to chalkbrood. However, researchers and beekeepers cannot assume that the ability of a colony to remove dead brood within a certain time will ensure colony-level resistance to chalkbrood. It is very important, especially for breeding purposes, to directly challenge colonies with A. apis in addition to the freeze-kill or pin-kill brood assay (See paragraph 1.5.1. Infection bioassay of colonies) and subsequent observation of the bees’ response to the challenged brood (Palacio et al., 2010).
1.8. Olfactory detection
Bees from hygienic colonies are particularly responsive to olfactory-based stimuli associated with diseased brood. All bees can perform uncapping and removal behaviours, but bees that detect abnormal brood odours at a low stimulus level may rapidly initiate uncapping behaviour, resulting in the removal of diseased brood before it becomes infectious (Wilson-Rich et al., 2009). Individual bees from rapid-hygienic line breeds exhibited significantly increased sensitivity to the odour of chalkbrood disease at lower concentrations compared with bees from the slow-hygienic line, based on electrophysiological recordings of nerve impulses from the antennae (EAG), by proboscis-extension response conditioning (PER), and by isolation of volatiles from chalkbrood–infected larvae for use in field bioassays (see detailed methods in: Masterman et al., 2000, 2001; Gramacho and Spivak, 2003; Spivak et al., 2003; Swanson et al., 2009; and general methods of EAG and collection of volatiles in the BEEBOOK paper on chemical ecology methods (Torto et al., 2013) and of PER in the BEEBOOK paper on behavioural methods (Scheiner et al., 2013)).
1.9. Inhibitory assays against chalkbrood
For both fundamental and applied research it can be important to test the inhibitory effects of certain chemicals, plant extracts, propolis, probiotic bacteria or hemolymph against chalkbrood. These substances can be tested for their direct effect on spore germination and on hyphal growth, or they can be used to test their effect on the ability of the fungus to infect individual bees in vitro or in a colony context.
1.9.1. Inhibition of spore germination and hyphae (zone of inhibition)
Prepare MY20 medium (see paragraph 1.4.1.1.) in petri dishes (15 cm)
Spread 2 ml of a spore solution (9.0 ×107 spores/ml) on the surface of the medium, approximately 106 spores/cm2
Make a central hole (7 mm) in the MY20 medium (Fig. 7A)
Place 0.5 ml of the test product into the hole
Incubate the cultures (30°C, 12% CO2, 65% relative humidity)
Measure the fungal growth (or the zone of inhibition) (Fig. 7B)
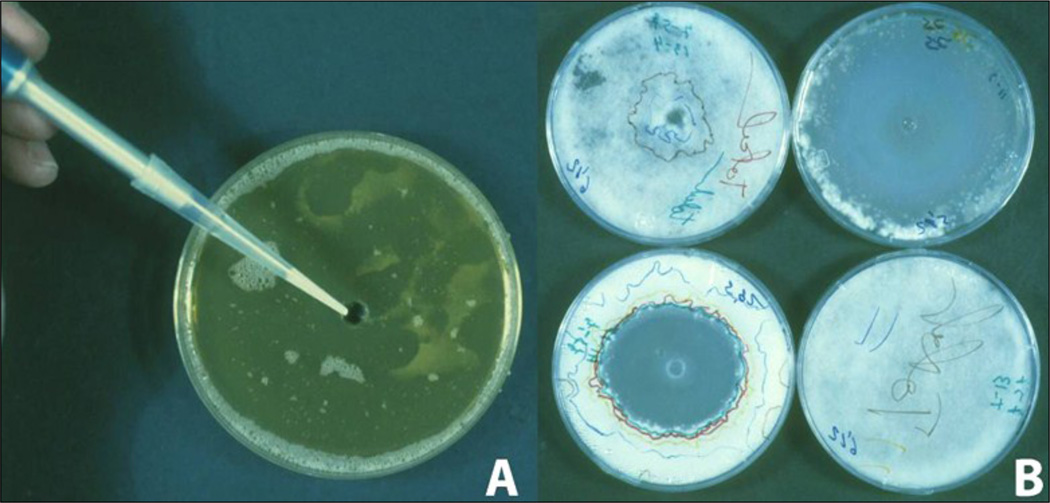
Fig. 7.
Zone of inhibition assay of different compounds against Ascosphaera apisA. MY-20 medium with spores spread on the entire surface and with a central hole for the test compound; B. The zone of inhibition was recorded daily by drawing on the Petri dish, a line over the border of the mycelium (each day a different colour) and the antifungal property was assessed as daily inhibition zone using an image analyser.
Photos: J M Flores
Measure the fungal growth (or the zone of inhibition) daily using a stereoscopic microscope magnification (× 20 to 40). Mycelium growth can be seen within the medium before emerging at the surface (Puerta et al., 1990). An Image Analyzer can be used to measure the diameter of the zone of inhibition.
1.9.2. Inhibition in colonies
Assess the degree of infection in colonies before treatment, as described in paragraph 1.3.1. (Quantify infection degree in colonies)
Increase colony infection with A. apis as described in paragraph 1.5.2. (Infection bioassay of colonies)
Apply the test item to the colony
Assess the level of infection in the colony, as described in paragraph 1.3.1. (Quantify infection level in colonies) the next day after treatment and repeat the assessment depending of the aim of the experiment.
We propose an indirect assessment of in vivo treatments because quantifying the disease by counting mummies is very difficult and inaccurate. If the disease is increasing in the colony, the spore numbers will also increase; likewise, if the disease is decreasing, the spore number will be lower due to the natural behaviours of the bees such as hygienic removal of larvae. The degree of infection would most probably be related to the dynamics of A. apis spore load in the colony. Evaluation of the level of infection in the colony before and after treatment is therefore recommended.
1.10. Minimizing chalkbrood in experimental colonies
The main techniques for chalkbrood control lie in management practices that reduce spore concentration in the colonies and that avoid stress (in particular chilling) of susceptible brood.
1.10.1. Breeding for resistance
Gilliam et al., (1983) and Taber (1986) demonstrated that it is possible to select and breed honey bees for resistance to chalkbrood disease. Spivak and Reuter (2001) demonstrated that colonies selected for rapid removal of freeze-killed brood showed resistance to chalkbrood in field experiments. Palacio et al. (2000) observed that hygienic colonies had a lower frequency of brood diseases including chalkbrood. Commercial queen breeders in the US and Denmark have found that if they have “zero-tolerance” for chalkbrood; i.e. they never raise queens from a colony that has had clinical symptoms of chalkbrood and they simultaneously select for rapid hygienic behaviour then they get rather chalkbrood resistant lines (Spivak and Jensen, unpublished).
1.10.2. Management and treatment
There are a number of management’s techniques that can be used to minimize the effects of chalkbrood in infected colonies:
Reduce volume of the brood chamber for the overwintering (Seal, 1957)
Enlarge colony entrance to aid ventilation (Gochnauer et al., 1975)
Replace old combs (Betts, 1951)
Heat treatment of the wax (Flores et al., 2005a)
Requeen affected colonies (Lunder, 1972)
To date, there is no effective chemical treatment against this disease. Nevertheless in vitro (Puerta et al., 1990; Flores et al., 1996b) and in vivo (Flores et al., 2001) techniques have been used to evaluate chemical treatments for chalkbrood control.
2. Stonebrood
2.1. Introduction
Stonebrood is a very rare honey bee brood disease caused by several fungi from the genus Aspergillus. The disease was first described by Massen (1906) and has since then been found worldwide. Aspergillus flavus has most frequently been reported, followed by Asp. fumigatus, but also Asp. niger and other species can kill honey bees (Gilliam and Vandenberg, 1997). Aspergillus is able to infect the host through the gut if the spores are ingested, but also through the cuticle. Therefore, adults as well as larvae and pupae can become infected. In addition, most species of Aspergillus produce aflatoxins that have been suggested to be the primary cause of death in stonebrood infected honey bees (Burnside, 1930). However, a non-aflatoxin producing Asp. flavus strain has been observed to induce stonebrood symptoms equally well in in vitro reared honey bee larvae as afaltoxin producing strains (Vojvodic, unpublished).
Aspergillus spp. are cosmopolitan filamentous fungi often found in soil, where they thrive as saprophytes, but occasionally they do infect living hosts including, plants, insects and mammals. Aspergillus can infect human lungs, eyes, pharynx, skin and open wounds, but most commonly this has been observed in immune deprived individuals (Gefter, 1992; Germaud and Tuchais, 1995; Denning, 1998; Galimberti et al., 1998; Garret et al., 1999). In addition, the alfatoxins are carcinogenic if inhaled or ingested; therefore precautions need to be taken when stonebrood disease occurs in honey bees principally to protect beekeepers and consumers. In several countries stonebrood is a notifiable disease that has to be reported to the authorities if it occurs.
2.2. Biohazards
Working with fungi requires good microbiological practice and containment, irrespective of whether they possess a potential risk for the environment or human, since proliferation on the growth medium of contaminants always poses a potential risk. Good microbiological practice is in principle the handling of a microorganism in a "test tube" without any other organisms entering and contaminating it. Containment is in principle the handling of a microorganism with emphasis on safety of the laboratory worker and the environment. Good microbiological practice and containment involves:
Aseptic techniques
Limit the open time of “test tube”
Open tubes or plates only in bio-safe cabinet/bench to avoid
contamination
Only use sterile tools (e.g. pipette tips or loops)
Avoid casual contact with the bench, fingers, or outside of the bottle
Dispose or decontaminate tools immediately after use
Personal hygiene and dress
Wash hands prior to and following manipulations
Wear appropriate personal protective equipment (gloves, clean lab coats)
Do not touch the skin, face, or unclean non-sterile surfaces
Confine loose or long hair and keep fingernails short
Area cleanliness and organization
Disinfect work area before and after work with 70% Ethanol
Immediately clean spills, and then disinfect the work surface
Keep only items important for the task in progress in the bench
Plan and lay out work in a logical order so the work in the bench becomes efficient
Minimize personnel traffic and unnecessary movements around the work area
Routine cleaning of difficult-to-access areas to prevent build up of dust and debris
Aspergillus spp. produce airborne conidia which represent a potential risk for the experimenter, but no additional attention has to be taken as long as good microbiological practice and containment are followed. In particular, it is important that the cultures grown on agar plates are only opened in a sterile bench and that .conidia are only handled outside the bench if they are in a liquid suspension.
If conducting a bioassay with Aspergillus spp. on in vitro reared larvae or in cages on adult honey bees, it is important that the assessment is done in a sterile bench or a fume hood once the Aspergillus start to sporulate. Dead infected bees can be removed with forceps to avoid production of numerous new conidia.
In several countries, permission from the authorities is required to work experimentally with stonebrood fungi, in particular if it is an outdoor experiment. If Aspergillus spp. are used for experiments in bee colonies we recommend using a mask and safety glasses for protection while conducting the experiments.
2.3. Diagnostics and qualitative detection
2.3.1 Morphological description
The typical symptoms observed in a colony affected by stonebrood are not very different from chalkbrood symptoms and includes irregular capping of the brood. Infected brood, also called "mummies”, can be seen in the combs. Stonebrood mummies turn hard and they resemble small stones, not sponge-like as chalkbrood mummies. Stonebrood mummies are difficult to remove from the cells with forceps and removal by the worker bees is also difficult. Infected brood becomes covered with powdery yellow, brown, green or black fungal spores depending on the species. In some cases infected or deceased larvae looked dry, but they do not produce visible conidia within a 48 hrs after pathogen inoculation (Vojvodic, unpublished)
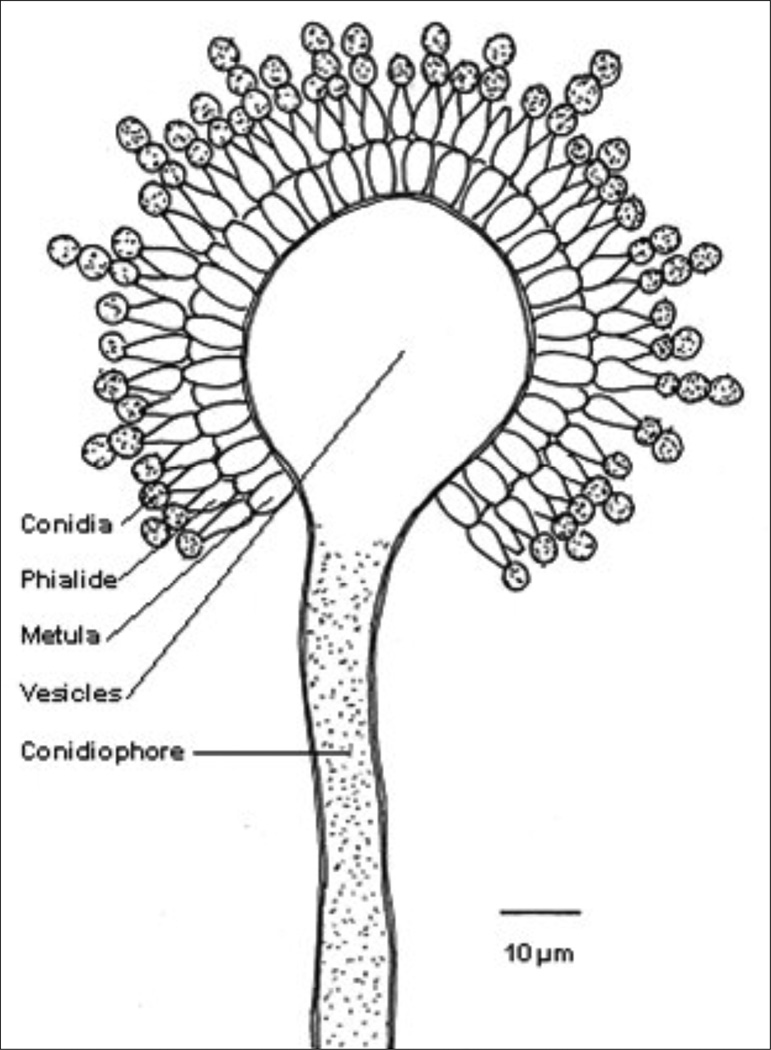
Stonebrood can be diagnosed by its gross symptoms, but positive identification requires its cultivation in the laboratory and subsequent microscopic examination. Structures of the conidiophores (spore forming structures) are very important for identification of Aspergillus spp. The conidiophores originate from a basal cell located on the supporting hyphae and terminate in a vesicle (Fig. 8). The morphology, colour and roughness of the conidiophores vary from species to species. Additionally, the position of the flask-shaped phialides (spore producing cells) on the vesicle is an important character. The phialides can cover the vesicle surface entirely ("radiate" head) or partially ("columnar" head) and the phialids can be attached to the vesicle directly (uniseriate) or attached via a supporting cell, called metula (biseriate). The phialides produce round conidia (2–5 µm in diameter) that form radial chains (Fig. 8). The conidia of the different species can have different colours (Table 4) (See Fig. 9 for Asp. flavus in vitro infected larvae). We however recommend contacting a mycologist for correct species identification of the Aspergillus specimens.
Fig. 8.
Structures of importance for identification of Aspergillus species.
Table 4.
Microscopic characters of three Aspergillus species most often reported to cause stonebrood
| Species | Conidia colour | Conidiophores | Phialides | Vesicle |
|---|---|---|---|---|
| Asp. flavus | Yellow-green | Colourless. Rough | Uni/biseriate | Round, radiate head |
| Asp. fumigatus | Blue-green to grey | Short (<300µm), smooth, colourless-qreen | Uniseriate | Round, columnar head |
| Asp. niger | Black | Long, smooth, colourless or brown | Biseriate | Round, radiate head |
Fig. 9.
In vitro reared honey bee larvae infected with Aspergillus flavus: the bottom left cell contains larvae in an early stage of disease visible by the change in larval colour; bottom right cell contains later stage of the diseases with the visible fungal body and conidia protruding out of the larval cuticle. The two upper cells contain healthy larvae.
Photo: S Vojvodic
2.3.2 Molecular methods
There is no single method (morphological, physiological or molecular) that can be used to recognize all of the approximately 250 Aspergillus species. Using a multi-locus approach will give a lot of information, but it also requires certain skills and equipment not always present in diagnostic laboratories (Geiser et al., 2007). A two-step barcoding has been suggested for identification of Aspergillus species in a clinical setting. The ITS regions can be used for inter-section level identification and the β-tubulin for identification of individual species within the various Aspergillus sections (Balajee et al., 2007) (see primers in Table 5).
Table 5.
List of primers used to be used in the two-step DNA barcoding of Aspergillus spp. for the ITS region and part of the β-tubulin gene.
| Primer name | Annealing temp (°C) |
Annealing temperature (°C) | Citation |
|---|---|---|---|
| ITS 1 | 55 | TCCGTAGGTGAACCTGCGG | White et al., 1990 |
| ITS 4 | 55 | TCCTCCGCTTATTGATATGC | White et al., 1990 |
| Bt2a (β-tubulin) | 58 | GGTAACCAAATCGGTGCTGCTTTC | Glass and Donaldson, 1995 |
| Bt2b (β-tubulin) | 58 | ACCCTCAGTGTAGTGACCCTTGGC | Glass and Donaldson, 1995 |
Aspergillus DNA can, as for Ascosphaera, be extracted with standard kits (see also the BEEBOOK paper on molecular methods (Evans et al., 2013)
2.4. Production and quality of inoculums
2.4.1. Isolation techniques
Aspergillus grows readily on many different standard media e.g. SDA, PDA, but Czapek-Dox medium, which contain sucrose as carbon source and nitrate as the nitrogen source should be very suitable. Czapek-Dox medium with addition of yeast extract (5.0 g/l) is recommended (Frisvad, pers. comm.). All these three media can be purchased premixed and easily prepared. E.g. to prepare 1 l of SDA medium, suspend 65 g SDA (Merck) in 1 l of demineralized water and autoclave 15 min at 121°C.
2.4.1.1. Aspergillus spp. can be isolated from sporulating mummies
Put a sterile microbial loop in an area with many conidia
Streak on agar plates
If the plates become contaminated with other microbes, repeat the procedure. Once a clean culture is established, proceed with single spore isolation described below.
2.4.1.2. Single spore isolation
Add 10 ml 0.05% Triton-X on the culture agar plate
Rub its surface gently with a sterile Drigalski spatula to loosen the conidia
Transfer the suspension conidia into a 15–50 ml sterile tube
- Wash the suspension twice (to remove agar and hyphal fragments)
-
4.1.Centrifuge 3 min at 7000 g for 3 min
-
4.2.Discharge the supernatant and add 10 ml 0.005% Triton-X
-
4.3.Centrifuge 3 min at 7000 g for 3 min
-
4.4.Discharge the supernatant and add 10 ml 0.005% Triton-X
-
4.1.
Prepare a serial dilution (remember to whirl mix before pipetting)
Count the spore concentration in a haemocytometer (described in the BEEBOOK paper on miscellaneous methods (Human et al., 2013))
Transfer 100 µl of a spore solution at 5 × 102 spore per ml to a new plate
Incubate at 25°C for two-four days
Transfer a single small colony to a new plate
To harvest conidia for experimental purposes the above procedure can be used (minus step 6–8). It is important to use Triton-X or another detergent to avoid spore clumping.
2.4.2. Preservation of in vitro cultures
Aspergillus has been observed to lose sporulation capacity and virulence after a couple of transfers on a standard medium (Scully and Bidochka, 2006). Therefore we recommend long-term storage of the isolates during the first transfers. Several long-term storage methods can be used: frozen in skim milk, refrigerated with silica gel, or freeze dried. It is possible to successfully recover various Aspergillus species (e.g. Asp. flavus, Asp. parasiticus and Asp. niger), stored in 10% glycerol at −80°C for more than a year, using the same method which we recommend for A. apis (see paragraph 1.4.3.1. Cryopreservation in glycerol at −80°C).
2.5. Quality test of inoculums
Viability testing of Aspergillus spp. can be performed using the standard insect pathology methods, which are briefly described below.
2.5.1. Spore viability and germination test
Transfer 10 µl of a spore solution at 5 × 105 spore per ml to an agar plate
Spread the spore solution over the entire ager plate with a sterile Digralski spatula
Incubate at 25°C for 24 hour
Place three cover slips on the plate
Place the agar plate under the microscope and count the proportion of germinated spores out of 100 spores under each of the three cover slips
It is recommended to use only new fresh spore solution for each experiment because the shelf life of Aspergillus spores in a water solution is about 3 days (Vojvodic, unpublished).
2.5.2. Availability and recommended reference isolates
Aspergillus has been deposited in various fungal culture collections, such as USDA-ARSEF, ATCC and CBS (see link below). None of the isolates deposited so far have been isolated from honey bees, neither larvae nor adults, but from sources such as plant material, soil, vertebrates and invertebrates. USDA-ARSEF is a collection of Entomopathogenic fungi, thus the majority of isolated deposited there originate from insects or other arthropods. It is possible to retrieve Aspergillus isolates from hymenoptera; solitary megachilid bees (Osmia lignaria and Megachile rotundata) and formicine ants (Anoplolepsis longipes and Solenopsis invicta).
Limited work has been performed with stonebrood, thus it is difficult to recommend a specific reference strain. A reference strain can be chosen based on its pathobiological properties (to honey bees or at least Hymenoptera); as in Vojvodic et al. (2011a) where an Asp.flavus strain isolated from an infected honey bee larvae was used to infect in vitro reared honey bee larvae. This particular strain is unfortunately lost. However, the reference strain could be based on its geographical origin or type specimen.
- USDA-ARSEF; ARS Collection of Entomopathogenic Fungi (83 isolates deposited, 15 Asp. flavus, 7 Asp. niger April 2012)
- ATCC; American Type Culture Collection (1618 isolates deposited, 180 Asp. favus, 111 Asp. fumingatus, 117 Asp. niger; April 2012)
- CBS; Centraalbureau voor Schimmelcultures (1055 isolates deposited, 47 Asp. flavus, 147 Asp. fumigatus, 7 Asp. niger; April 2012)
Aspergillus is classified as an opportunistic human pathogen, and thus some countries will need an import permit and proof that the laboratory is accredited to handle it.
2.6. Infection bioassays
Exposure bioassays with Aspergillus spp. can potentially be carried out, using in vitro reared larvae (Fig. 9), caged bees or colonies, due to its ability to infect larvae, pupae and adults. A bioassay with in vitro reared larvae has been conducted showing that stonebrood and chalkbrood had opposite temperature-dependent responses in virulence. Chilling of chalkbrood exposed larvae increased the pathogen virulence; whereas chilling of stonebrood exposed larvae increased larval survival (Vojvodic et al., 2011a). For infection of in vitro bee larvae with Aspergillus spp. the same precautions must be taken as described in paragraph 1.5.1. for A. apis.
Few experiments with caged bees have been conducted (Gilliam and Vandenberg, 1997). Infecting individual larvae while still in the brood frame is possible by placing the spore suspensions to be ingested in front of the larvae (Vojvodic unpublished).
3. Future perspectives
Chalkbrood and stonebrood diseases have been recognized for more than a century, but there is still much that remains to be discovered regarding these diseases and their impact on the general health status of honey bees. The numbers of studies of the two diseases reflects their frequency and abundance, with a magnitude difference in favour of chalkbrood.
The genome of A. apis was published in 2006 (Qin et al., 2006), and although the full annotation is still lacking, it will be useful for future research investigations on the expression of genes important for the infection and virulence of A. apis. Chalkbrood is a stress related disease and a recent longitudinal cohort study based on monitoring data collected over six years indicated that colonies with high numbers of varroa mites in the same season or Nosema ceranae infection in the spring had significantly higher chances of chalkbrood outbreaks (Hedtke et al., 2011). Such a correlation needs experimental confirmation, but it elucidates the complexity of the host-pathogen-interaction in honey bee colonies. Research on the interaction with other pathogens and stressors, such as sublethal concentration of various chemicals, is also warranted.
Aspergillus research is mostly focused on human health and food spoilage due to aflatoxin contamination of grains. Stonebrood outbreaks are rarely observed in honey bee colonies, but should not be underestimated. Their rarity could be a result of honey bees removing the stonebrood infected individuals very quickly, an area of research that has not been previously investigated. Furthermore, the basic biology of stonebrood is still poorly understood, and several studies elucidating stonebrood etiology are still to be performed. Aspergillus spp. spores are present everywhere and a high virulence towards honey bee larvae have been shown. Even though stonebrood is rarely reported, it would be interesting to understand which factors and mechanisms might play a role in the establishment and resistance of this disease.
References
- Anderson DL, Gibson NL. New species and isolates of spore-cyst fungi (Plectomycetes: Ascosphaerales) from Australia. Australian Systematic Botany. 1998;11:53–72. http://dx.doi.org/10.1071/SB96026. [Google Scholar]
- Anderson DL, Gibbs AJ, Gibson NL. Identification and phylogeny of sporecyst fungi (Ascosphaera spp ) using ribosomal DNA sequences. Mycological Research. 1998;102:541–547. http://dx.doi.org/10.1017/S0953756297005261. [Google Scholar]
- Aronstein KA, Murray KD, De Leon J, Jqin X, Weinstock G. High mobility group (HMG-box) genes in the honey bee fungal pathogen Ascosphaera apis . Mycologia. 2007;99:553–561. doi: 10.3852/mycologia.99.4.553. http://dx.doi.org/10.3852/mycologia.99.4.553. [DOI] [PubMed] [Google Scholar]
- Aronstein KA, Murray KD. Chalkbrood disease in honey bees. Journal of Invertebrate Pathology. 2010;103:20–29. doi: 10.1016/j.jip.2009.06.018. http://dx.doi.org/10.1016/j.jip.2009.06.018. [DOI] [PubMed] [Google Scholar]
- Aronstein KA, Murray KD, Saldivar E. Transcriptional responses in Honey bee larvae infected with chalkbrood fungus. BMC Genomics. 2010;11:391. doi: 10.1186/1471-2164-11-391. http://dx.doi.org/10.1186/1471-2164-11-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L. Honey bee pathology. Vol. 11. London, UK: Academic Press; 1981. pp. 40–44. [Google Scholar]
- Bamford S, Heath LAF. The effects of temperature and pH on the germination of spores of the chalkbrood fungus, Ascosphaera apis . Journal of Apicultural Research. 1989;28:36–40. [Google Scholar]
- Balajee SA, Houbraken J, Verweij PE, Hong S-B, Yaguchi T, Varga J, Samson RA. Aspergillus species identification in the clinical setting. Studies in Mycology. 2007;59:39–46. doi: 10.3114/sim.2007.59.05. http://dx.doi.org/10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts AD. The diseases of bees: their signs, causes and treatment. Camberley, UK: Hickmott; 1951. [Google Scholar]
- Bissett J. Contribution toward a monograph of the genus Ascosphaera . Canadian Journal of Botany. 1988;66:2541–2560. [Google Scholar]
- Burnside CE. Fungous diseases of the honey bee. US Department of Agriculture Technical Bulletin. 1930;149 [Google Scholar]
- Büchler R, Andonov S, Bienefeld K, Costa C, Hatjina F, Kezic N, Kryger P, Spivak M, Uzunov A, Wilde J. Standard methods for rearing and selection of Apis mellifera queens. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 1. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.1.07. [Google Scholar]
- Crailsheim K, Brodschneider R, Aupinel P, Behrens D, Genersch E, Vollmann J, Riessbergergallé U. Standard methods for artificial rearing of Apis mellifera larvae. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 1. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.1.05. [Google Scholar]
- De Miranda JR, Bailey L, Ball BV, Blanchard P, Budge G, Chejanovsky N, Chen Y-P, Van Dooremalen C, Gauthier L, Genersch E, De Graaf D, Kramer M, Ribière M, Ryabov E, De Smet L, Van Der Steen JJM. Standard methods for virus research in Apis mellifera . In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume II: standard methods for Apis mellifera pest and pathogen research. Journal of Apicultural Research. 4. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.4.22. [Google Scholar]
- Denning DW. Invasive aspergillosis. Clinical Infectious Diseases. 1998;26:781–803. doi: 10.1086/513943. http://dx.doi.org/10.1086/513943. [DOI] [PubMed] [Google Scholar]
- Evans JD, Chen YP, Cornman RS, De La Rua P, Foret S, Foster L, Genersch E, Gisder S, Jarosch A, Kucharski R, Lopez D, Lun CM, Moritz RFA, Maleszka R, Muñoz I, Pinto MA, Schwarz RS. Standard methodologies for molecular research in Apis mellifera . In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 4. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.4.11. [Google Scholar]
- Flores JM, Ruiz JA, Ruz JM, Puerta F, Bustos M, Padilla F, Campano F. Effect of temperature and humidity of sealed brood on chalkbrood development under controlled conditions. Apidologie. 1996a;27:185–192. http://dx.doi.org/10.1051/apido:19960401. [Google Scholar]
- Flores JM, Ruíz JA, Ruz JM, Puerta F, Bustos M, Padilla F, Campano F. Estudio in vivo e in vitro para el control de la ascosferiosis en apis mellifera . Revista Iberoamericana de Micología. 1996b;13:292–295. [Google Scholar]
- Flores JM, Puerta F, Gutiérrez I, Arrebola F. Estudio de la eficacia del Apimicos-b® en el control y la prevención de la ascosferiosis en la abeja de la miel. Revista Iberoamericana de Micología. 2001;18:187–190. [PubMed] [Google Scholar]
- Flores JM, Gutiérrez I, Puerta F. A comparison of methods to experimentally induce chalkbrood disease in honey bees. Spanish Journal of Agricultural Research. 2004a;2:79–83. [Google Scholar]
- Flores JM, Gutiérrez I, Puerta F. Oxytetracycline as a predisposing condition for chalkbrood in honey bee. Veterinary Microbiology. 2004b;103:195–199. doi: 10.1016/j.vetmic.2004.07.012. http://dx.doi.org/10.1016/j.vetmic.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Flores JM, Spivak M, Gutiérrez I. Spores of Ascosphaera apis contained in wax foundation can infect honey bee brood. Veterinary Microbiology. 2005a;108:141–144. doi: 10.1016/j.vetmic.2005.03.005. http://dx.doi.org/10.1016/j.vetmic.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Forsgren E, Budge GE, Charrière J-D, Hornitzky MAZ. Standard methods for European foulbrood research. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK: Volume II: Standard methods for Apis mellifera pest and pathogen research. Journal of Apicultural Research. 1. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.1.12. [Google Scholar]
- Fries I, Chauzat M-P, Chen Y-P, Doublet V, Genersch E, Gisder S, Higes M, Mcmahon DP, Martín-Hernández R, Natsopoulou M, Paxton RJ, Tanner G, Webster TC, Williams GR. Standard methods for nosema research. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK: Volume II: Standard methods for Apis mellifera pest and pathogen research. Journal of Apicultural Research. 1. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.1.14. [Google Scholar]
- Galimberti R, Kowalczuk A, Parra IH, Ramos MG, Flores V. Cutaneous aspergillosis: a report of six cases. British Journal of Dermatology. 1998;139:522–526. doi: 10.1046/j.1365-2133.1998.02424.x. http://dx.doi.org/10.1046/j.1365-2133.1998.02424.x. [DOI] [PubMed] [Google Scholar]
- Garrett DO, Jochimsen E, Jarvis W. Invasive Aspergillus spp. infections in rheumatology patients. Journal of Rheumatology. 1999;26:146–149. [PubMed] [Google Scholar]
- Gefter WB. The spectrum of pulmonary aspergillosis. Journal of Thoracic Imaging. 1992;7:56–74. doi: 10.1097/00005382-199209000-00009. [DOI] [PubMed] [Google Scholar]
- Germaud P, Tuchais E. Allergic bronchopulmonary aspergillosis treated with itraconazole. Chest. 1995;107:883. doi: 10.1378/chest.107.3.883. http://dx.doi.org/10.1378/chest.107.3.883. [DOI] [PubMed] [Google Scholar]
- Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA. The current status of species recognition and identification in Aspergillus . Studies in Mycology. 2007;59:1–10. doi: 10.3114/sim.2007.59.01. http://dx.doi.org/10.3114/sim.2007.59.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam M, Taber S, Richardson GV. Hygienic behaviour of honey bees in relation to chalkbrood disease. Apidologie. 1983;14:29–39. http://dx.doi.org/10.1051/apido:19830103. [Google Scholar]
- Gilliam M, Vandenberg J. Fungi. In: Morse RA, Flottum K, editors. Honey bee pests, predators, & diseases. 3rd Edition. Medina, USA: A I Root Co; 1997. pp. 81–110. [Google Scholar]
- Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochnauer TA, Furgala B, Shimanuki H. Diseases and enemies of the honey bee. In: Grout R, editor. The hive and the honey bee. 3rd Ed. Hamilton, USA: Dadant and Sons; 1975. pp. 615–621. [Google Scholar]
- Gramacho KP, Spivak M. Differences in olfactory sensitivity and behavioural responses among honey bees bred for hygienic behaviour. Behavioural Ecology and Sociobiology. 2003;54:472–479. http://dx.doi.org/10.1007/s00265-003-0643-y. [Google Scholar]
- Heath LAF. Development of chalkbrood in a honey bee colony: a review. Bee World. 1982;63:119–130. [Google Scholar]
- Heath LAF, Gaze BM. Carbon dioxide activation of spores of the chalkbrood fungus Ascosphaera apis . Journal of Apicultural Research. 1987;26(4):243–246. [Google Scholar]
- Hedtke K, Jensen PM, Jensen AB, Genersch E. Evidence for emerging parasites and pathogens influencing outbreaks of stress-related diseases like chalkbrood. Journal of Invertebrate Pathology. 2011;108:167–173. doi: 10.1016/j.jip.2011.08.006. http://dx.doi.org/10.1016/j.jip.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Hornitzky M. Literature review of chalkbrood. A report for the RIRDC. Publication No. 01/150. Australia: Kingston, ACT; 2001. [Google Scholar]
- Human H, Brodschneider R, Dietemann V, Dively G, Ellis J, Forsgren E, Fries I, Hatjina F, Hu F-L, Jaffé R, Köler A, Pirk CWW, Rose R, Strauss U, Tanner G, Tarpy DR, Van Der Steen JJM, Vejsnæs F, Williams GR, Zheng H-Q. Miscellaneous standard methods for Apis mellifera research. In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 4. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.4.10. [Google Scholar]
- Humber RA. Evolution of entomopathogenicity in fungi. Journal of Invertebrate Pathology. 2008;98:262–266. doi: 10.1016/j.jip.2008.02.017. http://dx.doi.org/10.1016/j.jip.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Invernizzi C, Penagaricano F, Tomasco IH. Intracolonial genetic variability in honey bee larval resistance to the chalkbrood and American foulbrood parasites. Insectes Sociaux. 2009;56:233–240. http://dx.doi.org/10.1007/s00040-009-0016-2. [Google Scholar]
- James RR, Skinner JS. PCR diagnostic methods for Ascosphaera infections in bees. Journal of Invertebrate Pathology. 2005;90:98–103. doi: 10.1016/j.jip.2005.08.004. http://dx.doi.org/10.1016/j.jip.2005.08.004. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch C, Matheny PB, Hofstetter V, Cox CJ, Celio G, Geuidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amroft A, Stajich JE, Hosaka K, Sung G-H, Johnson D, O’rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schüβler A, Longcore JE, O’donnell K, Mozley-Standridge S, Orter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman A, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lücking R, Büdel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, Mclaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. http://dx.doi.org/10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Jensen AB, James RR, Eilenberg J. Long-term storage of Ascosphaera aggregata and A. apis, pathogens of the leafcutting bee (Megachile rotundata) and the honey bee (Apis mellifera) Journal of Invertebrate Pathology. 2009a;101:157–160. doi: 10.1016/j.jip.2009.03.004. http://dx.doi.org/10.1016/j.jip.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Jensen AB, Pedersen BV, Eilenberg J. Differential susceptibility across honey bee colonies in larval to chalkbrood resistance. Apidologie. 2009b;40:524–534. http://dx.doi.org/10.1051/apido/2009029. [Google Scholar]
- Jensen AB, Welker DL, Kryger P, James RR. Polymorphic DNA sequences of the fungal honey bee pathogen Ascosphaera apis . FEMS Microbiology Letters. 2012;330:17–22. doi: 10.1111/j.1574-6968.2012.02515.x. http://dx.doi.org/10.1111/j.1574-6968.2012.02515.x. [DOI] [PubMed] [Google Scholar]
- Lunder R. Undersøkelse av kalkyngel i 1971 [Investigation on chalkbrood in 1971] Birøkteren. 1972;88:55–60. [Google Scholar]
- Maassen A. Weitere Mitteilungen uber der seuchenhaften Brutkrankheiten der Bienen [Further communication on the epidemic brood disease of bees] Mitteilungen aus der Kaiserlichen Biologischen Anstalt fur Land- und Forstwirtscshaft. 1913;14:48–58. [Google Scholar]
- Masterman R, Smith BH, Spivak M. Brood odour discrimination abilities in hygienic honey bees (Apis mellifera L) using proboscis extension reflex conditioning. Journal of Insect Behaviour. 2000;13(1):87–101. http://dx.doi.org/10.1023/A:1007767626594. [Google Scholar]
- Masterman R, Ross R, Mesce K, Spivak M. Olfactory and behavioural response thresholds to odours of diseased brood differ between hygienic and non-hygienic honey bees (Apis mellifera L.) Journal of Comparable Physiology A. 2001;187:441–452. doi: 10.1007/s003590100216. http://dx.doi.org/10.1007/s003590100216. [DOI] [PubMed] [Google Scholar]
- Maurizio A. Uber die Kaltbrut (Pericystis-Mykose) der Bienen. Archiv Bienenkunde. 1934;15:165–193. [Google Scholar]
- Moffett JO, Wilson WT. Feeding commercially purchased pollen containing mummies caused chalkbrood. American Bee Journal. 1978;118:412–414. [Google Scholar]
- Momot JP, Rothenbuhler WC. Behaviour genetics of nest cleaning in honey bees. VI. Interactions of age and genotype of bees, and nectar flow. Journal of Apicultural Research. 1971;10:11–21. [Google Scholar]
- Murray KD, Aronstein KA, Jones WA. A molecular diagnostic method for selected Ascosphaera species using PCR amplification of internal transcribed spacer regions of rDNA. Journal of Apicultural Research. 2005;44:61–64. [Google Scholar]
- Nilsson H, Kristiansson E, Ryberg M, Hallenberg N, Larsson KH. Intraspecific ITS variability in the kingdom Fungi as expressed in the International Sequence Databases and its implications for molecular species identification. Evolutionary Bioinformatics Online. 2008;4:193–201. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacio MA, Figini E, Rodriguez E, Ruffinengo S, Bedascarrasbure E, Delhoyo M. Changes in a population of Apis mellifera selected for its hygienic behaviour. Apidologie. 2000;31:471–478. http://dx.doi.org/10.1051/apido:2000139. [Google Scholar]
- Palacio MA, PEñA N, Clemente G, Ruffinengo S, Escande A. Viability and pathogenicity of Ascosphaera apis preserved in integral rice culture. Journal of Agricultural Research. 2007;5(4):481–486. [Google Scholar]
- Palacio MA, Rodriguez E, Goncalves L, Bedascarrasbure E, Spivak M. Hygienic behaviours of honey bees in response to brood experimentally pin-killed or infected with Ascosphaera apis . Apidologie. 2010;41:602–612. http://dx.doi.org/10.1051/apido/2010022. [Google Scholar]
- Puerta F, Flores JM, TaríN R, Bustos M, Padilla F, Hermoso De Mendoza M. Actividad antifúngica de productos seleccionados contra Ascosphaera Apis. Estudios in vitro . Revista Iberoamericana de Micología. 1990;7:103–106. [Google Scholar]
- Qin X, Evans JD, Aronstein KA, Murray KD, Weinstock GM. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis . Insect Molecular Biology. 2006;15(5):715–718. doi: 10.1111/j.1365-2583.2006.00694.x. http://dx.doi.org/10.1111/j.1365-2583.2006.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Evans JD. Microsatellite loci for the fungus Ascosphaera apis: cause of honey bee chalkbrood disease. Molecular Ecology Resources. 2009;9:855–858. doi: 10.1111/j.1755-0998.2008.02455.x. [DOI] [PubMed] [Google Scholar]
- Reynaldi FJ, Lopez AC, Albo GN, Alippi AM. Differentiation of Ascosphaera apis isolates by rep-PCR fingerprinting and determination of chalkbrood incidence in Argentinean honey samples. Journal of Apicultural Research. 2003;42:68–76. [Google Scholar]
- Rothenbuhler WC. Behaviour genetics of nest cleaning in honey bees. IV. Responses of F1 and backcross generations to disease-killed brood. American Zoologist. 1964;4(2):111–123. doi: 10.1093/icb/4.2.111. http://dx.doi.org/10.1016/0003-3472(64)90082-X. [DOI] [PubMed] [Google Scholar]
- Ruffinengo S, Peña NI, Clemente G, Palacio MA, Escande R. Suitability of culture media for the production of ascospores and maintenance of Ascosphaera apis . Journal of Apicultural Research. 2000;39(3/4):143–148. [Google Scholar]
- Scheiner R, Abramson CI, Brodschneider R, Crailsheim K, Farina W, Fuchs S, Grünewald B, Hahshold S, Karrer M, Koeniger G, Koeniger N, Menzel R, Mujagic S, Radspieler G, Schmickll T, Schneider C, Siegel AJ, Szopek M, Thenius R. Standard methods for behavioural studies of Apis mellifera . In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 4. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.4.04. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorfc S, Robert V, Spouge JL, Levesqueb CA, Chen W (FUNGAL BARCODING CONSORTIUM) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. www.pnas.org/cgi/doi/10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully LR, Bidochka MJ. A cysteine/methionine auxotroph of the opportunistic fungus Aspergillus flavus is associated with host-range restriction: a model for emerging diseases. Microbiology. 2006;152:223–232. doi: 10.1099/mic.0.28452-0. http://dx.doi.org/10.1099/mic.0.28452-0. [DOI] [PubMed] [Google Scholar]
- Seal DWA. Chalk brood disease of bees. New Zealand Journal of Agriculture. 1957;6:562. [Google Scholar]
- Skou JP. Ascosphaerales. Friesia. 1972;10(1):1–24. [Google Scholar]
- Skou JP. More details in support of the class Ascosphaeromycetes. Mycotaxon. 1988;31(1):191–198. [Google Scholar]
- Spiltoir CF. Life cycle of Ascosphaera apis (Pericystis apis) American Journal of Botany. 1955;42(6):501–508. http://dx.doi.org/10.2307/2438686. [Google Scholar]
- Spivak M, Gilliam M. Facultative expression of hygienic behaviour of honey bees in relation to disease resistance. Journal of Apicultural Research. 1993;32:147–157. [Google Scholar]
- Spivak M, Reuter GS. Resistance to American foulbrood disease by honey bee colonies, Apis mellifera, bred for hygienic behaviour. Apidologie. 2001;32:555–565. http://dx.doi.org/10.1051/apido:2001103. [Google Scholar]
- Spivak M, Masterman R, Ross R, Mesce KA. Hygienic behaviour in the honey bee (Apis mellifera L.) and the modulatory role of octopamine. Journal of Neurobiology. 2003;55:341–354. doi: 10.1002/neu.10219. http://dx.doi.org/10.1002/neu.10219. [DOI] [PubMed] [Google Scholar]
- Swanson J, Torto B, Kells S, Mesce K, Tumlinson J, Spivak M. Odorants that induce hygienic behaviour in honey bees: Identification of volatile compounds in chalkbrood-infected honey bee larvae. Journal of Chemical Ecology. 2009;35:1108–1116. doi: 10.1007/s10886-009-9683-8. http://dx.doi.org/10.1007/s10886-009-9683-8. [DOI] [PubMed] [Google Scholar]
- Taber S. Breeding bees for resistance to chalkbrood disease. American Bee Journal. 1986;126:823–825. [Google Scholar]
- Thompson VC. Behaviour genetics of nest cleaning in honey bees. III. Effect of age of bees of a resistant line on their response to disease-killed brood. Journal of Apicultural Research. 1964;3:25–30. [Google Scholar]
- Torto B, Carroll MJ, Duehl A, Fombong AT, Nazzi F, Gozansky KT, Soroker V, Teal PEA. Standard methods for chemical ecology research in Apis mellifera . In: Dietemann V, Ellis JD, Neumann P, editors. The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research Journal of Apicultural Research. 4. Vol. 52. 2013. http://dx.doi.org/10.3896/IBRA.1.52.4.06. [Google Scholar]
- Toumanoff C. Les Maladies des Abeilles [The diseases of bees] Revue Française d’Apicculture numéro spécial. 1951;68:1–325. [Google Scholar]
- Udagawa S, Horie Y. A new species of Emericella . Mycotaxon. 1976;4:535–539. [Google Scholar]
- Vojvodic S, Boomsma JJ, Eilenberg J, Jensen AB. Virulence of mixed fungal infections in honey bee brood. Frontiers in Zoology. 2012;9:5. doi: 10.1186/1742-9994-9-5. http://dx.doi.org/10.1186/1742-9994-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojvodic S, Jensen AB, James RR, Boomsma JJ, Eilenberg J. Temperature dependent virulence of obligate and facultative fungal pathogens of honey bee brood. Veterinary Microbiology. 2011a;149:200–205. doi: 10.1016/j.vetmic.2010.10.001. http://dx.doi.org/10.1016j.vetmic.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Vojvodic S, Jensen AB, Markussen B, Eilenberg J, Boomsma JJ. Genetic variation in virulence among chalkbrood strains infecting honey bees. PLoS ONE. 2011b;6(9):e25035. doi: 10.1371/journal.pone.0025035. http://dx.doi.org/10.1371/journal.pone.0025035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. A guide to methods and applications. Sand Diego, USA: Academic Press, Inc.; 1990. pp. 315–322. [Google Scholar]
- Wilson-Rich N, Spivak M, Fefferman NH, Starks PT. Genetic, individual, and group facilitation of disease resistance in insect societies. Annual Review of Entomology. 2009;54:405–423. doi: 10.1146/annurev.ento.53.103106.093301. http://dx.doi.org/10.1146/annurev.ento.53.103106.093301. [DOI] [PubMed] [Google Scholar]