Abstract
Phosphorylation of the α subunit of eukaryotic translation initiation factor 2 (eIF2α) at serine 51 inhibits protein synthesis in cells subjected to various forms of stress including virus infection. The human papillomavirus (HPV) E6 oncoprotein contributes to virus-induced pathogenicity through multiple mechanisms including the inhibition of apoptosis and the blockade of interferon (IFN) action. We have investigated a possible functional relationship between the E6 oncoprotein and eIF2α phosphorylation by an inducible-dimerization form of the IFN-inducible protein kinase PKR. Herein, we demonstrate that HPV type 18 E6 protein synthesis is rapidly repressed upon eIF2α phosphorylation caused by the conditional activation of the kinase. The remainder of E6, however, can rescue cells from PKR-mediated inhibition of protein synthesis and induction of apoptosis. E6 physically associates with GADD34/PP1 holophosphatase complex, which mediates translational recovery, and facilitates eIF2α dephosphorylation. Inhibition of eIF2α phosphorylation by E6 mitigates eIF2α-dependent responses to transcription and translation of proapoptotic genes. These findings demonstrate, for the first time, a role of the oncogenic E6 in apoptotic signaling induced by PKR and eIF2α phosphorylation. The functional interaction between E6 and the eIF2α phosphorylation pathway may have important implications for HPV infection and associated pathogenesis.
Eukaryotic cells respond to various stress conditions, including virus infection, in part by downregulating protein synthesis (12). This translation response is mediated largely through the phosphorylation of the α subunit of the eukaryotic translation initiation factor 2 (eIF2α) at serine 51 (28). Phosphorylated eIF2 has increased affinity for the translation initiation factor eIF2B, a guanine nucleotide exchange factor required for the recycling of eIF2-GDP to eIF2-GTP (28). Phosphorylation at serine 51 traps eIF2-GDP and eIF2B in a complex with reduced guanine nucleotide exchange factor activity. The resulting reduction in eIF2-GTP levels leads to the inhibition of the overall rate of protein synthesis (28). To date, there are four distinct eIF2α kinases that play a role in translational control by modulating eIF2 function; these are the heme-regulated inhibitor (9), the double-stranded RNA (dsRNA)-activated protein kinase PKR (36), the homologue of the Saccharomyces cerevisiae protein kinase GCN2 (38), and the endoplasmic reticulum (ER)-resident protein kinase PERK (64). Functional analyses of the eIF2α kinases have indicated that each enzyme provides the cell with a unique ability to modulate translation in response to specific types of stress (16). For example, heme-regulated inhibitor responds to heme depletion (9) and PERK mediates translational control in response to ER stress (64), whereas GCN2 is activated in response to amino acid starvation (38).
PKR is unique among the eIF2α kinase family for its ability to respond to virus infection (13, 36). The kinase is ubiquitously expressed in all cells at low levels but is transcriptionally induced by alpha/beta interferon (IFN-α/β), a family of cytokines with antiviral and antiproliferative actions that are secreted from infected cells (73). PKR consists of an N-terminal dsRNA-binding domain (dsRBD) and a C-terminal kinase domain (KD) (36). The dsRBD contains two dsRNA-binding motifs (dsRBMI and dsRBMII), which are essential for RNA binding, whereas the KD contains all 11 catalytic subdomains that are highly conserved among protein kinases (13, 36). Binding of PKR to dsRNA produced during virus replication induces dimerization and conformational changes that result in kinase activation by interphosphorylation on multiple sites (13, 36). Activated PKR then phosphorylates eIF2α at serine 51, causing the inhibition of protein synthesis. Through this capacity, the kinase functions as a mediator of the antiviral and antiproliferative actions of IFNs (73) and as an inducer of apoptosis (32). Because of the deleterious effects of the host's protein synthesis inhibition, many viruses have evolved distinct mechanisms to counteract PKR activation and eIF2α phosphorylation as a means to avoid, at least in part, the antiviral action of IFNs (21). These mechanisms include the direct inhibition of the kinase by viral proteins and/or RNAs, the downregulation of PKR protein, the regulation of eIF2α phosphorylation levels, or the control of translational pathways downstream of eIF2α phosphorylation (34, 35). In addition to translational control, PKR is implicated in signaling pathways that induce gene transcription in response to various cytokines and growth factors, virus infection, or various forms of environmental stress (12, 13, 36).
Infection with human papillomavirus (HPV) is associated with tumors in various tissues and organs and is clearly implicated in the development of cervical cancer, which has an incidence rate of ∼500,000 cases per year globally (15, 49, 87). To date, a total of almost 100 subtypes of this virus have been identified, which are divided into two groups: the low-risk HPVs, such as subtypes 6 and 11, which are rarely found in malignant tumors but induce benign genital warts, and the high-risk subtypes, such as 16 and 18, which are frequently found in cervical carcinoma (15, 49, 87). Among the several HPV proteins, E6 plays a major role in virus-mediated oncogenesis. It is a small basic protein of 151 amino acids (aa), whose major structural characteristic is the presence of two hypothetical zinc fingers (48). The best-characterized function of the high-risk E6 is the degradation of tumor suppressor p53 through ubiquitin-mediated proteolysis, a process that is dependent on the presence of a cellular protein termed E6-associated protein (48, 50). Degradation of p53 is an important mechanism by which E6 prevents apoptosis upon virus infection or exposure to ionizing radiation (48, 50). E6 can interact with and inactivate several other cellular proteins, including c-Myc, Bak, or the human homologue of the Drosophila melanogaster disks large tumor suppressor protein (48). E6 has also been shown to disrupt the transcriptional machinery through its association with the transactivator CBP/p300 protein or to induce cellular telomerase activity, a function with important implications in cellular immortalization and transformation (48, 50). Furthermore, E6 can efficiently immortalize human mammary epithelial cells and induce epithelial hyperplasia and skin tumors in transgenic mice (48, 50).
In addition to oncogenesis, E6 has a significant contribution to altering the immune response through its ability to inhibit apoptosis and suppress IFN action (41). This may account, at least in part, for the poor responsiveness of HPV-infected cells to IFN treatment in vitro and in vivo (41). Given the demonstrated role of eIF2α phosphorylation in the antiviral effects of IFNs, we were interested to examine a possible implication of E6 in eIF2α phosphorylation. Herein, we show that HPV type 18 (HPV-18) E6 protein synthesis is inhibited in response to IFN treatment or eIF2α phosphorylation by a conditionally active PKR. We also show that, despite its translational repression, E6 is capable of counteracting the inhibitory effect of eIF2α phosphorylation on cellular protein synthesis. We provide evidence that this is possible due to the ability of E6 to interact with the GADD34/PP1 holophosphatase complex and promote dephosphorylation of eIF2α. Furthermore, we demonstrate that inhibition of eIF2α phosphorylation by HPV-18 E6 prevents PKR-mediated apoptosis through the inhibition of expression of proapoptotic genes. The significance of these findings is underscored by our data showing that the antiapoptotic function is a property of the high-risk HPV-18 E6 but not of low-risk HPV-11 E6, thus supporting a role for the PKR-eIF2α phosphorylation pathway in virus-mediated tumorigenesis.
MATERIALS AND METHODS
Plasmids.
FLAG-tagged HPV-18 E6 or HPV-11 E6 in pcDNA3.1/Zeo vector (Invitrogen, Inc.) was constructed as described previously (46). The construction of GyrB.PKR cDNAs in pSG5 vector (Stratagene, Inc.) was previously described (78), and the generation and characterization of FLAG-GADD34 constructs were also described elsewhere (6).
Cell culture and IFN treatment.
The human fibrosarcoma HT1080 cells (ATCC CCL-121) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum and antibiotics. The generation and characterization of HT1080 cells expressing FLAG-HPV-18 E6 were previously described (46). To generate HT1080 cells expressing GyrB.PKR and E6 proteins, cells were transfected with either GyrB.PKR or GyrB.PKRK296H cDNA in pSG5 vector together with either FLAG-HPV-18 E6 or FLAG-HPV-11 E6 in pcDNA3.1/Zeo vector at a ratio of 5:1. In control transfections, GyrB.PKR cDNAs with pcDNA3.1/Zeo vector were used. Transfection was performed with Lipofectamine reagent (Invitrogen, Inc.) according to the manufacturer's instructions. Cells were selected in the presence of 200 μg of zeocin (Invitrogen)/ml, and clones were isolated, expanded, and characterized as described previously (46). For IFN treatment, 1,000 IU of human IFN-α2b (Intron A; Schering-Plough Corp.)/ml or 5 ng of human IFN-γ (Biosource International)/ml was used.
Microarray analysis.
For cDNA microarray analysis, total RNA (10) was used for hybridization of the Affymetrix human U133A cDNA chip, which covers 22,000 genes, as described previously (53). For each cell line, the values obtained after coumermycin treatment were normalized to those in the absence of the antibiotic. We focused on those genes whose expression was either induced or suppressed a minimum of fivefold in GyrB.PKR-expressing cells and remained unaffected in GyrB.PKRK296H-expressing cells (less than threefold induction or suppression) after coumermycin treatment.
Protein extraction, immunoblotting, immunoprecipitation, and pull-down assays.
Cells were washed twice with ice-cold phosphate-buffered saline (PBS) solution (140 mM NaCl, 15 mM KH2PO4 [pH 7.2], 2.7 mM KCl), and proteins were extracted with a lysis buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 2 mM MgCl2, 1% Triton X-100, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride, 3 μg of aprotinin/ml, 1 μg of leupeptin/ml, and 1 μg of pepstatin/ml. After incubation on ice for 20 min, the lysates were centrifuged at 14,000 × g for 10 min at 4°C. The supernatant was transferred to a fresh tube, and the protein concentration was measured by the Bradford assay (Bio-Rad). Samples were stored at −85°C.
Proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or two-dimensional (2D) gel electrophoresis and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore Corp.). Immunoblottings were performed according to the standard protocol (66). The primary antibodies (Abs) were as follows: anti-FLAG M2 mouse monoclonal Ab (Sigma; 2 μg/ml), anti-human PKR mouse monoclonal Ab (clone F9 or E8 [47], 1 μg/ml), anti-GyrB mouse monoclonal Ab (clone 7D3; John Innes Enterprises; 0.2 μg/ml), anti-human eIF2α rabbit polyclonal Ab (Cell Signaling; 1 μg/ml), rabbit serum to phosphoserine 51 of eIF2α (45) (1 μg/ml), anti-actin mouse monoclonal immunoglobulin G (IgG; ICN; 0.1 μg/ml), anti-Bik mouse monoclonal Ab (clone C33-1; BD Biosciences; 1 μg/ml), and anti-p53 mouse monoclonal Ab (Ab-2; Oncogene Science; 1 μg/ml). The following Abs were used from the APOPTOPAK miniature set from Upstate Biotechnology Inc.: anti-Bcl-2 mouse monoclonal Ab (clone 124; 1 μg/ml), anti-Bak rabbit polyclonal Ab (1 μg/ml), and anti-Bax rabbit polyclonal Ab (1 μg/ml). The following Abs were purchased from Santa Cruz Biotechnology: anti-GADD153 (C/EBP homologous protein [CHOP]) rabbit polyclonal Ab (sc-575; 1 μg/ml), anti-PP1 mouse monoclonal Ab (sc-7482; 2 μg/ml), and anti-FAS rabbit polyclonal Ab (sc-715; 1 μg/ml). The secondary Abs were horseradish peroxidase-conjugated anti-mouse Ab or horseradish peroxidase-conjugated anti-rabbit Ab (dilution, 1:1,000; Amersham Pharmacia Biotech). Proteins were visualized using the enhanced chemiluminescence detection system according to the instructions of the manufacturer (Perkin-Elmer Life Sciences, Inc.). Quantification of the bands in the linear range of exposure was performed by densitometry using NIH Image 1.54 software. Immunoprecipitations and glutathione S-transferase (GST) pull-down assays were performed as described previously (46). For GADD34 immunoprecipitation, 2 μg of anti-GADD34 rabbit polyclonal Ab (sc-8327) per 500 mg of protein extract was used.
In vivo 35S labeling, isoelectric focusing (IEF), and 2D gel electrophoresis.
Cells were treated with coumermycin in DMEM plus 10% calf serum for the appropriate time. Then, the medium was changed to DMEM lacking methionine and supplemented with 10% dialyzed fetal bovine serum for 2 h. Tran35S-label (ICN) was then added to the cells at a concentration of 100 μCi/106 cells, and culture was continued for an additional 2 h in the presence of coumermycin. Protein extracts prepared as described above were subjected to SDS-PAGE, and radioactive proteins were visualized by autoradiography.
For 2D gel electrophoresis, protein lysates were prepared using a specific lysis buffer {8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 65 mM DTT, 0.5% (vol/vol) immobilized pH gradient (IPG) buffer, (Amersham 17-6000-86) pH 4 to 7. The first dimension (IEF) was performed using the Ettan IPGphor IEF unit (Amersham) and 7-cm strips, pH 4 to 7 (14, 19). The isoelectric gels were first passively rehydrated in a total volume of 125 μl of rehydration solution (8 M urea, 2% [wt/vol] CHAPS, 10 mM DTT, 0.5% [vol/vol] IPG buffer [pH 4 to 7], trace of bromophenol blue) for 10 h (67). After the rehydration phase, IEF was performed at 150 V for 40 min, 500 V for 40 min, 1,000 V for 40 min, and 5,000 V for 3 h. After IEF, gels were incubated in equilibration buffer I (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, 1% [wt/vol] DTT, trace of bromophenol blue) for 12 min and in equilibration buffer II (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, 2.5% [wt/vol] iodoacetamide, trace of bromophenol blue) for 5 min prior to separation by SDS-PAGE (second dimension).
eIF2α dephosphorylation assay.
Purified recombinant histidine-tagged eIF2α was prepared as described previously (86) and phosphorylated by a purified GST fusion protein of human PKR in vitro based on a previously published protocol (86). The unincorporated [γ-32P]ATP was removed using MicroSpin G-25 columns (Amersham Pharmacia Biotech). An aliquot of 32P-eIF2α was then incubated with anti-FLAG immunoprecipitates and subjected to dephosphorylation as described previously (54). Phosphorylated eIF2α was detected by SDS-PAGE and autoradiography.
Cell staining and flow cytometry analysis.
Cells were prepared for flow cytometry analysis as described previously (81) with a few modifications. Briefly, approximately 106 cells per 10-cm-diameter dish were detached in PBS plus 0.1 mM EDTA and washed in ice-cold PBS. Following centrifugation at 900 × g for 5 min, cells were suspended in 0.5 ml of cold PBS and fixed by adding 4.5 ml of ice-cold ethanol dropwise with gentle mixing. Fixed cells were stored at −20°C for at least 8 h. For staining, pelleted cells were washed once with PBS and suspended in 0.5 ml of PBS containing 50 μg of propidium iodide (Sigma)/ml and 20 μg of RNase (Sigma)/ml. Cells were incubated at 37°C for 30 min and maintained at 4°C for 8 h before being subjected to flow cytometry analysis on a FACScan cell sorter.
RESULTS
Inhibition of HPV-18 E6 protein synthesis by IFN-α.
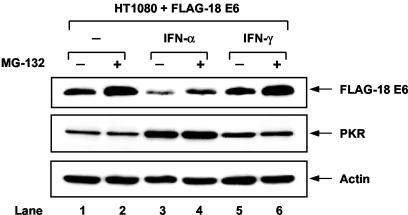
We previously generated a human epithelial cell-like fibrosarcoma HT1080 cell line carrying a FLAG-tagged form of the high-risk HPV-18 E6 (46). When HT1080 cells were treated with IFN-α, we noticed a decrease in FLAG-HPV-18 E6 expression levels compared to those in untreated cells (Fig. 1, top panel, compare lane 3 with lane 1). Inhibition of FLAG-HPV-18 E6 expression, however, was not observed in cells treated with IFN-γ (lane 5). Since E6 protein stability is controlled by the 26S proteasome (37), we tested whether inhibition of FLAG-HPV-18 E6 by IFN-α involved the proteasome-dependent degradation of the viral protein. Incubation of HT1080 cells with the proteasome inhibitor MG-132 increased FLAG-HPV-18 E6 levels equally in untreated and in IFN-α- or IFN-γ-treated cells (top panel, lanes 2, 4, and 6). However, MG-132 failed to completely restore FLAG-18E6 levels in IFN-α-treated cells (lane 4) as opposed to untreated (lane 2) or IFN-γ-treated (lane 6) cells, suggesting that the inhibitory effect of IFN-α is not exclusively based on the proteasome-mediated degradation of the viral protein. Since FLAG-HPV-18 E6 expression in HT1080 cells was mediated by a heterologous promoter, whose activity is not affected by IFNs (46), our findings implied a translational regulation of FLAG-HPV-18 E6 by IFN-α. This notion was supported by our observation that downregulation of FLAG-HPV-18 E6 was associated with an induction of the eIF2α kinase PKR protein in IFN-α-treated cells (Fig. 1, middle panel, lanes 3 and 4).
FIG. 1.
E6 protein levels are downregulated upon treatment with IFN-α. HT1080 cells expressing FLAG-tagged HPV-18 E6 were left untreated (lanes 1 and 2) or treated with either IFN-α (lanes 3 and 4) or IFN-γ (lanes 5 and 6) for 18 h followed by treatment with 40 μM MG-132 for an additional 2 h (lanes 2, 4, and 6). Protein extracts (50 μg) were subjected to immunoblot analysis with anti-FLAG monoclonal Ab (top panel), anti-human PKR monoclonal Ab (clone F9) (middle panel), and antiactin monoclonal Ab (bottom panel).
E6 protein synthesis is controlled by eIF2α phosphorylation.
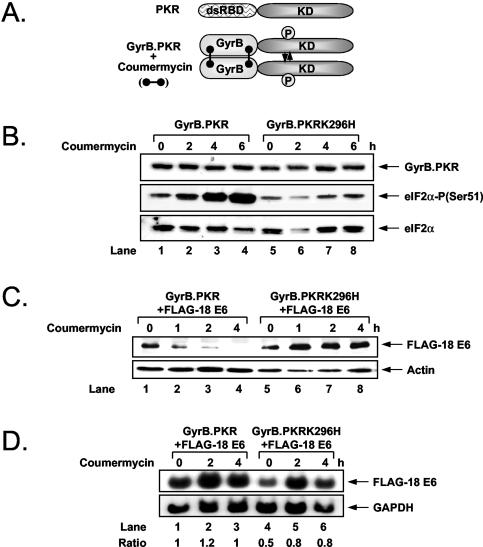
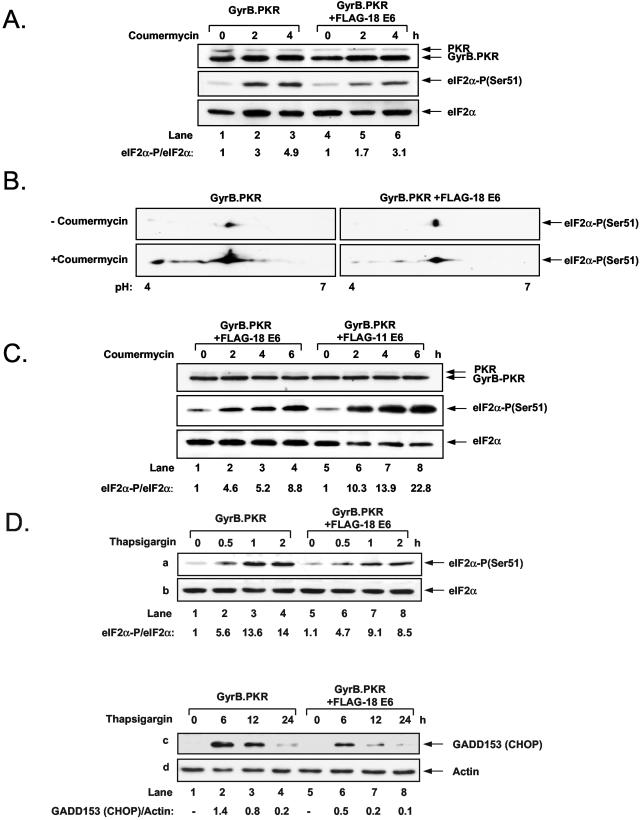
To establish a direct link between E6 protein synthesis and PKR, we employed an alternative approach to induce the activity of the kinase by expressing the KD of PKR as a fusion protein with the first 220 aa of Escherichia coli GyrB protein (Fig. 2A). Using this approach, Ung et al. previously demonstrated that chemical cross-linking of the GyrB domain within cells by the drug coumermycin resulted in the dimerization and activation of the fusion protein, leading to the induction of eIF2α phosphorylation and inhibition of protein synthesis (78). We generated HT1080 cells stably expressing either wild-type (wt) PKR or the catalytically inactive PKRK296H mutant as a fusion protein with GyrB. Immunoblot analysis with an anti-GyrB Ab showed equal expression of GyrB.PKR and GyrB.PKRK296H (Fig. 2B, top panel). When cells were treated with coumermycin, we found that phosphorylation of endogenous eIF2α was highly induced in GyrB.PKR-expressing cells but not in GyrB.PKRK296H-expressing cells (Fig. 2B, middle panel).
FIG. 2.
Inhibition of E6 protein synthesis by GyrB.PKR activation and eIF2α phosphorylation. (A) Schematic representation of PKR and GyrB.PKR proteins. The dsRBD of PKR was replaced by the GyrB domain, which mediates the dimerization of the chimera protein in the presence of coumermycin. This leads to the activation of GyrB.PKR, which in turn mimics the effects of wt PKR (78). (B) HT1080 cells expressing either GyrB.PKR (lanes 1 to 4) or GyrB.PKRK296H (lanes 5 to 8) were treated with 100 ng of coumermycin/ml for the indicated times. Protein extracts (50 μg) were subjected to immunoblotting with anti-GyrB monoclonal Ab (top panel), anti-eIF2α phosphoserine 51-specific rabbit Ab (middle panel), or anti-eIF2α rabbit polyclonal Ab (bottom panel). (C) HT1080 cells expressing FLAG-HPV-18 E6 (lanes 1 to 8) together with either GyrB.PKR (lanes 1 to 4) or GyrB.PKRK296H (lanes 5 to 8) were incubated with 100 ng of coumermycin/ml for up to 4 h. Immunoblot analysis of 50 μg of protein extracts with either anti-FLAG or antiactin monoclonal Ab is shown in the top or bottom panel, respectively. (D) HT1080 cells expressing FLAG-HPV-18 E6 and either GyrB.PKR (lanes 1 to 3) or GyrB.PKRK296H (lanes 4 to 6) were subjected to Northern blot analysis using 10 μg of total RNA followed by hybridization with either 32P-labeled HPV-18 E6 cDNA (top panel) or 32P-labeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA (bottom panel) as previously described (46). The radioactive bands were quantified by densitometry, and the ratio of FLAG-HPV-18 E6 to glyceraldehyde-3-phosphate dehydrogenase is shown.
Given that the GyrB.PKR system was functional in HT1080 cells, we further established cells expressing FLAG-HPV-18 E6 together with either GyrB.PKR or GyrB.PKRK296H (Fig. 2C). Treatment of these cells with coumermycin resulted in a rapid repression of FLAG-HPV-18 E6 levels in GyrB.PKR-expressing cells but not in GyrB.PKRK296H-expressing cells (Fig. 2C, top panel, compare lanes 1 to 4 with lanes 5 to 8). Northern blot analysis showed that 18E6 mRNA levels were not diminished by coumermycin treatment of GyrB.PKR- or GyrB.PKRK296H-expressing cells (Fig. 2D). Similar results were obtained with HT1080 cells expressing GyrB.PKR and a FLAG-tagged form of the low-risk HPV-11 E6 (data not shown). Collectively, these data suggested that E6 expression is translationally suppressed by PKR-mediated eIF2α phosphorylation.
High-risk 18E6 blocks PKR-mediated apoptosis.
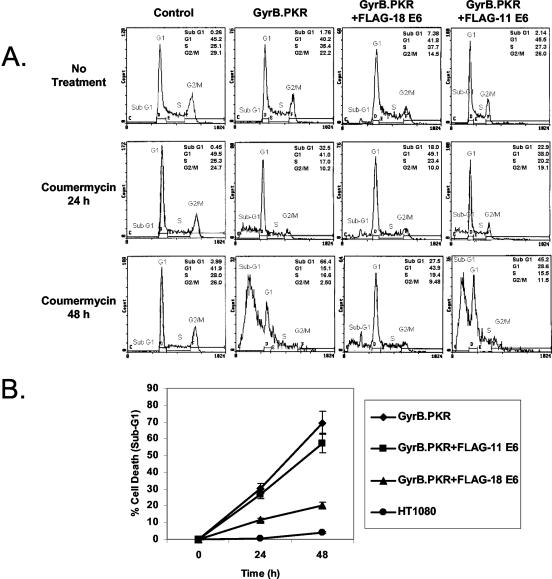
The inhibition of E6 protein synthesis by GyrB.PKR prompted us to examine a possible role of the viral protein in the biological effects of PKR activation. Considering that activation of PKR promotes apoptosis (75), we assessed the induction of death in HT1080 cells expressing GyrB.PKR in the absence or presence of either FLAG-HPV-18 E6 or FLAG-HPV-11 E6 (Fig. 3). When control HT1080 cells (i.e., cells transfected with the expression vector bearing the zeocin-resistant gene only) were treated with coumermycin, we observed that neither the growth (data not shown) nor the viability of the cells was affected by the presence of the antibiotic (Fig. 3A, leftmost panels). On the other hand, HT1080 cells expressing GyrB.PKR underwent massive death (see increased sub-G1 population) after treatment with coumermycin (panels second from the left). Interestingly, death was significantly lower in coumermycin-treated cells expressing GyrB.PKR and FLAG-HPV-18 E6 (Fig. 3A, panels third from the left). In contrast to this, the percentage of dead cells was not diminished in GyrB.PKR-expressing cells expressing FLAG-HPV-11 E6 compared to GyrB.PKR-expressing cells after treatment with coumermycin (Fig. 3A, rightmost panels). Quantitative analysis of cell death induced by GyrB.PKR activation in the absence or presence of E6 proteins is shown in Fig. 3B. Collectively, these findings clearly demonstrated the killing potential of activated GyrB.PKR and the ability of the high-risk HPV-18 E6 only to rescue cells from PKR-mediated death.
FIG. 3.
Control of GyrB.PKR-mediated cell death by E6. (A) HT1080 control cells (i.e., expressing the zeocin-resistant gene only) and cells expressing either GyrB.PKR alone or GyrB.PKR in the presence of either FLAG-HPV-18 E6 or FLAG-HPV-11 E6 were treated with 100 ng of coumermycin/ml for 24 or 48 h. Cells were harvested, fixed in ethanol, stained with propidium iodide, and subjected to flow cytometry analysis. The percentages of apoptotic cells or cells in various phases of the cell cycle are indicated. Data represent one of four reproducible experiments. (B) Quantification of cell death. The values represent the average percentages of cell death (sub-G1 population) for each cell line treated with coumermycin from three independent experiments.
High-risk 18E6 impairs translational control by eIF2α phosphorylation.
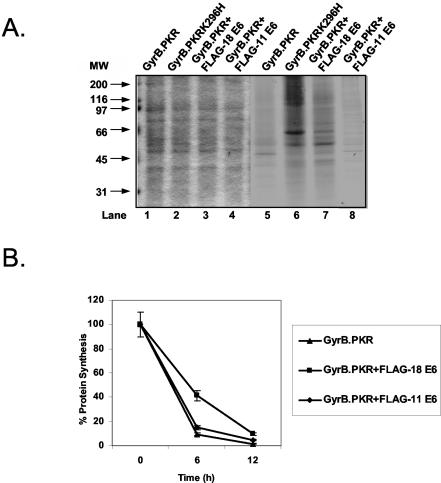
Since PKR-mediated cell death is tightly associated with protein synthesis inhibition (75), we next sought to examine the regulation of protein synthesis in HT1080 cells expressing GyrB.PKR in the absence or presence of the E6 proteins. Specifically, coumermycin-treated cells were labeled with [35S]methionine, and protein extracts were subjected to SDS-PAGE and autoradiography (Fig. 4A, lanes 5 to 8). We observed that, although total protein load measured by Coomassie blue staining was equal in all cells (Fig. 4A, lanes 1 to 4), treatment with coumermycin drastically inhibited protein synthesis in cells with GyrB.PKR (lane 5) as opposed to cells bearing the catalytic mutant GyrB.PKRK296H (lane 6). Interestingly, the presence of FLAG-HPV-18 E6 relieved the inhibition of protein synthesis by GyrB.PKR significantly (compare lane 7 with lane 5). In contrast to FLAG-HPV-18 E6, expression of FLAG-HPV-11 E6 did not affect inhibition of protein synthesis by GyrB.PKR (compare lane 8 with lane 5). To better assess the differences in protein synthesis, cells were labeled with [35S]methionine and radioactive proteins were extracted and quantified (Fig. 4B). We observed that protein synthesis was inhibited after 6 or 12 h of coumermycin treatment of GyrB.PKR-expressing cells or GyrB.PKR-expressing cells expressing FLAG-HPV-11 E6. On the other hand, protein synthesis was still reduced in GyrB.PKR-expressing cells expressing FLAG-HPV-18 E6 but to a lesser extent than in GyrB.PKR-expressing cells or GyrB.PKR-expressing cells containing FLAG-HPV-11 E6 particularly at 6 h after coumermycin treatment. These data showed the ability of FLAG-HPV-18 E6 to relieve the translational blockade induced by PKR-mediated eIF2α phosphorylation.
FIG. 4.
Regulation of GyrB.PKR-mediated inhibition of protein synthesis by E6. (A) HT1080 cells expressing GyrB.PKR alone, GyrB.PKRK296H alone, or GyrB.PKR together with either FLAG-HPV-18 E6 or FLAG-HPV-11 E6 were treated with 100 ng of coumermycin/ml for 10 h followed by labeling in vivo with [35S]methionine for an additional 2 h. Protein extracts (50 μg) were subjected to SDS-PAGE. Total protein was visualized by Coomassie blue staining (lanes 1 to 4), whereas radioactive proteins were detected by autoradiography (lanes 5 to 8). MW, molecular weight in thousands. (B) HT1080 cells were left untreated or treated with 100 ng of coumermycin/ml for 4 or 10 h followed by [35S]methionine labeling for an additional 2 h. The radioactive proteins were quantified in 10 μg of total protein extract after trichloroacetic acid precipitation and counting (66). Values represent the average percentages of protein synthesis (i.e., [35S]methionine incorporation) calculated from two independent experiments performed in triplicate.
To address the mechanism of translational control by E6, we assessed the phosphorylation levels of eIF2α in GyrB.PKR-expressing cells lacking or expressing FLAG-HPV-18 E6 (Fig. 5A). Immunoblot analysis with phosphospecific Abs to serine 51 of eIF2α showed that phosphorylation was significantly reduced in coumermycin-treated cells expressing GyrB.PKR and FLAG-HPV-18 E6 compared to cells expressing GyrB.PKR only (Fig. 5A, middle panel). We further verified this finding by testing eIF2α phosphorylation by IEF, 2D gel electrophoresis, and immunoblotting with phosphoserine 51-specific anti-eIF2α Abs (Fig. 5B). We found that eIF2α phosphorylation was highly induced in GyrB.PKR-expressing cells after coumermycin treatment. However, the levels of phosphorylated eIF2α induced in coumermycin-treated GyrB.PKR-expressing cells containing FLAG-HPV-18 E6 were lower than in GyrB.PKR-expressing cells lacking the viral oncoprotein. We also noticed that several species of phosphorylated eIF2α were recognized by the phosphospecific Ab based on their migration to acidic pH upon coumermycin treatment. These data indicated that activation of GyrB.PKR leads to hyperphosphorylation of eIF2α at multiple sites including serine 51. It is noteworthy that, although serine 51 is the only residue directly phosphorylated by PKR (22), hyperphosphorylation of eIF2α is indirect, most probably due to the ability of GyrB.PKR to activate pathways leading to multiple phosphorylation of eIF2α. Nevertheless, the above data clearly demonstrated the inhibitory effect of FLAG-HPV-18 E6 on GyrB.PKR-mediated eIF2α phosphorylation. We also compared eIF2α phosphorylation in GyrB.PKR-expressing cells containing either FLAG-HPV-18 E6 or FLAG-HPV-11 E6 (Fig. 5C). We found that the induction of eIF2α phosphorylation by activated GyrB.PKR was higher in cells expressing FLAG-HPV-11 E6 than in cells expressing FLAG-HPV-18 E6 (middle panel, compare lanes 5 to 8 with 1 to 4), indicating a higher capacity of FLAG-HPV-18 E6 than of FLAG-HPV-11 E6 to inhibit eIF2α phosphorylation.
FIG. 5.
E6 impairs eIF2α phosphorylation in response to GyrB.PKR activation or ER stress. (A) HT1080 cells expressing GyrB.PKR alone (lanes 1 to 3) or in the presence of FLAG-HPV-18 E6 (lanes 4 to 6) were treated by 100 ng of coumermycin/ml for 2 h (lanes 2 and 5) or 4 h (lanes 3 and 6). Protein extracts (50 μg) were subjected to immunoblot analyses with anti-human PKR monoclonal Ab (clone E8) (top panel), anti-eIF2α phosphoserine 51-specific rabbit polyclonal Ab (middle panel), or anti-eIF2α polyclonal Ab (bottom panel). The slower-migrating band recognized by the anti-PKR monoclonal Ab (top panel) is the fusion GyrB.PKR protein, which is slightly smaller in size than endogenous PKR. The ratio of phosphorylated to total eIF2α protein is indicated. (B) Detection of eIF2α phosphorylation by IEF and 2D gel electrophoresis. Protein extracts (100 μg) of untreated or coumermycin-treated HT1080 cells (4 h, 100 ng/ml) expressing GyrB.PKR in the absence or presence of FLAG-HPV-18 E6 were subjected to IEF and 2D gel electrophoresis as described in Materials and Methods. The phosphorylated forms of eIF2α were detected by immunoblotting with anti-phosphoserine 51 eIF2α-specific Ab. The acidic (pH 4) and basic (pH 7) ends of the gels are indicated. (C) HT1080 cells containing GyrB.PKR in the presence of either FLAG-HPV-18 E6 (lanes 1 to 4) or FLAG-HPV-11 E6 (lanes 5 to 8) were treated with 100 ng of coumermycin/ml for the indicated times and subjected to immunoblotting as described for panel A. The ratio of phosphorylated to total eIF2α protein is indicated. (D) HT1080 cells expressing GyrB.PKR alone (lanes 1 to 4) or together with FLAG-HPV-18 E6 (lanes 5 to 8) were treated with 1 μM thapsigargin for short (panels a and b) or long (panels c and d) time periods. Protein extracts (50 μg) were used for immunoblot analysis with a rabbit polyclonal Ab to serine 51 of eIF2α (panel a), a rabbit polyclonal Ab to eIF2α (panel b), a rabbit polyclonal Ab to CHOP (panel c), or a mouse monoclonal Ab to actin (panel d). The values of quantified bands are indicated.
We next addressed the specificity of FLAG-HPV-18 E6 in inhibiting eIF2α phosphorylation. If eIF2α phosphorylation was generally inhibited by E6, this should also take place in cells subjected to ER stress, which induces eIF2α phosphorylation through the activation of the PKR-like ER-resident kinase PERK (25). When cells expressing either GyrB.PKR alone or GyrB.PKR and FLAG-HPV-18 E6 were treated with the ER stress inducer thapsigargin in the absence of coumermycin, induction of eIF2α phosphorylation was reduced in cells expressing the viral oncoprotein compared to cells lacking it (Fig. 5D, panel a, compare lane 2 with lane 6 and lane 3 with lane 7). It has been well established that induction of eIF2α phosphorylation in ER-stressed cells leads to the expression of CHOP, which is also known as growth arrest and DNA damage gene 153 (GADD153) (26, 69). When CHOP/GADD153 was used as a marker for responses to eIF2α phosphorylation in thapsigargin-treated cells, we found that CHOP/GADD153 protein levels were more highly induced in cells with GyrB.PKR alone than in cells with GyrB.PKR and the viral oncoprotein (Fig. 5D, panel c, compare lane 2 with lane 6 and lane 3 with lane 7). Taken together, these findings demonstrated that the ability of FLAG-HPV-18 E6 to impair eIF2α phosphorylation is not specific for the PKR pathway but can be seen in other eIF2α kinase pathways such as ER stress, which induces eIF2α phosphorylation through the activation of PERK.
HPV-18 E6 promotes eIF2α dephosphorylation.
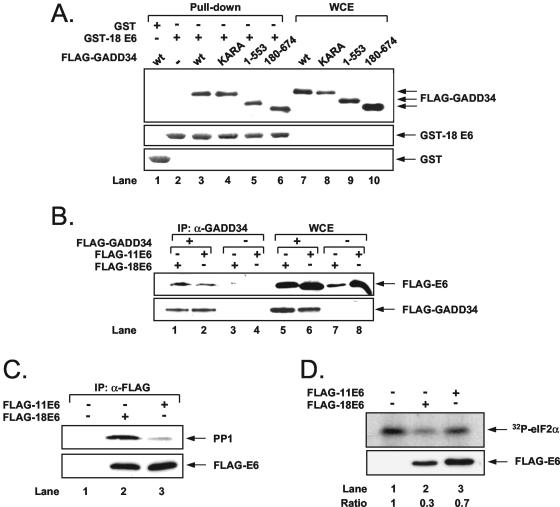
The growth arrest and DNA damage gene product 34 (GADD34) is a stress-inducible regulatory subunit of a holophosphatase complex, which contains the catalytic subunit of protein phosphatase 1 (PP1c) and specifically promotes the dephosphorylation of eIF2α in cells subjected to ER stress (6, 54, 55). Because eIF2α phosphorylation is reduced in ER-stressed cells expressing FLAG-HPV-18 E6, we hypothesized that E6 might play a role in eIF2α dephosphorylation via GADD34/PP1 (Fig. 6). To test this hypothesis, we first tested for a possible interaction between E6 and GADD34 or PP1. To this end, we used either FLAG-GADD34 or different mutants of FLAG-GADD34 with a deletion of the last 121 aa in the C terminus (1 to 553), deletion of the first 179 aa in the N terminus (180 to 674), or substitutions of the highly conserved KVRF sequence involved in PP1 binding (KARA mutant) (6). FLAG-GADD34 proteins were transiently expressed in HeLa cells, and binding to E6 was assessed in pull-down assays with a GST-18 E6 fusion protein (46) (Fig. 6A). Immunoblot analysis with anti-FLAG Ab revealed the interaction between E6 and the FLAG-GADD34 proteins independently of the type of mutation (top panel). The interaction of E6 with GADD34 was further tested in transient-transfection assays in HeLa cells. That is, coexpressed FLAG-GADD34 and FLAG-E6 proteins were subjected to immunoprecipitation with anti-GADD34 Ab followed by immunoblotting with anti-FLAG Ab (Fig. 6B). We found that both E6 subtypes were coimmunoprecipitated with GADD34 (lanes 1 and 2), thus confirming the interaction. We also examined the ability of E6 to interact with PP1. To this end, FLAG-E6 proteins transiently expressed in HeLa cells were immunoprecipitated with anti-FLAG Ab followed by immunoblotting with anti-PP1 Ab (Fig. 6C). We observed that a higher amount of endogenous PP1 was bound to FLAG-HPV-18 E6 than to FLAG-HPV-11 E6 (compare lanes 2 and 3). It is noteworthy that in this experiment the amount of transfected FLAG-HPV-18 E6 DNA was fivefold higher than that of FLAG-HPV-11 E6 DNA in order to achieve equal levels of expression of the two viral proteins. These data suggested that PP1 interacts more efficiently with FLAG-HPV-18 E6 than with FLAG-HPV-11 E6. To verify the significance of the interaction, we performed an eIF2α dephosphorylation assay by incubating FLAG-E6 immunoprecipitates with 32P-labeled eIF2α in vitro (54). We found that a higher amount of 32P-eIF2α was dephosphorylated by immunoprecipitated FLAG-HPV-18 E6 than by FLAG-HPV-11 E6 (Fig. 6D, lanes 2 and 3), suggesting that 18E6 promotes eIF2α dephosphorylation.
FIG. 6.
E6 recruits the GADD34/PP1 complex and enhances eIF2α dephosphorylation. (A) HeLa cells were transiently transfected with FLAG-GADD34 (lanes 1, 3, and 7) or FLAG-GADD34 mutants bearing either deletions (lanes 5, 6, 9, and 10) or the KARA mutation in the PP1 binding site (lanes 4 and 8). Protein extracts (500 μg) were used in pull-down assays with 1 μg of purified GST alone (lane 1) or GST-18E6 (lanes 2 to 6). Whole-cell extracts (WCE; 25 μg of protein) from each transfection were used as positive controls (lanes 7 to 10). GADD34 proteins were detected by immunoblotting with anti-FLAG monoclonal Ab (top panel) whereas GST proteins were visualized by Coomassie blue staining of SDS-polyacrylamide gels (middle and bottom panels). (B) HeLa cells were transfected with 1 μg of either FLAG-HPV-18 E6 DNA (lanes 1, 3, 5, and 7) or FLAG-HPV-11 E6 (lanes 2, 4, 6, and 8) in the presence of 1 μg of pcDNA3 vector DNA (lanes 3, 4, 7, and 8) or 1 μg of FLAG-GADD34 cDNA (lanes 1, 2, 5, and 6). Protein extracts (500 μg) were then subjected to immunoprecipitation with anti-GADD34 polyclonal Ab followed by immunoblotting with anti-FLAG monoclonal Ab to detect the levels of the viral proteins (top panel) or GADD34 (bottom panel). (C) HeLa cells were transiently transfected with 2 μg of pcDNA3 vector DNA (lane 1), 2 μg of FLAG-HPV-18 E6 DNA (lane 2), or 0.4 μg of FLAG-HPV-11 E6 DNA and 1.6 μg of pcDNA3 vector (lane 3). Protein extracts (500 μg) were subjected to immunoprecipitation with anti-FLAG monoclonal Ab. Half of the immunoprecipitates were immunoblotted for endogenous PP1 with a mouse monoclonal Ab to the catalytic subunit of the phosphatase (top panel) whereas the other half were immunoblotted with an anti-FLAG monoclonal Ab for detection of viral protein levels (bottom panel). (D) Protein extracts (500 μg) from HeLa cells transfected as described for panel C were subjected to immunoprecipitation with anti-FLAG monoclonal Ab. The immunoprecipitates were then subjected to dephosphorylation of 32P-labeled histidine-tagged eIF2α as described in Materials and Methods. Half of the reaction mixture was used to detect eIF2α phosphorylation by autoradiography (top panel) whereas the other half was used to detect E6 protein levels in the immunoprecipitates by immunoblotting with anti-FLAG monoclonal Ab (bottom panel).
HPV-18 E6 inhibits Bax induction by activated PKR.
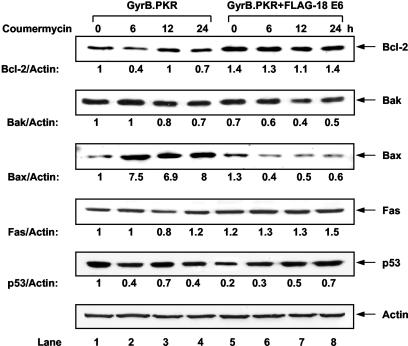
Considering the antiapoptotic role of FLAG-HPV-18 E6 in coumermycin-treated GyrB.PKR-expressing cells, we next sought to identify proteins implicated in the antiapoptotic function of E6 (Fig. 7). Oncogenic forms of E6 were shown to activate Bcl-2 and inactivate p53, Bak, or Bax (48), whereas induction of apoptosis by PKR was found to be associated with an increase in Bax and Fas protein synthesis (2, 17, 32). Immunoblot analysis showed that expression of the antiapoptotic Bcl-2 was resistant to GyrB.PKR activation (top panel, lanes 1 to 4), although its overall protein levels were elevated in cells expressing FLAG-HPV-18 E6 (top panel, lanes 5 to 8). On the other hand, expression of the proapoptotic Bak (panel second from the top) was not affected by the induction of GyrB.PKR (lanes 1 to 4), nor was its expression impaired by the presence of FLAG-HPV-18 E6 (lanes 5 to 8). In contrast to the above proteins, the proapoptotic Bax (third panel from the top) was highly induced upon GyrB.PKR activation (lanes 1 to 4). Significantly, Bax induction did not take place in cells expressing FLAG-HPV-18 E6 (lanes 5 to 8), suggesting a specific regulation of this protein in GyrB.PKR-mediated apoptosis. Unlike Bax, the proapoptotic Fas protein (fourth panel from the top) was not affected significantly by GyrB.PKR in either the absence or the presence of FLAG-HPV-18 E6. When we probed for p53, we found that its protein levels were reduced by 80% in cells expressing FLAG-HPV-18 E6 prior to GyrB.PKR activation (compare lane 1 with lane 5). This effect was most likely due to the proteasome-dependent degradation of the tumor suppressor protein by E6 in HT1080 cells (23). We also noticed that p53 protein levels were upregulated in coumermycin-treated GyrB.PKR-expressing cells containing FLAG-HPV-18 E6 (lanes 5 to 8). Since FLAG E6 protein synthesis is rapidly repressed by activated GyrB.PKR (Fig. 2C), the most conceivable explanation is that downregulation of FLAG-HPV-18 E6 contributes to stabilization of p53. From the above data, we concluded that FLAG-HPV-18 E6 specifically targets Bax protein in cells with activated PKR.
FIG. 7.
Expression of anti- or proapoptotic proteins in response to eIF2α phosphorylation. HT1080 cells expressing GyrB.wtPKR alone (lanes 1 to 4) or together with FLAG-HPV-18 E6 (lanes 5 to 8) were induced with 100 ng of coumermycin/ml for up to 24 h. Protein extracts (50 μg) were subjected to immunoblot analysis with Abs to the indicated proteins.
Transcriptional responses induced by eIF2α phosphorylation are mitigated by HPV-18 E6.
In addition to protein synthesis, induction of eIF2α phosphorylation can control gene transcription in response to diverse stressful conditions (12, 25). Based on this, we sought to identify genes that are transcriptionally regulated by GyrB.PKR in the absence or presence of E6. To this end, GyrB.PKR-expressing cells were subjected to cDNA microarray analysis using the human U133A DNA chip from Affymetrix, which contains 22,000 genes (53). Genes that were either induced or suppressed more than fivefold in coumermycin-treated GyrB.PKR-expressing cells are shown in Table 1. Among the nine genes suppressed by GyrB.PKR, some have been clearly implicated in cell cycle progression, such as the cyclins E1 and E2 (58) and the gene for centromere-associated protein E (CENPE) (83); in DNA repair, such as the radiation sensitivity protein RAD54L (44); or in cell signaling, such as the Rho GDP dissociation inhibitor beta (RhoGDIβ) (56), the regulator of G-protein signaling 4 (RSG4) (52), and peroxiredoxin 1 (PRDX1) (82). On the other hand, among the 22 genes induced by GyrB.PKR, some encode proteins involved in apoptosis, such as the growth arrest and DNA damage genes (GADD45) A and B (72), the natural-born killer and BH3-only Bcl-2 homologous protein (NBK/BIK) (61), and the IFN regulatory factor 1 (IRF1) (77) as well as the forkhead transcription factor FOXO3A/FKHRL1 (7). Interestingly, regulation of expression of these genes by GyrB.PKR was significantly mitigated by FLAG-HPV-18 E6 and to a much lesser extent by FLAG-HPV-11 E6 (Table 1). These data provided strong evidence for a role of E6 in gene transcription induced by the activation of the PKR-eIF2α phosphorylation pathway.
TABLE 1.
Genes regulated by coumermycin in GyrB.PKR-expressing cells
| Gene | Function | Regulation (fold) by coumermycin in cell line expressing:
|
Reference(s) | |||
|---|---|---|---|---|---|---|
| GyrB.PKR | GyrB.PKRK296H | GyrB.PKR + FLAG-HPV-18 E6 | GyrB.PKR + FLAG-HPV-11 E6 | |||
| Human homologue of yeast YA22 (HYA22) | Putative tumor suppressor | −29.3 | −1.5 | −1.9 | −10.1 | 31, 60 |
| Human homologue of rat pituitary tumor-transforming gene (PTTG3) | Proto-oncogene, regulator of cell proliferation and survival | −20.0 | 1.2 | −2.2 | −1.9 | 30, 63 |
| Cyclin E2 (CCNE2) | Cell cycle, overexpressed in human cancers | −13.5 | −1.4 | −2.5 | −2.6 | 58 |
| Radiation sensitivity 54L (RAD54L) | Double-stranded DNA-dependent ATPase, DNA repair, putative tumor suppressor | −12.8 | −1.8 | −1.2 | −3.3 | 44 |
| Rho GDP dissociation inhibitor (GDI) beta (ARHGDIB) | Extracellular signaling, cyto-skeleton organization, cell mobility | −12.0 | 1.0 | −1.3 | −3.4 | 56 |
| Regulator of G-protein signaling 4 (RGS4) | Regulation of G-protein function | −6.6 | 1.3 | −1.7 | −1.2 | 52 |
| Cyclin E1 (CCNE1) | Cell cycle, G/S transition | −5.6 | −2.0 | −1.4 | −1.3 | 58 |
| Peroxiredoxin 1 (PRDX1) | Redox regulation, cell signaling, and apoptosis | −5.3 | −1.0 | −1.1 | −1.9 | 82 |
| Centromere-associated protein E (CENPE) | Spindle microtubule organization, mitotic checkpoint signaling | −5.1 | 1.0 | −1.1 | −1.8 | 83 |
| Lymphotoxin beta (LT-beta; tumor necrosis factor superfamily, member 3) | Apoptosis, inflammatory responses | 5.4 | 1.4 | −1.9 | −1.4 | 79 |
| RNA polymerase III subunit 62 (RPC62) | Transcription of short mRNAs, involved in tRNA splicing | 5.5 | 1.4 | 1.4 | 2.9 | 70 |
| Methylthioadenosine phosphorylase (MTAP) | Putative tumor suppressor | 5.6 | −1.4 | −1.1 | 13.6 | 11, 18 |
| UV radiation resistance-associated gene (UVRAG) | Complement to UV sensitivity of xeroderma pigmentosum complementation group C | 5.7 | 1.1 | 1.1 | 1.5 | 59 |
| Protein kinase AMP-activated, beta-1 noncatalytic subunit (PRKAB1) | Adaptation to metabolic stress | 5.8 | −1.3 | 1.5 | 2.2 | 80 |
| Keratin 7 (KRT7) | Marker of epithelial differentiation | 6.2 | −1.1 | 1.6 | 1.9 | 40 |
| Thioredoxin reductase (TR) | Cellular defense against oxygen damage | 6.3 | 1.6 | −1.1 | 1.5 | 29 |
| Melanoma differentiation-associated gene 7 (mda-7)/interleukin 24 (IL-24) | Tumor suppressor | 6.4 | 1.2 | −2.7 | 4.1 | 68 |
| Retinoic acid receptor alpha (RARα) | Disruption of tumor progression | 7.0 | 1.1 | 1.1 | 1.7 | 33 |
| IRF1 | Transcription factor, cell cycle and apoptosis | 7.8 | 1.2 | 1.2 | 2.6 | 77 |
| Transcription factor RelB (RELB) | Regulation of development, cell cycle, cell survival | 8.0 | 1.4 | 1.5 | 1.6 | 22 |
| Growth arrest and DNA damage inducible 45A (GADD45A) | Regulator of DNA damage, apoptosis | 8.4 | 1.5 | 2.7 | 2.6 | 72 |
| Growth arrest and DNA damage gene 34 (GADD34) | Promoter of eIF2α dephosphorylation | 9.7 | 1.9 | 4.7 | 10.8 | 54, 55 |
| Growth arrest and DNA damage inducible 45B (GADD45B) | Regulator of DNA damage, apoptosis | 10.2 | 1.5 | −2.1 | 7.3 | 72 |
| Transcription factor JunB (JUNB) | Regulation of cell differentiation, proliferation, and apoptosis | 10.5 | 2.0 | 1.5 | 1.4 | 71 |
| Tumor necrosis factor alpha receptor-associated factor 1 (TRAF1) | Adaptor protein, negative regulator of tumor necrosis factor signaling | 11.9 | 1.2 | 1.7 | 2.6 | 85 |
| BH3-only Bcl-2 homologous protein BIK | Inducer of apoptosis | 13.0 | −1.4 | 6.6 | −1.1 | 61 |
| Tumor necrosis factor alpha-induced protein 3 (TNFAIP3) or A20 | Transcription-growth regulatory factor, involved in NF-κB pathway | 15.9 | 2.0 | −1.3 | 1.9 | 5 |
| Winged helix/forkhead transcription factor (FOXO3A) | Transcription factor, cell cycle, apoptosis, cell metabolism, and oxidative stress | 23.0 | −1.2 | 1.8 | 6.2 | 7 |
| IFN stimulatory gene 20 (ISG20) | IFN-induced RNase, 3′ to 5′ exonuclease, antiviral activity | 26.9 | 1.1 | 3.3 | 3.6 | 20 |
| Vitamin D3-upregulated protein (VDUP1/TXNIP) | Major regulator of cellular redox state, inhibitor of thioredoxin and tumor metastasis | 27.8 | 1.7 | 3.7 | 3.5 | 84 |
| Early growth response 1 (EGR1) transcription factor | Regulator of tumorigenesis | 42.8 | 2.8 | 2.0 | 5.2 | 1 |
DISCUSSION
Regulation of protein synthesis by eIF2α phosphorylation plays an important role in the cell's defense against viral infection (12, 34, 35). During virus replication, production of dsRNA generated by symmetric transcription can bind and induce PKR activity. This results in eIF2α phosphorylation, which in turn compromises the translational machinery and contains virus replication (Fig. 8). The significance of regulating PKR activity is emphasized by the distinct mechanisms evolved by DNA tumor viruses to circumvent PKR activation and eIF2α phosphorylation. For example, adenovirus and Epstein-Barr virus express VAI and EBER-1/2 RNA, respectively, which bind and block PKR activation (8, 51). Moreover, the herpes simplex virus encodes a protein, γ134.5, which complexes with the protein phosphatase 1α (PP1α) to dephosphorylate eIF2α (43). Interestingly, γ134.5 shares sequence homology with GADD34 (27), which functions as the regulatory subunit of the PP1 holoenzyme complex that targets eIF2α dephosphorylation (6, 54, 55). Furthermore, the simian virus 40 large T antigen prevents PKR-mediated translational shutoff at a step downstream of PKR (62, 74).
FIG. 8.
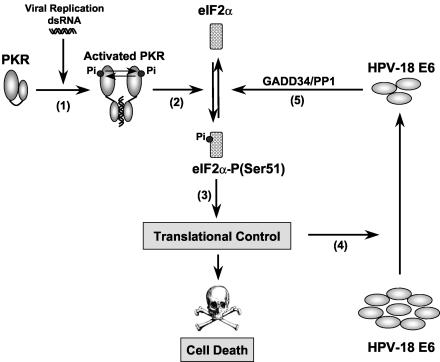
A model for regulation of the PKR-eIF2α phosphorylation pathway by E6. During infection, HPV gene expression produces transcripts containing dsRNA structures able to activate PKR by autophosphorylation (step 1). Activated PKR then catalyzes the phosphorylation of eIF2α at serine 51 (step 2), an event that leads to the translational inhibition and induction of apoptosis (step 3). Although most of the genes are translationally repressed by eIF2α phosphorylation, translation of specific mRNAs is likely to escape from this general translational blockade. These mRNAs may encode proteins that are involved in the inhibition of cell proliferation and induction of apoptosis. Translational inhibition by PKR rapidly downregulates HPV-18 E6 protein synthesis (step 4). However, the remainder of HPV-18 E6 is capable of counteracting this translational blockade by facilitating the dephosphorylation of eIF2α through the recruitment of GADD34/PP1 holophosphatase complex (step 5). This results in a translational relief that permits the expression of proteins with antiapoptotic properties. This may represent an important mechanism utilized by the high-risk HPVs to counteract the antiviral properties of PKR activation and promote virus-mediated oncogenesis.
Our findings further extend the above observations and demonstrate a functional relationship between E6 and the eIF2α pathway. Specifically, E6 protein synthesis rapidly decreases in cells with activated PKR. However, despite the significant downregulation of the viral protein, the remainder of E6 is able to partially rescue cells from the translational blockade posed by eIF2α phosphorylation. This is possibly mediated, at least in part, by the ability of E6 to promote the dephosphorylation of eIF2α through the GADD34/PP1 holophosphatase complex. This notion is based on our observations that high-risk E6 interacts with both GADD34 and PP1 and E6 promotes the dephosphorylation of eIF2α in vitro. In fact, a higher amount of PP1 was bound to HPV-18 E6 than to HPV-11 E6, consistent with the higher degree of eIF2α dephosphorylation by the high-risk viral protein. Although the precise mechanism utilized by E6 to promote eIF2α dephosphorylation through GADD34/PP1 is currently not known, this may be facilitated, at least in part, by the ability of high-risk E6 protein to be localized in both the nucleus and the cytoplasm, as opposed to low-risk viral protein, which exhibits predominantly nuclear localization (24). Mapping of the interaction between E6 and GADD34 showed that the domain of GADD34 required for binding to the viral protein is within the region between residues 180 and 553. The interaction is not mediated by the KVRF PP1-binding sequence of GADD34, since the KARA mutation, which abolishes PP1 binding to GADD34 (6), did not affect the interaction between GADD34 and HPV-18 E6. These findings also indicate that binding of HPV-18 E6 to GADD34 does not interfere with GADD34/PP1 complex formation. Interestingly, PP1 was previously found to directly bind and nullify PKR activity through the dephosphorylation of the activated kinase (76). However, GyrB.PKR is unlikely to be affected by PP1 since the N-terminal regulatory domain of the kinase, which is required for PP1 binding (76), is missing from the fusion protein. In addition, PKR autophosphorylation is not affected in E6-expressing cells (data not shown), further supporting the notion that the viral protein exerts its effects downstream of the activated kinase. A role of the GADD34/PP1 complex in eIF2α dephosphorylation is further supported by the cDNA microarray analysis data showing the regulation of GADD34 gene expression by GyrB.PKR. Specifically, the mRNA levels of GADD34 are induced almost 10-fold in coumermycin-treated GyrB.PKR-expressing cells and only 2-fold in GyrB.PKRK296H-expressing cells (Table 1). Interestingly, GADD34 mRNA expression was inhibited by 50% in GyrB.PKR-expressing cells expressing FLAG-18E6 and remained unaffected in GyrB.PKR-expressing cells with FLAG-HPV-11 E6 (Table 1). Since induction of GADD34 mRNA levels was previously shown to be dependent on eIF2α phosphorylation (55), its inhibition by the HPV-18 E6 further supports the inhibitory role of the viral oncoprotein in eIF2α phosphorylation.
The biological consequences of eIF2α phosphorylation and induction of apoptosis can be best explained in the context of virus infection. The ability of viruses to exert total control over the apoptotic response in infected cells is critical to their replication and induction of pathogenicity (3). For example, inhibition of early apoptosis is a necessary step to ensure efficient viral replication and facilitate virus spread by supporting replication in a broad range of cells and tissues. As such, viruses have evolved sophisticated means to inhibit apoptosis in infected cells. For example, adenoviruses and herpesviruses contain homologues of the Bcl-2 family and antiapoptotic proteins, whereas other viruses encode inhibitors of caspases (65). In the case of HPV, it has become clear that E6 plays a prominent role in prevention of apoptosis through the proteolytic inactivation of the proapoptotic p53, Bak, or Bax (48). Our findings further substantiate the antiapoptotic function of the oncogenic E6 and provide strong evidence that it can be mediated through the regulation of eIF2α phosphorylation. Our data support the notion that the antiapoptotic activities of E6 are mediated by its ability to attenuate both transcriptional and translational responses induced by the PKR-eIF2α phosphorylation pathway. At the translational level, we show that inhibition of apoptosis is likely to be mediated, at least in part, by downregulating the proapoptotic Bax. Interestingly, a role for Bax in PKR-dependent apoptosis was previously described for mouse cells expressing a tetracycline-inducible PKR (2). Mechanistically, it was proposed that increased Bax protein synthesis resembles the translational regulation of yeast GCN4 mRNA (32). That is, translation of GCN4 is controlled by the presence of four upstream open reading frames within the 5′ untranslated region (UTR) of its mRNA (16). Induction of eIF2α phosphorylation facilitates the correct initiation at the GCN4 AUG codon, leading to increased GCN4 protein synthesis (16). In analogy to GCN4, Bax mRNA possesses three upstream AUGs, all in frame with the authentic initiation codon, the first and third of which are followed by a termination codon (32). This striking similarity between GCN4 and Bax mRNAs has led to the hypothesis that the unusual 5′ UTR of Bax plays a role in its translational induction upon eIF2α phosphorylation (32). It is also possible that translation of antiapoptotic genes is facilitated in FLAG-HPV-18 E6 cells and this may play a role in the inhibition of cell death by PKR activation. Identification of genes that are specifically regulated by 18E6 at the translational level upon PKR activation by functional proteomics is the focus of our future experimentation.
In addition to translation, we provide evidence for a role of E6 in PKR-mediated gene transcription and apoptosis. For example, transcription of several proapoptotic genes is induced in cells with activated GyrB.PKR (Table 1). Expression of these genes is strongly suppressed by HPV-18 E6 and to a lesser degree by HPV-11 E6. These genes include GADD45, whose dependency on eIF2α phosphorylation was previously demonstrated in mouse cells containing a homozygous mutation at the serine 51 phosphorylation site of eIF2α (eIF2αS51A) (69). Specifically, GADD45 transcription was induced 15-fold in ER-stressed cells from wt mice but was completely abolished in knock-in cells with the eIF2αS51A mutation (69). Among the genes induced by GyrB.PKR, the melanoma differentiation-associated gene 7 (Mda-7) is an interesting target because of its strong apoptotic functions (68). It was recently shown that Mda-7 induces and activates PKR in lung cancer cells, leading to the destruction of the tumor cells by apoptosis (57). IRF1 is another gene induced by GyrB.PKR activation (Table 1). IRF1 is a protein with antiviral and tumor suppressor activities (77). Previous data provided evidence that transcription of IRF1 is defective in a PKR-null mouse (42) whereas the antiviral and antiproliferative effects of IRF1 are mediated, at least in part, by the activation of PKR (4, 39). It should be emphasized, however, that we do not as yet know whether transcription of all the genes in Table 1 is solely dependent on eIF2α phosphorylation. Inasmuch as PKR has been implicated in signaling to gene transcription through its functional interaction with transcriptional factors (12), it is conceivable that transcriptional control of some of the above genes could be exerted independently of eIF2α phosphorylation.
Collectively, our findings provide strong evidence for a role of the oncogenic HPV-18 E6 in gene translation and transcription modulated by the PKR-eIF2α phosphorylation pathway. Our data reveal a novel mechanism utilized by HPVs to bypass the translational blockade of eIF2α phosphorylation and the induction of an antiviral response by activated PKR. Although the role of eIF2α phosphorylation in virus-mediated tumorigenesis has already been established (12), the possibility that E6 affects various levels of translation in addition to eIF2α phosphorylation cannot be ruled out. Further understanding of the molecular functions of HPV oncoproteins in translational control and the identification of genes that are translationally regulated in HPV-infected cells may prove helpful in the design of strategies to combat HPV infection and associated disease.
Acknowledgments
We thank D. Moraitis for technical assistance; D. C. Tkachuk, M. H. Brush, and S. Shenolikar for FLAG-GADD34 constructs; R. Wek for histidine-tagged eIF2α cDNA; Genome Quebec-Chip Expression Laboratory, McGill University, for the cDNA microarray analysis; and K. Pantopoulos for helpful comments.
This work has been supported by a grant from the Cancer Research Society Inc. with the support of the Institute for Cancer Research of the Canadian Institutes of Health Research (CIHR) to A.E.K. A.E.K. is a recipient of a Scientist Award from CIHR.
REFERENCES
- 1.Adamson, E. D., and D. Mercola. 2002. Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol. 23:93-102. [DOI] [PubMed] [Google Scholar]
- 2.Balachandran, S., C. N. Kim, W. C. Yeh, T. W. Mak, K. Bhalla, and G. N. Barber. 1998. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 17:6888-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benedict, C. A., P. S. Norris, and C. F. Ware. 2002. To kill or be killed: viral evasion of apoptosis. Nat. Immunol. 3:1013-1018. [DOI] [PubMed] [Google Scholar]
- 4.Beretta, L., M. Gabbay, R. Berger, S. M. Hanash, and N. Sonenberg. 1996. Expression of the protein kinase PKR in modulated by IRF-1 and is reduced in 5q-associated leukemias. Oncogene 12:1593-1596. [PubMed] [Google Scholar]
- 5.Beyaert, R., K. Heyninck, and S. Van Huffel. 2000. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem. Pharmacol. 60:1143-1151. [DOI] [PubMed] [Google Scholar]
- 6.Brush, M. H., D. C. Weiser, and S. Shenolikar. 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1 alpha to the endoplasmic reticulum and promotes dephosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 23:1292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgering, B. M., and G. J. Kops. 2002. Cell cycle and death control: long live Forkheads. Trends Biochem. Sci. 27:352-360. [DOI] [PubMed] [Google Scholar]
- 8.Burgert, H. G., Z. Ruzsics, S. Obermeier, A. Hilgendorf, M. Windheim, and A. Elsing. 2002. Subversion of host defense mechanisms by adenoviruses. Curr. Top. Microbiol. Immunol. 269:273-318. [DOI] [PubMed] [Google Scholar]
- 9.Chen, J. J. 2000. Heme-regulated eIF2α kinase, p. 529-546. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Christopher, S. A., P. Diegelman, C. W. Porter, and W. D. Kruger. 2002. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16(cdkN2a/ARF), acts as a tumor suppressor in a breast cancer cell line. Cancer Res. 62:6639-6644. [PubMed] [Google Scholar]
- 12.Clemens, M. J. 2001. Initiation factor eIF2 alpha phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell. Biol. 27:57-89. [DOI] [PubMed] [Google Scholar]
- 13.Clemens, M. J., and A. Elia. 1997. The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res. 17:503-524. [DOI] [PubMed] [Google Scholar]
- 14.Corbett, J. M., M. J. Dunn, A. Posch, and A. Gorg. 1994. Positional reproducibility of protein spots in two-dimensional polyacrylamide gel electrophoresis using immobilised pH gradient isoelectric focusing in the first dimension: an interlaboratory comparison. Electrophoresis 15:1205-1211. [DOI] [PubMed] [Google Scholar]
- 15.Dell, G., and K. Gaston. 2001. Human papillomaviruses and their role in cervical cancer. Cell. Mol. Life Sci. 58:1923-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dever, T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545-556. [DOI] [PubMed] [Google Scholar]
- 17.Donze, O., J. Dostie, and N. Sonenberg. 1999. Regulatable expression of the interferon-induced double-stranded RNA dependent protein kinase PKR induces apoptosis and fas receptor expression. Virology 256:322-329. [DOI] [PubMed] [Google Scholar]
- 18.Dreyling, M. H., D. Roulston, S. K. Bohlander, J. Vardiman, and O. I. Olopade. 1998. Codeletion of CDKN2 and MTAP genes in a subset of non-Hodgkin's lymphoma may be associated with histologic transformation from low-grade to diffuse large-cell lymphoma. Genes Chromosomes Cancer 22:72-78. [PubMed] [Google Scholar]
- 19.Dunn, M. J., and J. M. Corbett. 1996. Two-dimensional polyacrylamide gel electrophoresis. Methods Enzymol. 271:177-203. [DOI] [PubMed] [Google Scholar]
- 20.Espert, L., G. Degols, C. Gongora, D. Blondel, B. R. Williams, R. H. Silverman, and N. Mechti. 2003. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278:16151-16158. [DOI] [PubMed] [Google Scholar]
- 21.Gale, M., Jr., S. L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 23.Gu, Z., D. Pim, S. Labrecque, L. Banks, and G. Matlashewski. 1994. DNA damage induced p53 mediated transcription is inhibited by human papillomavirus type 18 E6. Oncogene 9:629-633. [PubMed] [Google Scholar]
- 24.Guccione, E., P. Massimi, A. Bernat, and L. Banks. 2002. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology 293:20-25. [DOI] [PubMed] [Google Scholar]
- 25.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 26.Harding, H. P., I. Novoa, Y. Zhang, H. Zeng, R. Wek, M. Schapira, and D. Ron. 2000. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6:1099-1108. [DOI] [PubMed] [Google Scholar]
- 27.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hershey, J. W. 1991. Translational control in mammalian cells. Annu. Rev. Biochem. 60:717-755. [DOI] [PubMed] [Google Scholar]
- 29.Hirt, R. P., S. Muller, T. M. Embley, and G. H. Coombs. 2002. The diversity and evolution of thioredoxin reductase: new perspectives. Trends Parasitol. 18:302-308. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa, H., A. P. Heaney, R. Yu, G. A. Horwitz, and S. Melmed. 2001. Human pituitary tumor-transforming gene induces angiogenesis. J. Clin. Endocrinol. Metab. 86:867-874. [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa, S., M. Kai, M. Tamari, Y. Takei, K. Takeuchi, H. Bandou, Y. Yamane, M. Ogawa, and Y. Nakamura. 1997. Sequence analysis of a 685-kb genomic region on chromosome 3p22-p21.3 that is homozygously deleted in a lung carcinoma cell line. DNA Res. 4:35-43. [DOI] [PubMed] [Google Scholar]
- 32.Jagus, R., B. Joshi, and G. N. Barber. 1999. PKR, apoptosis and cancer. Int. J. Biochem. Cell Biol. 31:123-138. [DOI] [PubMed] [Google Scholar]
- 33.Kastner, P., and S. Chan. 2001. Function of RARα during the maturation of neutrophils. Oncogene 20:7178-7185. [DOI] [PubMed] [Google Scholar]
- 34.Katze, M. G. 1995. Regulation of the interferon-induced PKR: can viruses cope? Trends Microbiol. 3:75-78. [DOI] [PubMed] [Google Scholar]
- 35.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 36.Kaufman, R. J. 2000. Double-stranded RNA-activated protein kinase PKR, p. 503-527. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Kehmeier, E., H. Ruhl, B. Voland, M. C. Stoppler, E. Androphy, and H. Stoppler. 2002. Cellular steady-state levels of “high risk” but not “low risk” human papillomavirus (HPV) E6 proteins are increased by inhibition of proteasome-dependent degradation independent of their p53- and E6AP-binding capabilities. Virology 299:72-87. [DOI] [PubMed] [Google Scholar]
- 38.Kimball, S. R. 2001. Regulation of translation initiation by amino acids in eukaryotic cells. Prog. Mol. Subcell. Biol. 26:155-184. [DOI] [PubMed] [Google Scholar]
- 39.Kirchhoff, S., A. E. Koromilas, F. Schaper, M. Grashoff, N. Sonenberg, and H. Hauser. 1995. IRF-1 induced cell growth inhibition and interferon induction requires the activity of the protein kinase PKR. Oncogene 11:439-445. [PubMed] [Google Scholar]
- 40.Kirfel, J., T. M. Magin, and J. Reichelt. 2003. Keratins: a structural scaffold with emerging functions. Cell. Mol. Life Sci. 60:56-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koromilas, A. E., S. Li, and G. Matlashewski. 2001. Control of interferon signaling in human papillomavirus infection. Cytokine Growth Factor Rev. 12:157-170. [DOI] [PubMed] [Google Scholar]
- 42.Kumar, A., Y. L. Yang, V. Flati, S. Der, S. Kadereit, A. Deb, J. Haque, L. Reis, C. Weissmann, and B. R. Williams. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leib, D. A. 2002. Counteraction of interferon-induced antiviral responses by herpes simplex viruses. Curr. Top. Microbiol. Immunol. 269:171-185. [DOI] [PubMed] [Google Scholar]
- 44.Leone, P. E., M. Mendiola, J. Alonso, C. Mino, and A. Pestana. 2003. Implications of a RAD54L polymorphism (2290C/T) in human meningiomas as a risk factor and/or a genetic marker. BMC Cancer 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, S., and A. E. Koromilas. 2001. Dominant negative function by an alternatively spliced form of the interferon-inducible protein kinase PKR. J. Biol. Chem. 276:13881-13890. [DOI] [PubMed] [Google Scholar]
- 46.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 47.Li, S., K. Nagai, and A. E. Koromilas. 2000. A diminished activation capacity of the interferon-inducible protein kinase PKR in human T lymphocytes. Eur. J. Biochem. 267:1598-1606. [DOI] [PubMed] [Google Scholar]
- 48.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 49.Munger, K. 2002. The role of human papillomaviruses in human cancers. Front. Biosci. 7:d641-d649. [DOI] [PubMed] [Google Scholar]
- 50.Munger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 51.Nanbo, A., and K. Takada. 2002. The role of Epstein-Barr virus-encoded small RNAs (EBERs) in oncogenesis. Rev. Med. Virol. 12:321-326. [DOI] [PubMed] [Google Scholar]
- 52.Neubig, R. R., and D. P. Siderovski. 2002. Regulators of G-protein signalling as new central nervous system drug targets. Nat. Rev. Drug Discov. 1:187-197. [DOI] [PubMed] [Google Scholar]
- 53.Novak, J. P., R. Sladek, and T. J. Hudson. 2002. Characterization of variability in large-scale gene expression data: implications for study design. Genomics 79:104-113. [DOI] [PubMed] [Google Scholar]
- 54.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olofsson, B. 1999. Rho guanine dissociation inhibitors: pivotal molecules in cellular signalling. Cell. Signal. 11:545-554. [DOI] [PubMed] [Google Scholar]
- 57.Pataer, A., S. A. Vorburger, G. N. Barber, S. Chada, A. M. Mhashilkar, H. Zou-Yang, A. L. Stewart, S. Balachandran, J. A. Roth, K. K. Hunt, and S. G. Swisher. 2002. Adenoviral transfer of the melanoma differentiation-associated gene 7 (mda7) induces apoptosis of lung cancer cells via up-regulation of the double-stranded RNA-dependent protein kinase (PKR). Cancer Res. 62:2239-2243. [PubMed] [Google Scholar]
- 58.Payton, M., and S. Coats. 2002. Cyclin E2, the cycle continues. Int. J. Biochem. Cell Biol. 34:315-320. [DOI] [PubMed] [Google Scholar]
- 59.Perelman, B., N. Dafni, T. Naiman, D. Eli, M. Yaakov, T. L. Feng, S. Sinha, G. Weber, S. Khodaei, A. Sancar, I. Dotan, and D. Canaani. 1997. Molecular cloning of a novel human gene encoding a 63-kDa protein and its sublocalization within the 11q13 locus. Genomics 41:397-405. [DOI] [PubMed] [Google Scholar]
- 60.Protopopov, A., V. Kashuba, V. I. Zabarovska, O. V. Muravenko, M. I. Lerman, G. Klein, and E. R. Zabarovsky. 2003. An integrated physical and gene map of the 3.5-Mb chromosome 3p21.3 (AP20) region implicated in major human epithelial malignancies. Cancer Res. 63:404-412. [PubMed] [Google Scholar]
- 61.Puthalakath, H., and A. Strasser. 2002. Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ. 9:505-512. [DOI] [PubMed] [Google Scholar]
- 62.Rajan, P., S. Swaminathan, J. Zhu, C. N. Cole, G. Barber, M. J. Tevethia, and B. Thimmapaya. 1995. A novel translational regulation function for the simian virus 40 large-T antigen gene. J. Virol. 69:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos-Morales, F., A. Dominguez, F. Romero, R. Luna, M. C. Multon, J. A. Pintor-Toro, and M. Tortolero. 2000. Cell cycle regulated expression and phosphorylation of hpttg proto-oncogene product. Oncogene 19:403-409. [DOI] [PubMed] [Google Scholar]
- 64.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 66.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 67.Sanchez, J. C., V. Rouge, M. Pisteur, F. Ravier, L. Tonella, M. Moosmayer, M. R. Wilkins, and D. F. Hochstrasser. 1997. Improved and simplified in-gel sample application using reswelling of dry immobilized pH gradients. Electrophoresis 18:324-327. [DOI] [PubMed] [Google Scholar]
- 68.Sauane, M., R. V. Gopalkrishnan, D. Sarkar, Z. Z. Su, I. V. Lebedeva, P. Dent, S. Pestka, and P. B. Fisher. 2003. MDA-7/IL-24: novel cancer growth suppressing and apoptosis inducing cytokine. Cytokine Growth Factor Rev. 14:35-51. [DOI] [PubMed] [Google Scholar]
- 69.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165-1176. [DOI] [PubMed] [Google Scholar]
- 70.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16:2593-2620. [DOI] [PubMed] [Google Scholar]
- 71.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 72.Sheikh, M. S., M. C. Hollander, and A. J. Fornance, Jr. 2000. Role of Gadd45 in apoptosis. Biochem. Pharmacol. 59:43-45. [DOI] [PubMed] [Google Scholar]
- 73.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 74.Swaminathan, S., P. Rajan, O. Savinova, R. Jagus, and B. Thimmapaya. 1996. Simian virus 40 large-T bypasses the translational block imposed by the phosphorylation of eIF-2 alpha. Virology 219:321-323. [DOI] [PubMed] [Google Scholar]
- 75.Tan, S. L., and M. G. Katze. 1999. The emerging role of the interferon-induced PKR protein kinase as an apoptotic effector: a new face of death? J. Interferon Cytokine Res. 19:543-554. [DOI] [PubMed] [Google Scholar]
- 76.Tan, S. L., S. U. Tareen, M. W. Melville, C. M. Blakely, and M. G. Katze. 2002. The direct binding of the catalytic subunit of protein phosphatase 1 to the PKR protein kinase is necessary but not sufficient for inactivation and disruption of enzyme dimer formation. J. Biol. Chem. 277:36109-36117. [DOI] [PubMed] [Google Scholar]
- 77.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 78.Ung, T. L., C. Cao, J. Lu, K. Ozato, and T. E. Dever. 2001. Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 20:3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 80.Warden, S. M., C. Richardson, J. O'Donnell, Jr., D. Stapleton, B. E. Kemp, and L. A. Witters. 2001. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem. J. 354:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong, A. H., J. E. Durbin, S. Li, T. E. Dever, T. Decker, and A. E. Koromilas. 2001. Enhanced antiviral and antiproliferative properties of a STAT1 mutant unable to interact with the protein kinase PKR. J. Biol. Chem. 276:13727-13737. [DOI] [PubMed] [Google Scholar]
- 82.Wood, Z. A., E. Schroder, H. J. Robin, and L. B. Poole. 2003. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 28:32-40. [DOI] [PubMed] [Google Scholar]
- 83.Yao, X., A. Abrieu, Y. Zheng, K. F. Sullivan, and D. W. Cleveland. 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2:484-491. [DOI] [PubMed] [Google Scholar]
- 84.Yodoi, J., H. Nakamura, and H. Masutani. 2002. Redox regulation of stress signals: possible roles of dendritic stellate TRX producer cells (DST cell types). Biol. Chem. 383:585-590. [DOI] [PubMed] [Google Scholar]
- 85.Zapata, J. M., and J. C. Reed. 2002. TRAF1: lord without a RING. Sci. STKE 2002:PE27. [DOI] [PubMed] [Google Scholar]
- 86.Zhu, S., A. Y. Sobolev, and R. C. Wek. 1996. Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem. 271:24989-24994. [DOI] [PubMed] [Google Scholar]
- 87.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]