Abstract
The CDK11p110 protein kinases are part of large-molecular-weight complexes that also contain RNA polymerase II, transcriptional elongation factors, and general pre-mRNA splicing factors. CDK11p110 isoforms may therefore couple transcription and pre-mRNA splicing by their effect(s) on certain proteins required for these processes. The CDK11p58 kinase isoform is generated from the CDK11p110 mRNA through the use of an internal ribosome entry site in a mitosis-specific manner, suggesting that this kinase may regulate the cell cycle during mitosis. The in vivo role and necessity of CDK11p110/p58 kinase function during mammalian development were examined by generating CDK11p110/p58-null mice through targeted disruption of the corresponding gene using homologous recombination. While heterozygous mice develop normally, disruption of both CDK11p110/p58 alleles results in early embryonic lethality due to apoptosis of the blastocyst cells between 3.5 and 4 days postcoitus. Cells within these embryos exhibit both proliferative defect(s) and a mitotic arrest. These results are consistent with the proposed cellular functions of the CDK11p110/p58 kinases and confirm that the CDK11p110/p58 kinases are essential for cellular viability as well as normal early embryonic development.
The cyclin-dependent kinases (CDK1 through CDK11) have diverse cellular functions, including regulation of cell cycle progression (e.g., CDKs 1, 2, 3, 4, and 6), neuronal development (e.g., CDK5), and regulation of transcription (e.g., CDKs 7, 8, and 9) (1-3, 7, 23, 34, 35, 39, 40, 47). Other potential CDK family members with as yet unknown functions have been identified in the human and mouse genomes (27, 46). These kinases exhibit high homology to CDK1, including the PSTAIRE sequence motif for which they were initially named. Once an in vivo cyclin partner is identified, the protein kinase is renamed as a CDK. Included in this latter group were the PITSLRE p110 and p58 protein kinase isoforms (48), which due to the identification of a putative cyclin regulatory partner, cyclin L, have been renamed CDK11p110 and CDK11p58 (4, 10, 20). The CDK11p110/p58 protein kinase isoforms in humans are encoded by two distinct but closely related genes (Cdc2L1 and Cdc2L2) (14). In contrast, a single CDK11p110/p58 gene (cdc2l) is found in a syntenic region of mouse chromosome 4 (30). Evolutionarily conserved CDK11p110/p58 homologues are found in chickens, Caenorhabditis elegans, Drosophila, and Xenopus, while the Schizosaccharomyces pombe and Saccharomyces cerevisiae genomes express only the CDK11p58 isoform (24, 37).
The Cdc2L1 and -2 genes, as well as cdc2l, express a 110-kDa protein kinase isoform which also contains the open reading frame of the smaller mitosis-specific isoform, CDK11p58. The smaller CDK11p58 isoform is generated by an internal ribosome entry site sequence (IRES) found in most species of mRNA encoding the CDK11p110 isoform. The presence or absence of the CDK11p58 isoform must be ascertained by determining whether the 58-kDa protein kinase species is present by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblot analysis (9). CDK11p110/p58 localizes to the nucleoplasm and nuclear speckle-staining regions (i.e., splicing factor centers) (20, 25, 44). The CDK11p110 kinase isoform interacts with the general pre-mRNA splicing factors RNPS1 and 9G8 (both of which are classified like other splicing factors as SR proteins), RNA polymerase II (RNAP II), and casein kinase 2 (CK2) (25, 26, 43). Furthermore, the larger CDK11 kinase isoform coimmunoprecipitates and/or copurifies with these same proteins and with multiple transcriptional elongation factors, including ELL2, TFIIF, TFIIS, and FACT (44).
A subfamily of cyclin-dependent protein kinases, including CDK1, -7, -8, and -9, regulates transcription through phosphorylation of specific amino acids in the carboxyl-terminal domain of the larger RNAP II subunit. The action of these various protein kinases results in the sequential recruitment of various transcriptional repressors and promoters as well as RNA processing factors to the differentially phosphorylated carboxyl-terminal domain (16, 31, 32). This group of protein kinases, perhaps in a cell cycle-specific manner, links various growth factor signaling pathways to transcription and RNA processing events. CDK11p110 associates with RNAP II, the cyclin L regulatory protein, several transcriptional elongation factors, and the pre-mRNA splicing proteins RNPS1 and 9G8 (20, 25, 44). The general splicing factor 9G8 is phosphorylated both in vivo and in vitro by the CDK11p110 kinase. The other known SR protein kinases (i.e., Clk/Sty LAMMER kinases and SRPK-1 and −2) (8, 11, 13, 29) phosphorylate the SR splicing factors almost exclusively in RS domains, thereby changing their subnuclear localization by releasing them from the splicing factor center storage compartments, changing their binding partners during the splicing reaction, changing splice site selection, or altering their ability to interact with nonspecific RNAs (28). Thus far, none has been shown to directly affect the splicing activity of the SR protein. Immunodepletion of CDK11p110 from nuclear extracts leads to a marked reduction of the in vitro splicing activity (20). These observations support the hypothesis that CDK11p110 may play an important role in transcript production and in regulation of RNA processing.
The CDK11p110 protein kinase isoform is detected in all phases of cell cycle, while the CDK11p58 isoform is expressed only during mitosis (9, 36). As mentioned earlier, CDK11p58 is produced by initiation of translation that is controlled by a mitosis-specific IRES present in the coding region of the CDK11p110 transcript. In addition, increased expression of CDK11p58 in Chinese hamster ovary (CHO) fibroblasts results in a late telophase delay due to abnormal cytokinesis and in increased cell death (22).
In summary, CDK11p110 protein kinase isoforms are involved in the regulation of RNA transcription and processing. However, the biological function of CDK11p58 in mammalian cells is unclear. To gain further insight into the function of the CDK11p110/p58 protein kinases in vivo, we disrupted the cdc2l gene encoding these products in embryonic stem (ES) cells. Heterozygous CDK11p110/p58+/− mice are viable and appear to develop normally. However, no homozygous CDK11p110/p58−/−knockout mice were obtained, since deletion of both cdc2l alleles during the early embryonic phase is lethal. CDK11p110/p58−/−blastocysts are not viable and die in utero between 3.5 and 4.0 days postcoitus, most likely due to apoptosis as a result of proliferative defects and mitotic arrest. In vitro culturing of these isolated blastocyst cells confirmed that embryonic cell death was due to apoptosis. The CDK11p110/p58 protein kinases are therefore important for proper cellular proliferation and cell cycle progression during early embryonic development.
MATERIALS AND METHODS
Targeted disruption of the mouse cdc2l gene.
A 129-Sv mouse bacterial artificial chromosome genomic library (Stratagene) was screened with the mouse CDK11p58 cDNA as a probe. One genomic clone containing the complete mouse cdc2l gene was identified. This clone was subjected to further restriction mapping, subcloning, and DNA sequencing. A targeting vector comprising a 3.1-kb 5′ homologous region from this genomic cdc2l BAC, followed by a thymidine kinase-Neo cassette (TK-Neo) and a 5-kb 3′ homologous genomic region containing the IRES region of CDK11p58 was generated for the gene targeting experiments. This construct was designed such that after homologous recombination, a 10.3-kb genomic sequence that contained exons 3, 4, and 5 of the CDK11p110 portion of the cdc2l gene would be replaced by TK-Neo. The construct also contained a frameshift mutation that results in the introduction of a translational termination sequence following the deleted cdc2l exons to potentially prevent translation of a CDK11p110 product. A TK cassette was also cloned into the targeting vector to select against nonhomologous insertions. Twenty-five micrograms of BstXI-linearized targeting vector DNA was transfected by electroporation into 1.0 × 107 129/SvEv (Specialty Media) ES cells. The transfected ES cells were cultured on mitomycin C-treated mouse embryonic fibroblast cells in media containing 350 μg of G418 per ml and 1.2 μM ganciclovir. ES cell clones exhibiting the correct homologous recombination event into the cdc2l gene locus were identified by Southern blotting. The chromosomes of the targeted ES cells were then karyotyped to ensure that the chromosomes were otherwise normal; the cells were then injected into the C57BL/6 blastocysts to generate chimeras. Six male chimeras from two independent ES clones (i.e., clone p110KO109 and clone p110KO153) showed a high level of chimerism, as determined by coat color. These chimeras were mated with C57BL/6 female mice to generate mice that were heterozygous for the mutant cdc2l allele. Germ line transmission of the mutant allele was tested by genomic PCR and verified by Southern blot analysis of tail DNA from the F1 offspring from this mating that had agouti coat color. cdc2l+/− mice (CDK11p110/p58+/−) were obtained from these two independent targeted ES clones, and the animals that resulted from both of these cell lines displayed identical phenotypes.
Southern blot analysis and PCR genotyping of the mouse cdc2l locus.
The G418 and ganciclovir-selected ES cell clones were screened for disruption of the cdc2l gene by Southern blot hybridization of ES cell genomic DNA completely digested with BamHI. A 240-bp genomic sequence that flanked the 5′ homologous region used in the disruption construct was labeled by PCR with [32P]dCTP and used as a probe.
The cdc2l genotypes were assessed by using genomic PCR. Genomic DNA was extracted from mouse tails and incubated overnight at 55°C in 200 μl of lysis buffer (200 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.1% Triton X-100, 1% NP-40, 5 mM EDTA, 400 μg of proteinase K/ml). The sample was then diluted 1:10, and 2 μl of this was used in a standard PCR with Ex Taq polymerase (Takara). A mixture of three different primers was used to detect the wild-type and targeted allele of cdc2l: a, forward (5′-CTTTGACCTCCATGGGCACATG-3′); b, wild-type (5′-CAGAGGCAAGTGGGTTTCTGAG-3′); and c, Neo (5′-CTATCGCCTTCTTGACGAGTTC-3′). Primers a and b will generate a 180-bp wild-type allele, whereas a 320-bp fragment would be expected for the mutant cdc2l allele generated with primers a and c. The PCR amplification protocol consisted of an initial incubation at 94°C for 3 min, followed by 35 cycles at 94°C for 45 s, 60°C for 1 min, and 72°C for 35 s. To confirm the results of PCR genotyping, genomic DNA extracted from mouse tails was digested with BamHI, transferred to a nylon membrane, and subjected to Southern blot hybridization using the 1.3-kb Neo internal probe.
Genotyping of the preimplantation embryos was assessed by nested PCR. Individual embryos and cells from outgrowths in culture were transferred into Eppendorf tubes containing 1 μl of sterile water, to which 3 μl of lysis buffer (0.05% SDS [wt/vol] and 0.035 N NaOH) was added. The samples were boiled for 5 min, and 2.0 μl of this mixture was subjected to the first round of PCR amplification in a total volume of 50 μl using a mixture of three primers: 1 (5′-CAACCATCTGTAAGTCCAATTCCAG-3′), 2 (5′-GCACACACCTTTAAATCCCAGCAC-3′), and 3 (5′-GGCGATGCGCTGCGAATCGGGAG-3′) for 20 cycles at 94°C for 45 s, 60°C for 1 min, and 72°C for 35 s. Next, 2.5 μl of the PCR products was used for the second round of PCR amplification for 35 cycles with the mixture of primers a, b, and c as described above.
Blastocyst culturing.
Heterozygous male and female mutant mice were bred to obtain cdc2l+/+ (CDK11p110/p58+/+), cdc2l+/− (CDK11p110/p58+/−), and cdc2l−/− mutant (CDK11p110/p58−/−) mouse embryos. The morning of the day on which a vaginal plug was detected was designated embryonic day 0.5 (E0.5). Embryos at different stages of development (E1.5 through E4.0) were collected by flushing oviducts or the uterus with HEPES-buffered medium 2 (M2; Sigma). In some experiments, embryos were fixed immediately for 20 min at 4°C with phosphate-buffered saline (PBS) containing 4% paraformaldehyde, and their nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (Vectashield; Vector). For in vitro culturing, blastocysts were cultured for 16 h to 3 days in tissue culture plates containing complete ES medium containing leukemia inhibitory factor; outgrowths were inspected daily and photographed to monitor their development.
Immunoblot analysis.
CDK11p110/p58 wild-type and mutant ES cells were lysed in a buffer containing 25 mM Tris-HCl (pH 8.5), 1 mM dithiothreitol, and 100 mM NaCl by sonication; lysates were then cleared by centrifugation. One hundred micrograms of cell lysate was separated by SDS-12.5% polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane. The CDK11p110 and CDK11p58 protein kinases were detected by using a polyclonal antibody against the amino-terminal domain of CDK11p110 (P2N100) or the carboxyl-terminal domain common to both kinases (P1C).
BrdU labeling of cultured blastocysts.
E3.5 blastocysts were cultured in ES cell medium containing 10 μM 5′-bromo-2′-deoxyuridine (BrdU) (Sigma) for 16 h. Cells were then fixed in 4% paraformaldehyde for 20 min at 4°C. After three washes with PBS, the cells were treated with 0.5 M HCl for 30 min at room temperature, after which immunofluorescence was performed as described below.
Immunocytochemistry and in situ detection of mitotic arrest and apoptosis.
For immunofluorescence experiments, we used the rabbit polyclonal antibodies P1C and P2N100, which recognize the carboxyl-terminal domains of CDK11p110/p58 and the amino-terminal domain of CDK11p110, respectively. To determine the percentage of CDK11p110/p58+/+, CDK11p110/p58+/−, and CDK11p110/p58−/− blastocystcells in mitosis, the E3.5 blastocysts corresponding to these genotypes were cultured for 16 h to 3 days, and the cells were then immunostained with a rabbit polyclonal antibody, Phosphohistone (Ser10) (Cell Signaling Technology, Beverly, Mass.), which detects histone H3 when it is phosphorylated at serine residue 10. Blastocysts were fixed for 20 min in 4% paraformaldehyde, followed by permeabilization with 0.5% Triton X-100 in PBS for 20 min and blocking for 30 min in 5% fetal calf serum in PBS. Primary and secondary antibodies were left on cells for 1 h at room temperature, followed by three 5-min washes with PBS, after which DNA was stained with DAPI for 5 min. Embryos were embedded in Vectashield mounting medium and examined by fluorescent and confocal microscopy. This same procedure was used to immunostain CDK11p110/p58+/− and CDK11p110/p58−/− blastocysts with the P1C polyclonal antisera specific for the kinase domain shared by both CDK11p110 and CDK11p58 isoforms to verify that neither protein kinase isoform was expressed in the CDK11-deficient cells. To determine whether blastocyst cells were undergoing caspase-dependent apoptosis, both DNA fragmentation and caspase 3 enzymatic activity were examined. DNA fragmentation associated with cell death was detected by using an in situ terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) apoptosis detection kit (Roche). In brief, to determine whether active caspase 3 was present, we labeled embryos with FAM-DEVD-FMK for 1 h at 37°C. To fluorescently label the DNA in the nuclei, we added DAPI, and the incubation continued for 5 min. Embryos were washed twice in 1× working dilution wash buffer. Caspase 3 enzyme activity was assayed by the processing of its fluorescent substrate, FAM-DEVD-FMK (Intergen), resulting in green fluorescence, while nuclear DNA staining by DAPI resulted in blue fluorescence. Both were analyzed with fluorescence microscopy. For the TUNEL analysis, E3.5 blastocysts were collected as described above, fixed with 4% paraformaldehyde in PBS for 20 min at 4°C, and subjected to permeabilization for 20 min at room temperature with PBS containing 0.2% Triton X-100. TUNEL assays were then performed by using the in situ detection kit (Roche). Briefly, the fixed and permeabilized blastocysts were labeled with the TUNEL reaction mixture for 60 min at 37°C. The nuclei of these blastocysts were also stained with DAPI. Fluorescein-labeled DNA, indicating DNA fragmentation, was then analyzed by using a Nikon fluorescence microscope.
RESULTS
Targeted disruption of the single mouse cdc2l gene.
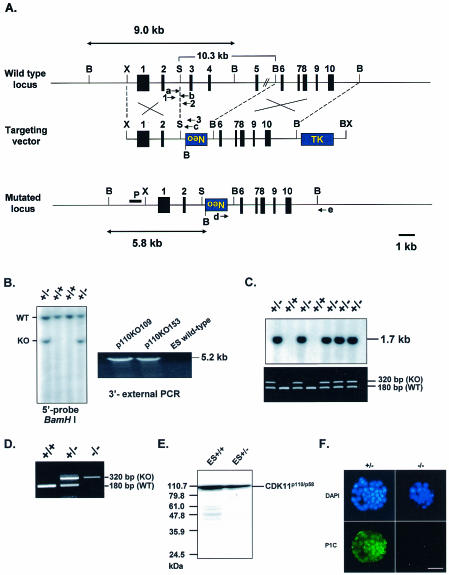
To determine the function of CDK11p110/p58 protein kinases during development and proliferation, we generated CDK11p110/p58-deficient mutant mice by gene targeting via homologous recombination in ES cells. The targeting vector was designed to replace 10.3 kb of the mouse genomic DNA containing exons 3, 4, and 5 of the cdc2l (CDK11p110/p58) gene with a TK-Neo cassette (Fig. 1A). The replacement of these three exons with the Neo cassette leads to a frameshift in the mRNA sequence and subsequent termination of translation, preventing any possible translation of CDK11p110 and possibly destabilizing the mRNA, thereby preventing the synthesis of the CDK11p58 protein kinase. Thus, functional CDK11p110, and more than likely CDK11p58, was not expected to be expressed from the mutated allele. The targeting construct was transfected by electroporation into ES cells. After 6 days of selection with G418 and ganciclovir, 5 of the original 240 colonies obtained were shown to be targeted correctly for the cdc2l (CDK11p110/p58) locus by Southern blot hybridization using a flanking 5′-external probe (Fig. 1B). This finding was further demonstrated by PCR using primers specific for a 3′-external genomic sequence and the Neo cassette (Fig. 1B). All five of these original clones were also karyotyped to ensure genomic integrity, and all were found to have 95% or greater normal chromosomes (data not shown). Two of these targeted ES clones were then injected into C57BL/6 blastocysts and transplanted into pseudopregnant C57BL/6 females. Six chimeric mice obtained from these two ES clones were found to transmit the mutated cdc2l allele through the germ line. Mice heterozygous for the cdc2l mutation (i.e., cdc2l+/− and CDK11p110/p58+/−) were generated by backcrossing male chimeras from the p110KO109 and p110KO153 clones to C57BL/6 females.
FIG. 1.
Targeted disruption of the mouse cdc2l gene locus by homologous recombination. (A) Structure of the targeting vector and partial restriction map of cdc2l gene locus in mouse before and after homologous recombination. Exons are represented by vertical black boxes, and the position of the Southern blot probe (P) is indicated by the horizontal black box. Restriction enzymes: B, BamHI; X, XbaI; S, SacI; BX, BstXI. The mutated cdc2l allele was detected by a set of PCR primers: a, b, and c. The primers 1, 2, and 3 were used for the first round of the nested PCR amplification for blastocyst genotyping. (B) Southern blot analysis of CDK11p110/p58 mutant ES cell lines. Genomic DNA isolated from ES cell clones following positive (G418) and negative (ganciclovir) selection were digested with BamHI and analyzed by Southern blotting using 5′ probe, yielding the predicted 5.8-kb band in addition to a 9.0-kb wild-type (WT) band. PCR amplification with primers d and e generated a 5.2-kb band containing the mutated allele. (C) Genotype analysis by Southern blot analysis (top) and PCR (bottom) of tail DNA of mice from heterozygote intercrosses. Tail DNA samples digested with BamHI were subjected to Southern blot hybridization with the 1.3-kb Neo probe, yielding a 1.7-kb band. The same DNA samples were also subjected to PCR with primers a and b for the WT allele and with primers a and c for the mutated (KO) allele, yielding amplification products of 180 and 320 bp, respectively. (D) The genotypes of the CDK11p110/p58 embryos from CDK11p110/p58+/− intercrosses were determined by nested PCR (see Materials and Methods). (E) Immunoblot analysis of CDK11p110/p58 expression in WT and heterozygous targeted ES cell lines. Whole-cell lysates from a WT ES cell line (+/+) and targeted ES cell line (+/−) were analyzed by immunoblotting with the CDK11p110-specific anti-P2N100 antibody. (F) Immunostaining of E3.5 CDK11p110/p58+/− and CDK11p110/p58−/− blastocysts with an affinity-purified C-terminal CDK11 polyclonal antibody, P1C, generated to a protein kinase domain shared by the CDK11p110 and CDK11p58 isoforms. The primary antibody was then detected with a fluorescein isothiocyanate-labeled secondary antibody (green, bottom panels) and indicates the presence of one or both of the protein kinases. The absence of a signal indicates that neither protein kinase is present in the blastocyst cells. To verify cellular integrity, we also stained blastocysts with DAPI, a reagent that specifically detects nuclear DNA (blue, top panels). White bar, 100 μm.
Disruption of CDK11p110/p58 results in early embryonic lethality.
We then intercrossed CDK11p110/p58+/− mice to ascertain the viability of CDK11p110/p58-null homozygotes (CDK11p110/p58−/−). The genotypes of the resulting offspring were determined by PCR analysis of tail DNA at age 3 weeks and further confirmed by Southern blot hybridization of the PCR products to an internal Neo probe (Fig. 1C), indicating the reliability of the PCR genotyping. A total of 326 offspring were genotyped. CDK11p110/p58+/+ and CDK11p110/p58+/− were present at near the expected frequencies, but no homozygote-null animals were observed (Table 1). These results indicated that CDK11p110/p58 deficiency causes embryonic death. To determine the stage of embryogenesis at which the CDK11p110/p58 disruption is lethal, we dissected embryos from the heterozygous intercrosses at different stages of embryonic development and genotyped them by PCR (Fig. 1D). No CDK11p110/p58−/− embryos were found at E9.5 or E7.5 of development (Table 1). In contrast, while at E3.5, CDK11p110/p58−/− embryos were found at a normal Mendelian frequency for all genotypes. These showed abnormal morphology (see below).
TABLE 1.
Genotype of offspring from CDK11p110/p58+/− heterozygote intercrossesa
| Age | No. of offspring with genotype:
|
Total no. | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Weaning | 107 | 219 | 0 | 326 |
| E9.5 | 12 | 22 | 0 | 34 |
| E7.5 | 8 | 16 | 0 | 24 |
| E3.5 | 18 | 39 | 14 | 71 |
Embryos from CDK11p110/p58+/− heterozygote intercrosses were collected at the indicated embryonic times, and the genotypes were determined by PCR with primers a, b, and c or by nested PCR with two sets of primers (1, 2, and 3; a, b, and c) (Fig. 1A).
Since both the CDK11p110 and CDK11p58 protein kinases are expressed ubiquitously in various cell types, including ES cells, it was essential to ensure that neither a truncated CDK11p110 protein nor the CDK11p58 isoform was being synthesized by the targeted allele in which exons 3, 4, and 5 of cdc2l (CDK11p110/p58) were replaced by the Neo cassette. To demonstrate that the targeted mutation did in fact affect CDK11p110/p58 protein kinase expression, we performed immunoblot analysis on whole-cell lysates obtained from ES cells that were heterozygous for the targeted allele by using antibodies against the amino-terminal domain of CDK11p110 and the carboxyl-terminal domain common to both CDK11p110/p58 kinases (i.e., P2N100 and P1C antibodies, respectively). As shown in Fig. 1E, expression of the CDK11p110 protein in wild-type ES cells was approximately 20 to 30% greater than that in heterozygous ES cells. Also, no truncated CDK11p110 or CDK11p58 proteins were detected in the ES cell lysates, suggesting that the targeted allele has a true null mutation. Due to the rather restricted window of CDK11p58 expression during mitosis (9), the results of the immunoblot analysis were not unanticipated and suggested that CDK11p58 was not expressed. The absence of both CDK11p110 and CDK11p58 protein kinase isoforms was confirmed by analysis of CDK11p110/p58−/−, CDK11p110/p58+/+, and CDK11p110/p58+/− blastocysts by indirect immunofluorescence using the P2N100 antiserum, which detects only the CDK11p110 kinase isoform, and the P1C antiserum, which detects both kinase isoforms. The P1C immunofluorescence microscopy studies demonstrated robust CDK11p110/p58 protein staining in the CDK11p110/p58+/− blastocyst cells but no significant CDK11p110 or CDK11p58 kinase in the CDK11p110/p58−/− blastocyst cells (Fig. 1F). These experiments established that neither the CDK11p110 nor the CDK11p58 kinase were expressed in the CDK11p110/p58−/− blastocysts, since any immunostaining in these cells would by necessity be the result of the expression of one or both of these protein kinases. No immunostaining of the CDK11p110/p58−/− blastocysts was observed with the P2N100 antisera, either, further verifying that the CDK11p110 protein was not expressed (data not shown). These data are consistent with the loss of both protein kinase isoforms due to the destabilization of the CDK11p110 mRNA transcript that produces the CDK11p110 and CDK11p58 kinases.
To further evaluate the role of CDK11p110/p58 in cell viability, we also attempted to generate ES cells with both cdc2l (CDK11p110/p58) alleles mutated by sequential gene targeting using CDK11p110/p58+/− ES cell clones. The mouse cdc2l gene was disrupted as described in detail in Materials and Methods. Nine targeted ES cell clones were isolated from 378 clones analyzed by PCR. All homologous recombination events occurred only at the previously targeted allele, and there were no CDK11p110/p58−/− ES cell clones obtained. All of the above data suggest that the CDK11p110/p58 protein kinases are essential for cell survival, and not for development, and that disruption of the cdc2l locus as well as the resulting loss of the CDK11p110/p58 kinases leads to an early embryonic lethality. Embryos lacking the CDK11p110/p58 kinases cease to develop before implantation, most likely just after the depletion of their maternal CDK11p58 mRNA and CDK11p110 mRNA and protein supplies. This difference is proposed due to the extremely short half-life of the CDK11p58 protein, which is degraded rapidly in late mitosis compared to the much longer half-life of the CDK11p110 protein, which is expressed throughout the cell cycle.
Aberrant cellular morphology and defective growth associated with CDK11p110/p58−/− embryonic cells.
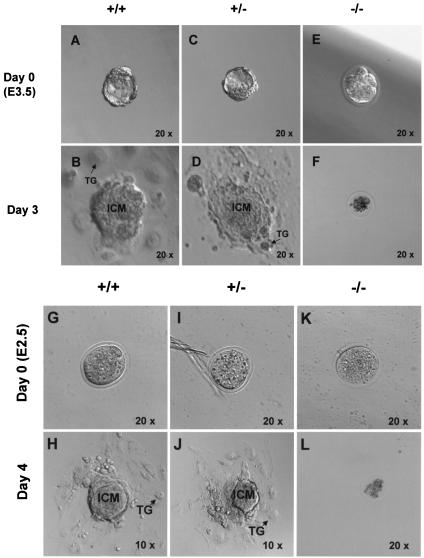
To more directly analyze the early embryonic lethality associated with the CDK11p110/p58−/− mice, we isolated blastocysts at different stages of development from intercrosses between heterozygous mutant mice, cultured them, and then examined the morphological differences among all types of embryos found. We observed that at the E3.5 stage of development, homozygous mutant (CDK11p110/p58−/−) blastocysts were morphologically distinguishable from their wild-type (CDK11p110/p58+/+) and heterozygous (CDK11p110/p58+/−) counterparts (Fig. 2A through F). Both CDK11p110/p58+/+ and CDK11p110/p58+/− blastocysts appeared normal, whereas the homozygous CDK11p110/p58−/− cells failed to form fully expanded blastocysts, which are composed of the hollow vesicle of the trophectoderm surrounding a fluid-filled cavity and a small group of internal cavity mass cells. The E3.5 CDK11p110/p58−/− blastocysts also appeared to be growth retarded at the morula stage of development (Fig. 2E). To further define the precise time point at which the CDK11p110/p58−/− embryos stop developing, we collected embryos from the E2.5 morula stage of heterozygous intercrosses and found that CDK11p110/p58−/− embryos at this stage appeared to be normal and did not have any morphological differences from those of the other genotypes (Fig. 2G through L), suggesting that embryos deficient in the CDK11p110/p58 kinases stop further development between E2.5 and E3.5. In an attempt to better understand the growth capacity of the CDK11p110/p58−/− embryos, we collected E3.5 and E2.5 blastocysts and cultured them individually in vitro in the presence of recombinant leukemia inhibitor for 16 h to 3 days. Unlike wild-type and heterozygous blastocysts that hatched from the zona pellucida, attached to the culture dish, generated apparently normal trophoblast giant cells (TG), and then continued to proliferate, the CDK11p110/p58−/− embryos never hatched and in fact gradually became smaller after in vitro culturing for several days and finally collapsed. This finding suggests that the CDK11p110/p58−/− homozygous blastocysts were incapable of implanting in the uterus and so died during the culture period (Fig. 2G through L). These results demonstrated that the development of CDK11p110/p58−/− embryos is impaired during in vitro culturing and reinforced in vivo observations that the growth of the CDK11p110/p58−/− blastocysts is arrested putatively after the maternally provided CDK11p110/p58 mRNA and protein stores are depleted.
FIG. 2.
Morphological analysis of the outgrowths derived from the CDK11p110/p58 blastocysts. Blastocysts were derived from CDK11p110/p58+/− intercrosses, explanted at E3.5 (day 0), and grown in culture for 3 days, during which time they developed outgrowths. The CDK11p110/p58+/+ (A and B) and CDK11p110/p58+/− (C and D) blastocysts grew normally. In contrast, CDK11p110/p58−/− (E and F) blastocysts stop further growth and development after E3.5. Magnification, ×11.2 for panels A through F. Embryos at E2.5 and their outgrowths after 4 days in culture in panels G through L. CDK11p110/p58+/+ (G and H), CDK11p110/p58+/− (I and J), and CDK11p110/p58−/− (K and L) embryos are indistinguishable at E2.5. All images are not shown to scale (magnification, ×6.5 for day 4 wild-type and heterozygous outgrowth [panels H and J]; ×13 for panels G, I, K, and L). ICM, inner cell mass; TG, trophoblast giant cell.
CDK11p110/58 deficiency leads to apoptotic cell death in blastocysts.
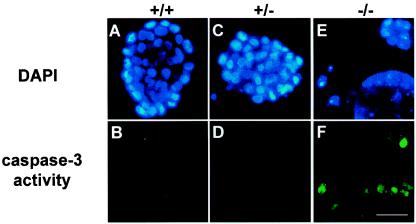
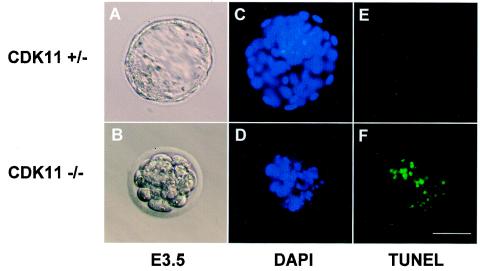
CDK11p110/p58 protein kinases have been reported to be involved in signaling pathway(s) that may help to coordinate transcription and RNA processing events during the cell cycle (20, 43, 44) as well as regulation of mitotic progression (9). At the early morula stage of blastocyst development, transcription and RNA processing of embryonic genes start to synthesize the proteins necessary for further embryonic development after the depletion of the maternally provided proteins. Failure to make the essential factors for cell proliferation within the embryos and/or mitotic arrest due to the CDK11p110p58−/− mutation could lead to extensive programmed cell death and contribute to a rapid collapse of CDK11p110/p58−/− blastocysts in vivo and in vitro. To assess whether the deficiency of the CDK11p110/p58 kinase isoforms resulted in apoptosis in CDK11p110/p58−/− homozygous embryos, we examined the cells present in the blastocysts isolated from the intercrosses between CDK11p110/p58+/− heterozygous mice by using a CaspaTag caspase 3 activity assay to detect active caspases normally associated with caspase 3-dependent cell death (see Materials and Methods) (21). The DAPI-stained nuclei of the CDK11p110/p58+/+, CDK11p110/p58+/−, and CDK11p110/p58−/− blastocysts (Fig. 3A, C, and E) are shown. Rare apoptotic cells observed in the proliferating CDK11p110/p58+/+ and CDK11p110/p58+/− blastocysts can be seen (Fig. 3B and D), in contrast to the extensive apoptosis that was already occurring in the growth-arrested CDK11p110/p58−/− embryos after only 2 to 3 h in culture (Fig. 3F). This finding indicated that CDK11p110/p58−/− embryonic lethality is at least partially due to apoptotic cell death. The extent of caspase 3-positive blastocyst cells from the CDK11p110/p58−/− mouse embryos, but not in the blastocyst cells from the CDK11p110/p58+/+ or CDK11p110+/− mouse embryos, was significantly increased during in vitro cell culturing up to 3 days (data not shown). Furthermore, when these blastocysts were examined for DNA fragmentation (another hallmark of apoptosis) by TUNEL analysis, the CDK11p110/p58−/− blastocyst cells were TUNEL positive (Fig. 4F), whereas the CDK11p110/p58+/− blastocysts were not positive (Fig. 4E). The blastocysts corresponding to CDK11p110/p58−/− and CDK11p110/p58+/− mice are also shown (Fig. 4A and B), as is the DAPI-stained DNA corresponding to the nuclei of these cells (Fig. 4C and D).
FIG. 3.
CDK11p110/p58 protein kinase deficiency resulted in apoptosis of the blastocyst cells. CDK11p110/p58+/+, CDK11p110/p58+/−, and CDK11p110/p58−/− blastocysts were isolated at E3.5 from CDK11p110/p58+/− intercrosses. The caspase 3 activity of the blastocysts was detected by the fluorescence of its covalently bound inhibitor, FAM-DEVD-FKM. Caspase 3 activity (B, D, and F) and nuclear DNA staining by DAPI (A, C, and E) were both examined by fluorescence microscopy. Bar, 50 μm.
FIG. 4.
Analysis of blastocyst cell apoptosis by TUNEL. CDK11p110/p58+/− (A) and CDK11p110/p58−/− (B) blastocysts were isolated at E3.5 from CDK11p110/p58+/− intercrosses. After 12 h in culture, the fixed, permeabilized blastocysts were stained with TUNEL reagents (E and F) and DAPI (C and D). Bar, 50 μm.
CDK11p110/p58−/− embryonic blastocyst cells do not proliferate normally and are arrested during mitosis.
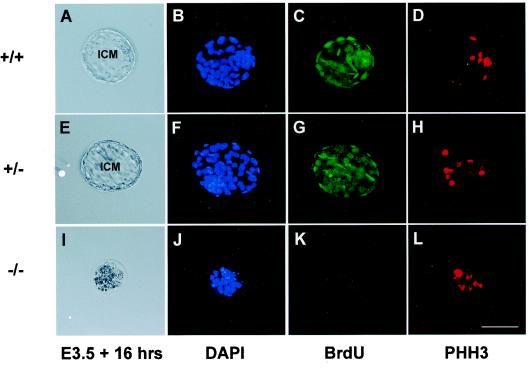
To examine the potential effect(s) of the CDK11p110/p58−/− mutation on the ability of these mouse embryos to proliferate as well as progress through the cell cycle appropriately, we cultured E3.5 blastocysts (Fig. 5A, E, and I) collected from the CDK11p110/p58+/− intercrosses in the presence of BrdU for 16 h. The nuclei of these cells were stained with DAPI for visualization by immunofluorescence microscopy also (Fig. 5B, F, and J). Incorporation of BrdU into the DNA during S phase of the cell cycle was assessed by indirect immunofluorescence with an antibody specific for BrdU. Both the CDK11p110/p58+/+ and heterozygous blastocysts showed strong incorporation of this DNA label (Fig. 5C and G), whereas the CDK11p110/p58−/− blastocysts showed very little BrdU incorporation (Fig. 5K), indicating that the proliferative capability of the cells from the CDK11p110/p58−/− mutant embryos is significantly impaired at E3.5.
FIG. 5.
Failure of the CDK11p110/p58−/− blastocyst cells to proliferate normally. CDK11p110/p58 blastocysts at E3.5 were recovered from CDK11p110/p58+/− intercrosses and grown in culture in the presence of 10 μM BrdU for 16 h. The fixed blastocysts (panels A, E, and I) were then stained with anti-BrdU antibody (panels C, G, and K), an antibody specific to the M-phase-specific marker histone H3 phosphorylated at Ser10 (panels D, H, and L), and DAPI (panels B, F, and J). The genotype of each embryo was determined by using nested PCR. Representative CDK11p110/p58+/+ (A to D), CDK11p110/p58+/− (E to H), and CDK11p110/p58−/− (I to L) blastocysts are shown. ICM, inner cell mass. Bar, 50 μm.
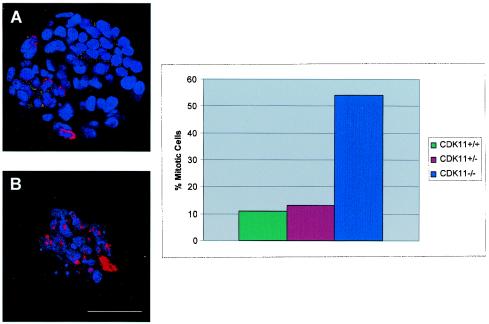
To further demonstrate that the cells from the CDK11p110/p58−/− blastocysts were arrested at a particular stage of the cell cycle, we immunostained the BrdU-labeled embryos by using an antibody specific for murine histone H3 phosphorylated on serine residue 10. The modification of histone H3 by phosphorylation at specific sites in the protein has been shown by numerous investigators to correlate with the events involved in chromosome condensation during mitosis (18, 19, 41). More specifically, the phosphorylation of histone H3 at serine residue 10 has been shown to coincide with the initiation of the induction of chromosome condensation, and it is therefore considered an excellent mitosis-specific marker. The presence of Ser10 histone H3 phosphorylation provides evidence for any possible cellular arrest occurring during mitosis. Our analyses by confocal microscopy showed that the percentage of phospho-Ser10-histone H3-positive cells is markedly higher in the blastocyst cells from the CDK11p110/p58−/− embryos (∼50 to 60%) than that in either their wild-type or heterozygous counterparts (∼10%) (Fig. 6A and B, inset). These results confirm that the majority of the cells from the CDK11p110/p58-null blastocysts were arrested during mitosis before E3.5. Taken together, the facts that (i) these blastocysts did not synthesize new DNA as shown by the BrdU-labeling experiments, (ii) the blastocysts consisted of a high percentage of cells positive for phosphorylated Ser10 on histone H3 (a mitotic marker), and (iii) the blastocysts were unable to produce CDK11p110/p58−/− ES cells after numerous attempts are all proof that the blastocyst cells from CDK11p110/p58−/− mouse embryos underwent cell death as a result of cell cycle arrest rather than of a specific developmental defect.
FIG. 6.
Representative confocal microscopy images of CDK11p110/p58+/+ (A) and CDK11p110/p58−/− mutant (B) embryos stained with DAPI (blue) and anti-PHH3 antibody (red). Bar,100 μm. Increased number of cells undergoing mitotic arrest observed in the CDK11p110/p58−/− mutant embryos at E3.5 are shown. Mitotic arrest was determined by counting the number of cells positive for phospho-Ser-histone H3 (PHH3), the M-phase-specific marker. The percentage of PHH3-positive blastocyst cells from among the total number of blastocyst cells (determined by DAPI staining of cellular nuclei) is represented in the inset bar graph.
DISCUSSION
The CDK family of protein kinases has been shown to directly regulate cell cycle progression through their control of mitosis and the G1/S-phase transition as well as regulate transcriptional events during the cell cycle (32, 46). The most studied members of this protein kinase family, CDK1 and CDK2, were believed to be essential for normal cell cycle progression (46). However, a number of recent studies, including those describing cdk2 gene knockouts in mice, have revealed that while CDK1 function is necessary for normal cell cycle progression (15, 42), CDK2 function is not (5, 33). The results of the cdk2 gene knockout studies were unexpected and reduce the number of CDK protein kinases that appear to be essential for regulating cell cycle progression normal to two, or possibly three (with the inclusion of CDK11). While CDK4 function is relevant to the G1-to-S-phase transition, it is not absolutely essential (45). However, the rather obscure CDK3 protein kinase, whose expression is extremely low and restricted, has been shown to be essential for normal embryonic development, yet its normal cellular function has not been resolved (46). In addition, the CDK family of protein kinases has been shown to be involved in the regulation of neuronal events (40).
Recently, an additional member of the CDK kinase family, CDK11p110, was identified as a regulator of RNA processing events and of transcription (20, 25, 26, 44), and a second CDK11 isoform, CDK11p58, has been shown to be expressed only during mitosis due to an IRES present in the CDK11p110 mRNA (9). While the exact function of the CDK11p58 isoform is not known at this time, its mitosis-specific expression strongly suggests that it regulates events during mitosis. In the present study, we have examined the effect(s) of the loss of CDK11p110/p58 function, due to the disruption of its corresponding cdc2l gene in mice, on normal embryonic development. Additionally, attempts to generate cdc2l−/− ES cells that do not express the CDK11p110/p58 kinases also proved to be unsuccessful after several different attempts (data not shown). While we cannot specifically attribute the resulting embryonic lethal phenotype to the specific loss of CDK11p110 or CDK11p58, the experiments reported here demonstrate that these particular CDK protein kinases are essential for the normal growth of a wide range of cell types. Additionally, these studies have shown that the loss of these protein kinase activities results in the failure of E3.5 CDK11p110/p58-null blastocyst cells to survive due to apoptosis induced by proliferative failure and mitotic arrest. Since E3.5 blastocyst cells are in the process of making the transition from early embryonic cell divisions that rely on a number of maternal mRNAs and proteins to promote their survival to cells whose survival is no longer dependent on such maternal products, it is likely that disruption of the cdc2l gene results in the complete loss of both CDK11p110 and CDK11p58 kinase function in postblastocyst cells. Therefore, we can conclude that the function of one or both of the CDK11 kinase isoforms is absolutely essential for the continued survival, and therefore development, of the postblastocyst murine cells. Additional conditional cdc2l gene knockouts, as well as cdc2l gene knock-in experiments, will now be needed to determine whether one or both of the CDK11 kinase isoforms is essential for normal cell growth and development. Furthermore, these results suggest that it may be of interest to examine the effects of crossing CDK11p110/p58 gene hypomorphs with well-known mouse tumor models to determine whether the decreased expression of one or both of these protein kinases might mediate tumorigenesis. Several recent reviews on the potential of pre-mRNA splicing factors to function as mediators of tumorigenesis suggest that this possibility should be considered (6, 12, 17). In addition, the phenotype of the CDK11p110/p58−/− blastocyst cells, which includes both proliferative failure and mitotic arrest, also supports the possible involvement of the CDK11 protein kinases in cancer and/or disease. Finally, the human Cdc2L genes are localized to human chromosome 1 band p36.3 (14), a chromosomal region that is frequently deleted or translocated in a number of different human tumors, including neuroblastoma, breast cancer, melanoma, and lymphoma, and the region has been proposed as the location of at least two tumor suppressor genes (38). Such studies will help us to determine whether the CDK11p110 and/or CDK11p58 protein kinases function as possible tumor suppressors or mediators of tumorigenesis, perhaps due to haploinsufficiency of one or both of the kinases.
Acknowledgments
We thank Gerard Grosveld, director of Transgenic/Knockout Core, and Christie Nagy for their help and assistance in generating the knockout mice; we also thank Nagy for help with the isolation of blastocysts and Madoka Inoue for maintaining the mouse colonies and performing tail DNA analysis. We also acknowledge Susan Ragsdale and the SJCRH Cancer Center Cytogenetics Shared Resource for performing the karyotype analysis of the ES cell clones, and we thank Ken Barnes in the SJCRH Cancer Center Scientific Imaging Shared Resource for assistance performing the confocal microscopy. Finally, we thank Dongli Hu and Pascal Loyer for helpful discussions.
This research was supported by a grant from the NIH (2RO1 GM44088-13) to V.J.K., from the NIH to SJCRH (P30 CA21765-25) and by the generous support of the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 2.Akoulitchev, S., T. P. Makela, R. A. Weinberg, and D. Reinberg. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377:557-560. [DOI] [PubMed] [Google Scholar]
- 3.Akoulitchev, S., and D. Reinberg. 1998. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12:3541-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berke, J. D., V. Sgambato, P. P. Zhu, B. Lavoie, M. Vincent, M. Krause, and S. E. Hyman. 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32:277-287. [DOI] [PubMed] [Google Scholar]
- 5.Berthet, C., E. Aleem, V. Coppola, L. Tessarollo, and P. Kaldis. 2003. Cdk2 knockout mice are viable. Curr. Biol. 13:1775-1785. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18:186-193. [DOI] [PubMed] [Google Scholar]
- 7.Cho, H., G. Orphanides, X. Sun, X. J. Yang, V. Ogryzko, E. Lees, Y. Nakatani, and D. Reinberg. 1998. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 18:5355-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colwill, K., L. L. Feng, J. M. Yeakley, G. D. Gish, J. F. Caceres, T. Pawson, and X.-D. Fu. 1996. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271:24569-24575. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, S., Y. Bruynooghe, G. Denecker, S. Van Huffel, S. Tinton, and R. Beyaert. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5:597-605. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, L. A., A. J. Edgar, J. Ehley, and J. M. Gottesfeld. 2002. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J. Biol. Chem. 277:25465-25473. [DOI] [PubMed] [Google Scholar]
- 11.Duncan, P. I., D. F. Stojdl, R. M. Marius, K. H. Scheit, and J. C. Bell. 1998. The Clk2 and Clk3 dual-specificity protein kinases regulate the intranuclear distribution of SR proteins and influence pre-mRNA splicing. Exp. Cell Res. 241:300-308. [DOI] [PubMed] [Google Scholar]
- 12.Faustino, N. A., and T. A. Cooper. 2003. Pre-mRNA splicing and human disease. Genes Dev. 17:419-437. [DOI] [PubMed] [Google Scholar]
- 13.Gui, J.-F., H. Tronchere, S. D. Chandler, and X.-D. Fu. 1994. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc. Natl. Acad. Sci. USA 91:10824-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gururajan, R., J. M. Lahti, J. Grenet, J. Easton, I. Gruber, P. F. Ambros, and V. J. Kidd. 1998. Duplication of a genomic region containing the Cdc2L1-2 and MMP21-22 genes on human chromosome 1p36.3 and their linkage to D1Z2. Genome Res. 8:929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamaguchi, J. R., R. A. Tobey, J. Pines, H. A. Crissman, T. Hunter, and E. M. Bradbury. 1992. Requirement for p34cdc2 kinase is restricted to mitosis in the mammalian cdc2 mutant FT210. J. Cell Biol. 117:1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hampsey, M., and D. Reinberg. 1999. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Gen. Dev. 9:132-139. [DOI] [PubMed] [Google Scholar]
- 17.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 18.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 19.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 20.Hu, D., A. Mayeda, J. H. Trembley, J. M. Lahti, and V. J. Kidd. 2003. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem. 278:8623-8629. [DOI] [PubMed] [Google Scholar]
- 21.Kidd, V. J. 1998. Proteolytic activities that mediate apoptosis. Annu. Rev. Physiol. 60:533-573. [DOI] [PubMed] [Google Scholar]
- 22.Lahti, J. M., J. Xiang, L. S. Heath, D. Campana, and V. J. Kidd. 1995. PITSLRE protein kinase activity is associated with apoptosis. Mol. Cell. Biol. 15:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lew, J., K. Beaudette, C. M. E. Litwin, and J. H. Wang. 1992. Purification and characterization of a novel proline-directed protein kinase from bovine brain. J. Biol. Chem. 267:13383-13390. [PubMed] [Google Scholar]
- 24.Li, H., J. Grenet, M. B. Valentine, J. M. Lahti, and V. J. Kidd. 1995. Structure and expression of the chicken PITSLRE protein kinase gene and mRNAs. Gene 153:237-242. [DOI] [PubMed] [Google Scholar]
- 25.Loyer, P., J. Trembley, J. M. Lahti, and V. J. Kidd. 1998. The RNP protein, RNPS1, associates with specific isoforms of the p34cdc2-related PITSLRE protein kinase in vivo. J. Cell Sci. 111:1495-1506. [DOI] [PubMed] [Google Scholar]
- 26.Mayeda, A., J. Badolato, R. Kobayashi, M. Q. Zhang, E. M. Gardiner, and A. R. Krainer. 1999. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 18:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyerson, M., G. H. Enders, C.-L. Wu, L.-K. Su, C. Gorka, C. Nelson, E. Harlow, and L.-H. Tsai. 1992. A family of human cdc2-related protein kinases. EMBO J. 11:2909-2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misteli, T. 1999. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 9:R198-R200. [DOI] [PubMed] [Google Scholar]
- 29.Misteli, T., and D. L. Spector. 1997. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 7:135-138. [DOI] [PubMed] [Google Scholar]
- 30.Mock, B. A., C. Padlan, C. A. Kozak, and V. J. Kidd. 1994. The gene for mouse p58cdc2L1 (Pitslre 1) protein kinase maps near Ski on mouse chromosome 4. Mamm. Genome 5:191-192. [DOI] [PubMed] [Google Scholar]
- 31.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 32.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 33.Ortega, S., I. Prieto, J. Odajima, A. Martin, P. Dubus, R. Sotillo, J. L. Barbero, M. Malumbres, and M. Barbacid. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25-31. [DOI] [PubMed] [Google Scholar]
- 34.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila p-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 35.Rickert, P., W. Seghezzi, F. Shanahan, H. Cho, and E. Lees. 1996. Cyclin/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 12:2631-2640. [PubMed] [Google Scholar]
- 36.Sachs, A. B. 2000. Cell cycle-dependent translation initiation: IRES elements prevail. Cell 101:243-245. [DOI] [PubMed] [Google Scholar]
- 37.Sauer, K., K. Weigmann, S. Sigrist, and C. F. Lehner. 1996. Novel members of the cdc2-related kinase family in Drosophila: cdk4/6, cdk5, PFTAIRE, and PITSLRE kinase. Mol. Biol. Cell 7:1759-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwab, M., C. Praml, and L. C. Amler. 1996. Genomic instability in 1p and human malignancies. Genes Chromosomes Cancer 16:211-229. [DOI] [PubMed] [Google Scholar]
- 39.Serizawa, H., T. P. Makela, J. W. Conaway, R. C. Conaway, R. A. Weinberg, and R. A. Young. 1995. Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374:280-282. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D. S., P. L. Greer, and L. H. Tsai. 2001. Cdk5 on the brain. Cell Growth Differ. 12:277-283. [PubMed] [Google Scholar]
- 41.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 42.Th'ng, J. P., P. S. Wright, J. Hamaguchi, M. G. Lee, C. J. Norbury, P. Nurse, and E. M. Bradbury. 1990. The FT210 cell line is a mouse G2 phase mutant with a temperature-sensitive CDC2 gene product. Cell 63:313-324. [DOI] [PubMed] [Google Scholar]
- 43.Trembley, J. H., D. Hu, C. A. Slaughter, J. M. Lahti, and V. J. Kidd. 2003. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J. Biol. Chem. 278:2265-2270. [DOI] [PubMed] [Google Scholar]
- 44.Trembley, J. H., D. Hu, L. C. Hsu, C. Y. Yeung, C. Slaughter, J. M. Lahti, and V. J. Kidd. 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277:2589-2596. [DOI] [PubMed] [Google Scholar]
- 45.Tsutsui, T., B. Hesabi, D. S. Moons, P. P. Pandolfi, K. S. Hansel, A. Koff, and H. Kiyokawa. 1999. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27Kip1 activity. Mol. Cell. Biol. 19:7011-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van den Heuvel, S., and E. Harlow. 1993. Distinct roles for cyclin-dependent kinases in cell cycle control. Science 262:2050-2054. [DOI] [PubMed] [Google Scholar]
- 47.Wei, P. F., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 48.Xiang, J., J. M. Lahti, J. Grenet, J. Easton, and V. J. Kidd. 1994. Molecular cloning and expression of alternatively spliced PITSLRE protein kinase isoforms. J. Biol. Chem. 269:15786-15794. [PubMed] [Google Scholar]