Summary
During CD4+ T cell activation, T cell receptor (TCR) signals impact T cell fate, including recruitment, expansion, differentiation, trafficking and survival. To determine the impact of TCR signals on the fate decision of activated CD4+ T cells to become end-stage effector or long-lived memory T-helper 1 (Th1) cells, we devised a deep sequencing-based approach that allowed us to track the evolution of TCR repertoires following acute infection. The transition of effector Th1 cells into the memory pool was associated with a significant decrease in repertoire diversity, and major histocompatibility complex (MHC) Class II tetramer off-rate, but not tetramer avidity, was a key predictive factor in the representation of individual clonal T cell populations at the memory stage. We conclude that stable and sustained interactions with antigen during the development of T-helper 1 (Th1) responses to acute infection are a determinative factor in promoting the differentiation of Th1 memory cells.
Introduction
Following their activation, CD4+ T cells undergo a period of clonal expansion that coincides with the acquisition of specific effector cell functions. Once the antigen is cleared, a small subset of effector CD4+ T cells survives and populates the long-lived memory T cell pool (van Leeuwen et al., 2009). The differentiation steps that lead to the formation of effector T helper-1 (Th1) cells have been studied extensively, but less is known regarding the signals that enable a subset of effector Th1 cells to differentiate into memory cells, although CD4+ T cells fated to become memory cells can be identified during the effector response to acute infection (Marshall et al., 2011). Identification of the signals that promote memory cell differentiation is key to understanding how activated T cells make fate decisions as well as to the design of better vaccination and immunotherapeutic strategies aimed at enhancing CD4+ memory T cell formation and function.
External environmental cues, including cytokines, control the expression of transcription factors that promote T helper subset differentiation, including T-bet, Blimp-1, STAT3, STAT4 and Bcl-6 in settings of Type I cell-mediated inflammation (Eto et al., 2011; Johnston et al., 2012; Johnston et al., 2009; Nakayamada et al., 2011; Pepper et al., 2011). The extent to which these factors promote effector or memory T cell fate decisions is less clear. Some recent articles have implied potential roles for Bcl-6 and IL-21 in the differentiation and formation of CD4+ central memory T cells, along with an opposing role for interleukin-2 (IL-2)-driven STAT5 activation in driving effector-memory Th1 cell differentiation (Crotty et al.; Johnston et al., 2012; Luthje et al., 2012; Pepper et al., 2011; Weber et al., 2012a).
Cell-intrinsic differentiation cues, in particular those dependent on T cell receptor (TCR) binding and signaling, also play a clear role in many aspects of CD4+ T cell differentiation. For CD4+ T cells, the strength of TCR-mediated signaling progressively drives effector differentiation and survival (Gett et al., 2003), and repeated stimulation selectively enriches for responding CD4+ T cells with high avidity TCRs (Savage et al., 1999). Additionally, several days of exposure to antigen in vivo are required for full differentiation of effector (Obst et al., 2005; Williams and Bevan, 2004) and memory (Jelley-Gibbs et al., 2005) CD4+ T cells. The nature of the TCR stimulus also influences the differentiation of T helper subsets, including Th1, T helper 2 (Th2), T follicular helper (Tfh) and regulatory T (Treg) cells (Brogdon et al., 2002; Fazilleau et al., 2009; Lee et al., 2012; Leitenberg and Bottomly, 1999; Moran et al., 2011; Olson et al., 2013). Low immunizing doses can result in the generation of CD4+ memory T cells with high affinity TCRs (Rees et al., 1999), and secondary responses are characterized by the emergence of secondary CD4+ T cell responders with high avidity for antigen (Savage et al., 1999). An additional study reports defects in memory cell formation related to naïve precursor frequency (Blair and Lefrancois, 2007). Based on the combined evidence, one can reasonably conclude that high avidity CD4+ T cells are progressively selected in the presence of antigen. However, it is unknown how TCR-mediated differentiation signals during the primary T cell response might influence long-term fate once antigen is cleared. The role of sustained TCR interactions with antigenic peptide bound to MHC Class II (pMHCII) in the specification of memory T cell fate has not been directly determined.
We previously showed that not all clones that participate in the effector Th1 response to acute infection are equally represented in the subsequent Th1 memory cell population (Williams et al., 2008). Instead, memory T cell differentiation potential corresponds to the development of high antigen sensitivity during the primary response, and stable maintenance of the memory state is associated with the emergence of Th1 memory cells with high functional avidity (Kim et al., 2012; Williams et al., 2008). These findings suggest the hypothesis that strong TCR-pMHCII interactions are a key element in Th1 memory cell fate decisions. To test this hypothesis, we generated a deep sequencing-based model system that allowed us to track TCR repertoire evolution during effector and memory Th1 cell differentiation, as well as characterize the binding of pMHCII by individual TCRs, thus relating the potential for memory differentiation to the kinetic and equilibrium binding properties of individual TCRs. TCR repertoire diversity substantially decreased as CD4+ memory T cell populations emerged following infection with either lymphocytic choriomeningitis virus (LCMV) or recombinant Listeria monocytogenes expressing the immunodominant MHC Class II-restricted epitope, GP61-80, derived from the LCMV Glycoprotein (Lm-gp61). However, when the binding properties of individual TCRs were assessed, memory T cell differentiation potential did not correspond to apparent Kd measurements as determined by MHC Class II tetramer binding. Instead, memory T cell differentiation was predicted by tetramer-binding off-rates, suggesting that in settings where antigen is not limiting, such as during a robust viral or bacterial infection, antigen off-rates may be a better predictor of the biological consequences of sustained TCR-pMHCII interactions. Furthermore, the differing potential of monoclonal populations of T cells to differentiate into memory T cells could be predicted by tetramer off-rates. Overall, we conclude that sustained TCR-mediated signaling during priming is a key element in the specification of CD4+ memory T cell fate.
Results
Generation and characterization of a fixed single-chain TCR transgenic mouse
To track virus-specific CD4+ T cell repertoires following acute viral or bacterial infection, we generated a singe chain TCR transgenic mouse that expressed the Tcra chain cloned from the SMARTA TCR (Oxenius et al., 1998). Because SMARTA TCR transgenic mice are specific for the immunodominant Class II-restricted epitope of lymphocytic choriomeningitis virus (LCMV) Glycoprotein, GP61-80, polyclonal expression of the TCRβ chain allowed for efficient tracking of antigen-specific TCRβ repertoires paired to a known TCRα during antigen-specific T cell responses. We crossed the fixed SMARTA Tcra chain transgenic mouse (SMα) to a TCRα-deficient background, ensuring the selection of only TCRβ chains that pair with the SMARTA TCRα (Fig. S1).
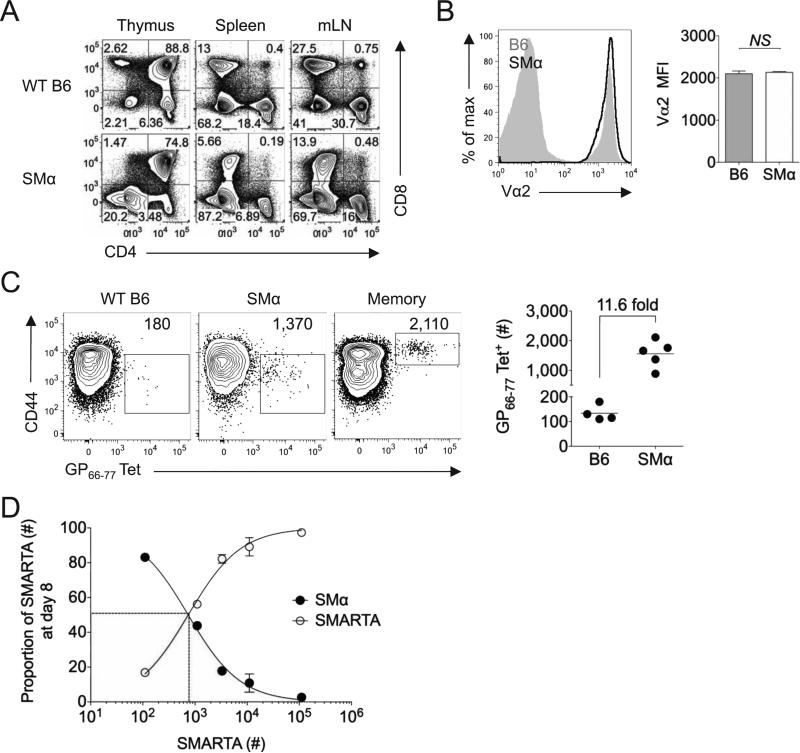
SMα mice generated readily detectable populations of CD4+ T cells in the spleen and lymph nodes, albeit at frequencies marginally lower than WT B6 mice. Surface expression of the SMARTA TCRα chain was comparable to that of polyclonal endogenous CD4+ T cells in WT B6 mice (Fig. 1A-B). As previously described (Moon et al., 2007), we used magnetic sorting to enrich CD4+ T cells from the spleens of B6, SMα, or B6 immune (>42 days post-infection with LCMV) mice that were capable of binding MHC Class II GP66-77 tetramers. We spiked the splenocyte population (Thy1.2+) with a known number (1 × 103) of congenically marked (Thy1.1+) SMARTA CD4+ T cells prior to enrichment as a positive control and to normalize total tetramer-binding cell counts. We also isolated tetramer-binding cells from the spleens of LCMV immune mice to control for the efficiency of tetramer enrichment. We calculated that the total number of GP66-77-specific naïve CD4+ T cells in the spleens of SMα mice ranged from 1-2 × 103, a 10-12-fold increase over naïve precursor frequencies in wildtype mice (Fig. 1C). As a second approach, we employed a previously described competitive inhibition assay (Whitmire et al., 2006) by transferring increasing numbers of Thy1.1+ SMARTA cells into Thy1.2+ SMα or B6 mice, followed by LCMV infection one day later. By measuring relative inhibition of the endogenous polyclonal response using either tetramers or ex vivo peptide stimulated IFN-γ production, we calculated naïve precursor frequencies in SMα mice to be ~1 × 103, 8-10-fold higher than those found in B6 mice (Fig. 1D and data not shown).
Figure 1.
SMα mice generate a readily detectable population of polyclonal naïve precursors specific for LCMV GP61-80. A) Representative flow plots indicate the frequency of CD4+ and CD8+ T cells in the spleen, mesenteric lymph nodes and thymus of B6 and SMα mice. B) Representative flow plot shows TCR surface expression (Vα2+) in B6 or SMα mice. Bar graph indicates the Vα2 MFI of Vα2+CD4+ cells in the spleens of B6 or SMα mice (n=4/group). C) Representative flow plots show tetramer staining after magnetic enrichment of tetramer-binding cells in B6, SMα or B6 immune (>6 weeks post-infection with LCMV) mice, with numbers indicating the estimated number of tetramer-binding cells per spleen after internal normalization to “spiked” Thy1.1+ SMARTA cells. (1 × 103) Graph indicates the estimated frequency of tetramer-binding CD4+ T cells in individual mouse spleens and the estimated fold difference in precursor frequency between B6 and SMα mice. D) SMARTA cells were adoptively transferred in various numbers into SMα mice, followed one day later by LCMV infection. Plot indicates the relative proportion of SMα and SMARTA responders at day 8 post-infection, compared to the estimated SMARTA precursor frequency in the spleen at day 0 (estimated as 10% take of the initial transfer). Error bars indicate the standard error of the mean (SEM)(n=4 mice/group). Results are representative of two independent experiments. See also Figure S1.
Although earlier studies have indicated that artificially elevating precursor frequency can lead to intra- and inter-clonal competition, these observations have typically taken place when precursor frequencies are an order of magnitude or more higher than those observed in our model system (Blair and Lefrancois, 2007; Foulds and Shen, 2006). Because we observed only modest increases in naïve precursor frequency in SMα mice, we analyzed their response following direct LCMV or Lm-gp61 infection. While SMα mice cleared the Lm-gp61 challenge with kinetics similar to WT mice, ~50% of SMα mice failed to clear the LCMV challenge (data not shown). Because all T cells in these mice bear a TCRα chain specific for a MHC Class II-restricted epitope, we hypothesized that MHC Class I-restricted responses were defective. Transfer of 5 × 106 naïve CD8+ T cells enabled SMα mice to clear acute LCMV infection with similar kinetics to WT mice even at early time points (days 3 and 5) post-infection (data not shown). Therefore, subsequent studies were conducted by infecting SMα mice that had received a CD8+ T cell adoptive transfer one day previously.
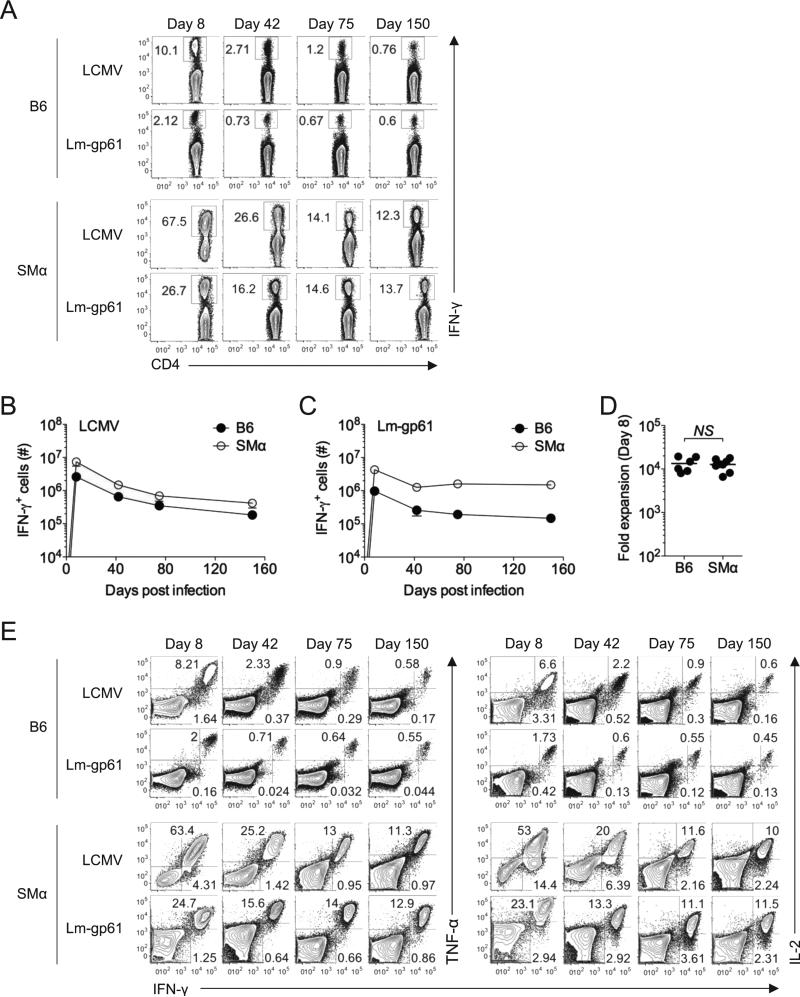
SMα mice generated robust CD4+ T cell responses to both LCMV and Lm-gp61 infection. By day 8 post-infection with LCMV, >60% of the CD4+ T cells in the spleen produced IFN-γ upon ex vivo restimulation with GP61-80 peptide. The response was also robust following Lm-gp61 infection of SMα mice, with ~25% of CD4+ T cells making IFNγ upon restimulation at the peak of the effector response (Fig. 2A). While the size of the responses was expected based on the elevated precursor frequencies in SMα mice, the overall kinetics and magnitude mirrored CD4+ T cell responses to the same epitope in B6 mice (Fig. 2B-C). The magnitude of primary expansion, estimated based upon our calculations of precursor frequency (Fig. 1C), was not significantly different in B6 and SMα mice (Fig. 2D). Furthermore, following LCMV infection, readily detectable memory T cell populations were generated in SMα mice and persisted with similar kinetics to B6 mice, although memory Th1 cells in SMα mice were more stably maintained following Lm-gp61 infection (Fig. 2A,C). Importantly, Th1 effector and memory cells induced in SMα mice displayed the same cytokine producing profile as polyclonal Th1 cells generated in B6 mice (Fig. 2E). In all, these data indicate that SMα mice are a robust model for analyzing the evolution and distribution of antigen-specific CD4+ T cell TCR repertoires following acute infection in vivo.
Figure 2.
Activated CD4+ T cells in SMα mice undergo physiological expansion and differentiation following infection with LCMV or Lm-gp61. B6 or SMα mice were injected with 5 × 106 naïve CD8+ T cells isolated from the spleens of B6 mice. One day later, mice were infected with LCMV or Lm-gp61. A) Representative plots indicate the frequency of IFN-γ-producing CD4+ T cells in the spleen at the indicated time points post-infection following ex vivo restimulation with GP61-80 peptide in the presence of Brefeldin A. B-C) Graphs indicate the number of IFN-γ-producing cells in the spleen of B6 or SMα mice in a time course post-infection with either LCMV or Lm-gp61. Error bars indicate SEM (n=4 mice/group). D) Plot displays estimated fold expansion of GP61-80-specific CD4+ T cells during the first 8 days post-infection based on our calculations of naïve precursor frequency in B6 or SMα mice. “NS” indicates “not significant”, as measured by a two-tailed student's T-test (p>.05). E) Representative flow plots indicate the frequency of IFNγ+TNFγ+ and IFNγ+IL-2+ CD4+ double producers in the spleen at the indicated time points post-infection for B6 and SMα mice. Results are representative of three independent experiments.
Skewed Vβ usage by Th1 memory cells corresponds to differences in functional avidity
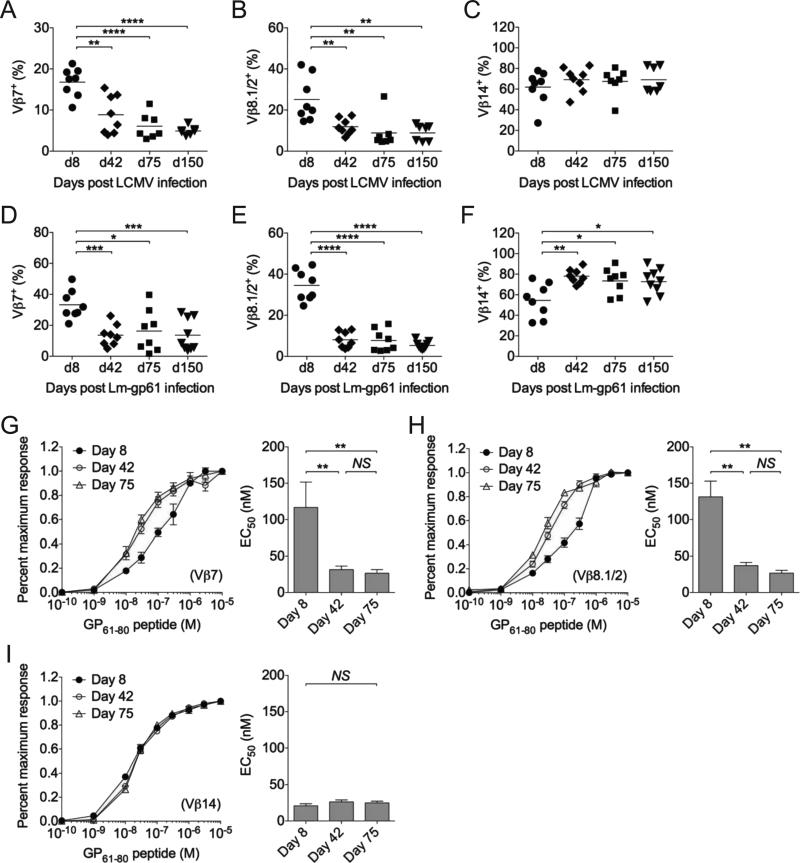
As an initial broad approach to measure TCR repertoire usage by effector and memory Th1 cells following LCMV or Lm-gp61 infection, we analyzed Vβ usage of antigen-specific responders by antibody staining, focusing primarily on the three pre-dominant Vβ subsets utilized by SMα CD4+ T cells during the response to the GP61-80 epitope: Vβ7, Vβ8.1 and Vβ14 (data not shown). MHC Class II tetramers may bind different TCRs with variable efficiency. Additionally, a recent report showed that a large proportion of IFN-γ-producing Th1 cells induced by LCMV fail to bind tetramers at all (Sabatino et al., 2011). Therefore, we measured Vβ expression by IFN-γ-producing cells. When comparing the peak of the effector response (day 8) to memory time points (≥42 days post-infection), the Vβ7 and Vβ8.1 subsets significantly decreased as a proportion of the overall antigen-specific repertoire following both LCMV and Lm-gp61 infection (Fig. 3A-B, D-E). We observed a corresponding increase in the representation of the most dominant subset, Vβ14, at memory time points (≥42 days post-infection) following Lm-gp61 infection (Fig. 3F). We did not observe the same increase in the Vβ14 subset following LCMV infection, (Fig. 3C), possibly reflecting variable and inconsistent participation of other Vβ subsets in the memory T cell pool, including Vβ8.3 and Vβ3 (data not shown). While antigen-specific polyclonal Th1 cells in B6 mice showed some changes in the distribution and usage of Vβ subsets, we observed a similar decrease in the proportion Vβ7 and Vβ8.1-expressing memory Th1 cells (Fig. S2A).
Figure 3.
TCR repertoires skew in the transition from effector to memory T cells. Graphs display the frequency of Vβ subsets among GP61-80-specific IFN-γ+CD4+ T cells in the spleen for Vβ7, Vβ8.1/2 and Vβ14 at the indicated time points after A-C) LCMV infection, or D-F) Lm-gp61 infection. G-I) Functional avidity, as measured by the percent maximal number of IFN-γ-producing cells at the indicated concentrations of peptide restimulation, was calculated for individual Vβ subsets following Lm-gp61 infection. Functional avidity peptide dose response curves are plotted at days 8, 42 and 75 post-infection for G) Vβ7, H) Vβ8.1 and I) Vβ14 subsets. Line plots display functional avidity maturation, and bar graphs indicate effective peptide concentration required to elicit a half maximal response (EC50) for each subset at the indicated time points. Error bars indicate SEM (n=4 mice/group). As determined by student's t-test: *p<.05; **p<.01; ***p<.001; ****p<.0001, NS=not significant (p>.05). See also Figure S2. Results are representative of two independent experiments.
We previously observed that the differentiation of memory Th1 cells is accompanied by an increase in their overall functional avidity, or antigen sensitivity, as measured by the production of IFN-γ in response to stimulation with decreasing concentrations of antigen (Williams et al., 2008). Similarly, effector Th1 cells in SMα mice displayed an increase in functional avidity as they transitioned to become memory Th1 cells (data not shown). We considered at least two possibilities to explain these observations. First, the population increase in functional avidity may represent a selective loss of low functional avidity responders in the formation of the memory T cell pool. Second, acquisition of higher antigen sensitivity may represent a normal facet of memory T cell differentiation that broadly applies to all individual clones within the response. To distinguish between these possibilities, we measured the functional avidity of the Vβ7, Vβ8.1 and Vβ14 subsets at the peak of their response (day 8) and at memory time points (days 42 and 75 post-infection) following Lm-gp61 infection. Functional avidity was assessed based on the ability of each subset to make IFN-γ, as measured by intracellular cytokine staining after ex vivo peptide restimulation. Importantly, while in vitro restimulation can result in TCR down-regulation, surface expression of TCR after restimulation was sufficient to readily detect with antibodies (Fig. S2B). Additionally, Vβ antibody staining of tetramer-binding Th1 cells at day 8 after LCMV infection in SMα mice was similar to that of IFN-γ-producing cells, (data not shown).
At day 8 post-infection, Vβ7+ and Vβ8.1+ IFN-γ-producing responders showed significantly lower functional avidity than did Vβ14+ responders. They required 4-5-fold higher concentrations of peptide to induce a half-maximal response as compared to the Vβ14 subset. Following the emergence of memory Th1 cells (≥42 days post-infection), both the Vβ7 and Vβ8.1 subsets showed an increase in functional avidity, whereas the Vβ14 subset, which began at higher functional avidity during the effector response, maintained its high functional avidity and did not demonstrate additional increase following memory Th1 cell differentiation (Fig. 3G-I). Overall these findings demonstrate that the TCR repertoire of memory Th1 cells shows broad skewing and highlight a role for TCR-driven differentiation events in the selection of the memory T cell pool. Additionally, the memory T cell differentiation potential of each subset corresponded with their functional avidity at the peak of the effector response, suggesting that the increased functional avidity of memory Th1 cells represents the selection of high functional avidity effector cells into the memory T cell pool and not broad functional avidity maturation of all responders.
Deep sequencing analysis reveals loss of repertoire diversity by memory Th1 cells
While Vβ analysis can provide a broad overview of TCR repertoires, we sought to analyze the evolution of SMα TCR repertoires in a more specific and comprehensive way. As an initial approach to study the evolution of TCR repertoires, we transferred large numbers of naïve CD4+ T cells (5 × 106) from SMα mice (Thy1.1+) into B6 hosts (Thy1.2+), followed by LCMV infection one day later. The response of adoptively transferred SMα Th1 cells resembled that of the B6 host in terms of estimated fold expansion, as well as the onset and kinetics of contraction (Fig. S3A-C). Following fluorescence-activated cells sorting (FACS) of IFN-γ-producing SMα Th1 cells and RT-PCR-based cloning and sequencing of expressed TCRβ transcripts within the Vβ14 subset, we observed the emergence of a variety of clones and an apparent loss of clonal diversity in the memory Th1 population (Fig. S3D-E). While these preliminary studies confirmed that CD4+ T cells derived from SMα mice differentiated normally in a wildtype setting, they also presented a key caveat. Because of the relatively low precursor frequency of antigen-specific T cells in SMα mice, even transfer of large numbers of CD4+ T cells did not guarantee adequate representation of the full naïve TCR repertoire in individual recipients. Therefore, we employed a more powerful deep sequencing approach for the characterization and analysis of TCR repertoires.
We purified IFN-γ-producing effector (day 8 post-infection) and memory (days 42, 75 and 150 post-infection) Th1 cells from the spleens of LCMV or Lm-gp61 infected SMα mice by FACS (>95% purity), followed by RT-PCR to amplify a small region of the Vβ7, Vβ8.1 or Vβ14 TCRβ chains encompassing the CDR3 region. Amplicons were multiplexed and sequenced using the Illumina HiSeq 2000 (Fig. S3F). Likely noise as a result of PCR error rate or contamination with non-specific T cells during FACS isolation was minimized by choosing a rate cut-off (0.1%) for the analysis that excluded all amino acid sequences that were observed only once (Fig. S3G). We analyzed 2.6-4.9 million nucleotide sequences per mouse and identified 275 unique CDR3 amino acid sequences across all mice, time points and infections. A number of these sequences (57) were shared by at least 75% of mice (3/4) at day 8 following either LCMV or Lm-gp61 infection (Table S1). We defined this group of TCR sequences as the “public” repertoire, with the remaining sequences, observed in fewer mice at day 8 post-infection, defined as the “private” repertoire.
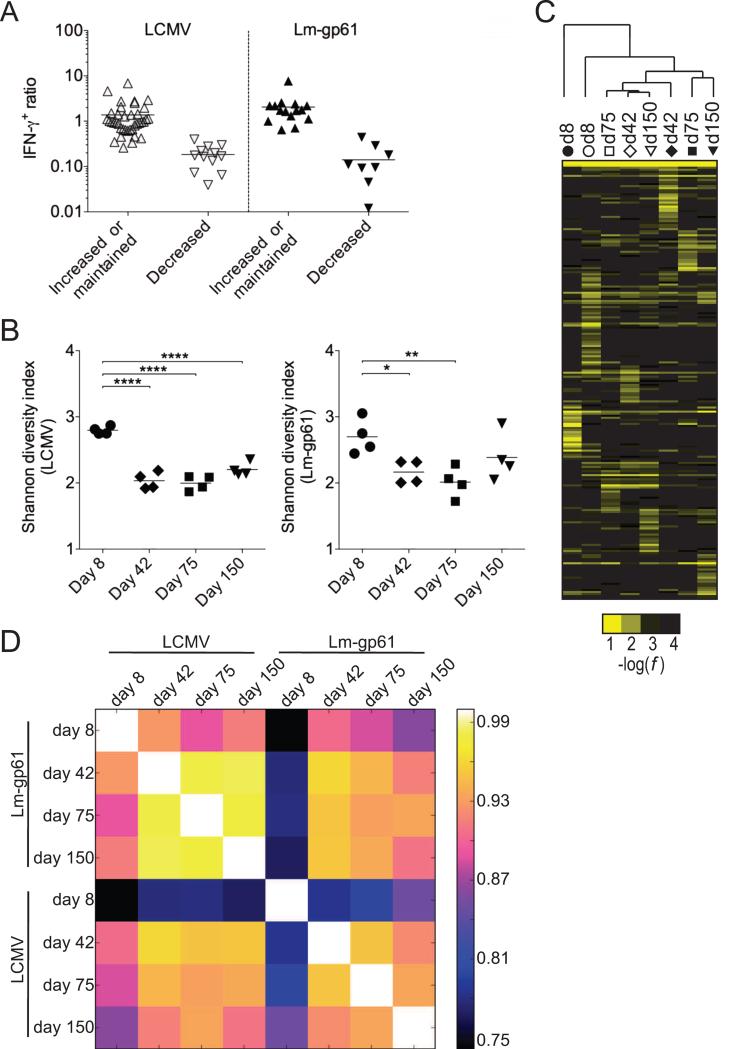
A substantial number of public TCRs (33) were unique to LCMV infection, while few (7) were unique to Lm-gp61 infection (Fig. S3H). These findings confirm that the public T cell repertoire recruited by Lm-gp61 is a subset of the repertoire recruited by LCMV and that the repertoire recruited by LCMV is broader overall. We next divided public clones into two groups: those whose representation within the overall T cell repertoire significantly declined between days 8 and 75 post-infection and those whose representation increased or remained unchanged (Fig. 4A, Table S1). Following LCMV infection 37% of public clones underwent at least 50% decrease in frequency during the transition to the memory state, and for 24% of public clones, this decrease was statistically significant (Fig. 4A, Table S1). Similarly, following Lm-gp61 infection 38% of public clones undergo >50% decrease in frequency as they enter the memory Th1 phase, and for 29% of public clones, this decrease was statistically significant. Overall, decreases in frequency ranged from 50% to almost 99% (Fig. 4A, Table S1).
Figure 4.
Deep sequencing TCR repertoire analysis reveals a loss of clonal diversity upon emergence of memory Th1 cells. After deep sequencing, public clones were identified as those CDR3 sequences present at frequencies of >0.1% in at least 75% of mice at effector time points. A) Public TCRs following LCMV or Lm-gp61 were separated into two groups: 1) those whose frequency within the antigen-specific repertoire increased or was maintained; and 2) those whose frequency within the antigen-specific repertoire decreased following memory Th1 cell differentiation. Plots indicate ratio of effector (day 8) to memory (day 75) Th1 cells. Clones were placed in each group based on the presence or absence of a statistically significant decrease in frequency within the total TCR repertoire between days 8 and 75 post-infection (p<.05), including only mice in which the clone was present (n=3-4/group). B) Shannon's diversity index was used to calculate changes in TCR distribution and diversity at effector (day 8) and memory (days 42, 75, 150) time points after infection with LCMV or Lm-gp61. C) Hierarchical cluster analysis of average rates (n=4 mice per column) for each CDR3 sequence. D) Pairwise Pearson correlation coefficients of average repertoire profiles. As measured by student's t-test, *p<.05; **p<.01; ****p<.0001. See also Figure S3.
To determine if changes in the frequency of individual clones within the memory cell pool could be observed on a global level, we calculated changes in clonal diversity amongst all public and private TCRs using Shannon entropy analysis (Singh et al., 2010; Stewart et al., 1997). We found that memory Th1 differentiation was accompanied by a significant decrease in overall diversity (Fig. 4B) for both LCMV and Lm-gp61 infection. In large part this decrease in diversity took place in between days 8 and 42 post-infection, whereas during the long-term maintenance of Th1 memory cells, TCR repertoire diversity remained stable (Fig. 4B). Additionally, we performed hierarchical cluster analysis based on the average frequency of TCR sequences at each time point. In general, memory T cell populations were more similar to each other than to the effector T cell populations from which they arose, even if they arose from disparate infections (Fig. 4C). We also calculated the Pearson correlation coefficient for the sets of sequence frequencies from each time point versus every other time point. In this analysis, the higher the coefficient, the greater the degree of similarity. Again, whereas memory T cell populations were similar to each other regardless of infection model, they diverged dramatically from the effector T cell populations that they arose from (Fig. 4D). Effector T cell populations also diverged strongly from each other, highlighting the diverse nature of the effector Th1 cell repertoire (Fig. 4D). These data demonstrate selective representation of some effector T cell clones but not others within the memory T cell pool and suggest a role for TCR signals in memory T cell fate specification.
Entry into the memory T cell pool corresponds with MHC Class II tetramer off-rates
We next sought to determine the characteristics of TCR binding that corresponded to enhanced memory T cell differentiation. Based on the public TCR sequences obtained in our deep sequencing (Table S1), we cloned the Tcrb gene of 16 different public clones (Table I). Tcrb chains were cloned with the SMARTA Tcra chain by fusion PCR into a retroviral expression vector with a GFP reporter. The Tcra and Tcrb genes were separated by a P2A sequence to allow for bicistronic expression, as previously described (Holst et al., 2006; Osborn et al., 2005).
Table I.
16 TCRβ sequences were cloned into a retroviral expression vector and paired to the SMARTA TCRα by fusion PCR. The CDR3β amino acid sequence, length, Vβ usage and Jβ usage for each clone are displayed. Retroviral vectors expressing cloned TCRs were transfected into 293T cells, followed by tetramer staining and calculation of apparent Kd and tetramer off-rates. See also Table S1.
| Clone ID | CDR3β | Length | Vβ | Jβ | Kd (nM) | off-rate (min) |
|---|---|---|---|---|---|---|
| clone 1 | ALQGDNNQAPL | 11 | TRBV31*01 | TRBJ1-5*01 | 5.52 | 824.4 |
| clone 2 | AWRPGTANSDYT | 12 | TRBV31*01 | TRBJ1-2*01 | 88.34 | 709.4 |
| clone 5 | AWSRDSSDYT | 10 | TRBV31*01 | TRBJ1-2*01 | 7.43 | 803.3 |
| clone 7 | AWSLPNYAEQF | 11 | TRBV31*01 | TRBJ2-1*01 | 7.93 | 591.9 |
| clone 10 | ASSDFGQGAERLF | 13 | TRBV13-3*01 | TRBJ1-4*02 | 2.77 | 54.2 |
| clone 13 | ASSDQNNQAPL | 11 | TRBV13-3*01 | TRBJ1-5*01 | 15.58 | 620.5 |
| clone 18 | AWSLWEYAEQF | 11 | TRBV31*01 | TRBJ2-1*01 | 92.37 | 212.0 |
| clone 19 | AWSPGLGVNYAEQF | 14 | TRBV31*01 | TRBJ2-1*01 | 116.20 | 501.6 |
| clone 20 | AWSLIEVF | 8 | TRBV31*01 | TRBJ1-1*01 | 2.33 | 466.5 |
| clone 22 | ASSDHNQANTEVF | 13 | TRBV13-3*01 | TRBJ1-1*01 | 10.86 | 379.0 |
| clone 24 | ASSEMGTGIETLY | 13 | TRBV13-3*01 | TRBJ2-3*01 | 3.31 | 124.4 |
| clone 25 | ASSLAGTGGYEQY | 13 | TRBV29*01 | TRBJ2-7*01 | 6.32 | 151.3 |
| clone 26 | ASSSPGTANYAEQF | 14 | TRBV29*01 | TRBJ2-1*01 | 4.50 | 38.2 |
| clone 27 | ASSPSGTGGYEQY | 13 | TRBV29*01 | TRBJ2-7*01 | 3.57 | 43.3 |
| clone 28 | ASSLHNSGNTLY | 12 | TRBV29*01 | TRBJ1-3*01 | 8.71 | 288.8 |
| clone 29 | AWSLPNSYEQY | 11 | TRBV31*01 | TRBJ2-7*01 | 5.46 | 265.7 |
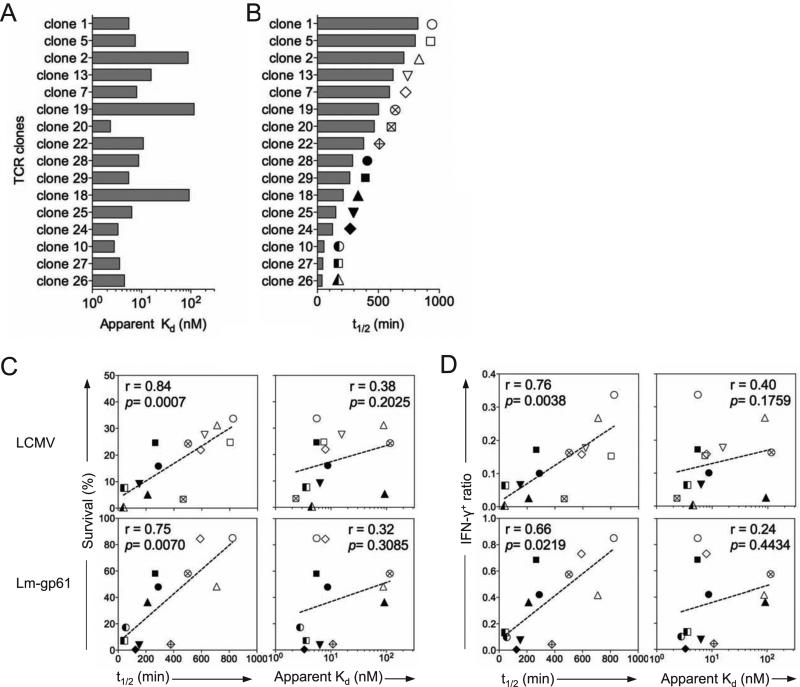
We transfected 293T cells with TCR retroviral expression vectors, along with multi-cistronic retroviral vectors encoding Cd3d, Cd3e, Cd3g and Cd247, in order to permit surface TCR expression (Holst et al., 2006). As the absence of CD4 does not enhance the avidity of tetramer binding to MHC Class II (Crawford et al., 1998; Hamad et al., 1998), we directly stained transfected 293T cells with decreasing concentrations of GP66-77 tetramer, normalized to GFP and surface TCR expression in equlilibrium binding assays,. We then generated Scatchard plots and calculated apparent Kd values (Fig. S4A-C). The panel of TCRs displayed Kd values across a 50-100-fold range (Fig. 5A). Next, we performed tetramer decay assays to determine the tetramer dissociation rate for each TCR (Fig. 5B, Fig. S4D). Tetramer off-rates and apparent Kd values for each TCR were discordant (Fig. 5A-B). Some clones demonstrated high avidity binding to tetramer but quick off-rates, while others demonstrated low avidity binding but extremely slow off-rates (Table I, Fig. S4E). When each of these parameters was compared to the survival of individual TCR clones between days 8 and 42 post-infection, the only significant predictor of memory T cell potential was the tetramer off-rate (Fig. 5C). Similar results were found when we compared tetramer off-rates to the total numbers of IFN-γ-producing cells in the spleen as a ratio between days 8 and 42 post-infection (Fig. 5D). One example of this phenomenon was TCR clone 2, a dominant Vβ14+ clone present at enriched frequencies within the memory Th1 population. While clone 2 TCR bound tetramer with low avidity at equilibrium binding concentrations, it maintained extremely slow off-rates in tetramer decay assays (Table I, Fig. S4D-E).
Figure 5.
CD4+ memory T cell differentiation is driven by slow TCR-pMHCII dissociation rates. 293T cells were transfected with retroviral vectors expressing the indicated TCR clone and a GFP reporter. Samples were subsequently stained with tetramer and normalized to GFP and cell surface TCR expression. A) Bar graph displays apparent Kd for each clone, as determined by intensity of tetramer binding under equilibrium binding conditions for varying tetramer concentrations. B) For each clone bar graphs display tetramer binding half-life by measuring tetramer binding decay (normalized fluorescence) following addition of high concentrations of MHC Class II blocking antibody. Apparent Kd and half-life measurements are representative of 2-3 separate transfections for each clone. C) For each clone, plots display tetramer-binding half-life or apparent Kd on the x-axis and the percent survival between day 8 and day 42 or D) the ratio of the total number of IFNγ-producing cells in the spleen at memory (day 42) versus effector (day 8) time points of individual TCR clones observed by deep sequencing on the y-axis. Dotted lines indicate the best fit by linear regression. Correlation and significance were calculated by two-tailed Spearman's rank correlation. See also Figure S4.
Sustained TCR-pMHCII interactions promote memory fate specification
To directly assess the role of sustained TCR-pMHCII interactions in promoting the differentiation of Th1 memory cells, we measured the impact of individual TCRs on Th1 memory cell differentiation potential in the context of infection in a wildtype mouse. We used the above-described TCR retroviral vectors to transduce RAG-deficient bone marrow and to subsequently generate TCR retrogenic bone marrow chimeras, as previously described (Holst et al., 2006). Eight-to-ten weeks after bone marrow transplantation, naïve CD4+GFP+ T cells were harvested from their spleens, transferred into naïve B6 mice in small numbers (1 × 104 cells per recipient) and stimulated by LCMV infection one day later. All TCRs that we tested expanded potently and produced cytokines in response to GP61-80 peptide stimulation following LCMV infection (Fig. S5 and data not shown), verifying that our model system and our criteria for establishing cut-offs for individual sequences effectively identified antigen-specific TCR clones.
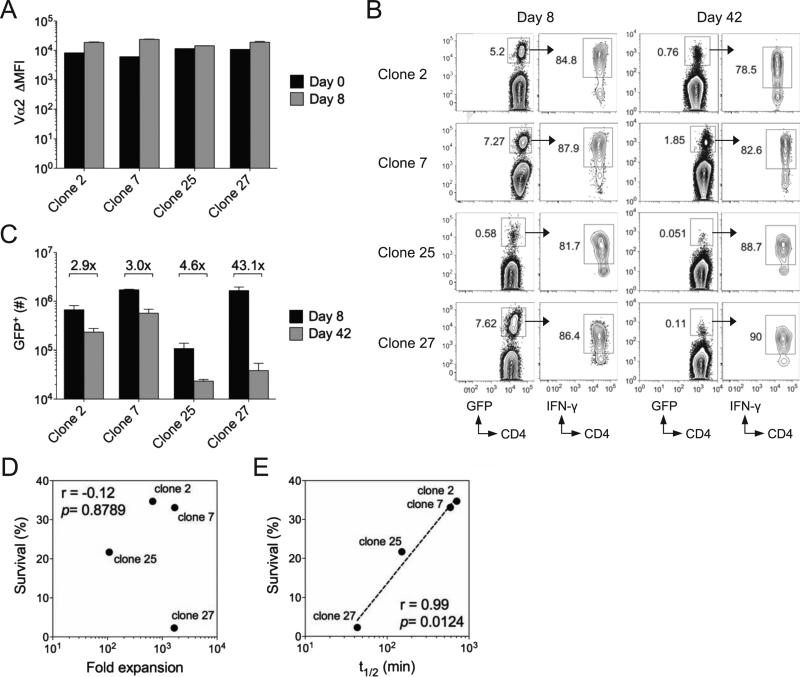
We selected four clones for further analysis on the basis of their similar surface TCR expression both before and after activation (Fig. 6A). These clones (clones 2, 7, 25, 27) exhibited a range of tetramer off-rates and tetramer binding avidity (Table I). Following LCMV infection of host B6 mice, each clone expanded and formed memory T cells, albeit to varying extents, and produced IFN-γ upon restimulation at both day 8 and day 42 post-infection in the spleen (Fig. 6B-C). The memory potential of each effector Th1 cell population, as measured by the overall decline in numbers in the spleen between days 8 and 42 post-infection, varied widely, with two clones (2 and 7) undergoing minimal contraction, a third clone (25) undergoing moderate contraction and a fourth clone (27) undergoing extensive contraction (Fig. 6C). The memory potential of each clone did not correspond to primary expansion (Fig. 6D), but to tetramer off-rates (Fig. 6E). These findings validate and recapitulate the results for individual clones derived from our deep sequencing data. We conclude that sustained TCR-pMHCII interactions are a key component in promoting Th1 memory cell fate decisions in vivo.
Figure 6.
Sustained TCR-pMHCII interactions promote CD4+ memory T cell differentiation. TCR “retrogenic” T cell clones (GFP+) were adoptively transferred (1 × 104) into B6 mice, followed by LCMV infection one day later. A) Bar graph indicates Vα2 surface expression as determined by antibody staining, calculated as the shift in mean fluorescence intensity as compared to Vα2-negative CD4+ T cells in the same host. B) Representative flow plots indicate the frequency of GFP+ retrogenic T cells within the CD4+ T cell population in the spleen, as well as IFN-γ production by gated GFP+ T cells at days 8 and 42 post-infection with LCMV. C) Bar graph indicates the total number of GFP+ retrogenic T cells in the spleen at days 8 and 42 post-infection for the indicated clones. Numbers indicate the fold difference in absolute numbers between days 8 and 42. Error bars indicate the SEM (n=4-5 mice/group). D) Plot indicates fold expansion between day 0 and 8 for each clone on the x-axis compared to percent survival between day 8 and 42 on the y-axis. E) Plot indicates tetramer off-rate for each clone on the x-axis compared to percent survival between day 8 and 42 on the y-axis. Dotted line indicates the best fit by linear regression. Correlation and their significance were calculated by two-tailed Spearman's rank correlation. Results are representative of two experiments. See also Figure S5.
Discussion
Overall, these findings demonstrate that sustained TCR-pMHCII interactions are a key component of the memory T cell differentiation signal for CD4+ T cells. While TCR-pMHCII interactions play important roles in T cell activation, function and survival, we report here that the kinetics of TCR-pMHCII interactions can differentially discriminate between end-stage effector and memory differentiation programs in T cells. Previous studies of the role of antigen in the emergence of high avidity secondary responders have largely concluded that this event occurs due to antigen-driven selection of high avidity clones throughout the primary and secondary response (Savage et al., 1999). In contrast, we found that specific TCR binding properties and the signals they deliver promote a CD4+ memory T cell differentiation program that takes place once antigen is cleared.
A simple model of TCR-pMHCII interaction would suggest that long-lived interactions between a single TCR and its MHC-restricted antigen are a key step in the initiation and amplification of the T cell signaling cascade required for robust activation and differentiation. In support of this, different occupation rates of phosphorylation sites of CD3 subunits have been associated with peptides over a defined range of affinities and agonist activity for a fixed TCR (Kersh et al., 1998; Rabinowitz et al., 1996). However, a strictly quantitative model of TCR signaling does not fully predict biological outcomes following T cell activation as they relate to memory T cell development and function. Recent studies have found that the polyclonal CD4+ T cell response to LCMV is populated with clones at the peak of the effector response and following viral clearance that are unable to bind pMHCII tetramers (Huang et al., 2010; Sabatino et al., 2011). Another study has found that the variable ability of monoclonal populations to either expand during the primary response or generate effective secondary responses is not necessarily determined by TCR affinity for cognate antigen, suggesting that the polyclonal response could be populated with clones that have highly variable and complex fates (Weber et al., 2012b). Our own findings suggest that representation within the Th1 effector cell compartment is not necessarily indicative of memory potential (Williams et al., 2008). Importantly, a recent report has also found that the duration of TCR-pMHCII interactions can influence CD4+ T cell responses during commitment to Th1 or Tfh cell differentiation (Tubo et al., 2013), providing additional evidence that sustained interactions between the TCR and antigen provide a unique signal for cellular differentiation independent of recruitment and expansion.
A variety of other factors likely influence biological outcomes related to TCR binding of pMHCII,, including TCR surface expression and clustering of TCRs and CD3 subunits on the cell surface, a factor that has previously been shown to enhance antigen sensitivity (Kumar et al., 2011). Deciphering the interplay of these factors, along with the actual kinetics of TCR-pMHCII interactions, is key for our understanding of how T cells incorporate activation signals to initiate distinct differentiation programs. For example, while quantitative differences in the magnitude of the TCR signal may play a role in differentiating between effector and memory T cell fate, TCR signals delivered in short bursts and with quick dissociation rates may also be qualitatively distinct from those characterized by more sustained signaling events and slow dissociation rates
Although we identify here a role for the TCR in promoting effector versus memory Th1 cell differentiation, the differentiation, function and survival of Th1 memory cells is driven by TCR-independent factors as well. For example, SMARTA TCR transgenic T cells exhibit a range of functional avidities at the peak of their effector response and an increase in functional avidity during the transition to the memory state (Williams et al., 2008). Factors such as inflammatory microenvironment, the activation status of the APC and amount of antigen presented could influence the acquisition and maintenance of high antigen sensitivity. One possibility is that effector Th1 cells that acquire higher antigen sensitivity during the primary response and are therefore better able to initiate sustained TCR activation compete more effectively for entrance into the memory T cell pool. A key focus of future studies will be to delineate how T cell intrinsic and extrinsic factors cooperate to initiate a memory differentiation program.
Most prior studies have relied on the analysis of a fixed TCR binding to altered peptide ligands, or on genetic alterations to the TCR itself to adjust binding properties. Here, however, we have relied on analysis of TCRs during a biological response to an infectious pathogen. All TCRs included in our analysis have passed thresholds of activation, differentiation and effector function, allowing us to compare differences in agonist-driven T cell activation. Given the availability of a large panel of naturally-derived TCRs with known antigen binding properties, our future studies will focus on the qualitative and quantitative nature of TCR signaling as it relates to off-rates and memory T cell fate determination.
Experimental Procedures
Mice and infections
C57BL/6, Rag1-deficient and TCRα-deficient (6-8 week) mice were purchased from Jackson Laboratories (Bar Harbor, ME). SMARTA mice (Oxenius et al., 1998) were maintained in our colony at the University of Utah. SMα mice were generated on a C57BL/6 background at the University of Utah Transgenic Core Facility by standard microinjection techniques using a T cell specific expression vector, VA-hCD2, in which the SMARTA Tcra gene was placed under the control of the human Cd2 promoter and a 3’ locus control region of the Cd2 gene (provided by M. Bevan, University of Washington, Seattle, WA) (Zhumabekov et al., 1995). LCMV Armstrong 53b was grown in BHK cells, titered in Vero cells as described (Ahmed et al., 1984) and injected i.p. into recipient mice at a dose of 2 × 105 plaque-forming units (PFU). Recombinant Listeria monocytogenes expressing the GP61-80 epitope of LCMV (Lm-gp61, provided by M. Kaja-Krishna, Emory University, Atlanta, GA) was grown to log phase in BHI broth and concentration determined by measuring the O.D. at 600 nm. Mice were injected i.v. with 2 × 105 colony forming units (CFU). All mouse experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Utah.
Cell preparations and flow cytometry
Splenocyte and lymph node cell suspensions were placed in DMEM containing 10% FBS, Penicillin, Streptomycin and L-glutamine. For cell surface stains, cells were incubated with fluorescently conjugated antibodies (eBiosciences, San Diego, CA or BD Biosciences, San Diego, CA) diluted in antibody staining buffer (PBS containing 1% fetal bovine serum) at 4°C. For intracellular cytokine assays, splenocytes were restimulated for 4 hours with 10 μM (or indicated dilutions in functional avidity assays) GP61-80 peptide from LCMV (GLKGPDIYKGVYQFKSVEFD) at 37°C in the presence of Brefeldin A (GolgiPlug, 1 μl/ml) per manufacturer's instructions (BD Biosciences). Samples were then stained with cell surface antibodies in antibody staining buffer, followed by permeabilization using a kit (BD Biosciences) and staining with fluorescently-labeled antibodies specific to the indicated cytokines.
Adoptive transfers and TCR sequencing
Untouched naïve (Thy1.1+CD44lo) CD4+ T cells were isolated from SMα mice using magnetic beads (Miltenyi) and injected i.v. into B6 mice, followed by LCMV infection one day later. For direct infection of SMα mice, untouched CD8+ T cells were isolated from the spleens of B6 mice using magnetic beads (Miltenyi) and injected i.v. into SMα mice one day prior to infection. MHC Class II-tetramer-based enrichment of naïve antigen-specific T cells was performed using magnetic beads (Miltenyi) as described {Moon, 2007 #80}. Live IFN-γ-producing antigen-specific CD4+ T cells were isolated from the spleens of infected mice at the indicated time points post-infection using a kit (Miltenyi), followed by FACS (FACSAria II, BDBiosciences). We purified RNA (RNEasy, Qiagen), generated a cDNA template (Superscript III, Invitrogen) and performed RT-PCR. For adoptive transfer experiments, we amplified the entire Tcrb molecule using primers specific for Vβ14, followed by TA cloning into the pCR2.1-TOPO vector using a kit (Invitrogen). We then isolated plasmids from individual colonies and sequenced Tcrb molecules at the University of Utah DNA Sequencing Core Facility. For deep sequencing studies of TCRs derived from direct infection of SMα mice, we used primers designed to amplify a small ~110 base pair portion of Tcrb encompassing the CDR3 region. The primer sets for each of Vβ subsets encompassed the following sequences: Vβ7: 5’-GACATCTGTGTACTTCTGTGC-3’, Vβ8.1: 5’-ACAGCTGTATATTTCTGTGCC-3’, Vβ14: 5’-TCTGGCTTCTACCTCTGTGCC-3’, and Cβ-specific reverse: 5’-CTTGGGTGGAGTCACATTTCTCAGATCC-3’. Amplicons were multiplexed and underwent single-end 50 base pair sequencing using the Illumina HiSeq 2000 at the University of Utah Microarray and Sequencing Core Facility. Data was segregated based on barcode as well as sequences corresponding to specific Vβ regions, and low quality reads (phred score <38 for 20 of 50 bases) were excluded from the analysis.
Analysis of TCR repertoires
After trimming the primer sequence from each read, nucleotide sequences were translated to determine the CDR3 amino-acid sequence, and intra-subset frequencies were calculated for each distinct amino-acid sequence for every mouse. These were converted to global frequencies for each CDR3 sequence in each mouse by multiplying by relative Vβ subset frequency, as determined by flow cytometry. To reduce the introduction of sequencing artifacts into the analysis, we required that an amino-acid sequence be present in at least two mice at a frequency greater than an empirically determined cut-off. Based on the distribution of sequence frequencies (Fig. S3G) a cut-off of 0.1% was chosen to exclude singleton observations from the analysis of public and private sequences. Hierarchical clustering was performed on average pathogen-specific subset-sequence frequencies, combining the four mice at each time point and applying the 0.1% cutoff to the average values. Clustering was performed with the Pearson correlation, using Cluster 3.0 (Eisen et al., 1998) and visualized with JavaTreeview (Saldanha, 2004). Pairwise correlations were calculated using NumPy and visualized with Matplotlib (Hunter, 2007). The IMGT database and the IMGT/V-QUEST tool (IMGT, The International Immunogenetics Information System; http://www.imgt.org) were used to identify and verify TRBV, TRBJ and CDR3 sequences. Shannon's diversity index, which reflects both abundance and richness, was used to evaluate TCR sequence diversity (Stewart et al., 1997). Shannon's diversity index was calculated as H′=−Σ[pi×ln(pi)], where pi is the proportion of TCR sequence i.
TCR cloning and retrogenic bone marrow chimeras
Tcrb genes were cloned by fusion PCR and expressed in a retroviral vector (MigR1) along with the SMARTA Tcra. In this vector, the Tcra and Tcrb coding regions were separated by the picornavirus-derived P2A sequence, a cis-acting hydrolase element that allows for bicistronic expression (Szymczak et al., 2004). The vector additionally contained a GFP reporter under the control of an IRES. To analyze TCR binding properties, TCR expression vectors, along with a retroviral vector driving expression of the Cd3d, Cd3e, Cd3g and Cd247 subunits (provided by D.A. Vignali, St. Jude Children's Research Hospital, Memphis, TN) (Holst et al., 2006) were transfected into 293T cells using FuGENE, (Promega). TCR-expressing retroviruses were transduced into Rag1-deficient bone marrow cells using described methods (Holst et al., 2006; Yun and Bevan, 2003), followed by i.v. injection of 1 × 106 bone marrow cells into irradiated (450 rads) Rag1-deficient hosts. Eight to ten weeks later, GFP+TCR+CD4+ T cells harvested from the spleens of the retrogenic chimeras were i.v. transferred (1 × 104 cells per recipient) into B6 hosts. Recipient mice were infected with LCMV one day later.
Tetramer staining and analysis
MHC Class II monomers bound to GP66-77 were expressed by stably transfected S2 cells, purified and converted into fluorescently-tagged tetramers using previously described methods (S2 cells provided by M. Pepper, U. Washington, Seattle, WA) (Pepper et al., 2011). Staining was performed at 25°C for 1 hour in RPMI containing 2% FCS and 0.1% sodium azide, followed by washing and cell surface staining. Tetramer fluorescence was normalized to samples stained with control hCLIP tetramer (NIH Tetramer Core Facility, Atlanta, GA). Scatchard plots and apparent Kd were calculated as described (Savage et al., 1999). Fluorescence units (bound) were plotted on the X-axis, and fluorescence units divided by tetramer concentration (bound/free) were plotted on the Y-axis. Kd was determined as the inverse of the slope. For tetramer decay assays, following extensive washing of tetramer-stained cells and cell surface staining, cells were incubated in high concentrations (100 μg/ml) I-Ab blocking antibody (BioLegend, San Diego, CA). To determine tetramer-binding half-life, total fluorescence of tetramer binding at various time points after MHC Class II blockade was normalized to the total fluorescence at the zero time point, as described (Savage et al., 1999).
Supplementary Material
Highlights.
CD4+ TCR repertoires skew following the emergence of memory T cells.
Memory T cell differentiation corresponds to a loss of TCR repertoire diversity.
MHC Class II tetramer off-rate, but not avidity, predicts memory T cell potential.
Sustained TCR interactions with antigen promote memory T cell fate decisions.
Acknowledgements
We thank J. Cassiano for technical assistance, M. Pepper (University of Washington) for providing GP66-77 tetramer constructs, D. Vignali (St. Jude Children's Research Hospital) for providing the retroviral plasmid expressing CD3 subunits, and B. Dalley (University of Utah Microarray Core Facility) for technical advice. This work was supported by NIH grant AI080830 (to M.A.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair DA, Lefrancois L. Increased competition for antigen during priming negatively impacts the generation of memory CD4 T cells. Proceedings of the National Academy of Sciences. 2007;104:15045–15050. doi: 10.1073/pnas.0703767104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogdon JL, Leitenberg D, Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. Journal of immunology. 2002;168:3825–3832. doi: 10.4049/jimmunol.168.8.3825. [DOI] [PubMed] [Google Scholar]
- Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nature immunology. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds KE, Shen H. Clonal Competition Inhibits the Proliferation and Differentiation of Adoptively Transferred TCR Transgenic CD4 T Cells in Response to Infection. Journal of immunology. 2006;176:3037–3043. doi: 10.4049/jimmunol.176.5.3037. [DOI] [PubMed] [Google Scholar]
- Gett AV, Sallusto F, Lanzavecchia A, Geginat J. T cell fitness determined by signal strength. Nat Immunol. 2003;4:355–360. doi: 10.1038/ni908. [DOI] [PubMed] [Google Scholar]
- Hamad AR, O'Herrin SM, Lebowitz MS, Srikrishnan A, Bieler J, Schneck J, Pardoll D. Potent T cell activation with dimeric peptide-major histocompatibility complex class II ligand: the role of CD4 coreceptor. J Exp Med. 1998;188:1633–1640. doi: 10.1084/jem.188.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, Szymczak-Workman AL, Vignali KM, Burton AR, Workman CJ, Vignali DAA. Generation of T-cell receptor retrogenic mice. Nat. Protocols. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, Zhu C. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: A 2D graphics environment. Computing in Science and Engineering. 2007;9:90–95. [Google Scholar]
- Jelley-Gibbs DM, Brown DM, Dibble JP, Haynes L, Eaton SM, Swain SL. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J. Exp. Med. 2005;202:697–706. doi: 10.1084/jem.20050227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. The Journal of Experimental Medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersh EN, Shaw AS, Allen PM. Fidelity of T cell activation through multistep T cell receptor zeta phosphorylation. Science. 1998;281:572–575. doi: 10.1126/science.281.5376.572. [DOI] [PubMed] [Google Scholar]
- Kim C, Jay DC, Williams MA. Stability and function of secondary Th1 memory cells are dependent on the nature of the secondary stimulus. Journal of immunology. 2012;189:2348–2355. doi: 10.4049/jimmunol.1200244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Ferez M, Swamy M, Arechaga I, Rejas MT, Valpuesta JM, Schamel WW, Alarcon B, van Santen HM. Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity. 2011;35:375–387. doi: 10.1016/j.immuni.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Lee HM, Bautista JL, Scott-Browne J, Mohan JF, Hsieh CS. A broad range of self-reactivity drives thymic regulatory T cell selection to limit responses to self. Immunity. 2012;37:475–486. doi: 10.1016/j.immuni.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitenberg D, Bottomly K. Regulation of naive T cell differentiation by varying the potency of TCR signal transduction. Seminars in immunology. 1999;11:283–292. doi: 10.1006/smim.1999.0184. [DOI] [PubMed] [Google Scholar]
- Luthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, Kaech SM. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. The Journal of Experimental Medicine. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst R, van Santen H-M, Mathis D, Benoist C. Antigen persistence is required throughout the expansion phase of a CD4+ T cell response. J. Exp. Med. 2005;201:1555–1565. doi: 10.1084/jem.20042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. Effector-like CD8(+) T Cells in the Memory Population Mediate Potent Protective Immunity. Immunity. 2013;38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Panoskaltsis-Mortari A, McElmurry RT, Bell SK, Vignali DAA, Ryan MD, Wilber AC, McIvor RS, Tolar J, Blazar BR. A picornaviral 2A-like Sequence-based Tricistronic Vector Allowing for High-level Therapeutic Gene Expression Coupled to a Dual-Reporter System. Mol Ther. 2005;12:569–574. doi: 10.1016/j.ymthe.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Oxenius A, Bachmann MF, Zinkernagel RM, Hengartner H. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur J Immunol. 1998;28:390–400. doi: 10.1002/(SICI)1521-4141(199801)28:01<390::AID-IMMU390>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Beeson C, Wulfing C, Tate K, Allen PM, Davis MM, McConnell HM. Altered T cell receptor ligands trigger a subset of early T cell signals. Immunity. 1996;5:125–135. doi: 10.1016/s1074-7613(00)80489-6. [DOI] [PubMed] [Google Scholar]
- Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4+ T cell responses in vivo and in vitro. Proceedings of the National Academy of Sciences. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatino JJ, Jr., Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Savage PA, Boniface JJ, Davis MM. A Kinetic Basis For T Cell Receptor Repertoire Selection during an Immune Response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- Singh Y, Ferreira C, Chan AC, Dyson J, Garden OA. Restricted TCR-alpha CDR3 diversity disadvantages natural regulatory T cell development in the B6.2.16 beta-chain transgenic mouse. Journal of immunology. 2010;185:3408–3416. doi: 10.4049/jimmunol.1001088. [DOI] [PubMed] [Google Scholar]
- Stewart JJ, Lee CY, Ibrahim S, Watts P, Shlomchik M, Weigert M, Litwin S. A Shannon entropy analysis of immunoglobulin and T cell receptor. Molecular immunology. 1997;34:1067–1082. doi: 10.1016/s0161-5890(97)00130-2. [DOI] [PubMed] [Google Scholar]
- Szymczak AL, Workman CJ, Wang Y, Vignali KM, Dilioglou S, Vanin EF, Vignali DA. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nature biotechnology. 2004;22:589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen EM, Sprent J, Surh CD. Generation and maintenance of memory CD4(+) T Cells. Curr Opin Immunol. 2009;21:167–172. doi: 10.1016/j.coi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JP, Fuhrmann F, Hutloff A. T-follicular helper cells survive as long-term memory cells. Eur J Immunol. 2012a;42:1981–1988. doi: 10.1002/eji.201242540. [DOI] [PubMed] [Google Scholar]
- Weber KS, Li QJ, Persaud SP, Campbell JD, Davis MM, Allen PM. Distinct CD4+ helper T cells involved in primary and secondary responses to infection. Proc Natl Acad Sci U S A. 2012b;109:9511–9516. doi: 10.1073/pnas.1202408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmire JK, Benning N, Whitton JL. Precursor Frequency, Nonlinear Proliferation, and Functional Maturation of Virus-Specific CD4+ T Cells. Journal of immunology. 2006;176:3028–3036. doi: 10.4049/jimmunol.176.5.3028. [DOI] [PubMed] [Google Scholar]
- Williams MA, Bevan MJ. Shortening the Infectious Period Does Not Alter Expansion of CD8 T Cells but Diminishes Their Capacity to Differentiate into Memory Cells. Journal of immunology. 2004;173:6694–6702. doi: 10.4049/jimmunol.173.11.6694. [DOI] [PubMed] [Google Scholar]
- Williams MA, Ravkov EV, Bevan MJ. Rapid culling of the CD4+ T cell repertoire in the transition from effector to memory. Immunity. 2008;28:533–545. doi: 10.1016/j.immuni.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun TJ, Bevan MJ. Notch-Regulated Ankyrin-Repeat Protein Inhibits Notch1 Signaling: Multiple Notch1 Signaling Pathways Involved In T Cell Development. Journal of immunology. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- Zhumabekov T, Corbella P, Tolaini M, Kioussis D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J Immunol Methods. 1995;185:133–140. doi: 10.1016/0022-1759(95)00124-s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.