Abstract
The anaphase-promoting complex (APC/C) is a large ubiquitin-protein ligase which controls progression through anaphase by triggering the degradation of cell cycle regulators such as securin and B-type cyclins. The APC/C is an unusually complex ligase containing at least 10 different, evolutionarily conserved components. In contrast to APC/C's role in cell cycle regulation little is known about the functions of individual subunits and how they might interact with each other. Here, we have analyzed Swm1/Apc13, a small subunit recently identified in the budding yeast complex. Database searches revealed proteins related to Swm1/Apc13 in various organisms including humans. Both the human and the fission yeast homologues are associated with APC/C subunits, and they complement the phenotype of an SWM1 deletion mutant of budding yeast. Swm1/Apc13 promotes the stable association with the APC/C of the essential subunits Cdc16 and Cdc27. Accordingly, Swm1/Apc13 is required for ubiquitin ligase activity in vitro and for the timely execution of APC/C-dependent cell cycle events in vivo.
Progression through mitosis depends on a multisubunit ubiquitin ligase called the anaphase-promoting complex or cyclosome (APC/C) (17, 19, 40). The APC/C catalyzes the transfer of ubiquitin molecules to cell cycle regulators and thereby marks them for proteolysis by the 26S proteasome (15, 32, 52). At the metaphase-to-anaphase transition, the APC/C activates the cysteine-protease separase by ubiquitinating its inhibitor Pds1/securin. The separase then destroys the cohesion between sister sequences of replicated chromosomes and enables the mitotic spindle to pull sister chromatids to opposite spindle poles (28). During anaphase, the APC/C ubiquitinates mitotic cyclins and thereby inactivates cyclin-dependent kinase 1 (Cdk1, called Cdc28 in budding yeast), which is required for exit from mitosis and cytokinesis (17, 19, 40, 51). During G1, APC/C activity prevents precocious accumulation of mitotic cyclins, which helps to create an extended period of low Cdk1 activity (16). Other APC/C substrates include mitotic kinases, proteins controlling spindle behavior, and regulators of DNA replication.
The temporal control and the substrate specificity of APC/C activity are mediated by the WD repeat proteins Cdc20 and Cdh1, which bind to the complex at different times during the cell cycle. Cdc20 and Cdh1 can also bind to substrates, suggesting that they activate ubiquitination by recruiting substrates to the catalytic center located in the complex (44). Binding of Cdc20 to the APC/C in mitosis triggers the degradation of Pds1 and initiates the destruction of Cdk1. The spindle assembly checkpoint blocks activation of APC/C-Cdc20 as long as sister kinetochores have not yet attached to microtubules emanating from opposite spindle poles (27). As cells exit from mitosis, Cdc20 disappears due to APC/C-dependent proteolysis and is replaced by Cdh1, which maintains APC/C activity during G1. In budding yeast APC/C-Cdc20 is required for activation of the phosphatase Cdc14, which dephosphorylates Cdh1 and thereby triggers binding of the activator to the APC/C as cells exit from mitosis. Dephosphorylation by Cdc14 also causes accumulation of the Cdk1-cyclin B inhibitor Sic1, whose phosphorylated form is subject to ubiquitin-dependent proteolysis. Together, Cdh1 and Sic1 suppress Cdk1 activity dependent on B-type cyclins until cells prepare to enter S phase (52).
While we are beginning to understand APC/C's role in cell cycle regulation, less is known about the functions of individual subunits and their interactions within the complex. This is in part due to the unusual size and complexity of this ligase (11). The budding yeast APC/C contains 11 subunits, most of which have homologues in the vertebrate particle (13, 50, 54). Mutations in yeast APC/C genes all reduce ligase activity in vitro and cause similar phenotypes in vivo, providing little clue to the specific functions of individual components (30, 48).
Insight into this problem came from the finding that the APC/C belongs to a large class of ligases characterized by a RING finger domain, which is thought to recruit ubiquitin-conjugating enzymes (18). APC/C's RING finger is located within the small subunit Apc11, which by itself has the properties of a minimal ubiquitin ligase (13, 22). Apc11 is anchored to the complex through Apc2, a subunit distantly related to cullins (50, 54). These proteins are subunits of SCF-type ligases, in which they link an Apc11-related RING finger protein to components concerned with substrate recruitment (9). A role in the catalytic mechanism was also proposed for the Doc1/Apc10 subunit, which stabilizes the interaction between APC/C and its substrates (6, 31). At least some APC/C subunits are expected to have structural functions and to provide binding sites for regulatory proteins. These functions may involve protein-protein interaction motifs such as the tetratricopeptide repeats (TPR) in the related subunits Cdc16, Cdc23, and Cdc27 (21).
Recently, two additional proteins, called Swm1/Apc13 and Mnd2, have been identified in the budding yeast APC/C, but their specific functions have not been reported (14, 31, 49). Here, we have analyzed Swm1/Apc13 and demonstrate that it is an evolutionarily conserved APC/C subunit. In budding yeast, Swm1/Apc13 mediates the association with high affinity of the TPR subunits Cdc16 and Cdc27 with the complex. Accordingly, Swm1/Apc13 is required for the proper timing of APC/C-dependent cell cycle events at elevated temperature.
MATERIALS AND METHODS
Strains and growth conditions.
Saccharomyces cerevisiae strains for experiments with vegetative cells were derivatives of W303 (ho ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1) and are listed in Table 1. Strains were grown, usually at 30°C, in yeast extract-peptone (YP) medium containing 2% glucose (YPD) or 2% raffinose (YPRaf). To induce the GAL promoter, 2% galactose was added to YPRaf medium (YPRafGal). Small G1 cells were isolated by centrifugal elutriation from strains grown in YPRaf and released into YPD. Meiosis was induced at 30°C in strains of the SK1 background (ho::LYS2 lys2 trp1::hisG leu2::hisG his3::hisG ura3) (Table 1) as described previously (4). Schizosaccharomyces pombe strains were derivatives of WSP20 (h− ura4-D18 leu1-32 ade6-M210) and were grown in YES medium at 30°C (24).
TABLE 1.
S. cerevisiae strains
| Strain | Relevant genotype |
|---|---|
| Z47 | MATapep4::URA3 bar1::hisG |
| Z652 | MATα |
| Z693 | MATaCLB2-myc12-URA3 |
| Z1104 | MATaura3::GAL1p-CLB2-ha3-URA3 pep4::LEU2 bar1::hisG |
| Z1780 | MATamyc18-CDH1-LEU2 pep4::URA3 bar1::hisG |
| Z1850 | MATα CDC16-TAP-KlTRP1 pep4::URA3 |
| Z1853 | MATα APC2-TAP-KlTRP1 pep4::URA3 |
| Z2075 | MATα swm1Δ::KanMX4 |
| Z2167 | MATα SWM1-TAP-KlTRP1 pep4::URA3 |
| Z2218 | MATaswm1Δ::KanMX4 ura3::GAL1p-CLB2-ha3-URA3 pep4::LEU2 bar1::hisG |
| Z2304 | MATα swm1Δ::KanMX4 CDC16-TAP-KlTRP1 pep4::URA3 |
| Z2314 | SK1 MATa/MATα |
| Z2328 | MATaswm1Δ::KanMX4 leu2::LEU2-GALLp-apc13+ |
| Z2351 | MATα APC1-myc18-HIS3 SWM1-ha3-KlTRP1 pep4::URA3 |
| Z2468 | MATaAPC11-ha3-TRP1 myc18-CDH1-LEU2 pep4::URA3 bar1::hisG |
| Z2591 | MATaswm1Δ::KanMX4 CLB2-myc12-URA3 |
| Z2620 | MATaswm1Δ::KanMX4 leu2::LEU2-GAL1p-hAPC13 |
| Z2622 | MATα swm1Δ::KanMX4 leu2::LEU2-GAL1p-hAPC13 |
| Z2625 | MATα swm1Δ::KanMX4 leu2::LEU2-GAL1p |
| Z2710 | MATacdc16-123 leu2::LEU2-GALLp-apc13+ |
| Z2711 | MATacdc16-123 leu2::LEU2-GAL1p-hAPC13 |
| Z2712 | MATaswm1Δ::KanMX4 cdc16-123 leu2::LEU2-GALLp-apc13+ |
| Z2714 | MATaswm1Δ::KanMX4 cdc16-123 leu2::LEU2-GAL1p-hAPC13 |
| Z2720 | MATaswm1Δ::KanMX4 APC2-TAP-KlTRP1 pep4::URA3 |
| Z2803 | MATα swm1Δ::KanMX4 cdc15-2 PDS1-myc18-LEU2 ura3::3xURA3-tetO112 leu2::LEU2-tetR-GFP |
| Z2804 | MATα cdc15-2 PDS1-myc18-LEU2 ura3::3xURA3-tetO112 leu2::LEU2-tetR-GFP |
| Z2827 | SK1 MATa/MATα PDS1-myc18-KlTRP1/PDS1-myc18-KlTRP1 |
| Z2894 | MATaCDC16-GFP-URA3 ADE2 |
| Z2922 | MATα swm1Δ::KanMX4 mad2Δ::KlTRP1 cdc15-2 PDS1-myc18-LEU2 ura3::3xURA3-tetO112 leu2::LEU2-tetR-GFP |
| Z2928 | MATα swm1Δ::KanMX4 ura3::URA3-SWM1-GFP ADE2 |
| Z2931 | MATaHa6-SWM1-LEU2 myc18-CDH1-LEU2 pep4::URA3 bar1::hisG |
| Z3089 | MATα cdc26Δ::KlURA3 APC2-TAP-KlTRP1 pep4::LEU2 |
| Z3182 | MATacdc28-4 ura3::8xGAL1p-CLB2-ha3-URA3 bar1::hisG |
| Z3186 | MATaswm1Δ::KanMX4 cdc28-4 ura3::GAL1p-CLB2-ha3-URA3 bar1::hisG |
| Z3190 | MATα APC4-TAP-KlTRP1 pep4::URA3 |
| Z3282 | MATα swm1Δ::KanMX4 APC4-TAP-KlTRP1 pep4::URA3 |
| Z3306 | SK1 MATa/MATα swm1Δ::KanMX4/swm1Δ::KanMX4 PDS1-myc18-KlTRP1/PDS1-myc18-KlTRP1 |
| Z3543 | MATα swm1Δ::KanMX4 ura3::URA3-ACE2 |
| Z3546 | MATα swm1Δ::KanMX4 ura3::URA3-ACE2-AAA |
| Z3616 | MATaAPC4-TAP-KlTRP1 |
| Z3885 | MATα swm1Δ::KanMX4 leu2::GPDp-hAPC13-LEU2 |
| Z3886 | MATα swm1Δ::KanMX4 leu2::GPDp-apc13+-LEU2 |
| Z4001 | SK1 MATa/MATα swm1Δ::KanMX4/swm1Δ::KanMX4 |
| Z4334 | MATα swm1Δ::KanMX4 leu2::SIC1-LEU2 |
| Z4359 | MATacdc23-54 APC4-TAP-KlTRP1 |
| Z4361 | MATα cdc23-51 APC4-TAP-KITRP1 |
| Z4363 | MATacdc23-39 APC4-TAP-KlTRP1 |
| Z4364 | MATα cdc27-1 SWM1-TAP-KlTRP1 pep4::URA3 |
| Z4367 | MATaapc9Δ::HIS3MX6 SWM1-TAP-KlTRP1 pep4::URA3 |
| Z4378 | MATacdc23-54 SWM1-TAP-KlTRP1 pep4::URA3 |
| Z4379 | MATacdc23-51 SWM1-TAP-KlTRP1 pep4::URA3 |
| Z4380 | MATacdc23-39 SWM1-TAP-KlTRP1 pep4::URA3 |
| Z4420 | SK1 MATa/MATα swm1Δ::KanMX4/swm1Δ::KanMX4 leu2::SIC1-LEU2/leu2::SIC1-LEU2 |
Construction of S. cerevisiae strains.
PCR-based methods were used for gene deletions (46) and C-terminal epitope tagging with HA3 (three hemagglutinin epitopes) (20) and a tandem affinity purification (TAP) tag (33). Tagged alleles of CLB2, PDS1, CDH1, and APC/C subunits have been described previously (38, 53, 55). Plasmids of the YIplac series (12) were used as integrative vectors. To create Ha6-SWM1 strains, SWM1 sequences (−216 to + 159, ATG = +1) containing six HA epitopes after the ATG were cloned into YIplac128, cut with BspEI, and integrated at the SWM1 locus. A HindIII-Asp718 fragment carrying a fusion of green fluorescent protein (GFP) to SWM1 under the control of the SWM1 promoter (43) was cloned into YIplac211 and integrated at the ura3 locus of swm1Δ cells. To construct CDC16-GFP strains, CDC16 sequences (+1970 to + 3037) containing GFP in front of the stop codon were cloned into YIplac211, cut with Eco47III, and integrated at the CDC16 locus. All strains containing tagged APC/C subunits grew indistinguishably from the wild type at different temperatures. APC13 homologues from S. pombe and human cells were amplified as BamHI-HindIII fragments from genomic DNA and IMAGE clone 4745731 (BG674084), respectively, and cloned into YIplac128 behind SacI-BamHI fragments carrying the GPD promoter, the GALL promoter, or the GAL1 promoter (25, 26). The plasmids were cut with EcoRV and integrated at the leu2 locus of swm1Δ cells. swm1Δ GALLp-apc13+ and swm1Δ GAL1p-hAPC13 strains were crossed to different apc mutants followed by tetrad dissection on YPRafGal plates. ACE2 and ACE2-AAA were isolated as XbaI-HindIII fragments from pMM2 and pMM2-TRIP (29), cloned into YIplac211, and integrated at the ura3 locus. To increase the gene dosage of SIC1, the gene (−425 to + 1895) was cloned into YIplac128 and integrated at the leu2 locus.
Construction of S. pombe strains.
Strain WSP20 was transformed with TAP-KanMX6 cassettes amplified from pDS1 (a gift from F. Stewart) to generate set1+-TAP (36), cut9+-TAP, and apc13+-TAP strains, which grew like WSP20. Diploid apc13Δ::KanMX4/apc13+ cells produced two viable, geneticin-sensitive spores and two spores that germinated but arrested as elongated cells after two or three divisions, demonstrating that apc13+ is essential. Haploid apc13Δ::KanMX4 cells were kept alive by an apc13+ plasmid but not by the vector.
Analysis of proteins.
Yeast cells were broken with glass beads in buffer B70 (54). To compare protein levels, lysates from 108 cells in 0.2 ml of buffer were boiled in sodium dodecyl sulfate (SDS) sample buffer and equal amounts of protein were analyzed by immunoblotting as described previously (51). For immunoprecipitations, lysates from 109 cells in 0.4 ml of buffer were centrifuged for 20 min at 18,000 × g and precleared with 0.1 ml of protein A-Sepharose. HA-tagged proteins were isolated from extracts (0.35 ml, 4 mg) with 5 μg of 12CA5 antibody as described previously (5). Extracts were separated in glycerol density gradients prepared in buffer B70 as described previously (55). Meiotic cells were broken in 10% trichloroacetic acid, pelleted, and resuspended in SDS sample buffer. HeLa cells grown under standard conditions were scraped from the plates in Tris-buffered saline, centrifuged, and lysed with glass beads in buffer A (11). Cleared extracts (0.4 ml, 6 mg) were subjected to immunoprecipitations, and beads were washed four times with buffer A.
Antibodies.
Rabbit antibodies were raised against His6-Apc11, His6-Cdc23, His6-Doc1, His6-human Apc13 (hApc13), and GST-hApc13 expressed in bacteria. His6-Cdh1 was expressed in baculovirus-infected insect cells. Antibodies to Apc1 and Apc4 were raised against C-terminal peptides. Antibodies to Apc2, Cct1, and Cdc20 have been described (5). Antibodies to the following proteins were provided by the indicated researchers: Cdc16 and Cdc27 (P. Hieter), Cin8 (A. Hyman), Cim5 (C. Mann), Clb3 (D. Drechsel), Sic1 (M. Tyers), Tub2 (W. Seufert), and hApc2 (J.-M. Peters). Commercial antibodies were used for the following proteins: HA tag (12CA5), myc tag (9E10), hCct1 and hCct2 (CTA-184 and CTA-200, respectively; Stressgen), hCdc16 and hCdc27 (sc-5615 and sc-9972, respectively; Santa Cruz), and Clb5 (sc-6704; Santa Cruz).
APC/C purification and mass spectrometry.
Strains were grown in parallel in 12-liter cultures yielding 30 to 40 g of cells, which were lysed by bead beating in 200 ml of buffer E (20 mM HEPES-NaOH [pH 8.0], 350 mM NaCl, 0.5 mM NaF, 10 mM β-glycerophosphate, 0.05% Tween 20, 10% glycerol, 1 mM dithiothreitol, protease inhibitors [Complete; Roche]). Extracts were centrifuged for 60 min at 100,000 × g, and TAP-tagged proteins were isolated by the TAP method (33). Purified proteins were precipitated with methanol-chloroform, separated on SDS-6 to 22% polyacrylamide gels, and stained with GelCode blue stain reagent (Pierce). Proteins were analyzed by matrix-assisted laser desorption/ionization peptide mass mapping and by nanoelectrospray tandem mass spectrometric sequencing (37).
Ubiquitination assay.
Substrates (Pds1 and Clb2) and activators (Cdh1, Cdh1-IK, Cdc20, and Cdc20-IA) were produced by coupled transcription/translation in reticulocyte lysates (TNT T7; Promega) from plasmids provided by D. Barford (31). Reactions were performed at 22°C in 10 μl of buffer (40 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 0.6 mM dithiothreitol, 1 mM ATP, 2 μM N-acetyl-Leu-Leu-Norleu-aldehyde [LLnL; Sigma]) containing 3 μg of ubiquitin (Sigma), 0.2 μg of ubiquitin-aldehyde (Affiniti), 0.6 μg of yeast His6-Ubc4, 1 μl of lysate with 35S-labeled substrate, 1.5 μl of lysate with activator, and 20 ng of APC/C. E1 activity was provided by the reticulocyte lysate. Reactions were stopped with SDS sample buffer, and mixtures were separated on SDS-8% polyacrylamide gels, with detection by fluorography.
Other techniques.
Flow cytometric analysis of cellular DNA and indirect immunofluorescence were performed as described previously (38). These procedures involve treating cells with pepsin and zymolyase, which dissociate the cell clusters produced by swm1Δ strains at 37°C. Sister chromatids were observed with the ura3::tetO/tetR-GFP system (23). The mean volume of 10,000 nonfixed cells was measured with a CASY1 Cell Counter Analyser (Schaerfe System, Reutlingen, Germany). Pictures of living CDC16-GFP and GFP-SWM1 cells were taken with a CCD (charge-coupled device) camera on an Olympus IX70 Delta Vision microscope. Database searches were performed at the National Center for Biotechnology Information with tblastn and blastp (1). Multiple sequence alignments were generated with Clustal X (41) and edited manually.
RESULTS
Identification of Swm1/Apc13 as an APC/C subunit.
Previous analysis of the budding yeast APC/C detected a protein of 19 kDa tentatively called Apc13 (54). To identify this protein, the APC/C was purified from cells in which the Cdc16 subunit was replaced by a version carrying a TAP tag (35). Mass spectrometric analysis of gel-separated proteins identified the 11 known APC/C subunits and two additional proteins (Fig. 1A). A band at 55 kDa contained the Mnd2 protein, which was discovered in a screen for genes required for meiosis but not for vegetative growth (34). Apc13 was identified as the product of the SWM1 gene, which is required for spore wall formation (43). Accordingly, purification of TAP-tagged versions of Swm1/Apc13 and Mnd2 yielded the same set of APC/C subunits as purification of Cdc16-TAP (Fig. 1A and data not shown). These findings confirm previous conclusions that Swm1/Apc13 and Mnd2 are stably associated with the APC/C (14, 31, 49). Here, we focus on the analysis of Swm1/Apc13.
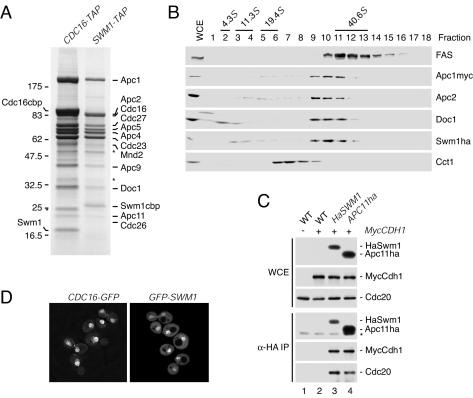
FIG. 1.
Swm1/Apc13 is a subunit of the APC/C. (A) Subunit composition of the S. cerevisiae APC/C. Proteins isolated by TAP from CDC16-TAP (Z1850) and SWM1-TAP (Z2167) cells were separated by SDS-PAGE, stained with Coomassie blue, and identified by mass spectrometry. Cdc26 is stained poorly with Coomassie. Asterisks mark nonspecific proteins, which were also purified with Stu2-TAP (results not shown). cbp, calmodulin-binding peptide remaining on the APC/C subunit. (B) Separation of a whole-cell extract (WCE) from APC1-myc18 SWM1-ha3 cells (Z2351) in a 10 to 35% glycerol density gradient. Fractions were analyzed by immunoblotting. Fatty acid synthetase (FAS) is a 40.6S complex, and Cct1 is a subunit of the 20S CCT chaperonin. Bovine serum albumin (4.3S), catalase (11.3S), and thyroglobulin (19.4S) were separated in parallel. (C) Immunoblot analysis of anti-HA immunoprecipitations from wild-type (−) and myc18-CDH1 (+) cells containing HA6-SWM1 or APC11-ha3. The asterisk marks antibody bands. Strains: lane 1, Z47; lane 2, Z1780; lane 3, Z2931; lane 4, Z2468. (D) Observation of living CDC16-GFP (Z2894) and GFP-SWM1 (Z2928) cells by fluorescence microscopy.
To determine what fraction of Swm1/Apc13 is associated with the APC/C, extracts from APC1-myc18 SWM1-ha3 cells were separated in glycerol density gradients. Most of Swm1/Apc13 cosedimented with other APC/C subunits as a 36S complex (Fig. 1B). Cdc20 and Cdh1 were not evident by Coomassie blue staining in our APC/C preparations. However, both activators were detected on immunoblots after immunoprecipitation of HA-tagged Swm1/Apc13 (Fig. 1C), demonstrating that Swm1/Apc13 is a component of the APC/C-Cdc20 and APC/C-Cdh1 holoenzymes. Swm1/Apc13 has been detected in the nuclei of cells undergoing meiosis I and II but not in mononucleated or vegetative cells (43). An integrative plasmid was used to express a GFP-Swm1/Apc13 fusion protein from the natural promoter as the sole source of Swm1/Apc13. The fusion protein accumulated in the nucleus during all stages of the mitotic cell cycle in vivo (Fig. 1D).
Homologues of Swm1/Apc13 in other organisms.
SWM1 was thought to encode a protein unique to budding yeast (14, 31, 43). However, BLAST searches with Swm1/Apc13 against the genome survey sequence database at the National Center for Biotechnology Information identified related sequences from the budding yeasts Zygosaccharomyces rouxii (AL394812), Candida glabrata (BZ299331), and Pichia farinosa (also called P. sorbitophila) (Fig. 2A and data not shown). Searches with the P. farinosa sequence uncovered related sequences from Candida albicans (AJ006641, starts at codon 20 of CaApc13) and S. pombe in the nonredundant database. The S. pombe sequence identified a protein from the fungus Glomus intraradices in the EST (expressed sequence tags) database. This sequence retrieved cDNAs from both yeasts and metazoan organisms, including plants, nematodes, insects, and vertebrates. The Swm1/Apc13-related sequences correspond to small proteins sharing a characteristic pattern of hydrophobic and negatively charged amino acids, which include highly conserved tryptophan and proline residues (Fig. 2A). These data suggest that Swm1/Apc13 is an evolutionarily conserved protein.
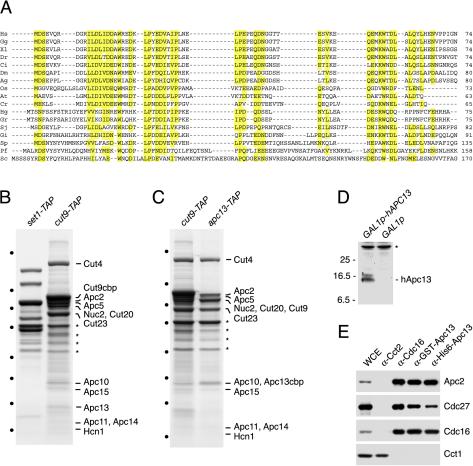
FIG. 2.
Swm1/Apc13 homologues in different organisms. (A) Multiple sequence alignment of Swm1/Apc13 homologues. All sequences start at the initiator methionine, and numbers give the total length of a protein. Hs, Homo sapiens (BI910334); Gg, Gallus gallus (AJ397343); Xl, Xenopus laevis (BF072239); Dr, Danio rerio (BM184675); Ci, Ciona intestinalis (BW016595); Dm, Drosophila melanogaster (AI062156); Ag, Anopheles gambiae (BM636962); Os, Oryza sativa (C25135); At, Arabidopsis thaliana (AY088589); Cr, Ceratopteris richardii (BQ087174); Hg, Heterodera glycines (BI749014); Gr, Globodera rostochiensis (BM354343); Sj, Schistosoma japonicum (BU715898); Gi, Glomus intraradices (BM027531); Sp, Schizosaccharomyces pombe (NP_595754); Pf, Pichia farinosa (AL415833); Sc, Saccharomyces cerevisiae Swm1. (B) Subunits of the fission yeast APC/C. Proteins purified from set1+-TAP (WSP21, control) and cut9+-TAP (WSP22) strains were gel separated, stained with Coomassie blue, and identified by mass spectrometry. Asterisks mark proteins identified in both purifications. Dots indicate molecular weight markers (see Fig. 1A). (C) Association of APC/C subunits with Apc13. Proteins were purified from cut9+-TAP (WSP22) and apc13+-TAP (WSP44) strains and analyzed as for panel B. (D) Detection of hApc13 expressed in budding yeast. Extracts were prepared from swm1Δ GAL1p-hAPC13 (Z2622) and swm1Δ GAL1p (Z2625) cells grown in YPRafGal medium for 120 min and immunoblotted with antibodies to GST-hApc13. (E) Immunoprecipitations from HeLa cell extracts with antibodies to GST-hApc13 (α-GST-hApc13), His6-hApc13, Cdc16, and the chaperonin component Cct2. Purified proteins were analyzed by immunoblotting with antibodies to APC/C subunits and Cct1, another chaperonin component.
To investigate whether the Swm1/Apc13-related proteins are APC/C subunits, we purified from fission yeast a TAP-tagged version of the Cdc16-homologue Cut9. The histone-methyltransferase complex Set1C was isolated as a control for specificity (36). Mass spectrometric analysis of the Cut9-TAP purification identified 10 proteins known or predicted to be APC/C subunits (48). In addition, three small proteins were identified as the products of the reading frames SPBC28E12.01c, SPAC27D7.05c, and SPBC83.04. These proteins were identified recently in purifications from lid1+-TAP cells and called Apc13, Apc14, and Apc15, respectively (49). Significantly, SPBC28E12.01c/Apc13 is identical to the Swm1/Apc13 homologue identified in our database searches (Fig. 2A). Accordingly, purification of a TAP-tagged version of Apc13 yielded the same set of APC/C subunits as Cut9-TAP (Fig. 2C), demonstrating that apc13+ encodes a subunit of the fission yeast APC/C. In accordance with previous results (49), we found that apc14+ and apc15+ are nonessential genes whereas apc13+ is essential (data not shown; see Materials and Methods).
To analyze the human Apc13 homologue, antibodies were raised against His6- and glutathione S-transferase (GST)-tagged versions expressed in bacteria. Both antibodies recognize a protein migrating at approximately 15 kDa after expression from the GAL1 promoter in S. cerevisiae (Fig. 2D). Human Apc13 is predicted to be a protein of 8.5 kDa; the low mobility may result from its acidic character (pI 4.0). The antibodies to human Apc13 immunoprecipitated different APC/C subunits but not the chaperonin component Cct1 from extracts of HeLa cells (Fig. 2E). These data show that the Apc13 homologue is associated with the APC/C in human cells.
Complementation of the SWM1 deletion with Swm1/Apc13 homologues.
The wild-type strain grew with a doubling time of 98 min at 37°C, whereas the swm1Δ strain grew more slowly, with a doubling time of 188 min (Fig. 3A). Microscopic examination of G1 cells growing on YPD plates at 37°C revealed that 30% of the mutant cells failed to produce colonies but arrested as large-budded cells in the first or second division. In contrast, more than 99% of wild-type cells produced colonies. Although SWM1 is formally called a nonessential gene, it is important for proliferation at 37°C because, on average, every third division fails in swm1Δ cells. Next, Swm1/Apc13 homologues under the control of the GPD promoter were integrated into swm1Δ cells. Expression of S. pombe apc13+ and human APC13 allowed swm1Δ cells to grow at 37°C with generation times close to that of wild-type cells (105 and 108 min, respectively) (Fig. 3A) and to form colonies with normal efficiency.
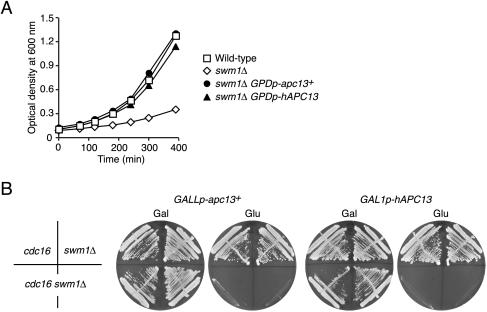
FIG. 3.
Complementation of the SWM1 deletion by Apc13 homologues. (A) Growth at 37°C of wild-type cells (squares, Z652), swm1Δ cells (diamonds, Z2075), and swm1Δ cells containing S. pombe apc13+ (filled circles, Z3886) or human APC13 (filled triangles, Z3885) under the control of the GPD promoter. (B) Rescue of the swm1Δ cdc16-123 double mutant by S. pombe apc13+ and human APC13. Strains were incubated for 2 days at 25°C on YPRafGal (Gal) and YPD (Glu) plates. S. pombe apc13+ was expressed from the GALL promoter in cdc16-123 (Z2710), swm1Δ (Z2328), and cdc16-123 swm1Δ (Z2712) cells. Human APC13 was expressed from the GAL1 promoter in cdc16-123 (Z2711), swm1Δ (Z2620), and cdc16-123 swm1Δ (Z2714) cells.
In order to test complementation more rigorously, we established conditions where SWM1 is absolutely essential for proliferation. The SWM1 deletion caused lethality at 25°C in combination with mutations in other APC/C genes (apc1-1, cdc16-123, cdc23-1, cdc27-1, doc1-22, apc9Δ, and cdc26Δ) and in CDC20 (cdc20-1 and cdc20-3). The double-mutant spores germinated but then arrested as large-budded cells after one to three divisions, whereas single-mutant spores formed colonies. This synthetic lethality was specific since mutations in other cell cycle genes (cdc28-4, cdc4-1, cdc53-1, cdc7-1, cdc15-2, cdc13-1, sic1Δ, mad2Δ, and cin8Δ) did not affect the growth of swm1Δ cells (data not shown). S. pombe apc13+ under the control of the weak GALL promoter or human APC13 fused to the GAL1 promoter was integrated into swm1Δ cells, and the resulting strains were crossed to different apc mutants. The GALLp-apc13+ and the GAL1p-hAPC13 plasmid allowed swm1Δ apc9Δ, swm1Δ cdc26Δ, and swm1Δ cdc16-123 double mutants to grow at 25°C on galactose- but not on glucose-containing medium (Fig. 3B). We conclude that the fission yeast and the human proteins can, at least partially, substitute for the function of Swm1/Apc13 in budding yeast.
Swm1/Apc13 promotes the structural integrity of the APC/C.
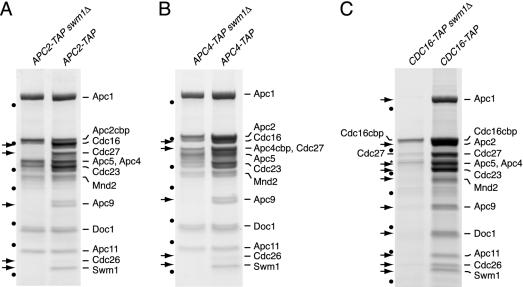
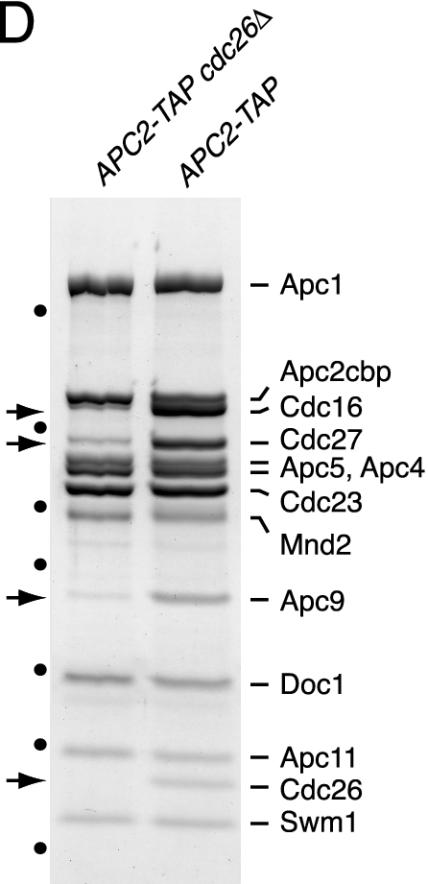
To investigate the function of S. cerevisiae Swm1/Apc13, the APC/C was purified in parallel from wild-type and swm1Δ cells grown at 25°C. Complexes isolated from APC2-TAP swm1Δ and APC4-TAP swm1Δ cells lacked Cdc16, Cdc27, Apc9, Cdc26, and Swm1/Apc13, whereas other subunits were present in normal stoichiometries (Fig. 4A and B). These data suggest that Swm1/Apc13 promotes the association with the APC/C of a group of subunits, including the essential components Cdc16 and Cdc27. Accordingly, purifications from CDC16-TAP swm1Δ cells yielded Cdc16 and trace amounts of Cdc27 and Apc9 but none of the other subunits (Fig. 4C). The consequences of deleting SWM1 resemble those resulting from a deletion of CDC26 (54). To analyze the relationship between these subunits, we purified the APC/C from APC2-TAP cdc26Δ cells grown at 25°C. The mutant APC/C contained reduced amounts of Cdc16, Cdc27, and Apc9 but normal levels of Swm1/Apc13 (Fig. 4D). This suggests that association of Cdc26 requires Swm1/Apc13 but not vice versa.
FIG. 4.
Subunit composition of the APC/C isolated from swm1Δ and cdc26Δ mutants. Proteins purified from strains grown at 25°C were gel separated, stained with Coomassie blue, and identified by mass spectrometry. Arrows mark subunits that were undetectable or present in reduced amounts. Dots indicate molecular weight markers (see Fig. 1A). The asterisk marks nonspecific proteins. The following strains were processed in parallel: (A) APC2-TAP swm1Δ (Z2720) and APC2-TAP (Z1853); (B) APC4-TAP swm1Δ (Z3282) and APC4-TAP (Z3190) (Apc4cbp migrated at the same position as Cdc27, which was absent from the mutant APC/C as revealed by mass spectrometry); (C) CDC16-TAP swm1Δ (Z2304) and CDC16-TAP (Z1850); (D) APC2-TAP cdc26Δ (Z3089) and APC2-TAP (Z1853).
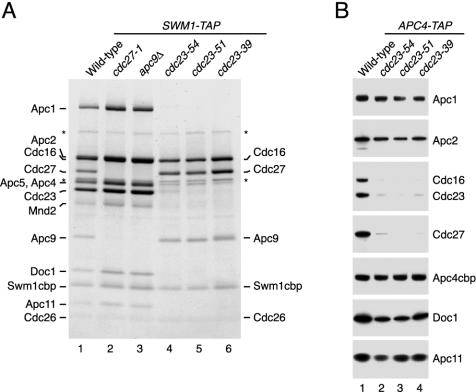
Next we sought to identify APC/C subunits located close to or even directly associated with Swm1/Apc13. TAP-tagged Swm1/Apc13 was purified from different apc mutants grown at 25°C and shifted to 37°C for 90 min (Fig. 5A). We reasoned that some of the temperature-sensitive (ts) mutations might result in fragmentation of the APC/C. Indeed, APC/C isolated from cdc27-1 mutant cells lacked both Cdc27 and Apc9. Interestingly, Swm1/Apc13 isolated from different cdc23-ts mutants was associated only with Cdc16, Cdc27, Apc9, and Cdc26, all of which were present in normal amounts (Fig. 5A). Accordingly, copurification of these subunits with Apc4-TAP was diminished specifically by cdc23-ts mutations (Fig. 5B). Our data suggest that Cdc16, Cdc27, Apc9, Swm1/Apc13, and Cdc26 form a stable subcomplex whose association with the APC/C requires Cdc23. In the absence of Swm1/Apc13, Cdc16 no longer associates efficiently with other subunits (Fig. 4C). This suggests that the subcomplex is less stable in swm1Δ cells, so that its components tend to dissociate from the APC/C.
FIG. 5.
Analysis of a subcomplex containing Swm1/Apc13. (A) SWM1-TAP strains containing the indicated mutations were grown at 25°C, shifted to 37°C for 90 min, and processed for TAP purification. Proteins were gel separated and stained with Coomassie blue. Proteins from SWM1-TAP cells are indicated on the left; those from SWM1-TAP cdc23-ts cells are marked on the right. Asterisks mark nonspecific proteins. Strains: lane 1, Z2167; lane 2, Z4364; lane 3, Z4367; lane 4, Z4378; lane 5, Z4379; lane 6, Z4380. (B) APC4-TAP strains containing cdc23-ts mutations or the wild-type allele were treated as in panel A. Purified proteins were detected by immunoblotting. Strains: lane 1, Z3616; lane 2, Z4359; lane 3, Z4361; lane 4, Z4363.
Swm1/Apc13 is required for ubiquitination.
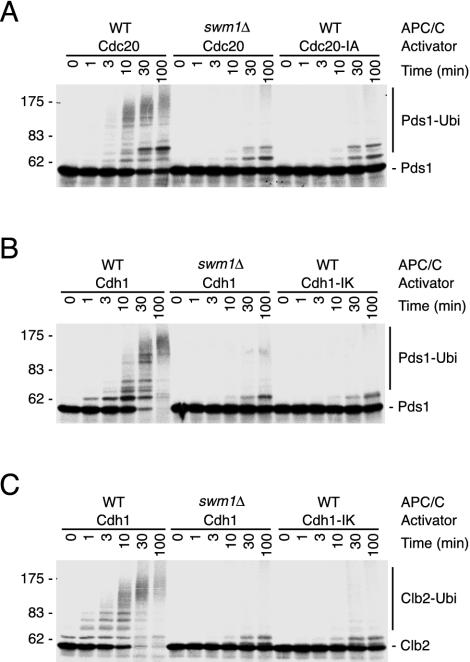
To measure ubiquitin ligase activity, complexes purified from APC4-TAP and APC4-TAP swm1Δ cells (Fig. 4B) were incubated with different activators and substrates generated by in vitro translation. Cdc20 activated wild-type APC/C to ubiquitinate Pds1 (Fig. 6A), whereas Cdh1 triggered ubiquitination of both Pds1 and Clb2 (Fig. 6B and C). Only background signals were generated in reaction mixtures lacking APC/C (not shown) or containing activators whose terminal, conserved arginine residue had been exchanged (Cdc20-IA and Cdh1-IK). Addition of Cdc20 or Cdh1 to APC/C from swm1Δ cells failed to result in significant ubiquitination of the substrates (Fig. 6). These data show that our purification conditions result in the isolation of active APC/C from wild-type cells. Complexes purified in parallel from swm1Δ cells contain structural defects that strongly reduce their ligase activity.
FIG. 6.
Swm1/Apc13 is required for APC/C-dependent ubiquitination. Equal amounts of APC/C purified from APC4-TAP (WT) and APC4-TAP swm1Δ (swm1Δ) cells (Fig. 4B) were incubated with in vitro-translated activator proteins and 35S-labeled substrates in the presence of Ubc4, ubiquitin, and ATP. Aliquots were withdrawn at the indicated times and analyzed by fluorography. Reaction mixtures contained 35S-Pds1 and Cdc20 or inactive Cdc20-IA (A), 35S-Pds1 and Cdh1 or inactive Cdh1-IK (B), and 35S-Clb2 and Cdh1 or inactive Cdh1-IK (C).
Defective cell cycle progression in swm1Δ mutant cells.
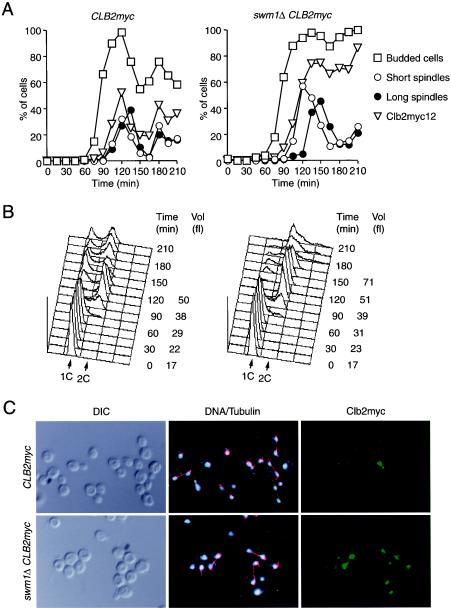
So far, the function of SWM1 in vivo has been analyzed only with respect to sporulation (43). Our results predict that SWM1 should also be important for the mitotic cell cycle. To analyze this, small G1 cells were isolated by centrifugal elutriation from wild-type and swm1Δ strains, both containing a myc-tagged version of the cyclin Clb2, and released into a synchronous cell cycle at 37°C. Bud formation, DNA replication, and spindle assembly occurred normally in the mutant cells (Fig. 7A and B). However, cells with elongated spindles appeared 30 min later and persisted longer in the swm1Δ culture, indicating a delay in both entry into and exit from anaphase (Fig. 7A), which is consistent with a defect in APC/C activity. Detection of Clb2-myc12 by indirect immunofluorescence revealed that the cyclin accumulated with similar kinetics in both strains (Fig. 7A). In wild-type cells Clb2-myc12 levels rapidly dropped below detectable levels during anaphase. In swm1Δ cells degradation of Clb2-myc12 was less efficient and the cyclin was still present in late anaphase, i.e., in cells containing a divided nucleus and a fully elongated spindle (Fig. 7C). Occasionally, it was even detectable in cells that had undergone cytokinesis. These data show that Swm1/Apc13 is required for efficient cyclin degradation during anaphase.
FIG. 7.
Cell cycle defects in swm1Δ mutant cells. Elutriated G1 cells from CLB2-myc12 (Z693) and swm1Δ CLB2-myc12 (Z2591) strains grown at 25°C were released at 37°C. (A) The percentages of budded cells (squares), of cells with a short spindle in an undivided nucleus (open circles), of cells with an elongated spindle and a divided nucleus (filled circles), and of cells with staining from Clb2-myc12 (triangles) were determined in samples taken every 15 min. (B) Cellular DNA content and mean cell volume. (C) Clb2-myc12 (green) and tubulin (red) were detected by indirect immunofluorescence, and DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI, blue) in samples taken 120 min (wild-type) and 135 min (swm1Δ) after release.
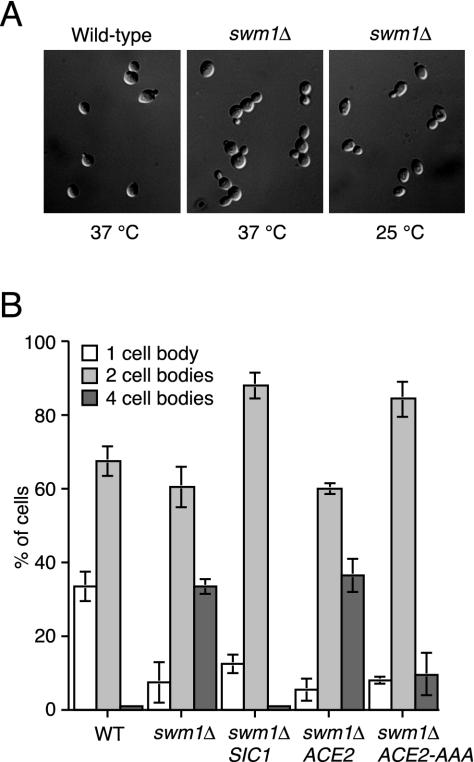
Cell separation defect of the swm1Δ mutant.
At 37°C most of the large-budded swm1Δ cells initiated a new round of budding before completing cell separation, thereby producing clusters of four cell bodies instead of unbudded cells (Fig. 8A). These clusters were resistant to sonication but were dissociated into intact, single-budded cells by a brief treatment with zymolyase. This suggests that swm1Δ cells underwent cytokinesis but failed to dissolve on time the cell wall material connecting mother and daughter cells. The cell separation defect was suppressed by increasing the gene dosage of the Cdk1-cyclin B inhibitor Sic1, suggesting that it resulted from inefficient inactivation of Cdk1 (Fig. 8B). Multiple copies of SIC1 also improved the growth of swm1Δ cells at 37°C (generation time, 108 min), but the fraction of budded cells was higher than in the wild-type culture. Cell separation requires expression of the chitinase Cts1, which is induced by translocation into the nucleus of the transcription factor Ace2 in late anaphase (10). Ace2's entry into the nucleus is inhibited by Cdk1 activity (29), raising the possibility that the phenotype of swm1Δ is caused by a deficiency in nuclear Ace2. To test this, we integrated plasmids into swm1Δ cells encoding Ace2 or the constitutively nuclear variant Ace2-AAA (29). Ace2-AAA, but not the wild-type protein, suppressed the cell separation defect of swm1Δ cells at 37°C (Fig. 8B). However, Ace2-AAA did not rescue the temperature-sensitive growth defect of the mutant cells.
FIG. 8.
Cell separation defect of swm1Δ mutants. (A) Wild-type and swm1Δ cells undergoing cytokinesis at 37°C. Mutant cells growing at 25°C are shown as a control. Pictures show fixed cells after sonication. (B) Suppression of the cell separation defect by additional copies of SIC1 and by a constitutively nuclear version of Ace2. Wild-type (Z652), swm1Δ (Z2075), and swm1Δ strains containing integrative plasmids with SIC1 (Z4334), ACE2 (Z3543), or ACE2-AAA (Z3546) were grown at 25°C and shifted to 37°C for 4 h. Cells were fixed, sonicated, and counted according to their morphology. Mean values from three independent experiments are shown. Bars represent standard deviations.
Swm1/Apc13 is required for timely sister chromatid separation.
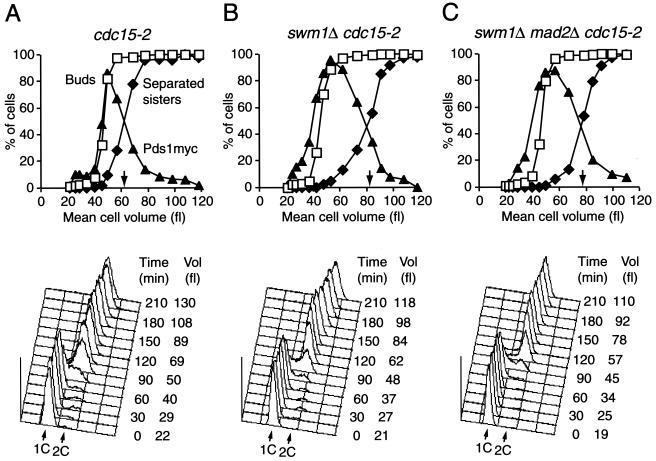
To test more rigorously whether Swm1/Apc13 was important for the timing of anaphase entry, we compared the kinetics of Pds1 degradation and sister chromatid separation after releasing small G1 cells of wild-type and swm1Δ strains at 37°C (Fig. 9). Sister chromatids were followed by observing fluorescent “dots” originating from GFP bound to the ura3 locus on chromosome V (23). To avoid difficulties associated with the cell separation defect of swm1Δ cells, we used strains containing a cdc15-2 mutation, which does not affect sister separation but blocks exit from mitosis at 37°C (7). cdc15-2 (i.e., wild-type) and cdc15-2 swm1Δ cells formed buds with similar kinetics, whereas Pds1-myc18 consistently accumulated earlier in the mutant cells, as would be expected if Pds1 degradation was defective during G1. Degradation of Pds1-myc18 and separation of sister chromatids were equally delayed in cdc15-2 swm1Δ cells (Fig. 9A and B). For example, sister chromatids were separated in 50% of the cells at a mean cell volume of 60 fl in the cdc15-2 culture but only at 80 fl in the cdc15-2 swm1Δ culture. This difference in cell volume corresponds to a delay in anaphase onset of 30 min. This delay was not caused by the mitotic checkpoint mechanism because deletion of the MAD2 gene in cdc15-2 swm1Δ cells failed to restore wild-type kinetics of Pds1-myc18 degradation and sister chromatid separation (Fig. 9C). We conclude that Swm1/Apc13 is required for the timely execution of cell cycle events dependent on APC/C-Cdc20.
FIG. 9.
Swm1/Apc13 is required for timely Pds1 degradation and sister chromatid separation. Elutriated G1 cells from cdc15-2 (wild-type, Z2804) (A), swm1Δ cdc15-2 (Z2803) (B), and swm1Δ mad2Δ cdc15-2 (Z2922) (C) strains grown at 22°C were released at 37°C. All strains contained PDS1-myc18 and a ura3 locus marked with GFP. Pds1-myc18 was detected by indirect immunofluorescence. Top panels, the percentages of budded cells (squares), of cells with staining from Pds1-myc18 (filled triangles), and of cells with separated sister chromatids (filled diamonds) were plotted over the mean cell volumes. Arrows mark separated sisters in 50% of cells. Bottom panels, cellular DNA content and the mean cell volume measured at different times after release.
Swm1/Apc13 is required for APC/C activity during G1.
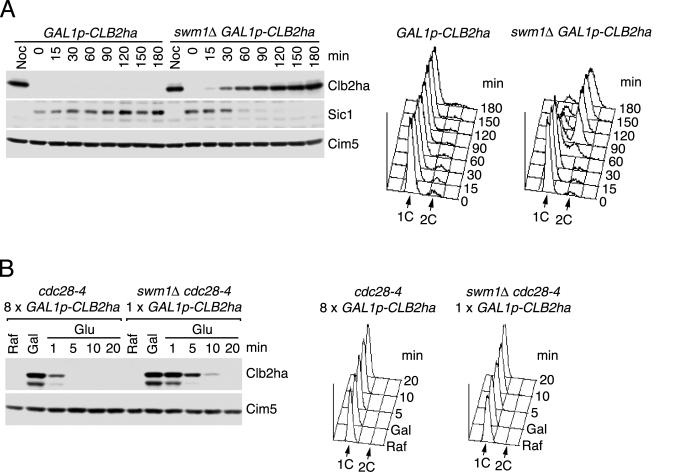
To test whether Swm1/Apc13 was required for cyclin degradation during G1, wild-type and swm1Δ strains were first arrested with α factor and then shifted to 37°C. Immunoblots revealed accumulation of the mitotic cyclins Clb2 and Clb3 after 60 min (data not shown). Expression of one copy of CLB2-ha3 from the GAL1 promoter at 37°C caused G1-arrested swm1Δ cells to accumulate Clb2-ha3 (after 30 min), to degrade Sic1, and to initiate DNA replication (after 60 min) (Fig. 10A). Similar to other apc mutants (16), swm1Δ cells did not form buds but elongated their mating projections. To measure Clb2's stability, the cyclin was expressed to detectable levels in G1-arrested cells at 37°C followed by “shutting-off” of further transcription and translation (Fig. 10B). Whereas the half-life of Clb2 was less than 1 min in the control cells, it increased to between 5 and 10 min in the swm1Δ cells. These data show that Swm1/Apc13 is required for efficient cyclin degradation also during G1, which depends on the activity of APC/C-Cdh1.
FIG. 10.
Swm1/Apc13 is important for APC/C activity during G1. (A) Expression of Clb2 in G1-arrested cells. Wild-type (Z1104) and swm1Δ (Z2218) cells of the genotype MATa GAL1p-CLB2-ha3 bar1Δ were grown at 25°C in YPRaf medium (GAL promoter off) and arrested in G1 with α factor (0.5 μg/ml). Cells were transferred to YPRafGal medium (GAL promoter on) with α factor at 35°C, and samples were withdrawn at different times. Noc, cells arrested with nocodazole and treated as before. (B) Half-life of Clb2 in G1-arrested cells. Wild-type (Z3182) and swm1Δ (Z3186) cells of the genotype MATa cdc28-4 GAL1p-CLB2-ha3 bar1Δ were arrested in G1 with α factor in YPRaf medium at 25°C (Raf) and then shifted to 35°C for 30 min. Galactose was added for 30 min (Gal), and cells were transferred to YPD medium with cycloheximide (Glu) at time zero. Samples were withdrawn at different times to analyze Clb2-ha3 levels and cellular DNA content. The cdc28-4 mutation prevents inactivation of Cdh1 by Cdk1. In wild-type cells eight copies of GAL1p-CLB2-ha3 were required to accumulate Clb2-ha3 to levels similar to those in swm1Δ cells.
Swm1/Apc13 is important for APC/C activity in meiosis.
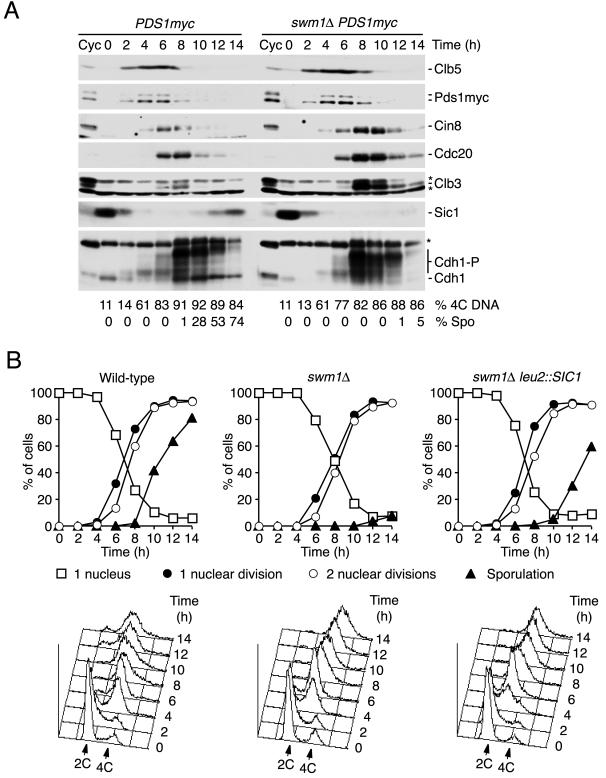
Since SWM1 was discovered as a gene required for sporulation (43), we analyzed whether Swm1/Apc13 was important for APC/C activity during meiosis. Both SWM1 alleles were deleted in diploid cells of the fast-sporulating SK1 strain. Upon transfer to sporulation medium at 30°C, the mutant cells underwent DNA replication and both nuclear divisions with normal kinetics whereas spore formation was strongly delayed (Fig. 11B), which agrees with previous results obtained with a different strain background (43). Consistent with normal nuclear divisions in swm1Δ cells, immunoblot analysis did not detect any defect in the degradation of Pds1. In contrast, the substrates Cin8, Cdc20, and especially Clb3 accumulated to higher levels than in the wild type and their degradation was delayed (Fig. 11A). Consistent with inefficient Cdk1 inactivation, reaccumulation of the Sic1 protein and dephosphorylation of Cdh1 both failed in the mutant cells. As shown in Fig. 11B, the sporulation defect of swm1Δ cells was partially rescued by increasing the gene dosage of SIC1. These data show that Swm1/Apc13 is required for APC/C activity in meiosis and suggest that the sporulation defect stems, at least in part, from persistent Cdk1 activity.
FIG. 11.
Swm1/Apc13 is required for APC/C activity in meiosis. (A) SK1 wild-type cells (Z2827) and swm1Δ/swm1Δ diploid cells (Z3306) homozygous for PDS1-myc18 were transferred to sporulation medium at 30°C at time zero and analyzed by immunoblotting. Cyc, extracts from growing cells. Asterisks mark nonspecific proteins. The percentages of cells with replicated (4C) DNA and of sporulated cells are indicated. (B) Suppression of the sporulation defect by multiple copies of SIC1. SK1 wild-type cells (Z2314) and swm1Δ/swm1Δ (Z4001) and swm1Δ/swm1Δ leu2::SIC1/leu2::SIC1 (Z4420) diploid cells were induced to enter meiosis at 30°C. Top panels, fractions of cells that contain one nucleus (squares) and that have undergone meiosis I (filled circles), meiosis II (open circles), and spore formation (filled triangles). Bottom panels, cellular DNA content.
DISCUSSION
Purification of the budding yeast APC/C in amounts readily detectable by Coomassie blue staining led to the identification of additional APC/C-associated proteins called Swm1/Apc13 and Mnd2 (14, 31, 49; this work). Our data show that most of Swm1/Apc13 is associated with the APC/C. Only APC/C subunits specifically copurified with TAP-tagged Swm1/Apc13, and the protein migrated together with other subunits as a 36S particle in glycerol density gradients. Accordingly, the SWM1 deletion caused lethality in combination with different apc mutations but not with mutations in other cell cycle genes. Swm1/Apc13 was proposed to be a substoichiometric subunit based on the finding that less APC/C is purified with Swm1/Apc13-TAP than with Cdc16-TAP (31). We note, however, that in different APC/C purifications the amount of untagged Swm1/Apc13 was similar, as judged from Coomassie blue staining, to that of other small subunits (Apc11, Doc1, and Apc9). It is possible that the TAP tag on Swm1/Apc13 is less accessible or that it reduces incorporation of the subunit into the complex. Ultimately, elucidating the stoichiometry of APC/C subunits has to await the determination of a high-resolution structure of the complex.
SWM1 was proposed to encode a protein unique to budding yeast (14, 31, 43). Our database searches, however, uncovered related sequences in many different species including fungi, plants, and animals, suggesting evolutionary conservation. Accordingly, the fission yeast and the human homologues are associated with APC/C subunits. Both homologues rescued the temperature-sensitive growth defect of swm1Δ single-mutant cells and the lethality of swm1Δ apc double mutants, demonstrating that different Swm1/Apc13 homologues have related functions. Yoon et al. have proposed other yeast and human proteins as homologues of Swm1/Apc13 (49). However, database searches revealed that S. cerevisiae YHR121w and C. albicans IPF8069 belong to an unrelated family of conserved fungal and animal proteins. The human sequence S47354 is a fragment of GAC-1 (AF317425), a predicted protein of 227 kDa containing ankyrin repeats.
Our data show that Swm1/Apc13 is important for APC/C's structural integrity. Complexes purified from APC2-TAP swm1Δ or APC4-TAP swm1Δ cells contained little Cdc16, Cdc27, Apc9, and Cdc26, whereas the remaining subunits were present at normal stoichiometries. Consistent with structural defects, the APC/C from swm1Δ cells has strongly reduced ubiquitin ligase activity. The five proteins Cdc16, Cdc27, Apc9, Swm1/Apc13, and Cdc26 form a stable subcomplex, which could be purified from cells containing ts mutations in Cdc23's N-terminal region (cdc23-51 and cdc23-54) or the TPR domain (cdc23-39) (39). Remarkably, the amounts of individual subunits in the subcomplex were similar to those in the intact APC/C. The subcomplex might recruit activator proteins since human Cdc27 was recently found to bind to Cdh1 (45). Cdc16 purified from swm1Δ cells yielded only trace amounts of Cdc27 and Apc9, suggesting that the subcomplex is less stable in the absence of Swm1/Apc13. We propose that in swm1Δ cells the subcomplex is sufficiently stable at low temperatures to maintain APC/C activity. At high temperatures, however, the subcomplex is less stable, so that subunits dissociate from the APC/C, which causes the cell cycle defects observed in vivo. The S. pombe subunit Lid1/Apc4 from wild-type cells sediments with the 20S APC/C but is shifted into a smaller complex upon depletion of Apc13 (49), which is consistent with our proposal for Apc13's function.
In addition, the small subunit Cdc26 is required to stabilize the interaction with the APC/C of Cdc16 and Cdc27 (54). This suggests that Swm1/Apc13 collaborates with Cdc26 in stabilizing the Cdc16/Cdc27 subcomplex. The functions of Swm1/Apc13 and Cdc26 are specific since deletions of Mnd2 (data not shown) or Doc1 (6) do not affect the association of other subunits. At low temperature in vivo either subunit is sufficient to promote formation of functional APC/C, whereas at high temperature both subunits are required. Homologues of Cdc26 have been identified in mammalian cells and in S. pombe (called Hcn1), and the yeast proteins were found to perform related functions (13, 47). Hcn1/Cdc26 and Apc13 are both essential in S. pombe (47, 49). We propose that in the fission yeast APC/C Hcn1/Cdc26 and Apc13 are both required for structural integrity already at low temperature.
Our results differ from those of a recent study reporting that APC/C purified from swm1Δ and cdc26Δ cells had normal subunit composition and ubiquitination activity (31). Conditional mutants must contain some APC/C activity at the permissive temperature. We speculate that the conditions of Passmore et al. (31) might preserve the mutant complexes whereas our conditions are more suitable to reveal the structural defects responsible for loss of APC/C function at elevated temperature in vivo. Nevertheless, the ubiquitination activity of our wild-type APC/C was comparable to that of the wild-type APC/C isolated by Passmore et al.
We show that Swm1/Apc13 is required for the timely execution of APC/C-dependent cell cycle events at 37°C. Degradation of Pds1, sister chromatid separation, and cyclin proteolysis during anaphase, all of which depend on APC/C-Cdc20, were delayed by approximately 30 min in swm1Δ cells. We propose that the cell separation defect is a consequence of inefficient Cdk1 inactivation, which interferes with nuclear accumulation of Ace2 and therefore reduces expression of the chitinase Cts1. Consistent with this idea, the cell separation defect was suppressed by a constitutively nuclear version of Ace2 or by increasing the gene dosage of the Cdk1 inhibitor Sic1. It might be important that translocation of Ace2 occurs abruptly because nuclear Ace2 is unstable (29). While this paper was being revised, Ufano et al. reported experiments showing that the cell separation phenotype stems from a defect in nuclear accumulation of Ace2 (42). The SWM1 deletion also reduced the activity of APC/C-Cdh1, causing a stabilization of Clb2 in G1-arrested cells.
In meiosis Swm1/Apc13 is required for spore formation at 30°C but not for nuclear divisions (43). Accordingly, degradation of Pds1 was not detectably affected in swm1Δ cells. However, degradation of the substrates Cin8 and Cdc20 and especially that of Clb3 was significantly delayed, demonstrating that Swm1/Apc13 is required for full APC/C activity during meiosis. Consistent with a defect in Cdk1 inactivation, swm1Δ cells failed to reaccumulate Sic1 and to dephosphorylate Cdh1 at late stages of meiosis. We propose that Swm1/Apc13 promotes sporulation, at least in part, through APC/C-dependent destruction of Cdk1 activity because the sporulation defect of swm1Δ cells could be rescued by multiple copies of SIC1. Consistent with this, Swm1/Apc13 is associated with the APC/C purified from cells synchronously progressing through meiosis (our unpublished results). Why is Swm1/Apc13 required specifically for sporulation, at least at 30°C? At this temperature, swm1Δ cells contain sufficient APC/C activity to proliferate almost normally. The sporulation process might be particularly sensitive to even a small reduction in APC/C function. In S. pombe moderate expression of a stabilized version of the B-type cyclin Cdc13 does not interfere with nuclear division but blocks sporulation (3). Another, mutually not exclusive, possibility is that Swm1/Apc13 is important for an APC/C form that specifically promotes sporulation. In both S. cerevisiae and S. pombe sporulation depends on meiosis-specific APC/C activators called Ama1 and Mfr1/Fzr1, respectively (2, 3, 8).
Acknowledgments
We thank David Barford, Geraldine Butler, David Drechsel, Anthony Hyman, Carl Mann, Kim Nasmyth, Jan-Michael Peters, Wolfgang Seufert, Francis Stewart, Mike Tyers, and Carlos Vazquez de Aldana for reagents. We thank Anna Shevchenko for tandem mass spectrometry and Carrie Cowan and Caroline Wilkinson for advice on fission yeast. We are grateful to Anthony Hyman and Elly Tanaka for valuable comments on the manuscript.
This work was supported by the Max Planck Gesellschaft and a grant from the Human Frontier Science Program Organization (RG0364/1999-M) to W.Z.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakawa, H., K. Kitamura, and C. Shimoda. 2001. A novel Cdc20-related WD-repeat protein, Fzr1, is required for spore formation in Schizosaccharomyces pombe. Mol. Genet. Genomics 265:424-435. [DOI] [PubMed] [Google Scholar]
- 3.Blanco, M. A., L. Pelloquin, and S. Moreno. 2001. Fission yeast mfr1 activates APC and coordinates meiotic nuclear division with sporulation. J. Cell Sci. 114:2135-2143. [DOI] [PubMed] [Google Scholar]
- 4.Buonomo, S. B., R. K. Clyne, J. Fuchs, J. Loidl, F. Uhlmann, and K. Nasmyth. 2000. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell 103:387-398. [DOI] [PubMed] [Google Scholar]
- 5.Camasses, A., A. Bogdanova, A. Shevchenko, and W. Zachariae. 2003. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol. Cell 12:87-100. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, C. W., and D. O. Morgan. 2002. The Doc1 subunit is a processivity factor for the anaphase-promoting complex. Nat. Cell Biol. 4:880-887. [DOI] [PubMed] [Google Scholar]
- 7.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067-1076. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, K. F., M. J. Mallory, D. B. Egeland, M. Jarnik, and R. Strich. 2000. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 97:14548-14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 10.Dohrmann, P. R., G. Butler, K. Tamai, S. Dorland, J. R. Greene, D. J. Thiele, and D. J. Stillman. 1992. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6:93-104. [DOI] [PubMed] [Google Scholar]
- 11.Gieffers, C., P. Dube, J. R. Harris, H. Stark, and J. M. Peters. 2001. Three-dimensional structure of the anaphase-promoting complex. Mol. Cell 7:907-913. [DOI] [PubMed] [Google Scholar]
- 12.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 13.Gmachl, M., C. Gieffers, A. V. Podtelejnikov, M. Mann, and J. M. Peters. 2000. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 97:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, M. C., M. P. Torres, G. K. Schroeder, and C. H. Borchers. 2003. Mnd2 and Swm1 are core subunits of the Saccharomyces cerevisiae anaphase-promoting complex. J. Biol. Chem. 278:16698-16705. [DOI] [PubMed] [Google Scholar]
- 15.Harper, J. W., J. L. Burton, and M. J. Solomon. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 16:2179-2206. [DOI] [PubMed] [Google Scholar]
- 16.Irniger, S., and K. Nasmyth. 1997. The anaphase-promoting complex is required in G1 arrested yeast cells to inhibit B-type cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110:1523-1531. [DOI] [PubMed] [Google Scholar]
- 17.Irniger, S., S. Piatti, C. Michaelis, and K. Nasmyth. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81:269-278. [DOI] [PubMed] [Google Scholar]
- 18.Joazeiro, C. A., and A. M. Weisman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 19.King, R. W., J. M. Peters, S. Tugendreich, M. Rolfe, P. Hieter, and M. W. Kirschner. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81:279-288. [DOI] [PubMed] [Google Scholar]
- 20.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 21.Lamb, J. R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257-259. [DOI] [PubMed] [Google Scholar]
- 22.Leverson, J. D., C. A. Joazeiro, A. M. Page, H. Huang, P. Hieter, and T. Hunter. 2000. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol. Biol. Cell 11:2315-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 24.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 25.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 27.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insight into dynamic signalling. Nat. Rev. Mol. Cell. Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 28.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 29.O'Conallain, C., M. T. Doolin, C. Taggart, F. Thornton, and G. Butler. 1999. Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262:275-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page, A. M., and P. Hieter. 1999. The anaphase-promoting complex: new subunits and regulators. Annu. Rev. Biochem. 68:583-609. [DOI] [PubMed] [Google Scholar]
- 31.Passmore, L. A., E. A. McCormack, S. W. Au, A. Paul, K. R. Willison, J. W. Harper, and D. Barford. 2003. Doc1 mediates the activity of the anaphase-promoting complex by contributing to substrate recognition. EMBO J. 22:786-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters, J. M. 2002. The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9:931-943. [DOI] [PubMed] [Google Scholar]
- 33.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 34.Rabitsch, K. P., A. Toth, M. Galova, A. Schleiffer, G. Schaffner, E. Aigner, C. Rupp, A. M. Penkner, A. C. Moreno-Borchart, M. Primig, R. E. Esposito, F. Klein, M. Knop, and K. Nasmyth. 2001. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr. Biol. 11:1001-1009. [DOI] [PubMed] [Google Scholar]
- 35.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 36.Roguev, A., D. Schaft, A. Shevchenko, R. Aasland, A. Shevchenko, and A. F. Stewart. 2003. High conservation of the Set1/Rad6 axis of histone 3 lysine 4 methylation in budding and fission yeasts. J. Biol. Chem. 278:8487-8493. [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko, A., D. Schaft, A. Roguev, W. W. Pijnappel, A. F. Stewart, and A. Shevchenko. 2002. Deciphering protein complexes and protein interaction networks by tandem affinity purification and mass spectrometry: analytical perspective. Mol. Cell. Proteomics 1:204-212. [DOI] [PubMed] [Google Scholar]
- 38.Shirayama, M., W. Zachariae, R. Ciosk, and K. Nasmyth. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski, R. S., W. A. Michaud, and P. Hieter. 1993. p62cdc23 of Saccharomyces cerevisiae: a nuclear tetratricopeptide repeat protein with two mutable domains. Mol. Cell. Biol. 13:1212-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudakin, V., D. Ganoth, A. Dahan, H. Heller, J. Hershko, F. C. Luca, J. V. Ruderman, and A. Hershko. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 6:185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ufano, S., M. E. Pablo, A. Calzada, F. Del Rey, and C. R. Vazquez De Aldana. 2004. Swm1p subunit of the APC/cyclosome is required for activation of the daughter-specific gene expression program mediated by Ace2p during growth at high temperature in Saccharomyces cerevisiae. J. Cell Sci. 117:545-557. [DOI] [PubMed] [Google Scholar]
- 43.Ufano, S., P. San-Segundo, F. del Rey, and C. R. Vazquez de Aldana. 1999. SWM1, a developmentally regulated gene, is required for spore wall assembly in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2118-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vodermaier, H. C. 2001. Cell cycle: waiters serving the destruction machinery. Curr. Biol. 11:R834-R837. [DOI] [PubMed] [Google Scholar]
- 45.Vodermaier, H. C., C. Gieffers, S. Maurer-Stroh, F. Eisenhaber, and J.-M. Peters. 2003. TPR Subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13:1459-1468. [DOI] [PubMed] [Google Scholar]
- 46.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, H., K. Kumada, and M. Yanagida. 1997. Distinct subunit functions and cell cycle regulated phosphorylation of 20S APC/cyclosome required for anaphase in fission yeast. J. Cell Sci. 110:1793-1804. [DOI] [PubMed] [Google Scholar]
- 48.Yanagida, M., Y. M. Yamashita, H. Tatebe, K. Ishii, K. Kumada, and Y. Nakaseko. 1999. Control of metaphase-anaphase progression by proteolysis: cyclosome function regulated by the protein kinase A pathway, ubiquitination and localization. Phil. Trans. R. Soc. Lond. B 354:1559-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoon, H. J., A. Feoktistova, B. A. Wolfe, J. L. Jennings, A. J. Link, and K. L. Gould. 2002. Proteomics analysis identifies new components of the fission and budding yeast anaphase-promoting complexes. Curr. Biol. 12:2048-2054. [DOI] [PubMed] [Google Scholar]
- 50.Yu, H., J. M. Peters, R. W. King, A. M. Page, P. Hieter, and M. W. Kirschner. 1998. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 279:1219-1222. [DOI] [PubMed] [Google Scholar]
- 51.Zachariae, W., and K. Nasmyth. 1996. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B-type cyclins in yeast. Mol. Biol. Cell 7:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]
- 53.Zachariae, W., M. Schwab, K. Nasmyth, and W. Seufert. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282:1721-1724. [DOI] [PubMed] [Google Scholar]
- 54.Zachariae, W., A. Shevchenko, P. D. Andrews, R. Ciosk, M. Galova, M. J. Stark, M. Mann, and K. Nasmyth. 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279:1216-1219. [DOI] [PubMed] [Google Scholar]
- 55.Zachariae, W., T. H. Shin, M. Galova, B. Obermaier, and K. Nasmyth. 1996. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science 274:1201-1204. [DOI] [PubMed] [Google Scholar]