Abstract
Epidermal growth factor receptor variant III (EGFRvIII) is a glycoprotein uniquely expressed in glioblastoma, but not in normal brain tissues. To develop targeted therapies for brain tumors, we selected RNA aptamers against the histidine-tagged EGFRvIII ectodomain, using an Escherichia coli system for protein expression and purification. Representative aptamer E21 has a dissociation constant (Kd) of 33×10−9 m, and exhibits high affinity and specificity for EGFRvIII in ELISA and surface plasmon resonance assays. However, selected aptamers cannot bind the same protein expressed from eukaryotic cells because glycosylation, a post-translational modification present only in eukaryotic systems, significantly alters the structure of the target protein. By transfecting EGFRvIII aptamers into cells, we find that membrane-bound, glycosylated EGFRvIII is reduced and the percentage of cells undergoing apoptosis is increased. We postulate that transfected aptamers can interact with newly synthesized EGFRvIII, disrupt proper glycosylation, and reduce the amount of mature EGFRvIII reaching the cell surface. Our work establishes the feasibility of disrupting protein post-translational modifications in situ with aptamers. This finding is useful for elucidating the function of proteins of interest with various modifications, as well as dissecting signal transduction pathways.
Keywords: EGFRvIII, glycosylation, RNA aptamer, SELEX
Introduction
Human epidermal growth factor receptor (EGFR) is a type I receptor tyrosine kinase expressed on the surface of a number of cell types (Baselga, 2002; Mendelsohn, 2002; Jorissen et al., 2003). EGFR consists of an ectodomain, a transmembrane domain, and an intracellular domain which contains a tyrosine kinase domain and a C-terminal tail. EGFR ectodomain can be further divided into four subdomains: I and III share high homology; II and IV are cystidine-rich. Domain II is crucial for EGFR homo-dimerization (Ogiso et al., 2002). Upon ligand binding, EGFR dimerizes, and cytoplasmic tyrosine kinase is activated, which triggers a set of signal transduction pathways, including the Ras/Raf/mitogen-activated protein kinase pathway [extracellular signal-regulated kinase (ERK) 1/2], the phosphatidylinositol 3-kinase/Akt pathway, etc., resulting in cell survival, proliferation, and differentiation (Jorissen et al., 2003).
EGFR overexpression has been detected in many human tumors. It is also the first cell surface receptor identified as being amplified and mutated in glioblastoma, the most frequently occurring and most malignant human brain tumor (Ohgaki and Kleihues, 2005). EGFR acts oncogenically to stimulate cancer cell growth and spread. In addition to overexpression, EGFR often undergoes rearrangements which make it constitutively active. The most common mutant is variant III (EGFRvIII), which has an in-frame deletion of 801 base pairs, resulting in absence of domain II. EGFRvIII has been detected in brain, breast, ovarian, and non-small cell lung cancers, but not in the corresponding non-malignant tissues (Garcia de Palazzo et al., 1993; Moscatello et al., 1995; Kuan et al., 2001).
EGFR is a highly glycosylated protein, as its ectodomain contains 12 N-glycosylation sites. Glycosylation is the most frequent protein modification in eukaryotic cells, proper glycosylation being crucial in protein folding and stability (Durand and Seta, 2000; Martin-Rendon and Blake, 2003). It is estimated that more than half of the proteins in eukaryotes undergo this modification. During glycosylation, glycans are synthesized in the endoplasmic reticulum, processed in the Golgi complex, and covalently linked to most plasma membrane and secretary proteins. EGFRvIII undergoes glycosylation as well. Despite the deletion of domain II, this variant retains eight glycosylation sites (Zhen et al., 2003), and proper glycosylation on these sites is required for EGFRvIII translocation to cell membrane, where it is constitutively activated.
The clear potential of EGFRvIII-targeted therapy for cancer has stimulated much research. Antibodies have been developed that specifically recognize EGFRvIII. Such antibodies bind EGFRvIII on the tumor cell and induce quick endocytosis. Once coupled with cytotoxic reagent, these antibodies kill tumor cells efficiently (Reist et al., 1995; Wikstrand et al., 1995). In practice, it is observed that EGFRvIII antibody injected intracranially may leak and cause systemic toxicity (personal communication). So far, there is no effective method to reverse antibody function. RNA aptamers can be viewed as oligonucleotide analogs of antibodies in that they have similar high affinity and specificity for their cognate proteins (Sullenger and Gilboa, 2002; Bunka and Stockley, 2006; Que-Gewirth and Sullenger, 2007). Yet owing to their nucleic acid nature, aptamers offer great advantages over antibodies. First, aptamers have smaller sizes (8–15 kDa), only half the molecular weight of the smallest single-chain fragment variant of IgG, which enables rapid tumor penetration and blood clearance. Second, aptamers have low to no immunogenicity. Third, aptamers can be bulk-synthesized and easily modified to increase stability. Most importantly, aptamers can be reversed by antidotes (Rusconi et al., 2002; Oney et al., 2007), which are typically 20–30 nucleotides long RNA oligonucleotides with complementary sequence to aptamers.
In this study, using the systematic evolution of ligands by exponential enrichment (SELEX) technique, we developed RNA aptamers against the ectodomain of EGFRvIII as a polyhistidine-tagged protein purified from Escherichia coli. We tested if these aptamers could recognize their eukaryotic targets with post-translational modifications. Finally, we explored the effects on cell viability of transfecting EGFRvIII aptamers into cells.
Results
Selecting RNA aptamers for the purified EGFRvIII ectodomain
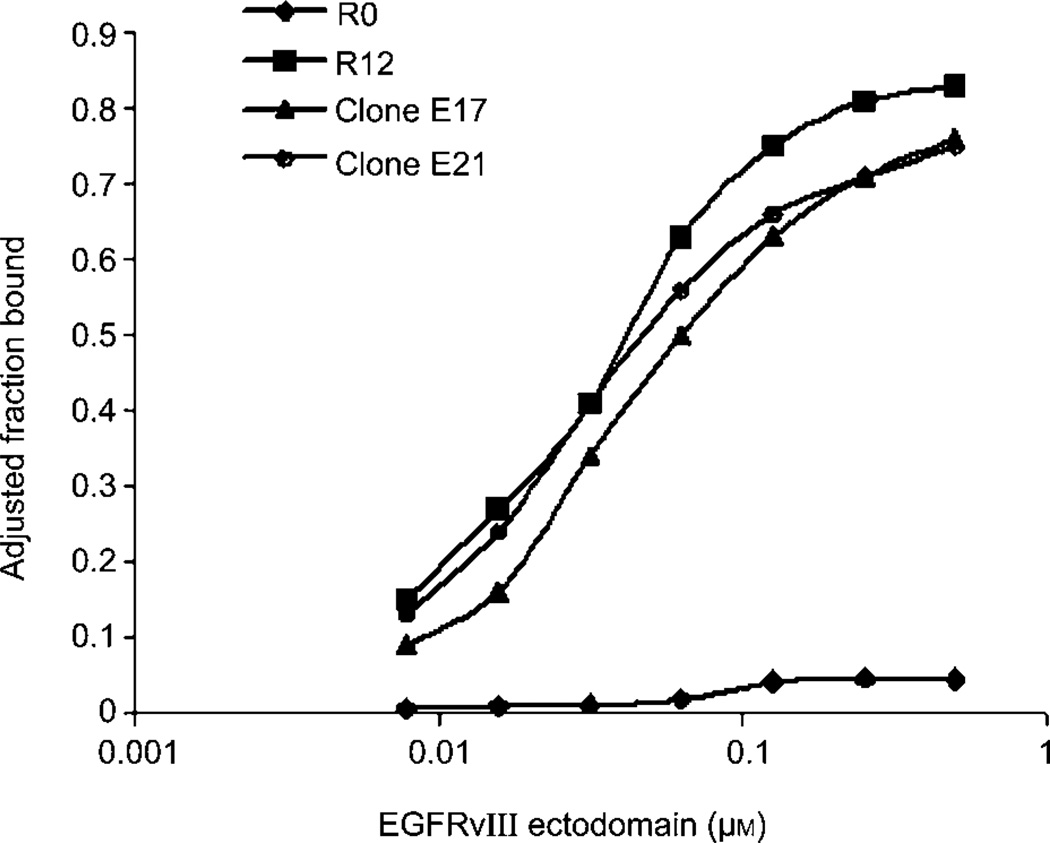
To obtain aptamers that specifically target EGFRvIII, we used E. coli-expressed, His-tagged EGFRvIII ectodomain as the target for in vitro selection. The initial RNA pool consists of 1014 oligonucleotides, each containing a 40-nucleotide random region flanked by fixed sequences. To improve stability, all the pyrimidines are 2′-fluoro modified. Initially, nitrocellular membrane filtration was used to trap aptamer-protein complexes. After four rounds, however, we found that aptamers were being inappropriately selected against the polyhistidine tag on the protein likely as a result of charge-charge interactions. Selections were then reinitiated via a bead-based approach, essentially consisting of pre-binding His-tagged EGFRvIII to Ni-NTA beads (Qiagen, Valencia, CA, USA), incubating protein-bead conjugate with oligonucleotides, washes to remove unbound oligonucleotides, and RT-PCR amplification of the bound oligonucleotides. After 12 rounds of such bead-based selection, the binding properties of the starting and final pools were compared. In sharp contrast to the starting pool, which essentially showed no binding to EGFRvIII, round 12 pool exhibited significantly increased affinity. The maximal binding was more than 80%, and the Kd value was around 30×10−9 m (Figure 1). We further excluded the possibility that the affinity was due to the His tag by showing that round 12 pool did not bind a His-tagged, but unrelated protein (data not shown).
Figure 1. Bead selection for EGFRvIII ectodomain significantly increased RNA pool affinity.
The affinity of the initial (R0) and final (R12) rounds RNA for EGFRvIII were compared in a double filter binding assay. Representative clones E17 and E21 for EGFRvIII are included, along with the pools.
Sequencing of round 12 identified two predominant clones, E17 and E27 (Table 1), each accounting for 54% and 10% of the pool, respectively. The rest of the aptamers can be grouped into one family, sharing the consensus TACCAAGATCCCACAACTAGCCGACCAN1–2-ATTGCCGGC. All EGFRvIII aptamers have a Kd ranging from 33 to 59×10−9 m, in the same range as the round 12 pool. For further study, we focused on E17, the predominant clone; and E21, a rare clone in the family, which has the highest affinity.
Table 1.
Sequences and affinities of EGFRvIII ectodomain aptamers.
| Aptamera | % of poolb | 5′-Sequence of random region-3′ | Kd (nm) |
|---|---|---|---|
| Predominant clones | |||
| E17 | 54 | ACCAAAATCAACGCAAAGAGCGCGCCTGCACGTCACCTCA | 50 |
| E27 | 10 | GCCAGCATCCCGACTGGGCTCCCTCGATAACGAGTGCCCG | 57 |
| Family | |||
| E1 | 12 | CGTACCAAGATCCCACAACTAGCCGACCACAATTGCCGGC | 41 |
| E40 | 12 | CGTACCAAGATCCCACAACTAGCCGACCAGGATTGCCGGC | 59 |
| E4 | 6 | CGTACCAAGATCCCACAACTAGCCGACCAG----ATTGCCGGC | N/D |
| E21 | 3 | TACCAAGATCCCACAACTAGCCGACCACAATTGCCGGCCA | 33 |
| Consensus | TACCAAGATCCCACAACTAGCCGACCA----ATTGCCGGC |
Aptamers from round 12 were grouped according to sequence similarity. For simplicity, the sequences at the 5′ and 3′ ends have been omitted. Affinities for EGFRvIII were measured by a nitrocellulose filter binding assay.
% of pool is the aptamer representation in the pool from which it was cloned. Kd, N/D: not determined.
When tested in the binding assay, E17 and E21 behaved quite similarly to the round 12 pool (Figure 1). E21 had a slightly higher affinity for EGFRvIII than E17 (Table 1).
Aptamer-EGFRvIII interactions confirmed by ELISA and Biacore
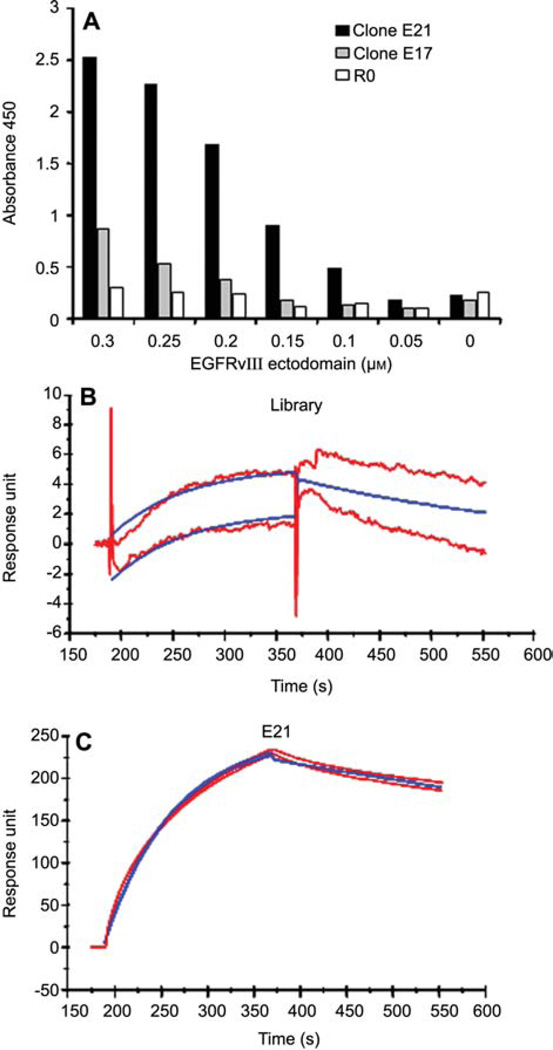
Since EGFRvIII is a membrane protein, we speculated that epitopes exposed in solution may not be accessible once the protein is immobilized. To determine whether EGFRvIII aptamers can bind immobilized EGFRvIII, we performed an ELISA assay by probing with the aptamers attached to a biotin-labeled oligonucleotide. Binding was monitored by addition of streptavidin-HRP-TMB substrate reaction. As shown in Figure 2A, compared to the initial pool, E21 shows more than an eight-fold increase in binding of EGFRvIII at 0.3 µm protein concentration. The enhanced affinity is protein concentration dependent. In the absence of EGFRvIII, no affinity difference between E21 and the starting pool was detected. Unlike E21, E17 exhibits only a moderate increase in affinity relative to the library control.
Figure 2. Aptamers bind their target proteins with high affinity.
(A) ELISA assay showing in vitro aptamer-protein interaction. (B, C) The interaction between protein EGFRvIII ectodomain and aptamer E21 is confirmed by surface plasmon resonance (Biacore 3000). For E21, the determined constants are ka = 1×104 m−1 s−1; kd = 4×10−3 s−1.
The interaction between aptamer E21 and EGFRvIII protein was further characterized by surface plasmon resonance analysis via Biacore 3000. As shown in Figure 2B and C, E21 binds EGFRvIII with a high Ka and a very low Kd, which indicates that once E21 binds EGFRvIII, it hardly dissociates. This binding characteristic is ideal for drug delivery or imaging purposes. In sharp contrast, library RNA showed a very low level of binding (<10 response units) to EGFRvIII (Figure 2B), which confirms the specificity of E21. Interestingly, although E17 did not work as well as E21 in previous tests, it functioned equally as well as E21 in the surface plasmon resonance assay (data not shown).
EGFRvIII aptamers distinguish glycosylated vs. unglycosylated EGFRvIII protein
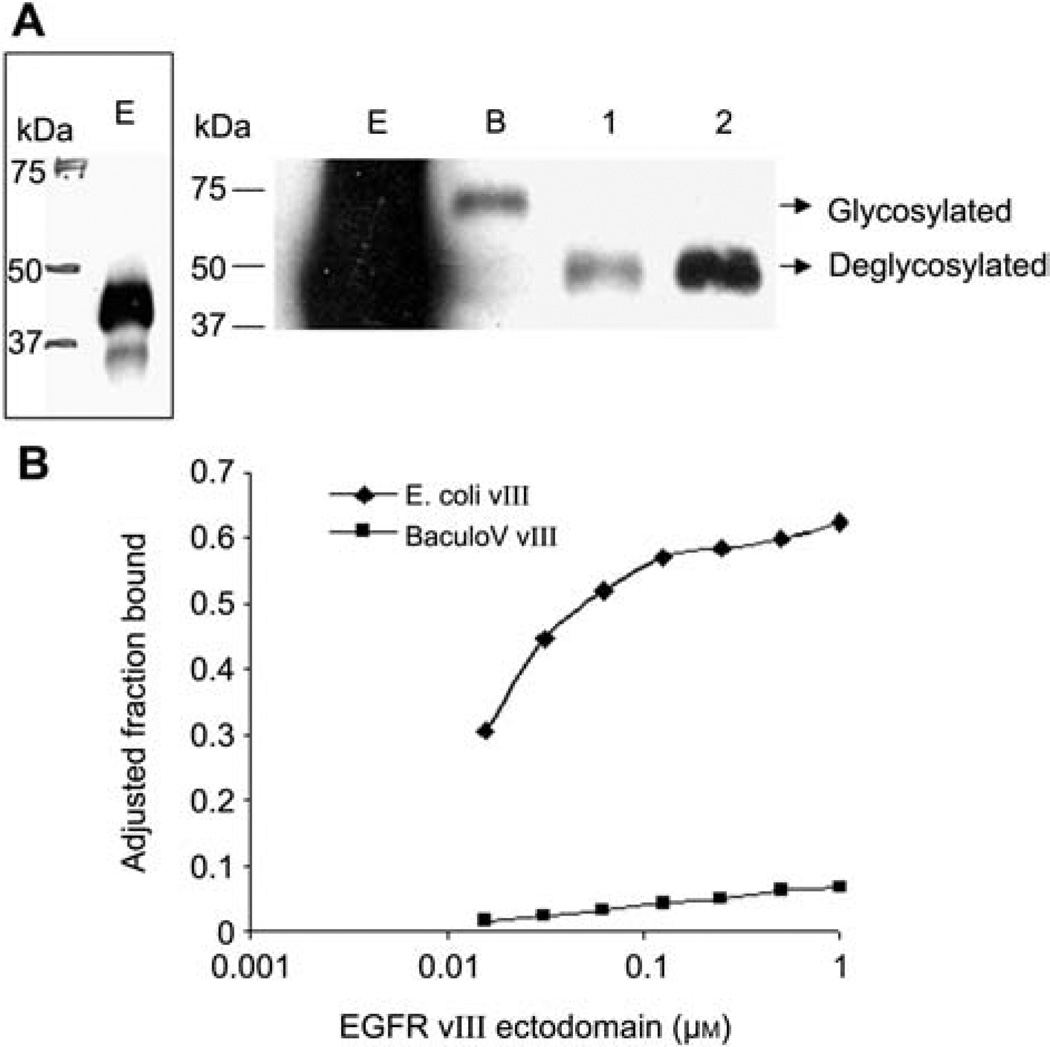
The EGFRvIII we used for SELEX is purified from E. coli, a prokaryotic system lacking post-translational modifications. The same protein was expressed and purified from baculovirus-infected insect SF9 cells, a eukaryotic system possessing post-translational modifications. We compared these two proteins and tested if post-translational modification exerts a negative effect on aptamer affinity. As expected, the same construct yields two proteins with distinct molecular weight, as shown in Figure 3A. E. coli-produced EGFRvIII is 40 kDa, as indicated by the bulky band below the 50 kDa marker. A better view is achieved with lighter exposure (Figure 3A, left panel). SF9-produced EGFRvIII is approximately 70 kDa, 30 kDa bigger than the one from E. coli. Since EGFRvIII is known to be a glycosylated protein, we tested if the molecular weight difference is due to glycosylation. Indeed, both the chemical removal of glycans by trifluoromethanesulfonic acid and enzymatic digestion by PNGase F of baculovirus-produced EGFRvIII resulted in a band of similar size of bacterial purified EGFRvIII, confirming that glycosylation accounts for the molecular weight difference.
Figure 3. EGFRvIII aptamers specifically bind unglycosylated EGFRvIII.
(A) Western blot showing the different molecular masses of EGFRvIII from E. coli (E), baculovirus (B), and two deglycosylated EGFRvIII. 1: chemical deglycosylation with trifluoromethanesulfonic acid, 2: enzymatic deglycosylation with PNGase F. Left panel: a light exposure to show EGFRvIII expressed from E. coli. (B) The binding assay revealed that aptamer E21 binds only E. coli-expressed EGFRvIII and not baculovirus-expressed, glycosylated EGFRvIII.
When tested for binding, aptamer E21 bound only E. coli-expressed EGFRvIII, and showed no affinity for baculovirus-produced EGFRvIII (Figure 3B), which demonstrates that glycosylation alters the binding surface of the aptamer on the protein and the interaction between aptamer and protein is disrupted.
EGFRvIII aptamers reduce membrane-bound EGFRvIII when introduced intracellularly
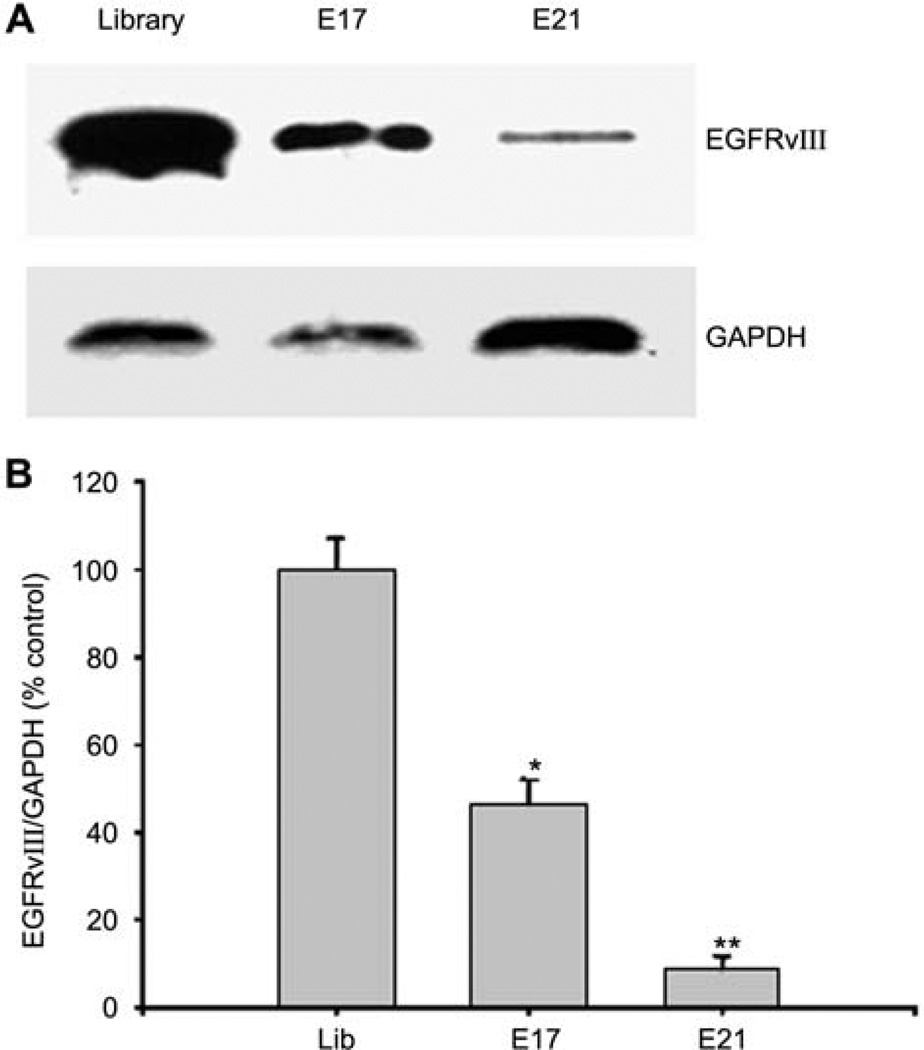
The maturation process of EGFRvIII involves synthesis at the rough endoplasmic reticulum, post-translational modification in the lumen of the Golgi complex, and export to the membrane (Johns et al., 2005). If the protein does not undergo proper glycosylation, it could be targeted for degradation (Ermonval et al., 2001). Since our aptamers specifically interact with unglycosylated EGFRvIII, we hypothesized that the aptamers may bind newly synthesized EGFRvIII in situ, block its glycosylation, cause its degradation, and reduce the expression of cell surface EGFRvIII. To test this hypothesis, we transfected EGFRvIII aptamers E17 and E21 into NR6M cells, a mouse cell line overexpressing EGFRvIII. Then, 28 h after transfection, membrane proteins were labeled with Sulfo-NHS-SS-biotin (Pierce, Rockford, IL, USA), purified on streptavidin beads, and analyzed by gel electrophoresis and Western blotting. Compared to control cells transfected with RNA library, membrane-bound EGFRvIII is decreased by 50% and 90% in E17- and E21-trans-fected cells, respectively (Figure 4), indicating that delivery of the aptamers into the cells can indeed reduce EGFRvIII surface expression.
Figure 4. EGFRvIII aptamers reduce membrane EGFRvIII after transient transfection.
(A) Representative Western blot to show that membrane EGFRvIII expression is reduced in response to aptamer transfection. The same blot was stripped, and reprobed with anti-GAPDH antibody to monitor loading. (B) Percent densitometric values of membrane EGFRvIII protein levels demonstrated that membrane EGFRvIII levels were significantly decreased in NR6M cells transfected with aptamer E17 or E21. *p<0.05 vs. library; **p<0.01 vs. library; n=3 experiments.
EGFRvIII aptamers induce apoptosis when introduced intracellularly
Cell surface expression of EGFRvIII confers cell growth advantage by constitutively activating downstream signaling pathways (Kuan et al., 2001). A crucial quality control exists at the Golgi complex to ensure properly glycosylated EGFRvIII reaches cell membrane, while improperly glyosylated ones become degraded. Since our aptamers specifically interact with unglycosylated EGFRvIII and reduce membrane-bound EGFRvIII, we tested whether the aptamers can abolish EGFRvIII-dependent growth advantage.
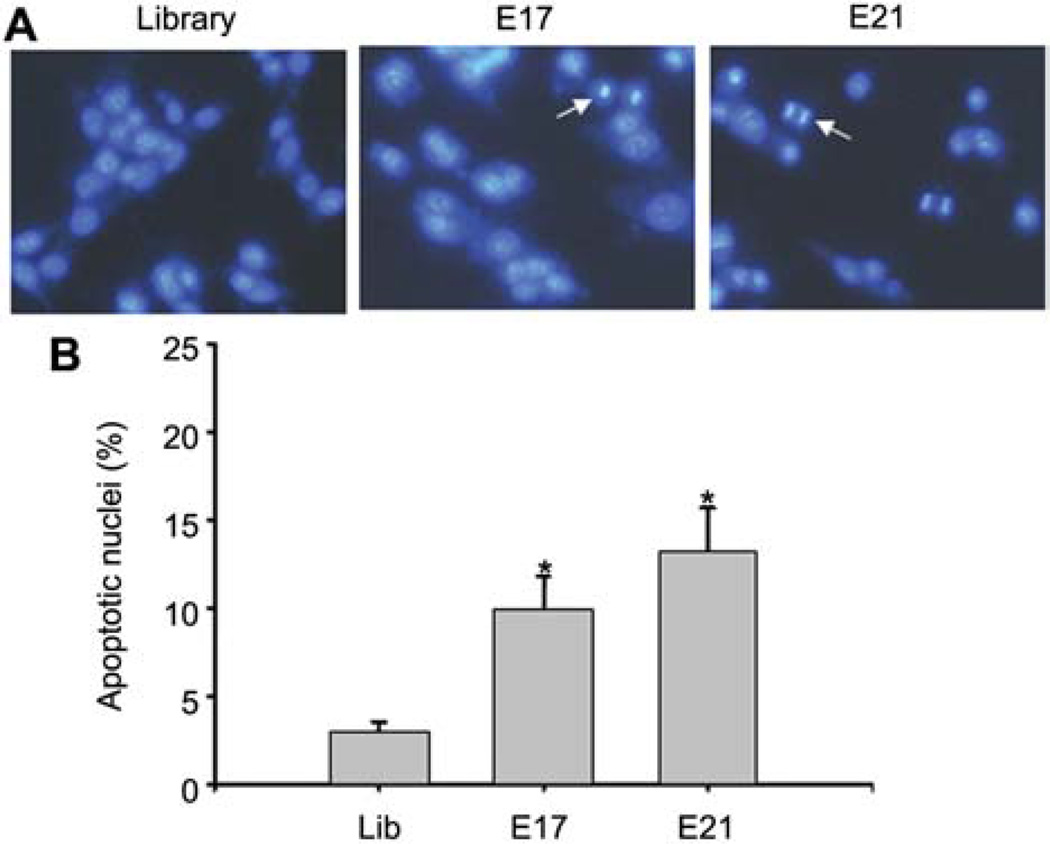
NR6M cells were transfected with EGFRvIII aptamers, as well as RNA library as control. Then, 28 h after transfection, cells were fixed and stained with Hoechst 33342. As shown in Figure 5A, both E17 and E21 transfected cells exhibit condensed nuclei, a characteristic morphological feature of apoptotic cells. Five different areas were then randomly picked, and the percentage of apoptotic cells was calculated (Figure 5B). Compared to cells transfected with RNA library, E17- and E21-trans-fected cells show a 4- and 5-fold increase in percentage of apoptotic cells, suggesting that transfected aptamers can indeed induce apoptosis in a subpopulation of cells.
Figure 5. EGFRvIII aptamers induce cell apoptosis after transient transfection.
(A) Hoechst 33342 staining of apoptotic nuclei. Condensed, bright nuclei exhibiting features of apoptosis are indicated by arrows. (B) Percentage of apoptotic nuclei. Apoptotic nuclei were counted from five different areas and averaged. *p<0.05 vs. library control. n=3 independent experiments.
Discussion
A practical SELEX strategy for histidine-tagged proteins
In a search for novel therapies for glioblastoma, we developed RNA aptamers against EGFRvIII ectodomain, a 40-kDa peptide that was polyhistidine tagged and expressed from E. coli. We found that positively charged His tag non-specifically attracts negatively charged RNA. After four rounds, selection is dominated by His tag binders that have no affinity for the target of interest. By prebinding His-tagged protein with Ni-NTA beads, we successfully blocked the His tag and directed selection toward the rest of the protein of interest. Since polyhistidine is a widely used tag for protein purification, our work provides a feasible SELEX strategy for His-tagged proteins.
Consideration of post-translational modification for successful SELEX
In the literature, there are reports of successful aptamers generation with prokaryotic system expressed proteins. However, our work presented here clearly demonstrates that such an approach is not feasible for EGFRvIII, a highly glycosylated protein. Glycosylation totals up to 30 kDa molecular mass, which accounts for three-quarters of the molecular weight of unmodified EGFRvIII ectodomain. This dramatic increase in molecular weight undoubtedly alters EGFRvIII conformation. Aptamers rely strictly on conformation complementary for target binding. When glycosylation adds significant molecular weight to a protein, the complementary surface an aptamer recognizes is unavoidably disrupted, and aptamers lose their affinity for the proteins. Our work on EGFRvIII reveals the pivotal role post-translational modification can play on aptamer-protein interactions, and demonstrates that it is not feasible to use prokaryotic system purified proteins for aptamer generation, especially when the target protein undergoes excessive post-translational modifications.
Although glycosylation is a universal modification in eukaryotic systems, glycans vary greatly among species. For our baculovirus-SF9 insect cell expressed EGFRvIII, glycans add 30 kDa to the molecular mass. Taking into account that EGFRvIII ectodomain has eight glycosylation sites, each glycan contributes 3.75 kDa of molecular mass. This glycan weight is quite different from the 1–2.3 kDa reported by other research groups (Kim et al., 2005). This might be due to different insect cells utilized for protein expression. We used SF9 cell, while the other group used S2 cells from Drosophila melanogaster. In addition, the glycan isoform changes according to whether conditions are normal or pathological (Durand and Seta, 2000; Martin-Rendon and Blake, 2003). Therefore, to obtain a functional aptamer, it is crucial to purify the target protein from the system where the aptamer will be applied eventually.
A new area for aptamer application: targeting cytoplasmic post-translational modifications
To date, the aptamer field has focused mainly on using aptamers to target plasma proteins and membrane receptors (Que-Gewirth and Sullenger, 2007). In 2004, the first aptamer-based drug Pegaptanib (Macugen) was approved by the Food and Drug Administration for the treatment of wet-age-related macular degeneration. A number of other aptamer drugs are in the pipeline (Que-Gewirth and Sullenger, 2007). In most cases, related antibodies have been produced earlier and stand in more advanced stages.
In contrast, targeting an intracellular protein is made possible only by aptamers. In 1999, the Famulok group reported intracellular expression of aptamers (which they termed as intramers) against integrin to block cell adhesion (Blind et al., 1999; Mayer et al., 2001). Later on, they simplified the expression procedure by directly transfecting intramers inside cells. Without any stabilization, the unmodified intramers persist inside cells for at least 6 h (Theis et al., 2004). They convincingly showed that by intramer technology, novel biological functions can be assigned to protein or protein domains, and targeted modulation of a signal transduction cascade can be achieved.
As a further advancement of intramer technology, our transfected EGFRvIII aptamers can reduce the amount of mature EGFRvIII that reached the cell surface, suggesting that transfected aptamers can reach cytoplasmic organelles and interfere with protein processing. Recently, we have also identified an aptamer that can specifically block GluR1, a neuron cell membrane receptor, site-specific phosphorylation and prevent its membrane translocation (Liu et al., 2008). Our finding broadens the frontier of aptamer applications, particularly for signal transduction studies. So far, more than 50 tyrosine kinase receptors have been identified (Robinson et al., 2000). They share a common activation step that involves their auto-phosphorylation, followed by the activity of a complex network of adaptors, effectors, etc. It is difficult to disrupt one particular pathway in its natural context to elucidate its function. Antibodies are inert under a cytoplasmic reducing environment. Small molecular inhibitors lack specificity. Aptamers may represent the only solution. We can develop aptamers against a specific cytoplasmic portion of a tyrosine kinase receptor and study the effect of aptamers. Our study has demonstrated that aptamers can efficiently interfere with a post-translational modification of protein, thereby its function, in an intact cell.
A novel strategy for EGFRvIII-targeted therapy against glioblastoma
Currently, the standard treatment for patients with glioblastoma includes surgery to remove the tumor, chemotherapy, and radiotherapy. Despite this effort, the median survival time is only around 6 months (Ohgaki and Kleihues, 2005). It is imperative to develop more specific, effective therapeutics. The fact that EGFRvIII is uniquely expressed only in brain tumor cells makes it an ideal target for therapeutic interventions (Kuan et al., 2001). Monoclonal antibody against EGFRvIII administered intracranially has effectively destroyed gliomas (Riva et al., 1999). EGFR tyrosine kinase inhibitors, erlotinib (OSI-774, Tarceva; Genentech, South San Francisco, CA, USA) and gefitinib (ZD1839, Iresssa; Astra-Zeneca, Wilmington, DE, USA) have been developed and tested for activity against tumors (Raizer, 2005). EGFRvIII is also being used as an antigen in a vaccine-based clinical trial for glioblastoma multiforme. In these trials, there are two concerns that need to be addressed. First, the blood-brain barrier represents a significant obstacle for successful treatment of intro-central nervous system (CNS) tumors. Consistent with our observation that intra-cranial administrated antibody leaks into the systemic circulation, there is a report now suggesting that the anti-EGFR antibody administrated systematically might cross from the systemic circulation into the CNS (Ramos et al., 2006). Moreover, EGFRvIII is resistant to degradation, allowing for prolonged activity at the cancer cell surface (Han et al., 2006). Resistance to the current generation of tyrosine kinase inhibitors may be mediated, in part, by this effect. Our aptamer provides a good solution for both concerns. Firstly, administrated with or without antidote, aptamer function can be refined in a desired region or in a sequential manner. For example, EGFRvIII-specific aptamers can be intracranially injected. Possible aptamer leakage through the blood-brain barrier can be neutralized by systematic antidote administration. Secondly, since our aptamers directly target EGFRvIII biogenesis and maturation, they represent a more potent inhibitor for a stable protein. With the development of novel techniques, such as nanoparticle or polymer-mediated intra-cellular delivery, our aptamers may provide potential for a novel therapy for glioblastoma.
Materials and methods
Selection procedure
In vitro selection was carried out as described previously (Ishizaki et al., 1996), with modifications. A random pool of RNA oligonucleotides of the sequence 5′-GGG AGG ACG ATG CGG (N40) CAG ACG ACT CGC TGA GGA TCC GAG A-3′ (N40 represents 40 random nucleotides with equimolar A, G, C, U) was generated by in vitro transcription with 2′-fluoro CTP and UTP (TriLink Biotech, San Diego, CA, USA), 2′-hydroxy GTP and ATP, and mutant T7 RNA polymerase that efficiently incorporates modified nucleotides (Sousa and Padilla, 1995). EGFRvIII ectodomain was histidine (His)-tagged and expressed in E. coli. Prior to each round of selection, His-tagged proteins were conjugated to Ni-NTA beads (Qiagen, Valencia, CA, USA) and then incubated with RNA pools at 37°C for 15 min. Bound RNA molecules were separated from unbound ones by centrifugation, phenol/chloroform extracted, ethanol precipitated, and amplified by RT-PCR. Selection stringency was increased by decreasing protein concentrations and increasing RNA-protein ratios. To prevent selection of RNA molecules that bind Ni-NTA beads and nitrocellulose membrane which is utilized in the binding assay, the pool was pre-bound to the beads and nitrocellulose disc in the absence of target proteins. RNA molecules that were retained on the beads and discs were discarded before each round of selection. For desired rounds, selected RNA molecules were reverse transcribed, amplified, cloned, and sequenced as described previously (Ishizaki et al., 1996).
Binding affinity assays
We determined RNA-protein equilibrium dissociation constants (Kd) by the double-filter, nitrocellulose-filter binding method as described previously (Wong and Lohman, 1993). For all binding assays, RNA molecules were dephosphorylated by using bacterial alkaline phosphatase (Gibco BRL, Gaithersburg, MD, USA) and 5′-end labeled by using T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) and [γ-32P]ATP. Binding assays were carried out by incubating [32P]-labeled RNA at a concentration of less than 0.2 nm and protein at concentrations ranging from 0.01 to 1 µm in selection buffer F (20 mm HEPES, pH 7.4; 150 mm NaCl; 2 mm CaCl2; 0.01% bovine serum albumin) at 37°C. The fraction of RNA bound was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA). Raw binding data were corrected for non-specific background binding of radiolabeled RNA to the nitrocellulose filter as described previously (Wong and Lohman, 1993).
ELISA
A 96-well plate was coated with EGFRvIII ectodomain at different concentrations (0–0.3 µm) in Dulbecco’s phosphate buffered saline (D-PBS) overnight. The plate was washed in 1× PBS/0.05% Tween-20 and blocked in 1% bovine serum albumin in 1× PBS for 1 h. Next, 100 pmol aptamers, which were annealed to a biotin-labeled 39 oligonucleotide (5′-[biotin]TCTCGGAT-CCTCAGC-3′) that recognize the fixed flanking region, were applied to the proteins and incubated at 37°C for 2 h. After extensive washing, streptavidin-horseradish peroxidase (HRP) was added and incubated for 20 min at room temperature in the dark. HRP substrate was then added and the reaction stopped by 2N H2SO4 20 min later. The optical density of each well was determined by a microplate reader at optical density 450 nm.
Affinity constant determination by surface plasmon resonance
Purified EGFRvIII ectodomain protein was immobilized on the surface of biosensor chips at a level of 2×104 response units for analysis with the BIAcore™3000 (BIAcore, Inc., Piscataway, NJ, USA). Coupling of antigen was achieved by using N-ethyl-N′-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide according to the manufacturers’ instructions. The running buffer was 10 mm HEPES/150 mm NaCl/3.4 mm EDTA, pH 7.4. The aptamer E21 and control samples were passed over the biosensor chip at concentrations from 25 to 200 nm. Starting RNA pool was included as a negative control. The association and dissociation rate constants (Ka and Kd) were determined by using the non-linear curve-fitting BIAevaluation software (BIAcore, Inc.).
Western blotting
For Western blotting, 0.2 µg of E. coli and baculovirus-expressed EGFRvIII ectodomain, as well as deglycosylated EGFRvIII were separated on a 10% Tris-HCl precast gel (Bio-Rad, Hercules, CA, USA), transferred to a polyvinylidene fluoride (PVDF) membrane, and probed as previously described (Mi et al., 2007). Deglycosylation was performed either by a chemical (trifluoromethanesulfonic acid, TMFS; Chemical deglycosylation kit; Sigma, St. Louis, MO, USA) or by an enzymatic digestion (PNGase F; New England Biolabs), following the manufacturer’s protocol.
Cell culture and transfection
NR6M, a mouse cell line overexpressing EGFRvIII (Batra et al., 1995), was grown in improved MEM Zinc option medium (Invitrogen Inc., Carlsbad, CA, USA) with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. For transfection, NR6M cells were plated on a 6-well plate at 8×105 cells/well, grown overnight, and 100 nm EGFRvIII aptamers or RNA library were used, together with siPORT lipid (Ambion, Austin, TX, USA). Then, 28 h after transfection, cells were analyzed as described below.
Membrane protein isolation and detection
Transfected NR6M cells were rinsed with cytostatic factor (CSF) buffer (150 mm NaCl, 3 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, 10 mm glucose, pH 7.4), and then incubated at 10°C with 1 mm sulfo-NHS-SS-biotin in CSF buffer for 30 min and lysed with RIPA buffer [0.15 mm NaCl; 0.05 mm Tris-HCl, pH 7.4; 10 µg/ml aprotinin; 0.5 mm phenylmethylsulfonyl fluoride (PMSF); 1% sodium deoxycholate; 1% Triton X-100; 0.1% SDS] after washing with ice-cold CSF (Man et al., 2007). Biotinylated surface proteins were precipitated with immobilized streptavidin beads, and the membrane EGFRvIII expression was probed with L8A4 antibody (Reist et al., 1995). GAPDH probing served as a loading control.
Hoechst 33342 staining for apoptotic morphology
Transfected NR6M cells were fixed in methanol:acetic acid (3:1) for 5 min at 4°C and washed three times with water. Subsequently, the cells were stained with Hoechst 33342 (5 µg/ml; Calbiochem, La Jolla, CA, USA) for 10 min at room temperature. Cells were washed three times with water, and apoptotic nuclei were visualized by fluorescence microscopy.
Acknowledgments
We thank Scott Szafranski and Jacoba G. Slagter-Jä ger for technique support and helpful discussions. This work is supported by NIH grant U54-CA-119343, NINDS Grant 5P50 NS20023-25, NIH SPORE Grant 5P50 CA108786-05, and NIH Merit Award R37 CA 011898-38.
References
- Baselga J. Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist. 2002;7(Suppl. 4):2–8. doi: 10.1634/theoncologist.7-suppl_4-2. [DOI] [PubMed] [Google Scholar]
- Batra SK, Castelino-Prabhu S, Wikstrand CJ, Zhu X, Humphrey PA, Friedman HS, Bigner DD. Epidermal growth factor ligand-independent, unregulated, cell-transforming potential of a naturally occurring human mutant EGFRvIII gene. Cell Growth Differ. 1995;6:1251–1259. [PubMed] [Google Scholar]
- Blind M, Kolanus W, Famulok M. Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. Proc. Natl. Acad. Sci. USA. 1999;96:3606–3610. doi: 10.1073/pnas.96.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunka DH, Stockley PG. Aptamers come of age – at last. Nat. Rev. Microbiol. 2006;4:588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Durand G, Seta N. Protein glycosylation and diseases: blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin. Chem. 2000;46:795–805. [PubMed] [Google Scholar]
- Ermonval M, Kitzmuller C, Mir AM, Cacan R, Ivessa NE. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 2001;11:565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- Garcia de Palazzo IE, Adams GP, Sundareshan P, Wong AJ, Testa JR, Bigner DD, Weiner LM. Expression of mutated epidermal growth factor receptor by non-small cell lung carcinomas. Cancer Res. 1993;53:3217–3220. [PubMed] [Google Scholar]
- Han W, Zhang T, Yu H, Foulke JG, Tang CK. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer Biol. Ther. 2006;5:1361–1368. doi: 10.4161/cbt.5.10.3226. [DOI] [PubMed] [Google Scholar]
- Ishizaki J, Nevins JR, Sullenger BA. Inhibition of cell proliferation by an RNA ligand that selectively blocks E2F function. Nat. Med. 1996;2:1386–1389. doi: 10.1038/nm1296-1386. [DOI] [PubMed] [Google Scholar]
- Johns TG, Mellman I, Cartwright GA, Ritter G, Old LJ, Burgess AW, Scott AM. The antitumor monoclonal antibody 806 recognizes a high-mannose form of the EGF receptor that reaches the cell surface when cells over-express the receptor. FASEB J. 2005;19:780–782. doi: 10.1096/fj.04-1766fje. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp. Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- Kim YK, Shin HS, Tomiya N, Lee YC, Betenbaugh MJ, Cha HJ. Production and N-glycan analysis of secreted human erythropoietin glycoprotein in stably trans-fected Drosophila S2 cells. Biotechnol. Bioeng. 2005;92:452–461. doi: 10.1002/bit.20605. [DOI] [PubMed] [Google Scholar]
- Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocr. Relat. Cancer. 2001;8:83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sun QA, Chen Q, Lee TH, Huang Y, Wetsel WC, Michelotti GA, Sullenger BA, Zhang X. Targeting inhibition of GluR1 Ser845 phosphorylation with an RNA aptamer that blocks AMPA receptor trafficking. J. Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05748.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man HY, Sekine-Aizawa Y, Huganir RL. Regulation of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor trafficking through PKA phosphorylation of the Glu receptor 1 subunit. Proc. Natl. Acad. Sci. USA. 2007;104:3579–3584. doi: 10.1073/pnas.0611698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Rendon E, Blake DJ. Protein glycosylation in disease: new insights into the congenital muscular dystrophies. Trends Pharmacol. Sci. 2003;24:178–183. doi: 10.1016/S0165-6147(03)00050-6. [DOI] [PubMed] [Google Scholar]
- Mayer G, Blind M, Nagel W, Bohm T, Knorr T, Jackson CL, Kolanus W, Famulok M. Controlling small guanine-nucleotide-exchange factor function through cytoplasmic RNA intramers. Proc. Natl. Acad. Sci. USA. 2001;98:4961–4965. doi: 10.1073/pnas.091100698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn J. Targeting the epidermal growth factor receptor for cancer therapy. J. Clin. Oncol. 2002;20:1S–13S. [PubMed] [Google Scholar]
- Mi J, Zhang X, Liu Y, Reddy SK, Rabbani ZN, Sullenger BA, Clary BM. NF-κB inhibition by an adenovirus expressed aptamer sensitizes TNFα-induced apoptosis. Biochem. Biophys. Res. Commun. 2007;359:475–480. doi: 10.1016/j.bbrc.2007.05.125. [DOI] [PubMed] [Google Scholar]
- Moscatello DK, Holgado-Madruga M, Godwin AK, Ramirez G, Gunn G, Zoltick PW, Biegel JA, Hayes RL, Wong AJ. Frequent expression of a mutant epidermal growth factor receptor in multiple human tumors. Cancer Res. 1995;55:5536–5539. [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- Oney S, Nimjee SM, Layzer J, Que-Gewirth N, Ginsburg D, Becker RC, Arepally G, Sullenger BA. Antidote-controlled platelet inhibition targeting von Willebrand factor with aptamers. Oligonucleotides. 2007;17:265–274. doi: 10.1089/oli.2007.0089. [DOI] [PubMed] [Google Scholar]
- Que-Gewirth NS, Sullenger BA. Gene therapy progress and prospects: RNA aptamers. Gene Ther. 2007;14:283–291. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- Raizer JJ. HER1/EGFR tyrosine kinase inhibitors for the treatment of glioblastoma multiforme. J. Neurooncol. 2005;74:77–86. doi: 10.1007/s11060-005-0603-7. [DOI] [PubMed] [Google Scholar]
- Ramos TC, Figueredo J, Catala M, Gonzalez S, Selva JC, Cruz TM, Toledo C, Silva S, Pestano Y, Ramos M, et al. Treatment of high-grade glioma patients with the humanized anti-epidermal growth factor receptor (EGFR) antibody h-R3: report from a phase I/II trial. Cancer Biol. Ther. 2006;5:375–379. doi: 10.4161/cbt.5.4.2522. [DOI] [PubMed] [Google Scholar]
- Reist CJ, Archer GE, Kurpad SN, Wikstrand CJ, Vaidyanathan G, Willingham MC, Moscatello DK, Wong AJ, Bigner DD, Zalutsky MR. Tumor-specific antiepidermal growth factor receptor variant III monoclonal antibodies: use of the tyramine-cellobiose radioiodination method enhances cellular retention and uptake in tumor xenografts. Cancer Res. 1995;55:4375–4382. [PubMed] [Google Scholar]
- Riva P, Franceschi G, Frattarelli M, Lazzari S, Riva N, Giuliani G, Casi M, Sarti G, Guiducci G, Giorgetti G, et al. Loco-regional radioimmunotherapy of high-grade malignant gliomas using specific monoclonal antibodies labeled with 90Y: a phase I study. Clin. Cancer Res. 1999;5:3275s–3280s. [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lin SF. The protein tyrosine kinase family of the human genome. Oncogene. 2000;19:5548–5557. doi: 10.1038/sj.onc.1203957. [DOI] [PubMed] [Google Scholar]
- Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger BA, Gilboa E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- Theis MG, Knorre A, Kellersch B, Moelleken J, Wieland F, Kolanus W, Famulok M. Discriminatory aptamer reveals serum response element transcription regulated by cytohesin-2. Proc. Natl. Acad. Sci. USA. 2004;101:11221–11226. doi: 10.1073/pnas.0402901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrand CJ, Hale LP, Batra SK, Hill ML, Humphrey PA, Kurpad SN, McLendon RE, Moscatello D, Pegram CN, Reist CJ, et al. Monoclonal antibodies against EGFRvIII are tumor specific and react with breast and lung carcinomas and malignant gliomas. Cancer Res. 1995;55:3140–3148. [PubMed] [Google Scholar]
- Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc. Natl. Acad. Sci. USA. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Caprioli RM, Staros JV. Characterization of glycosylation sites of the epidermal growth factor receptor. Biochemistry. 2003;42:5478–5492. doi: 10.1021/bi027101p. [DOI] [PMC free article] [PubMed] [Google Scholar]