Summary
The ability of cells to faithfully duplicate their two centrioles once per cell cycle is critical for proper mitotic progression and chromosome segregation. Multi-ciliated cells represent an interesting variation of centriole duplication in that these cells generate greater than 100 centrioles, which form the basal bodies of their motile cilia. This centriole amplification is proposed to require a structure termed the deuterosome, thought to be capable of promoting de novo centriole biogenesis. Here, we begin to molecularly characterize the deuterosome and identify it as a site for the localization of Cep152, Plk4, and SAS6. Additionally we identify CCDC78 as a centriole-associated and deuterosome protein that is essential for centriole amplification. Overexpression of Cep152, but not Plk4, SAS6 or CCDC78, drives over-amplification of centrioles. However, in CCDC78 morphants, Cep152 fails to localize to the deuterosome and centriole biogenesis is impaired, indicating CCDC78-mediated recruitment of Cep152 is required for deuterosome-mediated centriole biogenesis.
Introduction
The ability of ciliated epithelia to generate directed fluid flow is a critical component of tissue function in multiple contexts, such as proper neural development, egg migration through the oviduct and mucus clearance in the respiratory tract (Afzelius, 1976; Dirksen and Satir, 1972; Sawamoto et al., 2006). Loss of directional fluid flow leads to deleterious phenotypes including hydrocephaly, infertility and respiratory dysfunction (Bush et al., 1998). The skin of Xenopus embryos is comprised of a multi-ciliated epithelium similar to that found in the vertebrate airways, oviduct and ependyma (Werner and Mitchell, 2011). This tissue consists of various cell types including multi-ciliated cells (MCCs) that contain numerous motile cilia, which beat in a concerted manner to generate directed flow.
To generate multiple cilia, MCCs must first undergo a massive centriole amplification that will generate hundreds of centrioles (Dirksen, 1971; Kalnins and Porter, 1969; Sorokin, 1968). Centriole amplification occurs deep in the cell, and is followed by an apical migration of centrioles that ultimately dock with the apical surface and become the basal bodies of the motile cilia. Numerous groups have used Electron Microscopic (EM) techniques to describe two modes of centriole amplification in the MCCs of multiple systems including the Xenopus skin, mouse ependyma, and numerous vertebrate respiratory tracts and oviducts (Anderson and Brenner, 1971; Sorokin, 1968; Staprans and Dirksen, 1974; Steinman, 1968).
The first mode resembles typical centriole duplication in that daughter nucleation occurs orthogonal to the mother centriole. However, instead of a single daughter nucleating off the mother centriole, multiple daughters nucleate simultaneously. This is reminiscent of centriolar rosettes that have been observed around the mother centriole in cycling cells in which the critical centriolar components Polo like kinase 4 (Plk4), Cep152 or SAS6 have been manipulated (Dzhindzhev et al., 2010; Kleylein-Sohn et al., 2007; Peel et al., 2007). However, while it is common to have supernumerary centrioles, these mutant cells typically have less than 10 suggesting that mother centriole driven duplication alone cannot account for the approximately 150 centrioles found in MCCs.
A second ‘acentriolar’ mode of amplification has been described in which centrioles nucleate de novo without a mother centriole. Instead of forming clusters around a mother centriole, nascent centrioles bud off of a non-descript electron dense structure termed the deuterosome (Dirksen, 1971; Kalnins and Porter, 1969; Sorokin, 1968; Steinman, 1968). The deuterosome has also been called the precursor body as well as the condensation form, as it is thought to condense from amorphous filamentous material known as the fibrogranular complex (Anderson and Brenner, 1971; Staprans and Dirksen, 1974). In MCCs, the fibrogranular complex and deuterosomes are distinct from fibrous granules that are thought to be synonymous with the centriolar satellites that are found concentrated at the pericentriolar material (PCM) around centrosomes. While PCM-1 has been identified as a component of the fibrous granules it is not located at the deuterosome (Kubo et al., 1999; Vladar and Stearns, 2007).
Based on detailed EM analysis, the acentriolar mode of amplification has been proposed to account for the vast majority of centriole biogenesis in MCCs (Anderson and Brenner, 1971; Sorokin, 1968). Here we describe the fortuitous finding of a particular GFP fusion construct of CCDC78 that localizes specifically to an acentriolar structure at the center of centriolar clusters in MCCs undergoing amplification, allowing us to begin analyzing the molecular make up and regulation of deuterosome-mediated centriolar biogenesis.
Results
CCDC78 is a novel centriole associated/deuterosome protein
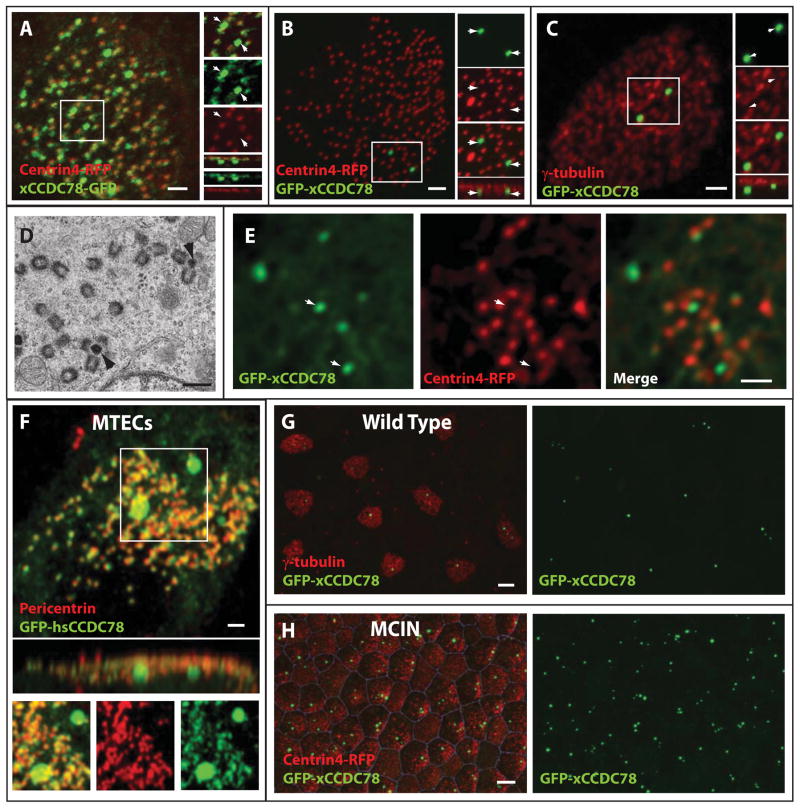
We initially identified coiled-coil domain containing protein 78 (xCCDC78) as a gene highly upregulated in the ciliated epithelia of Xenopus embryonic skin during the developmental window of centriole biogenesis (Stubbs et al., 2008). The xCCDC78 gene encodes a 559 amino acid protein containing multiple coiled-coil domains but no other predicted functional domains. CCDC78 has recently been genetically implicated in congenital myopathy although the biochemical nature of this remains obscure (Majczenko et al., 2012). In the ciliated epithelium of Xenopus embryos a C-terminal GFP tagged construct of xCCDC78 (xCCDC78-GFP) localizes weakly at the numerous centrioles in the MCCs (Figure 1A). We observe considerable association between xCCD78-GFP and centrioles marked with Centrin4-RFP (97%±0.65, n=9 cells)(Gavet et al., 2003). Furthermore, xCCDC78-GFP localizes strongly to additional foci specific to MCCs (Figure 1A). These foci do not co-localize with the core centriolar component Centrin4, indicating these xCCDC78-GFP foci are acentriolar structures (Figure 1A, inlay)(Gavet et al., 2003). The EM literature describes an acentriolar MCC specific structure, the deuterosome, involved in the de novo formation of nascent centrioles. Based on the data presented below we propose that xCCDC78 localizes to centrioles and the molecularly uncharacterized deuterosome. However, future work using immuno-EM based experiments will be required to definitively determine the precise subcellular localization of CCDC78.
Figure 1.
MCC-specific localization of xCCDC78. (A) Co-localization of C-terminally tagged xCCDC78-GFP with the centriole marker Centrin4-RFP (inlay shows a separate acentriolar foci of xCCDC78-GFP localization not co-localizing with Centrin4) (scale bar, 2μm). (B–C) Localization of N-terminal tagged GFP-xCCDC78 to an acentriolar structure below the apically fused centrioles (i.e. basal bodies) marked with either Centrin4-RFP (B) or γ-tubulin (C) (scale bars, 2μm). (D) TEM micrograph from MTECs depicting a deuterosome (arrowhead) with nascent centrioles forming around it (scale bar, 0.5μm). (E) Localization of GFP-xCCDCD78 to the center of nascent centriole clusters (arrowheads) marked by Centrin4-RFP during the process of centriole biogenesis (scale bar, 2μm). (F) Localization of GFP-hsCCDC78 to centrioles marked with pericentrin and to an acentriolar structure in MTECs (scale bar, 2μm). (G–H) MCC specific localization of GFP-xCCDC78 which is punctate in wild type ciliated epithelia (G) and becomes ubiquitous in MCIN induced ciliated epithelia (H)(scale bars, 10μm)(See also Figure S1 and S2).
N-terminal tagged CCDC78 is a unique identifier of the MCC specific deuterosome
Interestingly, an N-terminal tagged version of xCCDC78 (GFP-xCCDC78) failed to localize to centrioles as marked by either Centrin4-RFP or γ-tubulin but continued to localize to the acentriolar foci in MCCs (Figure 1B,C,E)(Muresan et al., 1993; Park et al., 2008). During centriole amplification we find these foci clustered at the site of centriole biogenesis adjacent to nascent centrioles, consistent with our interpretation that these foci represent the deuterosome (Figure 1E and S1B). We quantified the number of centrioles budding off of a given deuterosome in the MCCs of the mouse tracheal epithelium using EM and in Xenopus using Centrin4–RFP to mark the centrioles and GFP-xCCDC78 to mark the presumptive deuterosome (Figure S1A and B). In both cases, deuterosomes are often found unoccupied by growing centrioles. Furthermore, while they are capable of nucleating many centrioles simultaneously, it is more common for them to nucleate only 1 or 2 centrioles at a time (Figure S1A–C and S2A). Thus, GFP-xCCDC78 foci surrounded by Centrin4-RFP positive centrioles resemble the in vivo configuration of deuterosome/centriole complexes precisely, suggesting that GFP-xCCDC78, in fact marks the deuterosome. In mature MCCs that have completed centriole biogenesis, GFP-xCCDC78 is found in foci near the apical surface, but below the apically docked centrioles (i.e. basal bodies)(Figures 1B and C). We injected in vitro synthesized mRNA encoding GFP-xCCDC78 at the two-cell stage which results in ubiquitous protein synthesis in all epithelial cells (Werner and Mitchell, 2013). Interestingly, localization of the GFP-xCCDC78 protein is restricted to distinct foci specifically in MCCs, indicating that GFP-xCCDC78 localizes to a structure unique to these cells (Figure 1G).
We next wanted to test this MCC-specific protein localization in Xenopus skin. MCCs typically form a punctate pattern across the surface of the epithelium, but ectopic expression of the geminin-like protein Multicilin (MCIN) has been shown to convert all outer epithelial cells into MCCs (Stubbs et al., 2012). To test this MCC restricted localization we co-injected mRNA encoding GFP-xCCDC78 with mRNA encoding a Dexamethasone (Dex) inducible MCIN. Injected embryos treated with Dex contain epithelia comprised entirely of MCCs, marked by the presence of numerous Centrin4-RFP marked centrioles on the apical surface of every cell. We found that these cells now all contain foci of GFP-xCCDC78 indicating the subcellular localization of GFP-xCCDC78 protein is indeed restricted to MCCs and that deuterosome formation is downstream of MCIN in the MCC differentiation program (Figure 1H).
CCDC78 protein localization is conserved in vertebrates
The discrepancy between the C-terminal and N-terminal GFP constructs of xCCDC78 led us to test the localization of this protein in other systems. Consistent with the data from Xenopus, in the MCCs of mouse tracheal epithelial cells (MTECs), N- or C- terminal GFP fusions of xCCDC78 and hsCCDC78 as well as immunofluorescence (IF) of the endogenous protein using two CCDC78 antibodies reveals localization to centrioles and an acentriolar cluster (Figure 1F and S2B). The acentriolar clusters in MTECs are significantly larger than the foci in Xenopus or what would be predicted to be deuterosomes based on the EM literature, suggesting the CCDC78 localization may represent a less discreet structure in these cells, perhaps localizing to the fibrogranular material from which the deuterosome condenses (Figure S2A and S2B). Finally, both GFP fusions of hsCCDC78 and CCDC78 antibody labeling, reveals localization to the centrioles in human RPE-1 cells, with a bias for the mother centriole (Figure S2C and D). These results indicate that CCDC78 is a centriole associated protein that also localizes to a MCC-specific structure, the presumptive deuterosome. Our localization studies indicate a conserved commonality of CCDC78 protein enrichment to sites of centriole biogenesis. Additionally, the localization of GFP-xCCDC78 specifically to acentriolar sites of centriole biogenesis, but not centrioles, provides a powerful tool for the molecular characterization of this structure. These observations prompted us to use this construct as a tool to define the localization of other proteins involved in centriole duplication in relation to the GFP-xCCDC78 foci.
CCDC78 is required for proper centriole amplification in MCCs
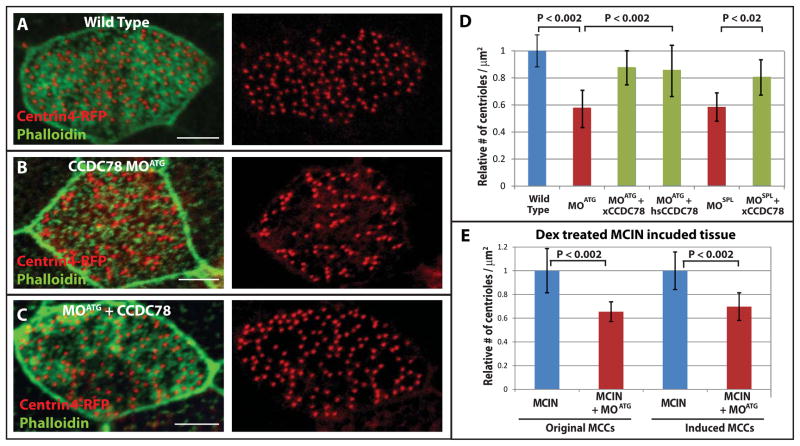
Localization of GFP-xCCDC78 to the acentriolar deuterosome led us to test whether centriole amplification in MCCs was regulated by xCCDC78 using morpholino oligonucleotide (MO) mediated knockdown. MCCs vary considerably in size yet maintain a constant ratio of centrioles per micron2 of apical surface. We quantified the number of centrioles per micron2 in wild type embryos and embryos injected with xCCDC78 MOs (Figure 2A,B,D). We injected 2–4 cell Xenopus embryos with mRNA encoding Centrin4-RFP together with either a start site MO (MOATG) or a splice site MO (MOSPL) targeting xCCDC78. Both the MOATG and the MOSPL resulted in a significant decrease in the number of centrioles generated (Figure 2D). Injection of both the MOATG (or MOSPL) and the GFP-xCCDC78 construct not targeted by the MO resulted in a significant rescue of centriole number (Figure 2C and D). Importantly, this rescue can occur with the GFP-xCCDC78, which only localizes to the deuterosome, suggesting this is the critical site of its function and indicating a role for CCDC78 in an acentriolar centriole amplification pathway (Figure 2C and D). Loss of centrioles is also rescued using RFP-hsCCDC78 suggesting the protein is functionally conserved throughout vertebrate evolution (Figure 2D). Importantly, morphant embryos are morphologically normal and we observe no significant defects in centriole duplication in non-MCCs (Figure S3A and D). This result suggests CCDC78 is not essential for cell cycle regulated centriole duplication, but is specifically required for the deuterosome mediated centriole amplification that is restricted to MCCs.
Figure 2.
xCCDC78 is required for proper centriole amplification in MCCs. (A–C) Representative images of a wild type MCC (A), CCDC78 morphant (B), and morphant rescued with GFP-xCCDC78 (C) that are co-injected with Centrin4-RFP to mark centrioles and stained with Phalloidin to mark cell boundaries (scale bar, 5μm). (D) Quantification of centrioles per micron2 surface area in MCCs of wild type embryos (n=75 cells) or embryos injected with CCDC78 MOATG (n=36, to wild type p<0.002), CCDC78 MOATG + GFP-xCCDC78 (n=64, to MOATG p<0.002), CCDC78 MOATG + RFP-hsCCDC78 (n=49, to MOATG p<0.002), CCDC78 MOSPL (n=58, to wild type p<0.002), or CCDC78 MOSPL + GFP-xCCDC78 (n=32, to MOSPL p<0.02) Error bars, SD. (E) Quantification of centrioles per micron2 surface area in Dex treated MCIN induced ciliated epithelia comparing cells that would normally have become MCCs (Original) and outer cells converted into MCCs (Induced) with (MCCs n=24, MCIN MCCs n=60) or without xCCDC78 MOATG (MCCs n=39 p<0.002, MCIN MCCs n=63 p<0.002). Error bars, SD. (See also Figure S3).
Localization of GFP-xCCDC78 in MCIN induced ectopic MCCs (Figure 1H) suggests CCDC78 is downstream of MCIN in the regulation of centriole biogenesis. To test this, we repeated our MO experiments in embryos expressing MCIN and found that ectopic MCCs also have a significant decrease in the number of centrioles generated (Figure 2E). Therefore, both wild type and ectopic MCCs require functional xCCDC78 to generate proper centriole numbers and CCDC78 is a critical downstream component of MCIN-induced MCC specification.
Depletion of CCDC78 leads to loss of tissue level cilia driven fluid flow
The normal gross morphology of xCCDC78 morphants led us to ask whether there was a physiological consequence of xCCDC78 depletion. The primary function of MCCs in numerous biological contexts is to generate directed fluid flow (Marshall and Kintner, 2008). The importance of cilia driven fluid flow is revealed in patients with primary ciliary dyskinesia (PCD) who present with numerous developmental and physiological problems including increased risk of hydrocephalus and respiratory infections (Bush et al., 1998). We found that xCCDC78 morphants have a significant decrease in cilia-generated fluid flow as measured by the displacement of fluorescent beads across the surface of the embryo (Figure S3B) (Werner and Mitchell, 2013). This loss of flow is consistent with the decrease in cilia that occurs as a result of the centriole biogenesis defect (Figure S3C). Importantly, the loss of flow is significantly restored in embryos where the centriole biogenesis defect is rescued by expression of GFP-xCCDC78 (Figure S3B).
GFP-CCDC78 transiently co-localizes with SAS6 but not Centrin4 or γ-tubulin
In an effort to identify components of the deuterosome we performed co-localization experiments with proteins known to be involved in centriole duplication. We performed these experiments both “early” when the deuterosomes are deep in the cell undergoing centriole biogenesis and “late” when mature MCCs have complete centriole amplification and the deuterosomes are just below the apical surface (Figure 3). In MCCs undergoing centriole amplification, as well as mature cells, we do not see co-localization of GFP-xCCDC78 with the MCC centriole markers Centrin4-RFP and γ-tubulin confirming that the foci are acentriolar (Figure 1B and C).
Figure 3.
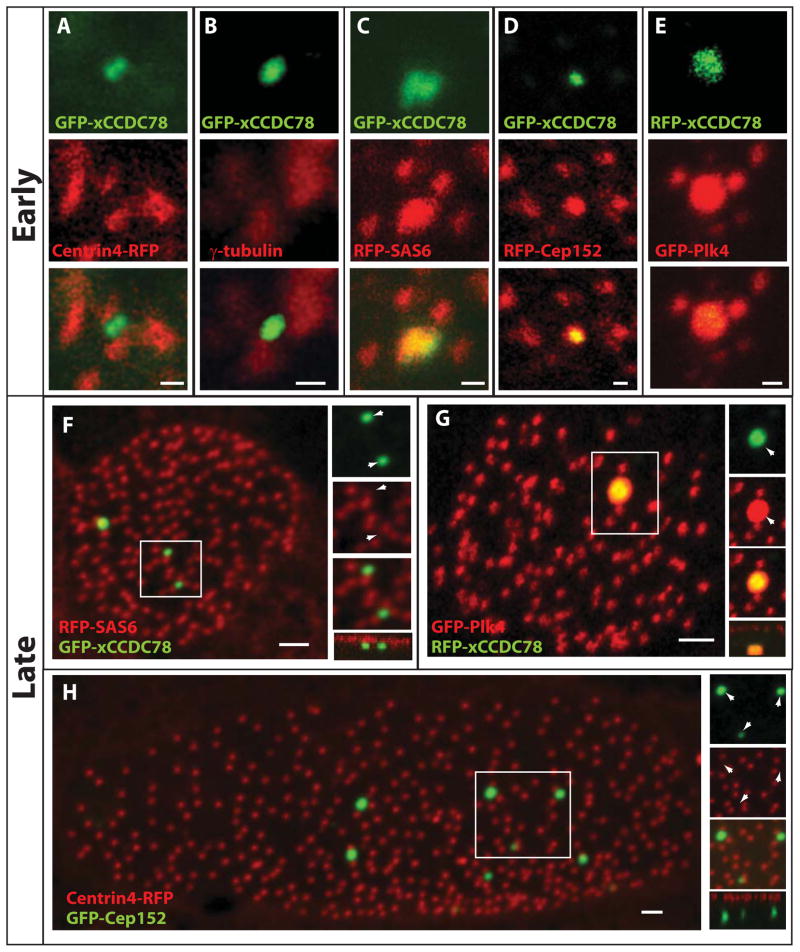
Localization of key components of centriole biogenesis to the deuterosome during centriole amplification. (A–E) Localization of GFP-xCCDC78 during centriole amplification with centriolar components centrin4-RFP (A), γ-tubulin (B), RFP-SAS6 (C), RFP-Cep152 (D) and GFP-Plk4 with RFP-xCCDC78 (E) (pseudo-colored for consistency) (Scale bars, 0.5 μm). (F) Localization of GFP-xCCDC78 in mature MCC that has completed centriole biogenesis with RFP-SAS6 (F). (G) Localization of GFP-Plk4 and RFP-xCCDC78 in a mature MCC (pseudo-colored for consistency) (scale bars, 2μm). (H) Localization of RFP-Cep152 in mature MCCs to the acentriolar structure not marked by GFP-Centrin4 (scale bar, 2μm)(See also Figure S4).
During centriole assembly there is a temporal hierarchy of structural proteins added to the growing centriole (Brito et al., 2012; Delattre et al., 2006). One of the earliest components, SAS6, is capable of establishing the initial 9 fold symmetric pattern of centriole structure (Gopalakrishnan et al., 2010; Kitagawa et al., 2011; van Breugel et al., 2011). Importantly, RFP-SAS6 localizes to centrioles in both non-ciliated cells (i.e. centrosomes) and in MCCs (i.e. basal bodies). As the MT based centriole grows, numerous other proteins become incorporated, including centrins and γ-tubulin (Brito et al., 2012). While we do not see co-localization of GFP-xCCDC78 with Centrin4-RFP or γ-tubulin we do see a transient co-localization with RFP-SAS6 in clusters of nascent centrioles suggesting that this early structural component is recruited to the deuterosome during the initiation of centriole formation (Figure 3A–C, S4A). In mature MCCs that have completed centriole biogenesis there is no co-localization of RFP-SAS6 and GFP-xCCDC78 despite the continued localization of RFP-SAS6 at centrioles (Figure 3F). This continued localization of SAS6 in centrioles but not the GFP-xCCDC78 foci supports our claim that this structure represents the acentriolar deuterosome.
Plk4 localizes to the deuterosome
Cell cycle regulated centriole duplication is well characterized and numerous regulators have been identified including Plk4 and Cep152 (Blachon et al., 2008; Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Habedanck et al., 2005; Hatch et al., 2010; Sillibourne et al., 2010). Plk4 is known in multiple systems to be the critical initial regulator of centriole biogenesis (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Pearson and Winey, 2010). In Xenopus embryos, injection of mRNA encoding GFP-Plk4 leads to early embryonic lethality consistent with the role of Plk4 in centriole duplication and the detrimental effects of its mis-regulation (Brownlee and Rogers, 2013). We therefore used plasmid DNA injections in which the α-tubulin promoter, which restricts expression to post-mitotic MCCs, was employed to drive expression of GFP-Plk4 (Stubbs et al., 2006). Using this construct we found that GFP-Plk4 strongly co-localized with the deuterosome as marked with RFP-xCCDC78 in clusters of forming centrioles and also co-localized with nascent centrioles as marked with Centrin4-BFP (Figure 3E and S4C). In mature cells, GFP-Plk4 is still strongly enriched at the deuterosome and continues to be localized at the centrioles (Figure 3G and S4D). Surprisingly, the over-expression of Plk4 in MCCs did not significantly alter the number of centrioles generated suggesting that Plk4 is not the rate-limiting factor in centriole formation in MCCs (147±65 vs. wt 165 ± 27, p=0.33, n=21 and 75 cells).
Cep152 localizes to the deuterosome and is rate-limiting for centriole amplification
The maintenance of Plk4 at mother centrioles is mediated by Cep152 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). We have observed in centriolar clusters that RFP-Cep152 co-localizes to both deuterosomes and the nascent centrioles during centriole amplification (Figure 3D and S4B). Additionally, we continue to observe RFP-Cep152 localization to the deuterosomes of mature MCCs, while we lose localization to the centrioles (Figure 3H, S4D and E).
While the overexpression of Plk4, SAS6 or CCDC78 did not alter centriole number, the overexpression of Cep152 leads to a dramatic increase in both MCC size and the number of centrioles generated (Figure 3H and 4C). The increase in centriole number could result from an increase in deuterosomes or higher activity at the existing deuterosomes. To address this we counted the number of CCDC78 positive foci in mature MCCs and found that wild type cells contained 1.5 (± 0.8) deuterosomes in contrast to Cep152 overexpressing cells that contained 9.6 (± 4.9) (p=4E-39) (e.g. Figure 1B, G and 3H). This indicates that Cep152 protein levels feedback to control deuterosome number and subsequently centriole biogenesis.
Figure 4.
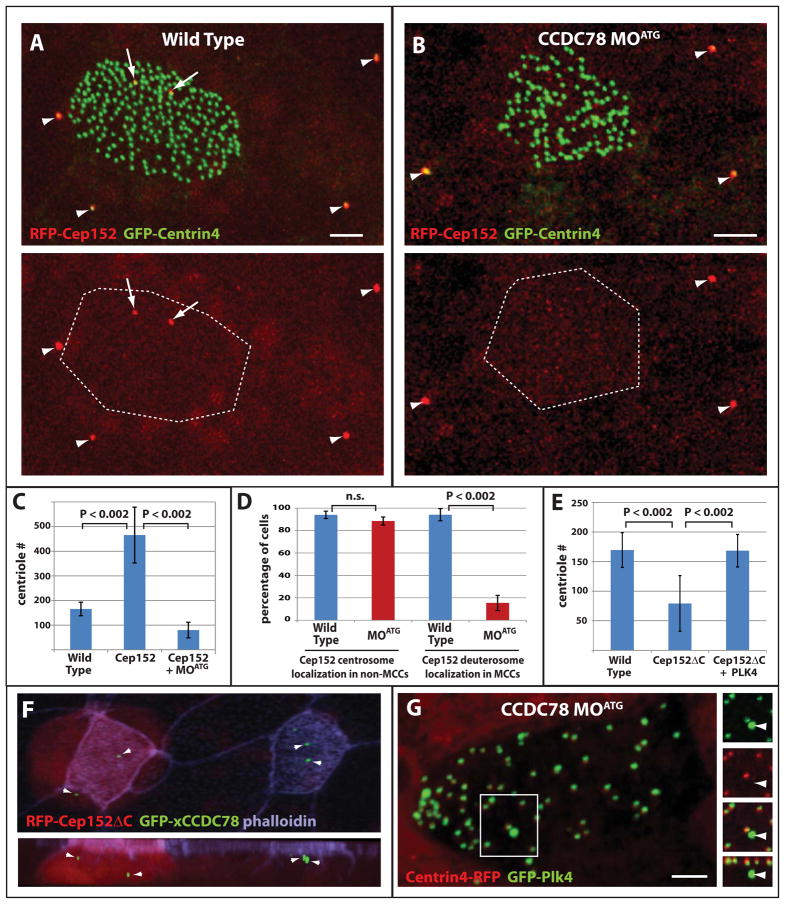
Role of Cep152 in centriole amplification. (A–B) Representative images of wild type (A) and CCDC78 morphant (B) ciliated epithelia showing co-localization of RFP-Cep152 with GFP-Centrin4 at centrosomes in non-ciliated epithelial cells (arrowheads) and the presence (A) or absence (B) of acentriolar localization in MCCs (arrows, scale bars, 5μm). (C) Quantification of centriole number in wild type embryos (n=75), embryos overexpressing Cep152 (n=42, p<0.002) and embryos overexpressing Cep152 in the presence of CCDC78 MOATG (n=86, p<0.002). Error bars, SD. (D) Percentage of cells containing RFP-Cep152 localization in the centrioles of non-ciliated epithelial cells and in the deuterosomes of MCCs in wild type (non-MCCs n=3 embryos, 335 cells, MCCs n=3 embryos, 72cells) and xCCDC78 MOATG morphant embryos (non-MCCs n=3 embryos, 396cells, MCCs n=embryos, 34cells)(Wt vs. MOATG non-MCCs, p=0.08, Wt vs. MOATG MCCs, p<0.002). Error bars, SD. (E) Quantification of centriole number in wild type cells (n=35 cells from 3 embryos), or cells expressing RFP-Cep152ΔC alone (n=34 cells from 4 embryos) or in combination with GFP-Plk4 (n=13 cells from 3 embryos). (F) Image from a mosaic embryo showing the localization of GFP-xCCDC78 in acentriolar foci in both wild type and Cep152ΔC expressing MCCs (red). (G) Image of CCDC78 morphant cell showing localization of GFP-Plk4 at both centrioles (marked with Centrin4-RFP) and at the deuterosome (no co-localization with Centrin4)(scale bars, 5μm).
Cep152ΔC acts as a dominant negative to disrupt centriole amplification
Cep152 is responsible for maintaining Plk4-localization at mother centrioles and it has been shown that an N-terminal fragment of Cep152 can bind to Plk4 but fails to localize to centrioles (Hatch et al., 2010; Sonnen et al., 2013). Overexpression of this construct in U2OS cells sequesters Plk4 away from centrioles disrupting centriole duplication (Sonnen et al., 2013). We drove expression of a similar construct (xCep152ΔC) specifically in post-mitotic MCCs using the α-tubulin promoter and found that it fails to localize to deuterosomes and leads to a significant decrease in the number of centrioles generated (Figure 4E and F).
If this phenotype is related to the ability of Cep152 to maintain Plk4 at the deuterosome, similar to its role at centrioles, one would predict that the overexpression of Plk4 would rescue the loss of centrioles. To test this we drove overexpression of both Cep152ΔC and Plk4 in the same cells. Co-expression of these constructs in MCCs resulted in a complete rescue of centriole biogenesis (Figure 4E). This result suggests that, similar to its role at mother centrioles, Cep152 is required for Plk4 function at the deuterosome. To test whether Cep152ΔC disrupted the deuterosome structure we generated mosaic embryos in which some MCCs received Cep152ΔC while others were wild type. We saw no difference in the accumulation of GFP-xCCDC78 positive foci in these two conditions indicating that the deuterosome is still present (Figure 4F).
Cep152 localization and function at the deuterosome requires CCDC78
The co-localization of Cep152 in mature MCCs to deuterosomes, but not centrioles allowed us to determine the relationship between these two proteins. We injected embryos with both mRNA encoding RFP-Cep152 and the MOATG to determine the effect of xCCDC78 depletion on Cep152 induced over-amplification of centrioles and the localization of Cep152 at the deuterosome. The average number of centrioles in a wild type embryo is 165 ± 27 (Figure 4C), and increases dramatically to 465 ± 113 in the presence of RFP-Cep152 (Figure 4C). This increase is completely abolished in the presence of xCCDC78 MOATG as centriole numbers fall to 95 ± 32 (Figure 4C), a number similar to when the xCCDC78 MOATG is injected alone (Figure 2D).
To explore the underlying mechanism of this result we tested the localization of Cep152 in CCDC78 morphants. A loss of Cep152 localization in MCCs could reflect a specific requirement for CCDC78 in the recruitment of Cep152 or a general loss of the deuterosome structure. To address this we took advantage of the fact that Cep152 localizes to both the centrosomes of non-MCCs (associated with Centrin4-RFP) and to the deuterosomes of MCCs (not associated with Centrin4-RFP e.g. Figure 3H). We found that RFP-Cep152 consistently marked the centrosomes of non-MCCs in both wild type and xCCDC78 morphant embryos indicating CCDC78 is not required for Cep152s localization to the centrosome (Figure 4A and B arrowheads, and D). In contrast, we saw a complete loss of Cep152 foci in morphant MCCs suggesting that CCDC78 is required for Cep152 recruitment to the deuterosome (Figure 4B and D). To determine if the deuterosome is present in the morphant MCCs we analyzed the localization of GFP-Plk4 in these cells. We found that GFP-Plk4 continues to localize to the deuterosome in CCDC78 morphants indicating that the structure remains intact and in normal numbers (Plk4 + MOATG=1.0 ± 0.8, n=21, vs. Plk4=1.4 ± 0.7, n=16, p=0.15, Figure 4G). Interestingly, over-expression of Plk4 did not rescue the morphant phenotype (MOATG = 0.40 centrioles/μm2, n=36 cells vs. MOATG + PLK4 =0.43 centrioles/μm2, n=18 cells p=0.63). We interpret this lack of rescue to be due to the lack of Cep152 at these deuterosomes (Figure 4D), indicating the ability of Cep152 to drive an increase in centriole biogenesis stems from its localization and function at the deuterosome, a feature that is lost in the absence of xCCDC78. The presence of Plk4 but not Cep152 at the deuterosome in CCDC78 morphants suggests that similar to cycling cells, Cep152 is likely involved in Plk4 maintenance or function rather than recruitment.
Discussion
Centriole biogenesis in MCCs has previously been proposed to involve an acentriolar structure termed the deuterosome. The identification of a GFP tagged construct of xCCDC78 that localizes to the presumptive deuterosome has allowed for the identification of other molecular components involved in deuterosome-driven centriole biogenesis. We have determined that the deuterosome contains the main regulators of centriole biogenesis Plk4 and Cep152, as well as the key structural component SAS6. These observations suggest deuterosome-mediated centriole biogenesis is molecularly quite similar to the canonical mother centriole driven process.
Multiple groups have performed comprehensive RNAi-based screens to identify regulators of centriole biogenesis (Nigg and Stearns, 2011). To our knowledge CCDC78 has not previously been identified in any of these screens, consistent with our observation that Xenopus morphant embryos develop normally with no defects in canonical centriole duplication. In contrast, we have identified CCDC78 as an important component for the deuterosome driven centriole amplification in MCCs. Interestingly, the depletion of xCCDC78 does not completely block centriole amplification. This could reflect an incomplete knockdown of xCCDC78 or compensation from the mother centriole driven mechanism, which we would predict to be intact in the absence of xCCDC78.
Mother centriole-mediated centriole duplication in cycling cells is generally restricted to S phase. However, deuterosome-mediated centriole amplification occurs in terminally differentiated MCCs (G1/0), suggesting that even though many of the critical regulators seem conserved, the cell cycle regulation has been modified. For example in cycling cells, the over-expression of Plk4 leads to the generation of supernumerary centrioles. In contrast, over-expression of Plk4 in post-mitotic MCCs did not alter the number of centrioles generated, however, it did rescue the loss of centrioles associated with the expression of a dominant negative Cep152. This result points to two conclusions: first, similar to cycling cells, an interaction between Plk4 and Cep152 is critical for the regulation of MCC centriole number; and second Plk4 function is distinct from its role in cell cycle regulated centriole duplication likely due to the G1/0 state of the MCCs.
The function of Plk4 in MCC centriole biogenesis coupled with the observation that CCDC78 is required for Cep152 localization to the deuterosome favors a model in which CCDC78 provides a scaffold, to which Cep152 is recruited to mediate regulation of Plk4 and centriole biogenesis. Together these proteins regulate the massive centriole amplification that occurs in MCCs. However, in contrast to cycling cells in which centriole number is precisely regulated, the number of centrioles generated in MCCs varies considerably, as does the size of the apical cell surface. Moving forward, a fascinating question remains unanswered: does centriole number dictate cell size or does cell size dictate centriole number?
Methods
Embryo Injections, Plasmids, RNA and MOs
The generation and injection of mRNA, plasmid DNA and MOs into Xenopus embryos was performed using standard techniques (Werner and Mitchell, 2013). Centrin4-RFP (Unigene: Xl.50473) was previously described (Werner et al., 2011). Full length xCCDC78 (Xl.4890), SAS6 (xl.33005) and Plk4 (xl.56605) were amplified from stage 17 Xenopus cDNA, and Cep152 (Xl.13956) and hsCCDC78 (hs.381943) were amplified from commercial clones (Open Biosystems) and cloned into the pCS2 vector containing GFP or RFP. Cep152ΔC was generated by PCR using the coding sequence of the first 306 amino acids (out of 1664) of xCep152.
Tissue Culture and Transfection
RPE-1 cells were transfected as previously described (Tanos et al., 2013), fixed in −20°C Methanol and stained with appropriate antibodies (See Supplemental Table 1) and DAPI (Invitrogen).
Immunofluorescence, Microscopy and Quantification
Embryo fixation, antibody staining and microscopy (Nikon-A1R confocal with 60X Plan Apo, 1.4NA objective) were performed as previously described (Werner and Mitchell, 2013). See Supplemental Table 1 for antibodies.. Quantification of centriole number, distance from deuterosome, cell size or presence of Cep152 was performed manually in Nikon Elements software. Quantification of flow velocities was performed as previously described (Werner and Mitchell, 2013). Statistics were performed using 2 tailed t-tests with the exception of Figure S1C and S3A in which we calculated the Chi square values to determine the p value.
MTEC Culture, Lentiviral Infection and Microscopy
Preparation of MTECs, lentiviral induction of hsCCDC78-GFP, IF, confocal microscopy and TEM were performed as previously described (Vladar and Stearns, 2007; You et al., 2002).
Supplementary Material
Highlights.
CCDC78 localizes to deuterosomes of multi-ciliated cells
Plk4, SAS6 and Cep152 localize to the deuterosome and facilitate centriole biogenesis
CCDC78 loss causes decreased centriole numbers and loss of Cep152 at deuterosomes
Cep152 is required to maintain Plk4 function at the deuterosome
Acknowledgments
This work was supported by the NIH/NIGMS (R01GM089970 to BJM), by a pilot grant from the NU-SDRC NIH/NIAMS (5P30AR057216-05). DAKD was supported by an ARRA NRSA (1F32AI08006901A1) and MEW was supported by postdoctoral fellowship from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- Anderson RG, Brenner RM. The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol. 1971;50:10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. Deconstructing the centriole: structure and number control. Curr Opin Cell Biol. 2012;24:4–13. doi: 10.1016/j.ceb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Brownlee CW, Rogers GC. Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell Mol Life Sci. 2013;70:1021–1034. doi: 10.1007/s00018-012-1102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush A, Cole P, Hariri M, Mackay I, Phillips G, O’Callaghan C, Wilson R, Warner JO. Primary ciliary dyskinesia: diagnosis and standards of care. Eur Respir J. 1998;12:982–988. doi: 10.1183/09031936.98.12040982. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Dirksen ER. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J Cell Biol. 1971;51:286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen ER, Satir P. Ciliary activity in the mouse oviduct as studied by transmission and scanning electron microscopy. Tissue Cell. 1972;4:389–403. doi: 10.1016/s0040-8166(72)80017-x. [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- Gavet O, Alvarez C, Gaspar P, Bornens M. Centrin4p, a novel mammalian centrin specifically expressed in ciliated cells. Mol Biol Cell. 2003;14:1818–1834. doi: 10.1091/mbc.E02-11-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T. Self-assembling SAS-6 multimer is a core centriole building block. J Biol Chem. 2010;285:8759–8770. doi: 10.1074/jbc.M109.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalnins VI, Porter KR. Centriole replication during ciliogenesis in the chick tracheal epithelium. Z Zellforsch Mikrosk Anat. 1969;100:1–30. doi: 10.1007/BF00343818. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majczenko K, Davidson AE, Camelo-Piragua S, Agrawal PB, Manfready RA, Li X, Joshi S, Xu J, Peng W, Beggs AH, et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am J Hum Genet. 2012;91:365–371. doi: 10.1016/j.ajhg.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Kintner C. Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol. 2008;20:48–52. doi: 10.1016/j.ceb.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Joshi HC, Besharse JC. Gamma-tubulin in differentiated cell types: localization in the vicinity of basal bodies in retinal photoreceptors and ciliated epithelia. J Cell Sci. 1993;104(Pt 4):1229–1237. doi: 10.1242/jcs.104.4.1229. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Winey M. Plk4/SAK/ZYG-1 in the regulation of centriole duplication. F1000 biology reports. 2010;2:58. doi: 10.3410/B2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamoto K, Wichterle H, Gonzalez-Perez O, Cholfin JA, Yamada M, Spassky N, Murcia NS, Garcia-Verdugo JM, Marin O, Rubenstein JL, et al. New neurons follow the flow of cerebrospinal fluid in the adult brain. Science. 2006;311:629–632. doi: 10.1126/science.1119133. [DOI] [PubMed] [Google Scholar]
- Sillibourne JE, Tack F, Vloemans N, Boeckx A, Thambirajah S, Bonnet P, Ramaekers FC, Bornens M, Grand-Perret T. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell. 2010;21:547–561. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Gabryjonczyk AM, Anselm E, Stierhof YD, Nigg EA. Human Cep192 and Cep152 cooperate in Plk4 recruitment and centriole duplication. J Cell Sci. 2013;126:3223–3233. doi: 10.1242/jcs.129502. [DOI] [PubMed] [Google Scholar]
- Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- Staprans I, Dirksen ER. Microtubule protein during ciliogenesis in the mouse oviduct. J Cell Biol. 1974;62:164–174. doi: 10.1083/jcb.62.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am J Anat. 1968;122:19–55. doi: 10.1002/aja.1001220103. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, Kintner C. Radial intercalation of ciliated cells during Xenopus skin development. Development. 2006;133:2507–2515. doi: 10.1242/dev.02417. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanos BE, Yang HJ, Soni R, Wang WJ, Macaluso FP, Asara JM, Tsou MF. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J Cell Biol. 2011;195:19–26. doi: 10.1083/jcb.201106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Understanding ciliated epithelia: The power of Xenopus. Genesis. 2011 doi: 10.1002/dvg.20824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Using Xenopus Skin to Study Cilia Development and Function. In: Cilia Part B, Marshall WF., editors. Methods in Enzymology. Vol. 525. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–1321. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.