Abstract
The leucine-rich acidic nuclear protein (LANP) belongs to a family of evolutionarily conserved proteins that are characterized by an amino-terminal domain rich in leucine residues followed by a carboxy-terminal acidic tail. LANP has been implicated in the regulation of a variety of cellular processes including RNA transport, transcription, apoptosis, vesicular trafficking, and intracellular signaling. Abundantly expressed in the developing cerebellum, this protein has also been hypothesized to play a role in cerebellar morphogenesis. LANP has been implicated in disease biology as well, both as a mediator of toxicity in spinocerebellar ataxia type 1 and as a tumor suppressor in cancers of the breast and prostate. To better understand the function of this multifaceted protein, we have generated mice lacking LANP. Surprisingly, these mice are viable and fertile. In addition we could not discern any derangements in any of the major organ systems, including the nervous system, which we have studied in detail. Overall our results point to a functional redundancy of LANP's function, most likely provided by its closely related family members.
Leucine-rich acidic nuclear protein (LANP or pp32 [for phosphoprotein with a molecular mass of 32 kDa]) is a member of the leucine-rich family of proteins (16-18). These proteins are involved in a variety of pathways including signaling, protein degradation, cytoskeletal dynamics, and morphogenesis, presumably based on the ability of the leucine-rich repeat (LRR) domains to serve as adapter sites for protein-protein interactions.
For LANP, the hydrophobic LRRs reside in its N-terminal domain, giving LANP a globular head domain. The C-terminal domain, on the other hand, is extended and hydrophilic, engaging interactors by virtue of its ionic interactions (23, 27, 30, 31).
Several proteins have extensive homology to LANP, both in displaying LRRs in their N terminal domains and in demonstrating extended acidic tail domains. These proteins represent true LANP family members that have probably arisen by gene duplication. The nomenclature of LANP family members is confusing because the same protein has been given more than one name based on the context of isolation. Thus APRIL, an acronym derived from A protein rich in leucines, is also known as PAL-31, for proliferation-associated leucine-rich protein, MW 31kDa, and SSP-29, for silver-stainable protein with a molecular mass of 29 kDa (24, 25, 41). CPD1 (cerebellar postnatal development protein 1), another LANP family member, has been called LANP-like protein (LANP-L) (21, 28). Two human homologues of LANP/pp32 have been called pp32r1 and pp32r2 to signify that they are related (14, 15). Besides having homology with these proteins, LANP family members also have limited homology to template-activating factor (TAF) proteins, TAF-1 alpha and beta (the latter being the product of the SET oncogene). The TAF-1 proteins, like the LANP family members, demonstrate long acidic C-terminal domains but, unlike LANP, do not display LRRs in their N-terminal domains (1, 22, 26, 38).
LANP and TAF family members have frequently been isolated together. This suggests that they have overlapping functions or work together as part of a complex. The putative functions and subcellular localizations of these proteins are almost bewildering in their variety. At the inner surface of the plasma membrane of lymphocytes, LANP and SET were observed to be HLA-associated proteins—hence the moniker putative HLA-associated protein 1 (PHAP1) for LANP and PHAP2 for SET. In this location, these proteins were postulated to play a role in signal transduction (37). In the cytoplasm, LANP binds to all classes of structural microtubule-associated proteins (MAPs) (11, 27, 35, 36). No other protein, besides tubulin, shares such an extensive repertoire of MAP binding capacity. Thus cytoplasmic LANP is well positioned to regulate microtubule function and microtubule-based vesicular trafficking and has been aptly referred to as mapmodulin in this context (11, 27, 35, 36). It has been postulated that, within the nucleus, LANP and TAF proteins are important members of the transcriptional repression machinery. This was based on the finding that these acidic proteins mask the positively charged histones, ultimately inhibiting histone acetylation. Chromatin is thus rendered less amenable to transcriptional activity and genes are silenced (30-32). Not surprisingly for proteins known to reside in both the nucleus and cytoplasm, LANP and SET family members have also been shown to shuttle between these two subcellular compartments. Their ability to shuttle, along with their ability to bind the RNA-binding protein HuR, is thought to contribute to RNA stability and transport (3, 10). Another seemingly unrelated function that appears to depend on the dual nucleocytoplasmic localization of these proteins is the modulation of apoptotic events at the level of apoptosome activity and DNA breakage (8, 9, 12). These functions taken as a group suggest that these proteins tie together vital cellular processes in metabolic loops that span from the cell surface to the nucleus and that they could therefore be key regulators of cellular homeostasis.
Another interesting feature of LANP is its well-regulated and distinct spatiotemporal pattern of expression during development (23). Although widely expressed and persisting into adulthood, LANP is particularly abundant in the developing cerebellum, leading to the speculation that LANP plays a role in cerebellar morphogenesis. LANP is also abundant in self-renewing stem cell-like populations and cell subtypes known for their rapid proliferative ability, such as intestinal crypt epithelial cells and prostatic epithelial cells competent for cell renewal (19, 39). There is also some evidence to suggest that LANP and its family members regulate the proliferation of tumor cells (2, 6, 13-15). Incidentally, the SET gene was originally identified as a putative oncogene, identified as the locus for a translocation breakpoint event in acute undifferentiated leukemia, although the cellular basis for oncogenesis is far from clear.
Our own interest in LANP stems from the observation that LANP interacts with ataxin-1, the protein mutated in the polyglutamine disease spinocerebellar ataxia type 1 (SCA1) (20). SCA1 is a neurodegenerative disease characterized by progressive incoordination, mainly because of progressive Purkinje cell dysfunction and loss. In this context, LANP, has a greater affinity for the mutated or polyglutamine-expanded version of ataxin-1. Thus one could imagine a scenario wherein LANP's functions are altered upon binding to mutant ataxin-1, triggering downstream neurodegenerative events. To begin to address what such events might be and to further understand the in vivo functions of this apparently versatile protein, we have generated mice lacking LANP. Since LANP is abundantly expressed in the nervous system, a site where all of its putative functions are well represented from development through adulthood, we have targeted our efforts to evaluating the neuronal consequences of loss of LANP. Despite morphological, histochemical, electrophysiological, and behavioral analysis of these mice, we could discern no differences between LANP null mice and their wild-type littermates. The most likely explanation for this apparent normality in the face of LANP's widespread cellular role could lie in the compensatory role of LANP's closely related family members that continue to be expressed in LANP null mice.
MATERIALS AND METHODS
Generation of LANP knockout mice.
We obtained genomic clones corresponding to the mouse Lanp locus from a genomic library prepared from the 129/SvEV mouse strain (Stratagene, La Jolla, Calif.). We generated a targeting vector by flanking a 2.2-kb region of genomic DNA containing exons 2 and 3 of the LANP gene with loxP sites. A selectable cassette containing the genes for thymidine kinase (TK) and neomycin resistance (TK-Neo cassette) was inserted downstream of the genomic fragment followed by a third loxP site. The LANP targeting construct was electroporated into embryonic stem (ES) cells that were subsequently grown in media containing G418 selection to select for homologous recombination. Targeted ES clones were subsequently electroporated with Cre recombinase, which mediates recombination between the loxP sites, thus allowing excision of the TK-Neo cassette and the generation of an edited version of the Lanp locus. ES cells with the targeted Lanp locus were injected into C57BL/6J blastocysts to generate chimeras. We then crossed the chimeras to C57BL/6J females for germ line transmission. Homologous and Cre-mediated recombination events in ES cell clones and germ line transmission of the correct construct in mice were confirmed by Southern blotting. Genomic DNA was digested with EcoRV, and a 5′ probe upstream of the targeted Lanp locus was used to probe the Southern blots. All protocols involving the use of animals were in compliance with the National Institutes of Health's Guide for the Care and Use of Laboratory Animals and the Baylor College of Medicine Institutional Animal Care and Use Committee.
Biochemical assays.
We prepared protein extracts from mouse brain tissue as previously described (4). Briefly tissues were homogenized in protein extraction buffer containing 100 mM Tris, pH 6.8, 2% sodium dodecyl sulfate (SDS), 25 mM dithiothreitol, and protease inhibitors (Complete; Roche Molecular Biochemicals). Proteins were separated on SDS-10% polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes (Schleicher & Schuell). Western blots were probed with an antibody against LANP (antibody 3118) (27) by using the ECL detection kit (Amersham Life Science). Antibodies generated against human APRIL and the rat APRIL/PAL31 antibody were kind gifts from J. Steitz and K. Shiota, respectively. Monoclonal antibodies against actin (A6-40; Sigma) and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; 6C5; Advanced Immunochemicals) were used to confirm equivalent loading of tissue samples.
Hippocampal-slice preparation and electrophysiology.
Hippocampal electrophysiology was performed as previously described (29). Briefly, adult mice were sacrificed and brains were rapidly removed and briefly submerged in ice-cold cutting saline (110 mM sucrose, 60 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 28 mM NaHCO3, 0.5 mM CaCl2, 5 mM d-Glucose, 0.6 mM ascorbate). All solutions were saturated with 95% O2 and 5% CO2. Whole brains were then dissected on a cutting solution-soaked filter paper mounted on a glass platform resting on ice. Hippocampal slices (400 μm) were prepared on a Vibratome and allowed to equilibrate in a 50% cutting saline-50% ACSF (125 mM NaCl, 2.5 mM KCl, 1.24 mM NaH2PO4, 25 mM NaHCO3, 10 mM d-glucose, 2 mM CaCl2, 1 mM MgCl2) solution at room temperature for a minimum of 30 min. Slices were transferred to an interface chamber supported by a nylon mesh and allowed to recover for a minimum of 1 h prior to recording. Extracellular field recordings were obtained from the area CA1 stratum radiatum. Stimulation was supplied with a bipolar Teflon-coated platinum electrode, and recording was obtained with a glass microelectrode filled with ACSF (resistance, 1 to 4 MΩ). Tetani used to evoke CA1 long-term potentiation (LTP) consisted of one set of high-frequency stimulation. Each set consisted of two trains of 100-Hz frequency stimulation for 1 s, with each train separated by a 20-s interval. Stimulus intensities were adjusted to give population excitatory postsynaptic potentials (pEPSPs) with slopes that were ≤50% of that corresponding to the maximum stimulus intensity determined from an input-output curve. The 50% maximum stimulus intensity was used for all LTP experiments. Experimental results were obtained only from those slices that exhibited stable baseline synaptic transmission for a minimum of 30 min before the LTP-inducing stimulus was given. The average slope of the pEPSP from six individual traces, standardized to baseline responses, was measured and graphed.
Behavioral analysis of LANP null mice. (i) Rotarod.
Mice at 5 and 11 weeks of age were placed on a rotating-rod apparatus that accelerates linearly from 4 to 40 rpm (Ugo Basile). Mice were tested in four consecutive trials (trials 1 through 4) on four consecutive days (days 1 through 4). The amount of time a mouse took to fall off the rotating rod to a maximum of time of 10 min, the duration of the trial, was noted. Two episodes of holding on to a rod rotating 360 degrees were also scored as a fall, and the time of the second rotation was recorded as such.
(ii) Open-field test.
Mice were placed in the center of an open field (40 by 40 by 30 cm) for 30 min monitored by a computer-operated optical system (RXYZCM; Acuscan) that monitors the movement of mice as they wander in the open field.
(iii) Wire test.
Mice were placed by their forepaws on an approximately 2-mm-diameter wire constructed from a coat hanger. The amount of time they remained suspended was recorded to a maximum time of 1 min.
(iv) Dowel test.
Mice were allowed to balance on the center of a long dowel (0.9 cm in diameter), and the time they remained on the dowel was recorded up to a maximum of 2 min. Mice that walked across the dowel before completion of the test were placed back onto the center of the dowel.
(v) Conditioned-fear test.
The conditioned-fear test was preceded by a 7-min training period (conditioning training). In the training the mice were placed in a chamber capable of delivering an electric shock to their feet. The mice were allowed to accustom themselves to their new chamber for 3 min, following which a conditioned stimulus (CS) of 80 dB of white noise was delivered for 30 s. The CS was followed by the unconditioned stimulus (US) of a mild (2-s, 0.5-mA) foot shock. Another CS-US pair was presented 2 min later (5 min from the start), and the mice were finally removed at 7 min. Throughout the testing period freezing behavior was recorded every 10 s. The mice were then tested for fear based on context (cage environment) and fear based on cue (the auditory stimulus) 1 and 24 h later. In the contextual-testing phase, each mouse was returned to the test chamber and freezing behavior was recorded every 10 s for 5 min. To test for cue conditioning, the environment of the mice was changed by placing mice in a novel box with a new vanilla odor. At 3 min the auditory stimulus was delivered over another 3 min. Once again, freezing was recorded every 10 s throughout the test.
Immunohistochemistry.
Immunohistochemistry was performed on mouse tissues fixed in formalin as previously described (7, 34). The anti-MAP1 antibody was commercially obtained (SC-8971; Santa Cruz Biotechnology). Other antibodies used were identical to those used in the Western blots.
RESULTS
Generation of LANP null mice.
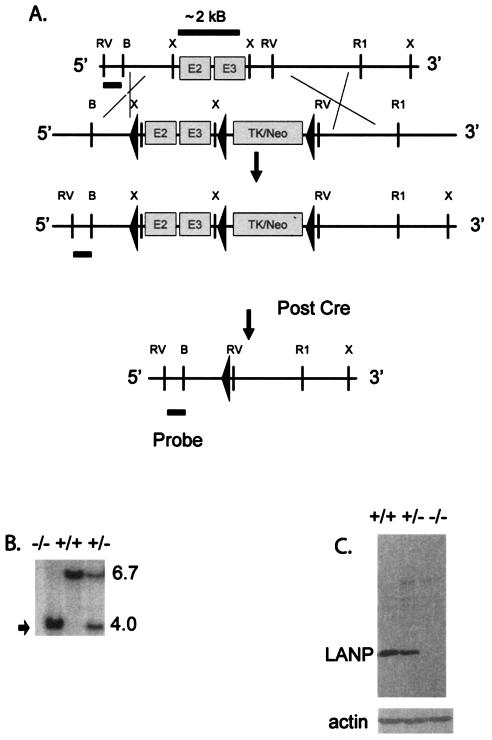
To generate mice lacking LANP, we used a homologous-recombination strategy to target the mouse Lanp locus in ES cells. A detailed scheme for the targeting construct is shown in Fig. 1A. Briefly, we flanked exons 2 and 3 of the LANP gene along with a selectable TK-Neo cassette with loxP sites. Exon 1 of the LANP gene is short and encodes only the first 18 residues. ES cells selected for homologous recombination were then electroporated with a plasmid expressing Cre recombinase. This resulted in two loxP-mediated recombination events, resulting in either of two constructs. In the first construct (type 1 deletion) only the exogenous TK-Neo selection cassette of the targeting construct was deleted; this was intended to make a conditional LANP knockout in the event that LANP null mice were either infertile or were not viable. In the second construct (type 2 deletion) the DNA sequence between loxP sites 1 and 3 was deleted; this construct was used to generate the LANP null mice described in detail in the rest of the report. In this construct, even if splicing were to occur between exon 1 and exon 4, the mRNA would encode only the first 18 residues of LANP; next in the coding sequence would be an out-of-frame exon four (a stop codon occurring after 10 codons). Using this strategy, we obtained 15 ES clones with the type 1 deletion and 76 ES clones with the type 2 deletion from 125 successfully targeted clones. We confirmed successful targeting by Southern blotting using probes that recognized genomic DNA outside the targeted region (Fig. 1B). Loss of protein expression was confirmed with an antibody generated against LANP (Fig. 1C).
FIG. 1.
Generation of LANP null mice. (A) Targeting strategy. The targeting vector was generated by flanking a 2.2-kb region of genomic DNA containing exons 2 and 3 of the LANP gene with loxP sites. The selectable cassette (TK-Neo) containing the genes for neomycin resistance and TK was inserted downstream of the genomic fragment, followed by a third loxP site. Targeted ES clones were subsequently electroporated with Cre recombinase, and the recombination event that deletes exons 2 and 3 and the selection cassette is shown. (B) Southern blotting of DNA isolated from mouse tails using EcoRV-digested DNA and a 5′ probe upstream of the targeted locus. Mice null for LANP display a 4-kb band (arrow) compared to the wild-type band of 6.7 kb. (C) Western blot analysis of cerebellar extracts using an anti-LANP antibody and an antiactin antibody. LANP null mice show no detectable LANP and equal intensity of the actin signal as a loading control.
LANP null mice are viable and fertile.
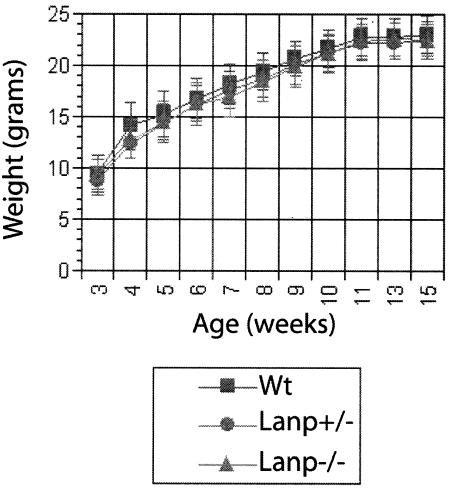
Mice lacking LANP are viable and fertile and display a normal growth curve (Fig. 2). We could not distinguish LANP null mice from their wild-type littermates based on home cage behavior including gait, general activity, and grooming. Breeding of LANP heterozygous mice resulted in approximately Mendelian ratios of progeny (38 LANP null mice, 42 wild-type mice, and 75 heterozygous mice out of a total of 155). We also could not observe any pathological derangements based on light-microscopic analysis of hematoxylin-eosin sections of any of the major organ systems (data not shown). Two of the mice lacking LANP that were sacrificed during our studies displayed hydronephrosis of the kidneys. Another mouse had to be sacrificed because of a urethral concretion that deformed the penis and caused an acute inflammatory reaction in the pudendal region. Both urethral concretions and hydronephrosis can be sporadic in C57BL6 mice, so it is unlikely that these low-frequency events are attributable to LANP deficiency. One mouse also developed a tumor at the base of the tail that appeared to be rich in fibrous tissue with abundant hyalinized vessels. This cancer did not appear to share any features reminiscent of cancer of the prostate, a cancer that might be caused by alterations of LANP gene expression (13, 14). Since there was no overall increase in the number of tumors, we believe that this pathology also is unlikely to be attributable to the fact that these mice are deficient in LANP.
FIG. 2.
LANP mice display normal growth curves. LANP null mice and wild-type (Wt) and heterozygote littermates show overlapping growth curves. Results are means ± standard errors of the means. Wild type, n = 13; LANP null mice, n = 9; heterozygous mice, n= 14.
Analysis of the nervous systems of mice lacking LANP. (i) Morphological analysis of the brains of LANP null mice.
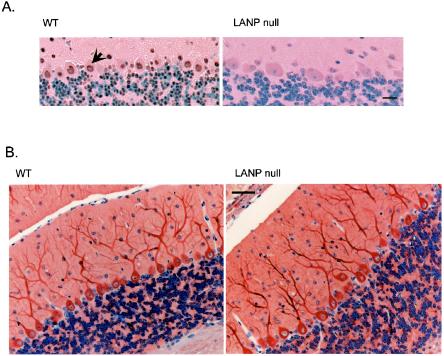
LANP null mice appeared to have normal brain structure. We could clearly observe by immunohistochemistry a depletion of LANP in null mice compared to their wild-type littermates using an LANP-specific antibody (Fig. 3A). Since LANP has been reported to interact with all structural MAPs, we performed immunohistochemistry against the light chain of MAP1; this MAP staining gives a good overview of dendritic arborization. Here as well our analysis did not reveal any variability in cerebellar morphology, Purkinje cell number and size, or dendritic arborization (Fig. 3B).
FIG. 3.
Morphological analysis of brains of LANP null mice. (A) LANP null mice show depletion of LANP by immunohistochemical staining (right). Note the predominantly nuclear staining (arrow) of LANP in a section from a wild-type littermate. Scale bar = 20 μm. (B) LANP null mice do not show any differences in cerebellar morphology, cell position, or dendritic arborization compared to wild-type littermates based on immunohistochemical analysis using an antibody against the light chain of MAP1B. Scale bar = 50 μm.
(ii) Behavior.
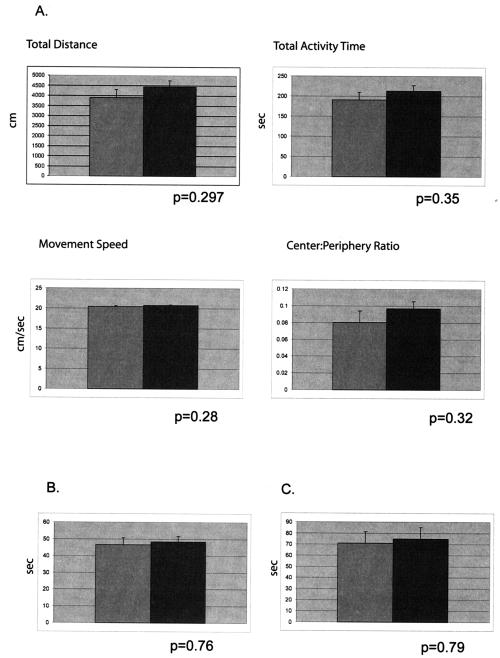
LANP null mice showed no evidence of gait abnormalities or ataxia. To evaluate their general exploratory behavior, we performed open-field testing (Fig. 4A). LANP mice and wild-type littermates explored similar distances (4,442.3 ± 314.6 versus 3,905.4 ± 403.3 cm; P = 0.297) in similar times (213.1 ± 14.0 versus 190.6 ± 19.3 s; P = 0.35), thus displaying similar speeds of movement (LANP null mice, 20.7 ± 0.17 cm/s; wild type, 20.4 ± 0.17 cm/s; P = 0.28). LANP null mice also spent approximately equivalent amounts of time in the center and periphery of the open-field box, an overall indicator of exploratory behavior and level of anxiety (center/periphery ratios: LANP null mice, 0.097 ± 0.01; wild type, 0.08 ± 0.01; P = 0.32).
FIG. 4.
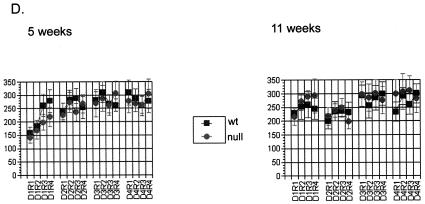
Behavioral profile of LANP null mice. (A) Open-field test. LANP null mice (n = 17; dark gray bar) are as active as wild-type mice (n = 15; light gray bar) in the open field and display no increase in anxiety. In this test LANP null mice travel distances similar to those traveled by wild-type mice and spend similar times moving; hence, they and wild-type mice exhibit similar speeds during the test. Moreover they and wild-type mice spend approximately similar amounts of time in the center versus the periphery, suggesting no sense of heightened anxiety. Results are means ± standard errors of the means (SEM). (B) Wire test. LANP null mice (n = 20) and wild-type littermates (n = 19) spend similar times suspended on the wire. Results are means ± SEM). (C) Dowel test. LANP null mice (n = 20) and wild-type littermates (n = 23) spend similar times balanced on the dowel. Results are means ± SEM. (D) Rotarod test. LANP null mice (n = 9) perform similarly to wild-type mice (n = 13) on the rotating-rod apparatus. Wild-type and LANP null mice were subjected to Rotarod testing of four trials per day for 4 days at 5 and 11 weeks of age. Results are means ± SEM. (E) Conditioned-fear testing. LANP null mice (n = 8) learn as well as wild-type animals (n = 10) to associate the context or the CS (cue) with a foot shock, as determined by measuring freezing behavior. Mice were tested for both short-term (1 h) and long-term (24 h) fear-conditioned behavior. Results are means ± SEM.
To test for general motor strength, we tested LANP null animals and control littermates for their ability to suspend themselves from a wire with their forepaws (Fig. 4B). In this test LANP null mice (48.3 ± 3.37 s) performed comparably to their wild-type counterparts (46.68 ± 4.03 s; P = 0.76). We then tested the mice for their ability to maintain their balance on a suspended dowel (Fig. 4C). This is a good test for both motor and cerebellar function. LANP null mice maintained themselves on the dowel for 74.9 ± 10.35 s, similar to the amount of time wild-type mice could remain balanced (71.04 ± 10.36 s; P = 0.79). To detect any defects in motor learning, we tested mice for their ability to remain on an accelerating Rotarod. This test is designed to test for subtle deficits in cerebellar function. Despite abundant LANP expression in Purkinje cells, LANP null mice performed well on this test. Moreover they showed no deterioration in their ability to perform this test as a function of age, which might have suggested a neurodegenerative phenotype (Fig. 4D).
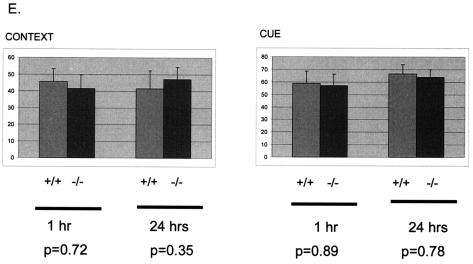
Because we also detected high expression of LANP in hippocampal pyramidal cells (Fig. 5), we subjected LANP null mice to conditioned-fear testing (Fig. 4E). In this test mice were placed into a test chamber and were trained to associate an 80-dB noise (CS) with a mild electric shock to the feet (US). One and 24 h later mice were tested for freezing behavior, which would demonstrate fear in response to either context (the cage that was used for delivering the US) or cue (by delivering the auditory CS in a novel environment). Both wild-type and LANP null mice performed similarly in context- and cue-based testing (context at 1 h: LANP null, 41.67 ± 8.04; wild type, 45.67 ± 7.62; P = 0.72; context at 24 h: LANP null, 47.08 ± 7.5; wild type, 41.7 ± 10.5; P = 0.35; cue at 1 h: LANP null, 56.94 ± 9.2; wild type, 58.89 ± 9.88; P = 0.89; cue at 24 h: LANP null, 63.89 ± 6.03; wild type, 66.67 ± 7.2; P = 0.78).
FIG. 5.
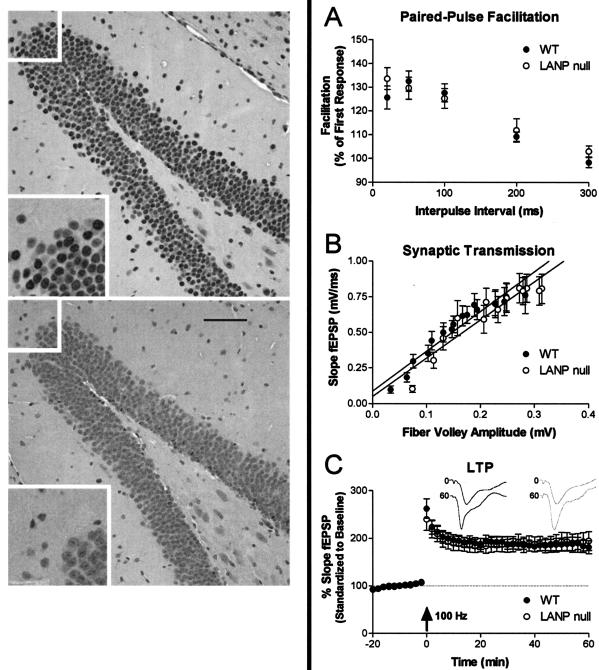
Electrophysiologic assessment of LANP null mutants compared to wild-type mice. (Left) Immunohistochemical analysis using a LANP-specific antibody demonstrates that LANP is abundantly expressed in hippocampal cells of wild-type (WT) mice (top); LANP null mice show no staining and serve as a staining control (bottom). In each image the upper left inset, magnified twofold, is displayed in the lower left corner. Scale bar = 50 μm. (A) The PPF paradigm was used to assess short-term plasticity in LANP null and WT mice. PPF in mutant mice was similar to that in WT mice (LANP null: 134% ± 4.6%, n = 19; WT: 126% ± 4.9%, n = 17; P = 0.1217). (B) Synaptic transmission in the stratum radiatum in area CA1 of the hippocampus was unaltered in LANP null mutants compared to WT mice. No significant differences in the slopes of the lines were seen when a linear regression was calculated with data from all stimulus intensities (LANP null mice: n = 19; slope, 2.68 ± 0.27; r2 = 0.8791; WT, n = 16; slope, 2.78 ± 0.24; r2 = 0.9107). (C) LTP was induced in LANP null and wild-type mice with two trains, each of 1 s, of 100-Hz stimulation separated by 20 s (arrow). No change in the amount of posttetanic potentiation was seen immediately following stimulation (LANP null: 240% ± 22.8%; n = 14; WT: 263% ± 20.5%, n = 12; P = 0.2350) or 60 min after stimulation (LANP null, 195% ± 19.7%; WT, 182 ± 13.9%; P = 0.3120). (Inset) Representative traces (means of six successive EPSPs) are shown for baseline (0) and 60 min after tetanic potentiation (60) for LANP null mice (gray) and wild types (black). Results (A to C) are means ± standard errors of the means.
In summary LANP null mice performed similarly to their wild-type littermates on a variety of behavioral tests. LANP null mice beyond 12 months of age continued to be indistinguishable from wild-type littermates based on home cage behavior.
Electrophysiology.
Since LANP is expressed in hippocampal cells (Fig. 5, left), we examined the well-defined Schaffer collateral synapses of the hippocampus to determine if there are any alterations in baseline synaptic transmission and/or short- or long-term synaptic plasticity.
Paired-pulse facilitation (PPF) is a short-lived and calcium-dependent form of plasticity likely to be involved in some forms of learning and memory (33, 40). Since PPF varies with the stimulus intensity and the size of the pEPSP slope for the first response, the stimulus intensity was adjusted so that the slopes of the pEPSP from the first stimulus for all slices were the same (data not shown). The percentage of PPF was determined at interpulse intervals of 20, 50, 100, 200, and 300 ms. LANP null mutants showed no significant difference in PPF at any of the interpulse intervals tested (P = 0.1217; Fig. 5A).
Loss of the LANP in the hippocampus appeared to have no deleterious effect on baseline synaptic transmission at Schaffer collateral synapses. Overall synaptic transmission and connectivity were determined from pEPSP of field recordings from the stratum radiatum in hippocampal area CA1. This was performed by determining the amplitude of the evoked fiber volley versus the slope of the field excitatory postsynaptic potentials at increasing stimulus intensities (Fig. 5B). Linear regression through these data points reveals no significant difference in the slope of the line. This suggests that in LANP null mutant mice the overall connectivity and synaptic transmission across the Schaffer collateral synapses are unaltered.
Hippocampal LTP is a use-dependent increase in synaptic efficacy involving biochemical processes similar to those underlying long-term memory formation in mammals. Hippocampal LTP was induced by stimulation of the Schaffer collateral pathway in the area CA1 with a modest protocol consisting of two trains of tetani (twice for 1 s at 100 Hz, separated by 20 s) performed while maintaining slices at 30°C. No differences in the induction or maintenance of LTP were observed (Fig. 5C). These results suggest that LANP is not necessary for normal LTP induction or maintenance across the Schaffer collateral synapse.
Expression of the LANP paralogue APRIL in LANP null mice is not altered.
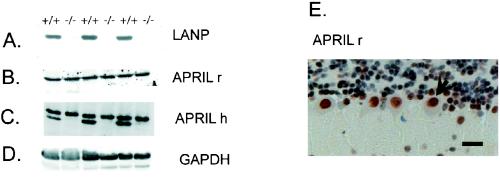
Because LANP has closely related family members, we were keen to test whether they were expressed in the same cell types and if they were up-regulated in mice lacking LANP. We focused on LANP's closest family member, APRIL (based on phylogenetic analysis; see Fig. 7), which is also expressed in the nervous system. Mouse LANP and APRIL demonstrate 63% identity. APRIL, however, is slightly larger, with an expected molecular mass of 31 kDa and a pI of 3.72 (in comparison with a molecular mass of 28.5 kDa and a pI of 3.84 for LANP). APRIL and LANP have similar properties. Like LANP, APRIL binds the RNA binding protein HuR, playing a role in RNA stability and transport (3); more recently, it has been demonstrated that both LANP and APRIL modulate apoptosis (12). Moreover by immunohistochemical staining (using the specific antibody on LANP null mice), it is clear that APRIL overlaps with LANP in the subtypes of neuronal cells that express these family members (Fig. 6E). Thus they are both abundant in the nuclei of Purkinje cells. We evaluated the abundance of APRIL in lysates prepared from knockout mice and wild-type littermates by immunoblotting to test if it is up-regulated. One of the antibodies (generated against rat PAL31/APRIL) is indeed specific for APRIL and shows similar levels in both LANP knockout and wild-type mice (Fig. 6B). Another antibody (generated against human APRIL) clearly cross-reacts with LANP, as seen by the disappearance of this band in the knockout mice (Fig. 6C). This cross-reaction both attests to the high level of homology between the two proteins and demonstrates the continued presence of this LANP family member in the cerebella of LANP null mice.
FIG. 7.
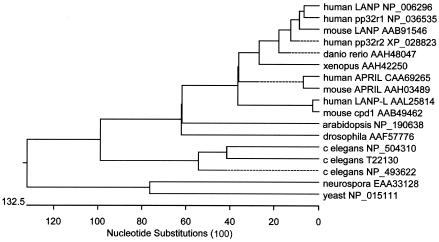
Phylogenetic tree of LANP and family members. LANP and representative family members from a variety of species were aligned in clustalV, and a phylogenetic tree was drawn to show their relationships.
FIG. 6.
Normal expression of APRIL/PAL31, an LANP family member, in LANP null mice. Western blotting of mouse cerebellar extracts from three pairs of wild-type and LANP null littermates reveals the absence of LANP staining in LANP null mice (A). Persistence of APRIL/PAL31 is observed with two different antibodies generated against rat (APRIL r) and human (APRIL h) versions of this protein (B and C, respectively). Note that APRIL h recognizes mouse APRIL and LANP, causing a doublet pattern in wild-type lanes (C). (D) A Western blot against GAPDH serves as a loading control. (E) Immunohistochemical localization of APRIL with antibody APRIL r from a LANP null cerebellum shows significant staining in the nuclei of Purkinje cells.
DISCUSSION
LANP has been found to have several important functions in the cytoplasm and nucleus. It belongs to an evolutionarily conserved family of proteins seen in all eukaryotes (Fig. 7). Its closest yeast homologue is Lea1 (GenBank accession number NP 015111), a specific component of the U2 snRNA particle required for prespliceosome formation (5). Although homologues of Lea1 persist in metazoans, LANP family members characterized by longer acidic domains are now observed in both the plant and animal kingdoms.
To begin to understand the role of this family of proteins, we generated mice lacking LANP. In addition to examining the general health and development of LANP null mice, we focused our efforts on evaluating the neuronal structure and function in these mice for a variety of reasons. LANP is abundant in the nervous system; moreover loss of LANP's putative cellular functions, particularly those involved in neuritogenesis, transcriptional regulation, and apoptosis, would be expected to play havoc, particularly in neurons. To take just a few instances, in the context of LANP's role as a modulator of MAPs, LANP null mice might be expected to display neuritic abnormalities, defects in vesicular transport, and/or electrophysiological defects, whereas LANP's role in RNA stability and transcriptional regulation means that loss of LANP should in effect significantly derange the neuronal proteome, resulting in alterations in neuronal structure and function. In addition LANP's role as an apoptotic regulator suggested to us a priori that loss of LANP might result in a developmental phenotype, or at least an alteration in the size and cellularity of various organs, including the brain. We did not see any of these abnormalities, nor did we see any neuronal loss despite looking for such evidence in vivo.
Indeed LANP mice for the most part are indistinguishable from their wild-type littermates. LANP mice perform well on comprehensive neurological tests, including those that test for strength, balance, motor coordination, and learning. Moreover detailed histological analysis and electrophysiological studies do not reveal any major abnormalities. The most likely explanation for the apparent normality of LANP null mice, in view of the multifaceted role of LANP, is the presence of proteins closely related to LANP. Based on spatiotemporal expression patterns, APRIL stands out as a prime candidate, although it is possible that rescue might be provided by other newly discovered and less-characterized family members. To test this hypothesis, one would have to engineer, mate, and characterize mice lacking two or more LANP family members. An alternative and perhaps more elegant approach might be to try to disrupt the function of the LANP complex by overexpressing LANP or by generating dominant negative disruptors, such as an LANP molecule with its LRR domain intact but lacking its C-terminal domain. Such an approach might be feasible, since LANP appears to function as a trimer in vivo and, complexed with other proteins in the SET/TAF complex, can be as large as 450 kDa (8). Another approach might be to use less-complex organisms, such as Drosophila melanogaster, which appears to have one LANP homologue, so as to understand its function at an organismal level. In view of LANP's role in rapidly dividing cells and possibly in mediation of neurodegeneration, it is interesting that, without LANP, mice develop normally and do not show an increased tendency to develop spontaneous tumors. Future studies exposing LANP null mice to ionizing radiation or mating them with other genetically engineered mice that have increased susceptibility to tumors might yet reveal a role for LANP in cancer.
What about LANP's role in SCA pathogenesis? Although at first glance the fact that depletion of LANP does not result in ataxia appears to speak against a loss of function of LANP in SCA1, this conclusion may be premature. It is possible that ataxin-1 binds all LANP family members in vivo, resulting in a phenotype of far greater magnitude than that resulting from loss of LANP alone. It is also possible that LANP might play a role in SCA1 by vesting mutant ataxin-1 with novel properties when the two interact. These scenarios are eminently testable by mating mice lacking LANP with ataxin-expressing mice to observe whether the spinocerebellar phenotype is ameliorated or worsened.
Acknowledgments
We thank members of the Zoghbi laboratory for their input throughout the course of this project, Bobby Antalffy for technical assistance, and Joan Steitz and K. Shiota for providing us valuable antibodies.
This work was funded in part by NIH grant K08 NS02246-03 (P.O.), a Molecular Medicine Scholar award from Baylor College of Medicine (P.O.), R01 NS27699-13 from NIH (H.Y.Z.), and Baylor Mental Retardation Research Center grant HD24064.
REFERENCES
- 1.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J. Biol. Chem. 269:2258-2262. [PubMed] [Google Scholar]
- 2.Bai, J., J. R. Brody, S. S. Kadkol, and G. R. Pasternack. 2001. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene 20:2153-2160. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burright, E. N., J. D. Davidson, L. A. Duvick, B. Koshy, H. Y. Zoghbi, and H. T. Orr. 1997. Identification of a self-association region within the SCA1 gene product, ataxin-1. Hum. Mol. Genet. 6:513-518. [DOI] [PubMed] [Google Scholar]
- 5.Caspary, F., and B. Seraphin. 1998. The yeast U2A′/U2B complex is required for pre-spliceosome formation. EMBO J. 17:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, T. H., J. R. Brody, F. E. Romantsev, J. G. Yu, A. E. Kayler, E. Voneiff, F. P. Kuhajda, and G. R. Pasternack. 1996. Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol. Biol. Cell 7:2045-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings, C. J., E. Reinstein, Y. Sun, B. Antalffy, Y. Jiang, A. Ciechanover, H. T. Orr, A. L. Beaudet, and H. Y. Zoghbi. 1999. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron 24:879-892. [DOI] [PubMed] [Google Scholar]
- 8.Fan, Z., P. J. Beresford, D. Y. Oh, D. Zhang, and J. Lieberman. 2003. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell 112:659-672. [DOI] [PubMed] [Google Scholar]
- 9.Fan, Z., P. J. Beresford, D. Zhang, and J. Lieberman. 2002. HMG2 interacts with the nucleosome assembly protein SET and is a target of the cytotoxic T-lymphocyte protease granzyme A. Mol. Cell. Biol. 22:2810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 11.Itin, C., N. Ulitzur, B. Muhlbauer, and S. R. Pfeffer. 1999. Mapmodulin, cytoplasmic dynein, and microtubules enhance the transport of mannose 6-phosphate receptors from endosomes to the trans-Golgi network. Mol. Biol. Cell 10:2191-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, X., H. E. Kim, H. Shu, Y. Zhao, H. Zhang, J. Kofron, J. Donnelly, D. Burns, S. C. Ng, S. Rosenberg, and X. Wang. 2003. Distinctive roles of PHAP proteins and prothymosin-alpha in a death regulatory pathway. Science 299:223-226. [DOI] [PubMed] [Google Scholar]
- 13.Kadkol, S. S., J. R. Brody, J. I. Epstein, F. P. Kuhajda, and G. R. Pasternack. 1998. Novel nuclear phosphoprotein pp32 is highly expressed in intermediate- and high-grade prostate cancer. Prostate 34:231-237. [DOI] [PubMed] [Google Scholar]
- 14.Kadkol, S. S., J. R. Brody, J. Pevsner, J. Bai, and G. R. Pasternack. 1999. Modulation of oncogenic potential by alternative gene use in human prostate cancer. Nat. Med. 5:275-279. [DOI] [PubMed] [Google Scholar]
- 15.Kadkol, S. S., G. A. El Naga, J. R. Brody, J. Bai, Y. Gusev, W. C. Dooley, and G. R. Pasternack. 2001. Expression of pp32 gene family members in breast cancer. Breast Cancer Res. Treat. 68:65-73. [DOI] [PubMed] [Google Scholar]
- 16.Kobe, B., and J. Deisenhofer. 1994. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 19:415-421. [DOI] [PubMed] [Google Scholar]
- 17.Kobe, B., and J. Deisenhofer. 1995. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5:409-416. [DOI] [PubMed] [Google Scholar]
- 18.Kobe, B., and J. Deisenhofer. 1995. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374:183-186. [DOI] [PubMed] [Google Scholar]
- 19.Malek, S. N., A. I. Katumuluwa, and G. R. Pasternack. 1990. Identification and preliminary characterization of two related proliferation-associated nuclear phosphoproteins. J. Biol. Chem. 265:13400-13409. [PubMed] [Google Scholar]
- 20.Matilla, A., B. Koshy, C. J. Cummings, T. Isobe, H. T. Orr, and H. Y. Zoghbi. 1997. The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1. Nature 389:974-978. [DOI] [PubMed] [Google Scholar]
- 21.Matsubae, M., T. Kurihara, T. Tachibana, N. Imamoto, and Y. Yoneda. 2000. Characterization of the nuclear transport of a novel leucine-rich acidic nuclear protein-like protein. FEBS Lett. 468:171-175. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto, K., K. Nagata, M. Ui, and F. Hanaoka. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582-10587. [PubMed] [Google Scholar]
- 23.Matsuoka, K., M. Taoka, N. Satozawa, H. Nakayama, T. Ichimura, N. Takahashi, T. Yamakuni, S.-Y. Song, and T. Isobe. 1994. A nuclear factor containing the leucine-rich repeats expresses in murine cerebellar neurons. Proc. Natl. Acad. Sci. USA 91:9670-9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mencinger, M., I. Panagopoulos, J. A. Contreras, F. Mitelman, and P. Aman. 1998. Expression analysis and chromosomal mapping of a novel human gene, APRIL, encoding an acidic protein rich in leucines. Biochim. Biophys. Acta 1395:176-180. [DOI] [PubMed] [Google Scholar]
- 25.Mutai, H., Y. Toyoshima, W. Sun, N. Hattori, S. Tanaka, and K. Shiota. 2000. PAL31, a novel nuclear protein, expressed in the developing brain. Biochem. Biophys. Res. Commun. 274:427-433. [DOI] [PubMed] [Google Scholar]
- 26.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opal, P., J. J. Garcia, F. Propst, A. Matilla, H. T. Orr, and H. Y. Zoghbi. 2003. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule associated protein 1B and modulates neuritogenesis. J. Biol. Chem. 275:34691-34699. [DOI] [PubMed] [Google Scholar]
- 28.Radrizzani, M., G. Vila-Ortiz, E. G. Cafferata, M. C. Di Tella, A. Gonzalez-Guerrico, C. Perandones, O. H. Pivetta, H. Carminatti, V. P. Idoyaga Vargas, and T. A. Santa-Coloma. 2001. Differential expression of CPD1 during postnatal development in the mouse cerebellum. Brain Res. 907:162-174. [DOI] [PubMed] [Google Scholar]
- 29.Roberson, E. D., and J. D. Sweatt. 1996. Transient activation of cyclic AMP-dependent protein kinase during hippocampal long-term potentiation. J. Biol. Chem. 271:30436-30441. [DOI] [PubMed] [Google Scholar]
- 30.Seo, S. B., T. MacFarlan, P. McNamara, R. Hong, Y. Mukai, S. Heo, and D. Chakravarti. 2002. Regulation of histone acetylation and transcription by nuclear protein pp32, a subunit of the INHAT complex. J. Biol. Chem. 277:14005-14010. [DOI] [PubMed] [Google Scholar]
- 31.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 32.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva, A. J., T. W. Rosahl, P. F. Chapman, Z. Marowitz, E. Friedman, P. W. Frankland, V. Cestari, D. Cioffi, T. C. Sudhof, and R. Bourtchuladze. 1996. Impaired learning in mice with abnormal short-lived plasticity. Curr. Biol. 6:1509-1518. [DOI] [PubMed] [Google Scholar]
- 34.Skinner, P. J., B. Koshy, C. Cummings, I. A. Klement, K. Helin, A. Servadio, H. Y. Zoghbi, and H. T. Orr. 1997. Ataxin-1 with extra glutamines induces alterations in nuclear matrix-associated structures. Nature 389:971-974. [DOI] [PubMed] [Google Scholar]
- 35.Ulitzur, N., M. Humbert, and S. R. Pfeffer. 1997. Mapmodulin: a possible modulator of the interaction of microtubule-associated proteins with microtubules. Proc. Natl. Acad. Sci. USA 94:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitzur, N., C. Rancaño, and S. R. Pfeffer. 1997. Biochemical characterization of mapmodulin, a protein that binds microtubule-associated proteins. J. Biol. Chem. 272:30577-30582. [DOI] [PubMed] [Google Scholar]
- 37.Vaesen, M., S. Barnikol-Watanabe, H. Gotz, L. A. Awni, T. Cole, B. Zimmermann, H. D. Kratzin, and N. Hilschmann. 1994. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol. Chem. Hoppe-Seyler 375:113-126. [DOI] [PubMed] [Google Scholar]
- 38.von Lindern, M., S. van Baal, J. Wiegant, A. Raap, A. Hagemeijer, and G. Grosveld. 1992. can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 12:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walensky, L. D., D. S. Coffey, T. H. Chen, T. C. Wu, and G. R. Pasternack. 1993. A novel Mr 32,000 nuclear phosphoprotein is selectively expressed in cells competent for self-renewal. Cancer Res. 53:4720-4726. [PubMed] [Google Scholar]
- 40.Wu, L. G., and P. Saggau. 1994. Presynaptic calcium is increased during normal synaptic transmission and paired-pulse facilitation, but not in long-term potentiation in area CA1 of hippocampus. J. Neurosci. 14:645-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, L., L. Perlaky, D. Henning, and B. C. Valdez. 1997. Cloning and characterization of a new silver-stainable protein SSP29, a member of the LRR family. Biochem. Mol. Biol. Int. 42:927-935. [DOI] [PubMed] [Google Scholar]