Abstract
In earlier studies, we identified the C-9 amido derivative 1 (9-(4'-hydroxy-4-biphenyl)acetamido-9-deoxy-Neu5Gcα2-6GalOMP) and the C-9 amino derivative 2 (9-(4'-hydroxy-4-biphenyl)methylamino-9-deoxy-Neu5Gcα2-6GalOMP) have the most promising affinity for mouse CD22 and human CD22, respectively. Replacing the subterminal galactose residue (2-6Gal-OMP) of 1 with benzyl (5) or biphenylmethyl (6) as aglycone led to even higher potency for mCD22. In this study, both compounds showed improved potency and selectivity for CD22 (IC50 70 nM) and 712-fold more selective for CD22 than for MAG. The corresponding derivatives of 2, compounds 8 and 9, showed comparable activity to 2 but lower potency and selectivity than 5 and 6. Although compounds 5-9 are simple and small molecular weight antagonists, they showed much high potency and selectivity than the corresponding compounds having α 2-6Gal linkage. Both biological and computational docking simulation studies suggest that the 2-6Gal-OMP residues of 1 and 2 are not critical for binding process and could be replaced with hydrophobic non carbohydrate moieties. The data presented herein has significant implications for the design and discovery of next-generation CD22-antagonists
Keywords: benzyl and biphenylmethyl sialosides, CD22, Hydrophobic interaction, Selectivity, MAG, ELISA
1. Introduction
Advances in the functional understanding of carbohydrate–protein interactions have enabled the development of a new class of small-molecule drugs, known as glycomimetics. These compounds mimic the bioactive function of carbohydrates and address the drawbacks of carbohydrate leads, namely their low activity and insufficient drug-like properties.1
CD22 (siglec-2) is the best characterized member of siglec family.2 It is an accessory molecule of the B-cell receptor complex (BCR) that exhibits both positive and negative effects on receptor signaling. The carbohydrate ligand recognized by CD22 is the sequence Siaα2-6Galβ1-4GlcNAc found on both neighboring glycoconjugate of the same cell (cis ligands) and on cells that interact with B cells (e.g. T cells, trans ligands). Interactions of CD22 with cis or trans ligands regulate aspects of B cell activation, proliferation and development. Therefore, CD22 is a regulatory protein that prevents the overactivation of the immune system and the development of autoimmune diseases.3 Our current understanding of the biological functions of CD22 is mostly based on in vitro experiments. The present challenge will be to transfer this knowledge to in vivo studies, in order to learn more about their role in regulating immune responses. To face this challenge, high affinity ligands suitable for such studies are required. In addition CD22 antagonists with sufficient avidity could find several applications, e.g. modulation of the immune response4 and targeting device for treatment of B cell related diseases.5 Chen et al 2010.
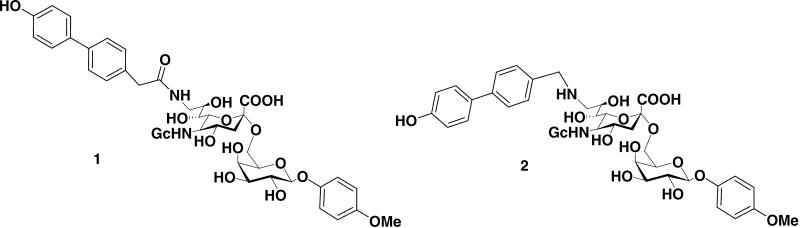
Recently,6 we have reported the dramatic improvement of binding affinity for CD22 achieved by the modifications at C-9 of Neu5Gcα2-6GalOMP core as exemplified by the 9-amido derivative 1 (9-(4′-hydroxy-4-biphenyl)acetamido-9-deoxy-Neu5Gcα2-6GalOMP) and the 9-amino derivative 2 (9-(4′-hydroxy-4-biphenyl)methylamino-9-deoxy-Neu5Gcα2-6GalOMP) which exhibited the highest potency for mouse CD22 (mCD22) and human CD22 (hCD22), respectively, (Fig. 1). Our previous docking studies revealed that C-9 amido or amino sialic acid derivatives linked with 2-6GalOMP moiety form extra interactions as compared with reference compound, which could account for their increased binding affinities for both hCD22 and mCD22.6
Figure 1.
Structures of the 9-amido (1) and 9-amino (2) sialosides.6
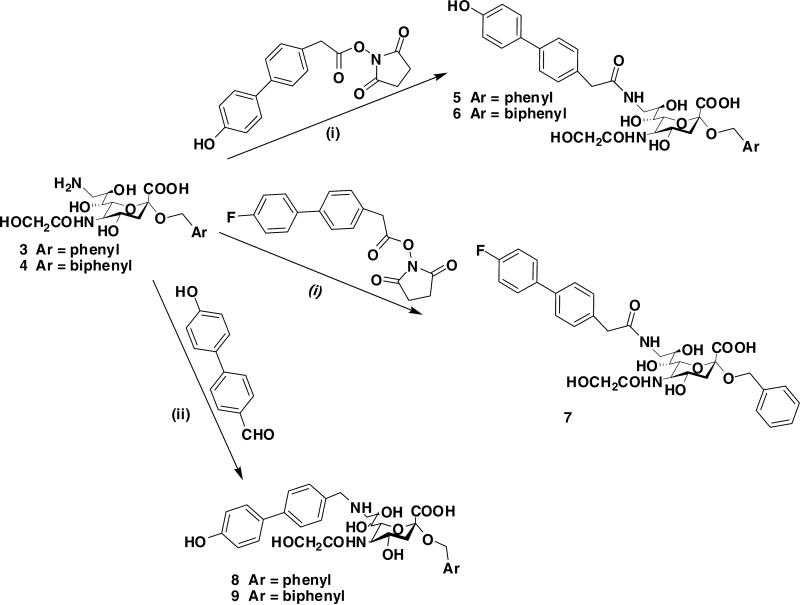
More recently, we have found that replacing the subterminal galactose residue of 1 (2-6GalOMP) with benzyl or biphenylmethyl as aglycone at the C-2 of sialic acid scaffold led to higher affinity for mCD22 (compounds 5 and 6; Scheme 1).7 The further analysis of binding affinity of the tested compounds guided that 2-6GalOMP group which is independent from the core sialic acid, could be replaced with more flexible hydrophobic groups. The hydrophobicity of benzyl or biphenylmethyl could compensate the desolvation penalty fetched by 2-6Gal-OMP (unfavorable desolvation enthalpy of OH groups in the 2-6Gal) and also gain in the entropy (the change in solvation entropy is more favorable if the surfaces are more hydrophobic) can make the binding affinity more favorable.
Scheme 1.
Synthesis of 9-amido (5-7) and 9-amino (8 and 9) benzyl and biphenylmethyl sialosides. Reagents and conditions: (i) NaHCO3, MeCN/H2O, 75%; (ii) NaBH3CN, AcOH, MeOH, r. t., 24 h, 68%.
Sequence comparison of Siglecs and inspection of the 2-3-sialyllactose/SnD1 crystal structure suggested that α-sialosides with modified substituents in the glycerol side chain could yield increased binding affinities8-10. Moreover, such studies indicated that these compounds would be likely to display enhanced specificity to individual family members and could potentially aid in the dissection of Siglec function10.
In continuation of our interest in investigating the molecular basis of the interaction of CD22 with sialosides, we report herein the synthesis of the new compounds 7-9 and the determination of the binding affinity of compounds 5-9 for CD22 and MAG. Manual docking and molecular dynamics simulations were carried out to investigate the structural basis of the observed affinity.
2. Results and Discussion
This work describes the novel small molecule hydrophobic sialosides as selective antagonists for the ligand binding domain of CD22.
2.1. Chemistry
As outlined in our preceding communication7 we have achieved the syntheses of benzyl 3, 5, 9- trideoxy-5-glycolamido-9-(4′-hydroxy-4-biphenyl)acetamido-d-glycero-α-d-galacto-2-nonulopyranosidonic acid (5) and the corresponding biphenylmethyl sialoside (6) by acylation of the 9-amino intermediates (3 and 4), respectively, with N-hydroxysuccinimide ester of 4′-hydroxybiphenyl-4-acetic acid (Scheme 1). Similarly compound 7 was prepared by acylation of 3 with the succinimidyl ester6 of 4'-fluorobiphenyl-4-acetic acid.11 Based on the higher affinity of 9-aminosialoside 2 for hCD22 than 9-amidosialosides 16 and encouraged by the promising activity of compounds 5 and 6; the corresponding 9-amino analogues (8 and 9) were prepared. They were synthesized by reductive alkylation of 3 and 4 with 4′-hydroxybiphenyl-4-carbaldehyde6 (Scheme 1).
2.2. Biological assay
A competition enzyme-linked immunosorbent assay (ELISA) based on a biotinylated form of 112 was used for measuring the inhibitory potency of the synthesized sialosides. The results are shown in Tables 1, 2 and 3.
Table 1.
Inhibitory potencies of 9-amido analogs (5-7) for mCD22 and hCD22.
| Compd. | Activity |
|||
|---|---|---|---|---|
| mCD227 | hCD22 | |||
| IC50 (μM)a | rIPb | IC50 (μM) | rIP | |
| 1 | 3.84 ± 0.32 | 1 | 0.90 ± 0.00 | 1 |
| 5 | 0.10 ± 0.00 | 38.4 | 0.13 ± 0.00 | 7 |
| 6 | 0.19 ± 0.00 | 20.2 | 0.07 ± 0.01 | 12 |
| 7 | 5.00 ± 0.6 | 0.77 | ND | |
Sialoside concentration which leads to 50% inhibition of binding. The values are the mean ± SD of triplicates.
The rIP of each sialoside was calculated by dividing the IC50 of the reference compound 1 by the IC50 of the compound of interest. The results in rIPs above 1.0 for derivatives binding better than the reference and rIPs below 1.0 for compounds with a lower affinity than the reference.
ND; not determined. The values for mCD22 and hCD22 cannot be directly compared
Table 2.
Inhibitory potencies of 9-amino analogs (8 and 9) for hCD22 and mCD22.
| Compd. | Activitya |
|||
|---|---|---|---|---|
| mCD22 | hCD22 | |||
| IC50 (μM) | rIPb | IC50 (μM) | rIP | |
| 2 | 6.43 ± 0.00 | 1 | 0.53 ± 0.00 | 1 |
| 8 | 5.03 ± 3.54 | 1.2 | 0.82 ± 0.02 | 0.6 |
| 9 | 23.33 ± 5.51 | 0.3 | 1.12 ± 0.06 | 0.5 |
Footnote of table 1.
Table 3.
Comparison of Inhibitory Potentials of the synthetic sialosides to CD22 and MAG
| Compd. No. | Activity |
Selectivity | |
|---|---|---|---|
| hCD22 IC50 (μM) |
MAG IC50 (μM) |
||
| 1 | 3.27 ± 0.62 | 136 ± 27 | 41.5 |
| 2 | 3.29 ± 0.33 | 37.4 ± 3.1 | 11.4 |
| 5 | 0.24 ± 0.05 | 171 ± 35 | 712.5 |
| 6 | 0.38 ± 0.00 | 164 ± 11 | 431.5 |
| 8 | 2.75 ± 1.3 | 9.27 ± 1.3 | 3.4 |
| 9 | 2.24 ± 0.28 | 16.6 ± 5.2 | 7.4 |
The selectivity of each sialoside was determined. They were calculated by dividing the IC50 value of each compound for MAG by the IC50 value of the same compound for CD22.
2.2.1 Comparison between the binding affinity of 9-amido (5 and 6) and 9-aminosialosides (8 and 9) for hCD22 and mCD22
The high affinity of 5 (IC50 100 nM, 38-fold higher potency) and 6 (IC50 190 nM, 20-fold higher potency) for mCD22 in comparison with 17, prompted us to investigate their potencies for hCD22. Accordingly, we found that both compounds 5 and 6 also showed improved affinity for hCD22; 7 and 12-fold more potent than 1, respectively. As observed for mCD22, these hydrophobic sialosides are the most potent antagonists for hCD22 (IC50 130 nM and 70 nM). In case of hCD22, biphenyl methyl sialoside (6) is more potent than benzyl sialoside (5), while in case of mCD22, benzyl sialoside is more potent. This result may be due to steric interaction with the binding site as will be discussed in the molecular modeling study. Unexpectedly, and in contrast with the improved potency of the analogs of 1, the benzyl and biphenylmethyl analogs of 2 (8 and 9) did not show any improvement of binding affinity (Table 2). They are nearly equipotent to 2. Since the analogs of 1 and 2 are different only in the substituent at C-9, it seems that the flexibility of amino group is responsible for such differences in binding affinity. Therefore, the greater flexibility of compounds 8 and 9 may result in entropic penalties upon binding. Alternatively, the flexibility may constrain multivalent interaction with CD22, while the 9-amido group of 5 and 6 is more rigid giving the correct orientation for multivalent interaction. Despite having a simpler structure the target compounds have either higher (5 and 6) or comparable (8 and 9) potency to more complex parent structures 1 and 2. These sialic acid derivatives combine the beneficial modifications at C-2 and C-9 positions in one molecule and are supposed to have improved pharmacokinetic properties due to the presence of aromatic moieties, a lower molecular weight, and a reduced number of polar hydroxy functions compared to the parent compounds. Compound 5 and 6 are the most potent inhibitors for hCD22 have ever been synthesized.
2.2.2 Comparison between the binding affinity of the test compounds for CD22 and MAG
At this stage of development of these potent inhibitors, it was crucial to address the issue of specificity of the synthetic sialosides for CD22. Therefore, appropriate control, a closely related Siglec-4 (myelin-associated glycoprotein, MAG), was included in the ELISA experiments to ascertain if the inhibitors are really specific to CD22.
Inhibitory potentials of the synthetic sialosides to CD22 and MAG were determined by competitive ELISA (Table 3). The most interesting results in our study are the high binding affinity and selectivity of compounds 5 and 6 (Table 3). Both of them contain the same hydrophobic moiety at C-9 and benzyl or biphenyl, respectively, at C-2. Compound 5 is 712-fold more selective for hCD22 than for MAG. Similarly compound 6 is 431-fold more selective. Surprisingly, they are much more selective than the corresponding compounds containing α2-6 linkage; 1 and 2. They are also much more potent and selective than 8 and 9 which contain 9-amino instead of 9-amido.
On the other hand, binding studies on MAG revealed that benzyl sialosides are 10-fold more potent than methyl sialosides.13 This effect was rationalized by the results of STD-NMR investigations, indicating a hydrophobic interaction with MAG.14 Moreover, chlorobenzamide was found to be the best substituent in the 9-position.15 Accordingly, we can conclude that the main source of selectivity of 5 and 6 is the presence of 9-(4'-biphenylacetamido) which contributes significantly to the improved affinity as well as selectivity.
2.2.3 The effect of replacing 4'-hydroxy by 4'-fluoro on the binding affinity of compound 5
The promising activity and selectivity of 5 encouraged us to study the effect of replacing the 4'-hydroxy group with 4'-fluoro. Accordingly, compound 7 was synthesized and its binding for CD22 was evaluated. The rationale for such strategy stems from similarities between fluorine and oxygen in polarity and close isosteric relationship.16 Consequently, a F atom is considered as a good substitute of a OH group. One important advantage of introducing fluoride is to render the compound relatively resistant to metabolic transformation due to the high energy of carbon-fluorine bond.16 Unexpectedly, we found that compound 7 has significantly lower activity for CD22 (IC50 5 μM; 50-fold less potent than 5). Obviously, a fluorine atom cannot donate hydrogen bonds, but it is electronegative and can accept them. This finding confirms our model of binding to CD22 and the involvement of 4'-hydroxy in H-bond formation.6
The solubility of the test compounds in aqueous media and the reproducibility of the assay results under different condition from different laboratories are thought to provide insight into the interactions with the binding site and exclude the possibility of micelle or multimeric formation as one possible mechanism of improved activities of such compounds. Moreover, the lower activity of compound 7 gives a good evidence for the dependence of binding on structural feature and not on physical interactions of molecules. In addition, closely related structures of MAG antagonists have been reported and the improved activities of such compounds were accounted for by increased lipophilicity as rationalized by the results of trNOE NMR and STD NMR investigations.13-15
2.3. Molecular modeling
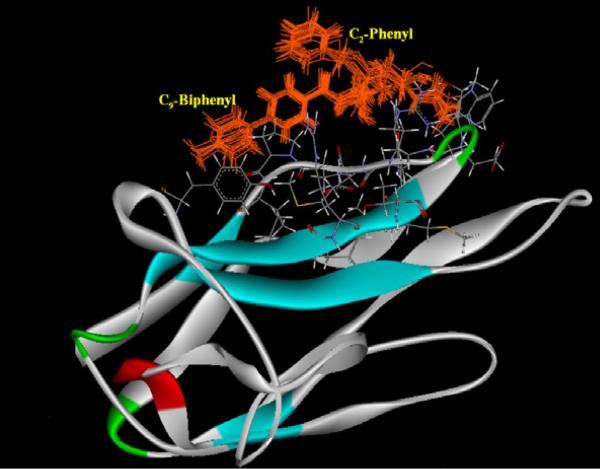
In order to investigate the structural basis of the observed affinity and interpret the unexpected differences in the inhibition potencies between 9-amido (5 and 6) and 9-amino (8 and 9) analogs, manual docking and molecular dynamics simulations were carried out against mCD22. The binding modes of the benzyl sialosides (5 and 8) apparently found to be different with the previously studied 2-6Gal-OMP binding mode.6 As can be seen from Figure 2, the benzyl portion of the representative structures obtained during simulations of 5 is slightly exposed out to solvent and partially interacts with the 9-biphenylacetamido portion of the same compound. These attractive intra-molecular interactions could be induced dipole– induced dipole (London) dispersion forces as both aromatic groups (C-2 phenyl and C-9 biphenyl of 5) are present in their proximities (~3.1Å to 5.0 Å). These momentary van der Waals attractions could also be aided by the solvophobic effects, in which desolvation can stabilize interactions of hydrophobic molecular surfaces. Therefore, they can stabilize protein ligand complex.17 In connection with this, the differential activities of benzyl or biphenylmethyl sialosides between the hCD22 and mCD22 can be inferred to subtle amino acid differences in the C-9-biphenyl binding region (Arginine in hCD22, Proline in mCD22).6
Figure 2.
View of the simulated representative structure of compound 5 bound to the mCD22. Structures colored in grey are protein residues.
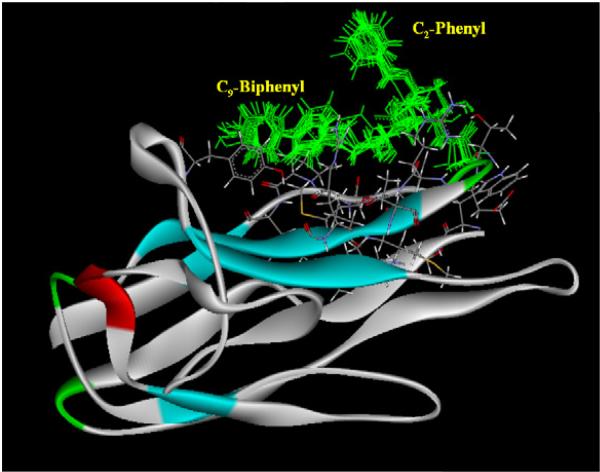
On the other hand, C-2 benzyl and C-9 biphenyl of 7 (an amino analogue of compound5) are stay farther distance than those of 5 (~4.6 Å to 8.5 Å; Figure 3). This could be due to the flexibility of amino group, which can disturb the intra-molecular interactions described above. At this point, we can speculate that the observed differences in the activity of compounds 5-9 as compared to the parent compounds (1 or 2) could be due to their ability to form the intra-molecular interactions with the head and tail hydrophobic portions.
Figure 3.
View of the simulated representative structure of compound 8 bound to the mCD22. Structures colored in grey are protein residues.
Previously reported crystal structures of the Siglec sialoadhesin (SnD1), in the absence of ligand, and in complex with sialic acid analogs, revealed that SnD1 undergoes very few structural changes on ligand binding which unexpectedly can induce dimerization of two SnD1 molecules.18 These results highlight the need to account for the possibility of ligand induced dimerization when measuring affinity.18 On the other hand, thermodynamic analysis of recently developed MAG antagonists, having hydrophobic substituents at C-2 of sialic acid moiety, revealed that the improved affinity predominantly results from an increased binding enthalpy and not from an entropy gain.19 Hydrophobic interactions with the side chains of Trp59, Tyr60, and Tyr69, lining the main hydrophobic pocket was proposed through homology modeling and docking.19
Both of the above mentioned facts could not sufficiently rationalize our results; though second one look more promising. However, the sequence alignment of some member of siglecs showed that the hydrophobic cavity of MAG is not relevant or not clearly known in CD22 and Sn.10
Based on our models of CD226, 2-benzyl or biphenyl group are slightly exposed to the solvent and also interacting the 9-biphenyl group in a shallow active of CD22s. The proposed intramolecular interactions, between C-2 and C-9 substituents, could be better in explaining the observed activity differences. Residues related to hydrophobic cavity of MAG in the CD22s are not in same of position or even close; if 2-benzyl of 5 takes similar conformation as seen in MAG antagonists, it can severely affect the conserved Arg-97 interaction in CD22s.
Taken together, the current experimental and computational studies support our previous finding that the subterminal sugar part (2-6Gal-OMP) could be replaced with non-carbohydrate moieties. Particularly, groups with optimal hydrophobicity could be exploited for improved binding affinity.
3. Conclusion
Structurally simplified and highly potent CD22-antagonists of high selectivity were synthesized. The improved affinity of the target compounds (5-9) might be due to desolvation and intra-molecular interactions of hydrophobic groups at C-2 and C-9 or due to dimerization of CD22 molecules. The higher binding affinity exhibited by the more rigid 9-amido derivatives (5 and 6) in comparison with the flexible 9-amino derivatives (8 and 9) may be due to the stronger intra-molecular forces between the hydrophobic moieties at C-2 and C-9. We are encouraged by these promising results and are moving forward regarding the optimization of the substituent at C-2 of sialic acid scaffold for further improvement of pharmacodynamic and pharmacokinetic parameters.
4. Experimental
4.1. Synthesis
4.1.1. General information
1H and 13C NMR spectra were recorded on JEOL JNM-EX-400 (400 MHz) or JEOL JNM-ECA-500 (600 MHz). Chemical shifts are expressed in ppm (δ) relative to the signal of Me4Si. MALDI-TOF MS spectra were recorded in positive ion mode on a Bruker Autoflex with the use of α-cyano-4-hydroxycinnamic acid (CHCA) as a matrix. Solvents and reagents were used as received from commercial distributors without further purification. Anhydrous reactions were conducted under an argon atmosphere. TLC analysis was carried out on Merck TLC (silica gel 60F254 on glass plate), and compounds were visualized by exposure to UV light and/or by charring with 10% H2SO4 in ethanol. Silica gel (80 mesh and 300 mesh) manufactured by Fuji Silysia Co. was used for flash column chromatography. Quantity of silica gel was usually estimated as 100 to 150-fold weight of sample to be charged. Reversed phase silica gel (Wakogel® 50C18) was purchased from Wako Pure Chemical Industries, Ltd. HPLC grade water and methanol (WAKO) were used for separation of the final compounds. Solvent systems in chromatography were specified in v/v. All evaporations and concentrations were carried out in vacuo.
4.1.2. Benzyl 3, 5, 9- trideoxy-5-glycolamido-9-(4′-fluoro-4-biphenyl)acetamido-d-glycero-α-d-galacto-2-nonulopyranosidonic acid (7)
To the amine 3 (49.7 mg, 0.12 mmol) in water (1.5 mL) was added solution of N-hydroxysuccinimidyl ester of 4′-fluorobiphenyl-4-acetic acid (52.4 mg, 0.16 mmol) in acetonitril (5 mL), while maintaining the pH between 8.0-9.0 with saturated sodium hydrogen carbonate. The mixture was stirred at room temperature for 48 h. The solvent was evaporated and the residue was reconstituted into water, and applied onto a silica reversed phase column pre-equilibrated in water. The compound was eluted with a gradient of methanol-water (0:1 to 40:100) to afford compound 9 in (52.6 mg, 70%) yields as a white fluffy solid after a final lyophilization from water. The solvent system (ethyl acetate/methanol/ water/acetic acid 10:3:3:1) was used for following up the reaction.
1H NMR (CD3OD): δ 7.60-7.45 (m, 4 H, Ar-H), 7.40-7.10 (m, 9 H, Ar-H), 4.78 (d, J = 11.4 Hz, 1 H, glycosidic CH2), 4.5 (d, J = 11.0 Hz, 1 H, glycosidic CH2), 4.03 (s, 2 H, glycolyl CH2CO), 3.96-3.92 (m, 1 H, H8b), 3.82-3.67 (m, 4 H, H4, H5, H6, H7), 3.57 (br. s, 2 H, biphCH2CONH), 3.37 (br. d, J = 9.8 Hz, 1 H, H9a), 3.20 (dd, J = 9.8, 13.7 Hz, 1 H, H9b), 2.90 (dd, J = 4.6, 12.0 Hz,1 H, H3eq), 1.64 (t, J = 12.0 Hz, 1 H, H3ax). 13C NMR (CD3OD): δ 180.2, 178.9, 174.3, 169.7, 158.12, 150.5, 141.0, 140.0, 137.1, 135.4, 131.5, 129.2, 129.1, 128.9, 128.3, 127.5, 116.7, 116.6, 114.1, 102.1, 74.0, 72.3, 71.3, 69.5, 67.4, 62.6, 61.4, 44.0, 43.4, 42.7, 26.3. MALDI-TOF MS calcd for C32H35FN2O10Na (M + Na)+, 626.23; found 626.22.
4.1.3. General procedures for reductive alkylation
To a mixture of the amine (3 or 4, 0.084 mmol) and 4′-hydroxybiphenyl-4-carbaldehyde6 (8.5 mg, 0.043 mmol) in dry MeOH (3 mL) was added acetic acid (0.1 mL), followed by NaBH3CN (1.0 M in THF, 60 μL, 0.060 mmol). After stirring at r. t. for 24 h, the reaction mixture was concentrated, reconstituted into water and loaded on a silica reversed phase column pre-equilibrated in water. The compounds were eluted with a gradient of methanol-water (0:1 to 60:100) to afford the title compounds 7 or 8 in about 68% yield as a white fluffy solid after a final lyophilization from water. The solvent system (ethyl acetate/methanol/water/acetic acid 10:3:3:1) was used for following up the reaction.
4.1.4. Benzyl 3, 5, 9- trideoxy-5-glycolamido-9-(4′-hydroxy-4-biphenyl)methylamino-d-glycero-α-d-galacto-2-nonulopyranosidonic acid (8)
1H NMR (CD3OD): δ 7.62-7.21 (m, 11 H, Ar-H), 6.86-6.83 (m, 2 H, Ar-H), 4.81 (d, J = 11.0 Hz, 1 H, glycosidic CH2), 4.71 (d, J = 11.0 Hz, 1 H, glycosidic CH2), 4.24-4.16 (m, 3 H, H8, glycolyl CH2CO), 4.05 (s, 2 H, biphCH2NH), 3.89-3.85 (m, 1 H, H4), 3.75-3.69 (m, 2 H, H5, H6), 3.40 (d d, J = 8.9, 1.8 Hz, 1 H, H7), 3.36 (dd, J = 2.8, 12.7 Hz, 1 H, H9a, ), 2.96 (br. d, J = 12.7 Hz, 1 H, H9b), 2.93 (dd, J = 4.8, 12.4 Hz,1 H, H3eq), 1.66 (t, J = 12.4 Hz, 1 H, H3ax). MALDI-TOF MS calcd for C31H37N2O10(M + H)+, 597.24; found 597.24.
4.1.5. (4-Biphenyl)methyl 3, 5, 9-trideoxy-5-glycolamido-9-(4′-hydroxy-4-biphenyl) methylamino-d-glycero-α-d-galacto-2-nonulopyranosidonic acid (9)
1H NMR (CD3OD): δ 7.58-7.38 (m, 11 H, Ar-H), 6.85-6.83 (m, 2 H, Ar-H), 4.88 (d, J = 11.7 Hz, 1 H, glycosidic CH2), 4.58 (d, J = 11.7 Hz, 1 H, glycosidic CH2), 4.15 (m, 1 H, H8), 4.04 (s, 2 H, glycolyl CH2CO), 3.99 (s, 2 H, biphCH2NH), 3.88-3.84 (m, 1 H, H4), 3.79-3.72 (m, 2 H, H5, H6), 3.40 (d d, J = 8.9, 1.4 Hz, 1 H, H7), 3.18 (dd, J = 4.8, 12.4 Hz, 1 H, H9a, ), 2.94 (d d, J = 4.8, 12.4 Hz, 1 H, H9b), 2.81 (dd, J = 8.9, 12.4 Hz,1 H, H3eq), 1.68 (t, J = 12.4 Hz, 1 H, H3ax). 13C NMR (CD3OD): δ 177.4, 158.7, 156.6, 153.3, 143.6, 132.6, 131.6, 130.3, 129.1, 128.0, 119.3, 116.8, 115.4, 104.0, 75.3, 74.7, 74.0, 72.7, 72.4, 69.9, 69.1, 68.4, 64.1, 62.6, 56.0, 53.8, 52.0, 51.2, 42.6. MALDI-TOF MS calcd for C37 H41N2O10 (M + H)+, 673.27; found 673.28.
4.2.1 Competition ELISA
The Fc region of human IgG1 fused to the N-terminal three Ig domains of murine and human CD22 (mCD22Fc and hCD22Fc, respectively) was prepared from transfectants of Lec2 cells, a mutant line of CHO cells deficient in protein sialylation, as described previously.20 Binding of synthetic sialosides to CD22 was analyzed by competition ELISA.
96-well plates (Greiner) were coated overnight with 50 μL of streptavidin (40 μg/mL) dissolved in 50 mM NaHCO3 (pH = 8 .5). Plates were then incubated with 50 μL of 4 μg/mL of biotinylated form of compound 112 in PBS for 1 h. After blocking of the wells with 0.5% BSA in PBS for 3 hr, various concentrations of synthetic sialosides were incubated for 1 h together with 0.5 μg of the CD22Fc fusion protein. After washing with PBS containing 0.05% Tween 20, CD22Fc bound to the plates was detected using alkaline phosphatase- conjugated anti-human IgG Fc antibody (Southernbiotech). Alternatively, high-binding 96-well flat bottom plates (Nunc) precoated with neutravidin (Pierce Biotechnology) at 1 μg/50 μL of PBS/well overnight at 4°C were washed with Hanks balanced salt solution (HBSS; 200 μL, 3x) and blocked with HBSS containing 0.5% BSA.
4.2.2 Competition ELISA (hCD22)
High-binding 96-well flat bottom microtiter plates (Nunc) precoated with neutravidin (Pierce Biotechnology) at 1 μg/50 μL of PBS/well overnight at 4°C were washed with Hanks balanced salt solution (HBSS; 200 μL, 3x) and blocked with HBSS containing 0.5% BSA for 30 min at 37°C. Wells were then washed with HBSS once (200 μL). Fifty microliters of the Siaα2-6LacNAc-PAA- biotinylated sialoside (0.625 μg/mL) was aliquoted into each well, and allowed to bind for 1 h at room temperature and free sialoside was washed away with HBSS (200 μL, 5x). Affinity purified F(ab')2 goat anti-human IgG (0.6 μg/mL; Jackson ImmunoResearch Laboratories), horseradish peroxidase (HRP), conjugated affinity purified F(ab')2 fragment rabbit anti-goat F(ab')2 specific (0.3 μL/ml; Jackson ImmunoResearch Laboratories) and hCD22-Fc21 (~1.25 μg/mL) were mixed in PBS and allowed to precomplex for 45 min. at room temperature. 50 μL of the inhibitor and 50 μL of the hCD22-Fc-antibody complex solution were added to each well and allowed to bind for 30 min at 37°C before washing five times with 200 μL of HBSS.Wells were developed with 100 μL of TMB solution (3,3′,5,5′-tetramethylbenzidine, Pierce). The reaction was stopped with 100 μl of 1 M H2SO4 and the OD was measured at 450 nm.
4.2.3 Competition ELISA (MAG)
As described for hCD22 but in this case fifty microliters of the Siaα2-3LacNAc-PAA-biotinylated sialoside (0.33 μM) was aliquoted into each well, and allowed to bind for 1 h at room temperature and free sialoside was washed away with HBSS (200 μL, 5x). Affinity purified F(ab')2 goat anti-human IgG (0.05 μg/mL) and HRP conjugated affinity purified F(ab')2 fragment rabbit anti-goat F(ab')2 specific (0.0167 μg/mL) were allowed to precomplex in HBSS for 45 min at room temperature. This was mixed with 1:10 sialidase treated MAC-Fc21 (~0.12 μg/ml) which had been pretreated with V. cholerae sialidase (0.4 mU/ml) for 45 min at room temperature), stopped by adding EDTA to 1mM and incubated for an additional 45 min at room temperature. The MAG-Fc-antibody complex solution was then diluted 3:2 with HBSS/BSA/EDTA, 50 μL of the inhibitor and 50 μL of the MAG-Fc-antibody complex were added to each well and allowed to bind for 30 min at 37°C before washing five times with 200 μL of HBSS. Wells were developed with 100 μL of TMB solution (3,3′,5,5′-tetramethylbenzidine, Pierce). The reaction was stopped with 100 μl of 1 M H2SO4 and the OD was measured at 450 nm.
4.3. Molecular Modeling
Manual docking and Molecular dynamics were carried out on an Intel P4 based Microsoft windows 2000 workstation using Discovery Studio Modeling 1.5 and 2.0 packages (Accelrys, Inc., San Diego, CA). The construction of the complex structure of mCD22 with the ligand was accomplished in three steps. First, the structures of 5 and 8 were built using BIP model complexes as a template as reported.6 The individual antagonists were superimposed into the BIP in the models, confirming that interactions essential for competitive binding are maintained. Then obtained complexes were then energetically minimized with 1000 iterations of “in-situ ligand minimization algorithm” using Smart Minimizer option using CHARMm force field. After the minimizations, molecular dynamics simulations were initiated using standard dynamics cascade module. Molecular dynamics cascade consisted of 1000 steps of steepest descent energy minimization and conjugate gradient energy minimization. Then, heating of 2000 steps (initial temperature 50K, final temperature 300K), equilibration of 120 ps (1fs time step, coordinates saved every 250 steps), production of 120ps (1fs time step, 300 K, NVT ensemble, nonbond cutoff 14A, switching function between 10 and 12A, coordinates saved every 150 steps) were applied. For all the above calculations, dielectric constants of 1 (interior) and 80 (exterior) were used. The representative ligand binding modes were analyzed to investigate the relative differences between interactions to understand experimental selectivity differences.
Supplementary Material
Acknowledgements
H. H. M. A. is grateful to the Egyptian Government for Ph.D. funding scholarship. This work was partly supported by the ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (Grant-in-Aid for Scientific Research to M. Kiso, No. 1710100 and No. 22380007) and CREST of JST (Japan Science and Technology Corporation) and NIH grants GM60938 and AI50143 to J. Paulson.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article can be found, in the online version, at...
References and notes
- 1.Ernst B, Magnani JL. Nature Rev. Drug Discov. 2009;8:661. doi: 10.1038/nrd2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crocker PR, Paulson JC, Varki A. Nat. Rev. Immunol. 2007;7:255. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 3.O'Reilly M, Paulson JC. Trends Pharmacol. Sci. 2009;30:240. doi: 10.1016/j.tips.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onodera T, Poe JC, Tedder TF, Tsubata T. J. Immunol. 2008;180:907. doi: 10.4049/jimmunol.180.2.907. [DOI] [PubMed] [Google Scholar]
- 5.Chen WC, Completo GC, Sigal DS, Crocker PR, Saven A, Paulson JC. Blood. 2010;115:4778. doi: 10.1182/blood-2009-12-257386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdu-Allah HHM, Tamanaka T, Yu J, Zhuoyuan Lu, Sadagopan M, b Adachi T, Tsubata T, Kelm S, Ishida H, Kiso M. J. Med. Chem. 2008;51:6665. doi: 10.1021/jm8000696. [DOI] [PubMed] [Google Scholar]
- 7.Abdu-Allah HHM, Watanabe K, Hayashizaki K, Takaku C, Tamanaka T, Takematsu H, Kozutsumi Y, Tsubata T, Ishida H, Kiso M. Bioorg. Med. Chem. Lett. 2009;19:5573. doi: 10.1016/j.bmcl.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 8.Kelm S. Results Probl. Cell Differ. 2001;33:153. doi: 10.1007/978-3-540-46410-5_9. [DOI] [PubMed] [Google Scholar]
- 9.Kelm S, Gerlach J, Brossmer R, Danzer CP, Nitschke L. J. Exp. Med. 2002;195:1207. doi: 10.1084/jem.20011783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaccai NR, Maenaka K, Maenaka T, Crocker PR, Brossmer R, Kelm S, Jones EY. Structure. 2003;11:557. doi: 10.1016/s0969-2126(03)00073-x. [DOI] [PubMed] [Google Scholar]
- 11.Gala P, Stamford A, Jenkins J, Kugelman M. Org. Process Res. Dev. 1997;1:163. [Google Scholar]
- 12.Abdu-Allah HHM, Watanabe K, Hayashizaki K, Iwayama Y, Takematsu H, Kozutsumi Y, Tsubata T, Ishida H, Kiso M. Tetrahedron Lett. 2009;50:4488. doi: 10.1016/j.bmcl.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 13.Kelm S, Brossmer R, Isecke R, Gross HJ, Strenge K, Schauer R. Eur. J. Biochem. 1998;255:663. doi: 10.1046/j.1432-1327.1998.2550663.x. [DOI] [PubMed] [Google Scholar]
- 14.Bhunia A, Schwardt O, Gaethje H, Gao G, Kelm S, Benie AJ, Hricovini M, Peters T, Ernst B. ChemBioChem. 2008;9:2941. doi: 10.1002/cbic.200800458. [DOI] [PubMed] [Google Scholar]
- 15.Shelke SV, Gao GP, Mesch S, Gathje H, Kelm S, Schwardt O, Ernst B. Bioorg. Med. Chem. 2007;15:4951. doi: 10.1016/j.bmc.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman M, Rychlewski J. Int. J. quantum chem. 2002;89:419. [Google Scholar]
- 17.Sims PA, Wong CF, Vuga D, McCammon JA, Sefton BM. J. Comput Chem. 2005;26:668. doi: 10.1002/jcc.20207. [DOI] [PubMed] [Google Scholar]
- 18.Zaccai NR, May AP, Robinson RC, Burtnick LD, Crocker PR, Brossmer R, Kelm S, Jones EY. J. Mol. Biol. 2007;365:1469. doi: 10.1016/j.jmb.2006.10.084. [DOI] [PubMed] [Google Scholar]
- 19.Mesch S, Moser D, Strasser DS, Kelm A, Cutting B, Rossato G, Vedani A, Koliwer-Brandl H, Wittwer M, Rabbani S, Schwardt O, Kelm S, Ernst B. J. Med. Chem. 2010;53:1597. doi: 10.1021/jm901517k. [DOI] [PubMed] [Google Scholar]
- 20.Naito Y, Takematsu H, Koyama S, Miyake S, Yamamoto H, Fujinawa R, Sugai M, Okuno Y, Tsujimoto G, Yamaji T, Hashimoto Y, Itohara S, Kawasaki T, Suzuki A, Kozutsumi Y. Mol. Cell Biol. 2007;27:3008. doi: 10.1128/MCB.02047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blixt O, Collins BE, van den Nieuwenhof IM, Crocker PR, Paulson JC. J. Biol. Chem. 2003;278:31007. doi: 10.1074/jbc.M304331200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.