Abstract
Allorecognition has been described in many metazoan phyla, from the sponges to the mammals. In vertebrates, allorecognition is a result of a MHC-based recognition event central to adaptive immunity. However, the origin of the adaptive immune system and the potential relationship to more primitive allorecognition systems is unclear. The colonial ascidian, Botryllus schlosseri, has been used as a model organism for the study of allorecognition for over a century, as it undergoes a natural transplantation reaction controlled by a single, highly polymorphic locus. Herein we will summarize our current understanding of the molecular mechanisms that underlie this innate allorecognition reaction.
Keywords: Allorecognition, ascidian, Botryllus schlosseri, innate immunity
The ability to discriminate self from non-self is a process found throughout the metazoa and is the basis of immune function. In the vertebrates, allorecognition is carried out mainly via the adaptive immune system, which is characterized by its capacity for somatic recombination of germ-line encoded immune receptors coupled to recognition of highly polymorphic MHC proteins. As one of the most complex processes that occur within the vertebrate body, it has been thought that the origins of the adaptive immune system may reside within the more primitive organisms. In fact, highly polymorphic allorecognition systems have been discovered across a wide evolutionary spectra of taxa, including sponges, cnidarians, echinoderms, tunicates and the jawless fishes [1], suggesting the presence of complex and discriminatory recognition systems in non-vertebrate organisms. However, to date the ligands and receptors used within these systems are unrelated to those in the adaptive immunity, [2-4], thus the evolutionary precursor to the adaptive immune system remains elusive.
The colonial ascidian, Botryllus schlosseri, belongs to the chordate subphylum, Tunicata, and occupies a key phylogenetic position in vertebrate evolution. Botryllus has also been used as model organism for the study of allorecognition for over a century, as it undergoes a natural transplantation reaction controlled by a single, highly polymorphic locus (the fuhc), semantically similar to the MHC. The allorecognition reaction in B. schlosseri is highly specific and capable of discriminating between 100s of naturally occurring allelic variants, but recent molecular evidence has shown that the fuhc is not the ancestral MHC [13]. Nevertheless, as the closest living invertebrate relatives to the vertebrate line [14], allorecognition in the ascidians represents a basal form of self/non-self recognition, and properties of this system may be the building blocks for the evolution of chordate immunity. We are just beginning to dissect and understand the underlying genetic architecture of this exquisitely specific allorecognition system, which evolved nearly 500 million years before the emergence of adaptive immunity. Here we will summarize our current understanding of the molecular mechanisms which underlie this innate allorecognition reaction.
Biology of Botryllus schlosseri
The life history of Botryllus schlosseri cycles between two distinct body plans, beginning with a sexually derived, free-swimming chordate tadpole larvae which hatches, swims away from its parents and searches for a suitable substratum, then settles, usually within 12-24 hours. The larva then undergoes metamorphosis, resorbing most of its chordate characteristics and transforming to an asexually propagating invertebrate bauplan, called an oozoid. The oozooid immediately begins a budding process called blastogenesis, eventually giving rise to a colony of genetically identical individuals (zooids) encased in a common tunic and sharing a common blood supply. Each individual is autonomous, possessing a gut, heart, gonads etc., and can be manually dissected from the colony and transferred to a new substratum, where it will continue to bud. Thus multiple naïve subclones can be generated for any particular genotype. A connecting, extracorporeal vasculature permeates throughout the entire colony and terminates at structures called ampullae, which are oval-shaped protrusions found throughout the periphery of the tunic. Functionally, ampullae adhere the animal to the substratum and are also the location of the allorecognition phenomenon we study.
Allorecognition Phenotype in Botryllus schlosseri
Botryllus schlosseri are typically found on man-made structures in shallow subtidal environments around the world, and competition for space in these habitats can be fierce. Since the colonies are continuously undergoing asexual reproduction and expanding outwards, they often come into physical contact with other Botryllus colonies. The first part of the colonies to touch each other are the ampullae, and two outcomes can result from this interaction: either the two ampullae will initiate a blood-based inflammatory rejection reaction causing the two colonies to no longer interact, or they will undergo vascular reorganization, fusing to form a chimeric colony which shares a common blood supply (Figure 1) [6,7,15].
Figure 1.

A. A dorsal view of a Botryllus schlosseri colony as described in the text. Adult individuals (zooids), the asexually developing progeny (buds) and the structures involved in allorecognition (ampullae) are highlighted. B. An allorecognition reaction occurring between three distinct colonies (a, b1 and b2). Colonies b1 and b2 are genetically identical subclones that are undergoing vascular reorganization (black arrow) which will eventually lead to a fusion event. Simultaneously, both colonies b1 and b2 are rejecting colony a, and have developed several points of rejection (red arrows) which prevent blood transfer.
The reaction begins when the ampullae of two adjacent colonies come into physical contact. The initial contact is typically tip to side, where the ampullar tip of one colony penetrates through the tunic matrix and touches the side of the ampullae on the adjacent individual. Following this, a characteristic set of morphological changes occur which eventually lead to fusion or rejection.
A rejection response is an active, blood-based inflammatory reaction that begins with the migration of a refractive cell called a morula [16] into the tips of the ampullae, causing swelling and the retreat of pigmented blood cells. Next, the epithelia of the ampullae lose their integrity and the morula cells leak into the periphery where they discharge their vacuoles, initiating a prophenoloxidase cascade which eventually forms dark melanin scars called points of rejection (PORs; Figure 1b, red arrows). The formation of PORs prevents any further blood exchange between the two colonies and often causes the ampullae to disintegrate and the colonies to no longer interact. In contrast, if two colonies are set to fuse, the tip of the ampullae rapidly penetrate through the tunic and come into contact with base of the peripheral blood vessel. The pigmented cells retreat and the ampullae fill with an unknown population of cells, which causes the ampullae to swell and pulse and eventually adjoin with the neighboring blood vessel (Figure 1b, black arrow).
From contact to a fusion or rejection reactions typically takes 24-48 hours and is limited to the ampullae that are interacting. As shown in figure 1b, a colony can simultaneously fuse and reject. The two colonies positioned on the right (labeled b1 and b2) are fusing, genetically identical subclones that are simultaneously rejecting with the colony on the left (labeled a). At the interface between colonies b1 and b2, the ampullae have migrated much further into the neighboring colony than the interface between the rejecting pairs. This observation suggests that allorecognition is due to spatially restricted interactions between juxtaposed ampullae and is not globally activated within the colony.
The Self Ligand: fuhc
Fusion or rejection is governed by a single, highly polymorphic locus called the fuhc [11,17,18]. Several criteria were put forth during the identification of the fuhc, the first being that the candidate must be highly polymorphic. Next, these polymorphisms must correlate to fusion and rejection outcomes in both wild-type and lab-reared colonies with fusing individuals sharing at least on fuhc allele and rejecting colonies none. Finally, the segregation of these polymorphisms in the crosses must also correlate 100% with histocompatibility outcomes. Genetic mapping of the fuhc locus in five independent crosses identified several candidate histocompatibility ligands, however only one satisfied all the genetic criteria to be the putative fuhc [13].
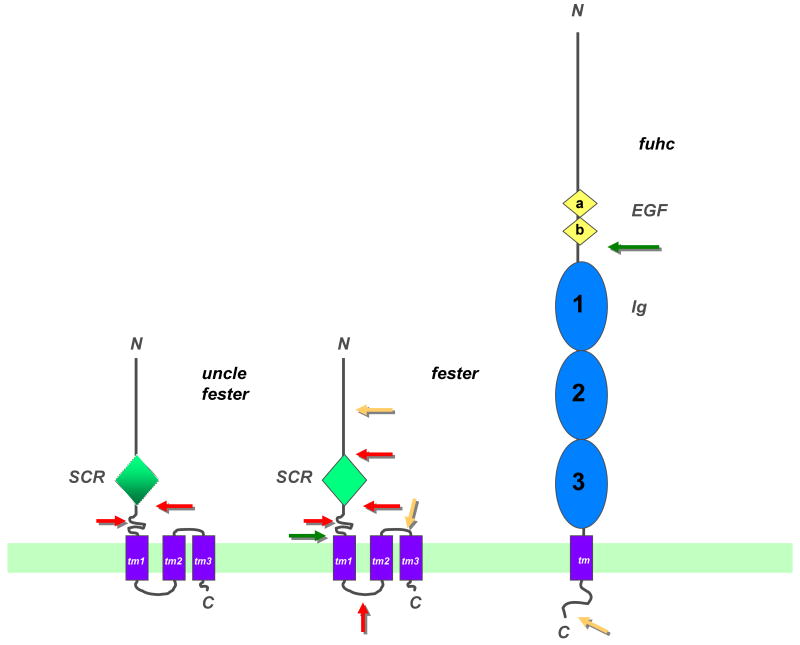
This candidate fuhc gene encodes a large (1008 aa) type I transmembrane protein containing a signal sequence, two and potentially three extracellular Ig domains and two EGF domains of about 750aa, followed by a TM domain and an intracellular tail which had no known signaling or other domains (Figure 2). Besides the full-length form, we identified two alternative splice variants. One removed the last half of the protein and added three new exons creating a putative secreted protein that consisted of about ½ the ectodomain. Another added a new exon within the tail. This was seen in about 5% of the TM containing clones but added no known domains. There is no clear homolog covering the length of this protein in any other genome, including three related ascidian genomes that have been sequenced (although they are all in different Orders). However, homology is seen with the vertebrates over the Ig domains by sequence similarity (similar to the protein IgSF4). 3D predictions of the Ig domain followed by a structural comparison done by PSSM revealed that the predicted Ig domains strongly resembled those of the poliovirus receptor, CD155. Finally, SMART searches reveal that this domain architecture (extracellular Ig and EGF domains) are only on the proteins Tie-1 and Tie-2 in the vertebrates, although the fuhc does not have fibronectin domains also found on these genes [13]. Wu-Kabat analysis of the distribution of polymorphisms revealed that the majority of alleles were different by about 25-50 amino acids spread throughout the ectodomain. At this point, we have not identified any hypervariable regions, and this result lends some insight into how fuhc polymorphism might be interpreted by its effector system (discussed below). In addition, results of in situ hybridization of the fuhc found that this transcript is expressed on ampullae in both adults and juveniles and at the anterior portion of the tadpole. Thus, the fuhc is found on all tissues known to be intimately involved in the allorecognition response. The expression profile in combination with the high degree of polymorphism and the 100% correlation with fusion rejection outcomes, led us to conclude that the fuhc is the putative self-ligand in this species.
Figure 2.
Topological models of uncle fester, fester and the fuhc proteins. Both fester and uncle fester encode an extracellular SCR domain (green diamond), although there is no amino acid homology between the two. This is followed by three predicted transmembrane domains and an intracellular COOH tail. Portions of the molecule deleted by alternative splicing are shown (arrows). Red arrows show regions which are spliced frequently, yellow arrows are regions which are spliced rarely. The green arrow on fester depicts a splice variant which removes all the transmembrane domains, creating a putatively secreted form of the protein (see text for details). The fuhc protein is shown on the right, with two tandem EGF domains (yellow diamonds), followed by three tandem immunoglobulin (C2 type) domains (blue ovals), followed by a TM region and short intracellular tail. Two splice variants of the fuhc have been detected; the green arrow shows a splice which creates a putative secreted form of the protein, containing ca. 50% of the ectodomain, while the yellow arrow indicates a region where a small exon is inserted in ca. 5% of the transcripts, however it encodes no obvious motifs. These structures are based on predictions from multiple algorithms, but have not been directly demonstrated [13]. For example, one program predicts an Ig domain at the N-terminus of fester, and two Ig domains at the N-terminus of the fuhc, preceding the EGF domains (F. Bazan, personal communication). In addition, Ig domain 1 is predicted by some, but not all programs.
It has been estimated that over 100 fuhc alleles exist within the Monterey Bay, CA population alone, suggesting that the fuhc is one of the most highly polymorphic loci ever described. How have these extraordinary levels of polymorphism been generated and maintained over evolutionary time? Interestingly, in our laboratory, we have observed a severe overdominant selection and heterozygote advantage of fuhc polymorphism in both new and established populations [19]. A consistent observation during derivation of inbred strains of Botryllus over the past 20 years was that few homozygotes survived to adulthood, as assayed by histocompatibility assays. During positional cloning of the fuhc locus, molecular genetic markers were developed that enabled analysis of fuhc segregation in both juveniles and adults. A severe, developmentally regulated over- dominant selection was seen in every F2 population using different combinations of five fuhc alleles. In each population, progeny were born in normal Mendelian ratios but in a period between hatching and the onset of sexual maturity, 80–95% of fuhc homozygotes died. The phenotype was identical in each cross: homozygotes seemed to be less fit overall and died at a greater rate during the first 12 weeks of life compared with their heterozygous siblings. However, homozygotes that survived to maturity were indistinguishable from heterozygotes. Both complementation and genetic mapping studies demonstrated that this was not due to the presence of common recessive lethal genes in each haplotype. Moreover, the probability of five fuhc alleles having linked recessive lethals that both complemented each other and produced the same phenotype seems improbable. We hypothesize that this is an epigenetic effect of the detection of homozygosity, such as homology dependent gene silencing [20].
A fusion event and the formation of a chimeric colony will occur if the two colonies share one or both alleles at the fuhc. Thus, in order for allorecognition specificity to be achieved, a system must be in place that can discriminate between hundreds of fuhc alleles. Given this high degree of polymorphism, it seems highly unlikely that recognition is occurring via homotypic interactions. Thus an effector system must exist and recognition is probably accomplished by receptors that learn to identify specific fuhc alleles. The rules of allorecognition are that fusion can occur if only a single fuhc allele is shared, so this effector system is clearly not directly recognizing non-self. The rules of fusibility are reminiscent of the “missing-self” recognition utilized by the effector arm of the vertebrate natural killer cells [21,22], whereby receptors are educated to self fuhc alleles, thus anything they do not recognize is, by default, non-self. We hypothesize that this discriminatory strategy, which is analogous to the recognition of polymorphic MHC residues by germline encoded receptors in Natural Killer (NK) cells, is achieved by an education process that takes place during development and involves differential expression of cell surface receptors, which continues until the correct affinity for self is achieved [23]. In Botryllus, this education process must be completed during the first 6 days of sexual development, since juvenile oozooids are competent to make allorecognition decisions upon settlement [24]. Thus far, we have identified two candidate receptors we believe to be involved in this process, both embedded within the Fuhc locus. These two structurally similar loci, called fester and uncle fester, exhibit markedly distinct functions and will be discussed in the following two sections.
The effector system: fester
One criterion for identifying candidate receptors to the fuhc was polymorphism, i.e., a gene product that could respond to the incredible amount of diversity exhibited by the fuhc itself. In 2006, Nyholm et. al identified fester, located over 100 kb from the fuhc. The full-length fester gene is 11 exons long and encodes a 368 residue, type I transmembrane protein with a signal sequence, 7 extracellular exons, an SCR or sushi domain located in the extracellular exon 5, three exons (8- 10) which each encode a predicted transmembrane α-helix, and exon 11, encoding a short intracellular tail (Figure 2). We have now identified over 45 alleles on both west and east coast populations; however these polymorphisms do not contribute to histocompatibility: colonies sharing alleles could reject, and those sharing neither allele could fuse in both lab and wild- type interactions.
The gene is polymorphic, is polygenic and is somatically diversified by alternative splicing. Six of the eleven exons (ectodomain exons 2,3,6,7 and transmembrane domain exons 9, 10) can be alternatively spliced in all combinations, creating 64 splice variants, all of which have been identified. In addition, another 16 splice variants are made that encoded potentially secreted proteins. In this case exons 8-10 were deleted, and a new exon (exon 12), which encoded two amino acids and a stop codon, was added, and this was found in combination with the different ectodomain exons spliced. All splice variants are productive and encode open reading frames with no change in amino acid sequence in any exon. The intracellular splice variants with exons 9, 10 spliced out are intriguing. If they truly encode TM domains, these would change the topology of the protein, and we are currently testing this in vitro.
To summarize, sequencing of multiple clones always revealed a varying number and type of splice variants, with a minimum of 8: each individual we have analyzed appears to express both the full-length fester gene, one of the splice variants (exons 6 and 7 removed) plus an individual-specific subset of alternatively spliced variants which encode proteins with different extracellular and intracellular coding regions, as well as putative secreted forms. Spatially, fester expression was analyzed using in situ hybridization using multiple probes and by immunohistochemistry (IHC) using a mAb, raised against a fusion protein containing exons 1-8 of an allele (fester A) carried in our lab-reared lines. In the adult, fester protein is expressed on the epithelia of the ampullae, as well as a subset of blood cells which were both inside the circulation, as well as migrated into the tunic. In the larvae, fester is expressed on the epithelia of the nascent ampullae, as well as a sensory structure on the anterior of the tadpole, called the adhesive papillae. The tadpole touches the adhesive papillae to the substrate as it makes decisions on where to settle [25-27]. The secreted form of the gene is also expressed in these tissues as assayed by RT-PCR. Thus, similar to the fuhc, fester expression is restricted to all tissues known to be involved in allorecognition.
Two experiments were done that implicated fester as playing a key role in the allorecognition response and as the putative receptor to the fuhc [28]. First, a monoclonal antibody (mAb) generated to the full-length A-clade fester allele was injected into two colonies as they came into contact. This had no effect on fusing colonies, however it caused rejecting colonies to fuse. However, this affect of the mAb was only observed if three A-clade fester alleles were present between the two genotypes. In other words, one colony had to be a homozozygote, and the other a heterozygote for the allele fester A allele that the mAb is specific for [28]. These results have important implications as to how fester functions. First, we have demonstrated that a threshold to the inhibition of a rejection response exists and that ampullae from both colonies are required to initiate a fusion event. Importantly, the injection of fester mAb functionally mimicked the recognition of self and initiated a fusion between histoincompatible colonies. This suggests that fester is acting as an inhibitory receptor, capable of discriminating fuhc polymorphisms and consequently inhibiting a rejection response.
Around the same time, a new technique had been developed in our laboratory that coupled siRNA-mediated knockdown with vascular regeneration [28-30]. In brief, we are able to deliver siRNA specific to the gene of interest by simply injecting into the vasculature and/or soaking the siRNA in the seawater. Simultaneously, we perform a micro-surgery removing all of the ampullae surrounding the colony, effectively removing the existing protein and subsequently forcing the animal to regenerate new ampullae under the effects of the siRNA. This process takes 48-72 hours, at which the newly regenerated ampullae are capable of making fuhc based allorecognition decisions. In contrast to the mAb functional data, siRNA mediated knockdown of all fester splice variants yielded a no reaction phenotype between both histocompatible and incompatible colonies. Neither colony received a signal initiating a response, suggesting that fester was acting as an activating receptor required for the initiation of both a fusion and rejection event.
Thus, fester appears to be playing a dual role in the allorecognition response, both as an activating receptor, required to initiate both fusion and rejection, and as an inhibitory receptor involved in the recognition of self and the inhibition of a rejection response. How could this dual functionality be accomplished? We hypothesize that this may be a result of different splice variants playing different roles in the response. As previously mentioned, each individual expresses a full-length form and two membrane bound forms swapping out the two exons located at the cell surface (exons 6 and 7). It is possible that these forms are involved in both the activation of a rejection and a fusion, and thus the recognition of the fuhc.
Are fusion and rejection mutually exclusive?
There are nine botryllid ascidian species which each appear to have a single locus allorecognition system. However, only in several combinations are xenorejections observed, and fusion between species have never been reported [31,32]. In addition, characteristic morphological changes which precede fusion or rejection, such as morula cell inflammation, do not occur when a colony grows into a rock or other non-botryllid species. In context of the disparate functional results from the fester experiments, it appears that the allorecognition response consists of both an activation step, alerting the colonies they are in contact with a conspecific, as well as an inhibitory step, which blocks a rejection reaction. But how are the fusion and rejection responses related? We have recently identified another member of the fester family, called uncle fester, which has allowed functional dissection of these two processes, suggesting that the fusion and rejection are independent pathways (McKitrick, et al., in preparation). Structurally and topologically, uncle fester resembles fester but shows much less alternative splicing and polymorphism (Fig. 2).
Working hypothesis: How is allorecognition specificity achieved?
A colony can pinpoint a self fuhc allele from hundreds of competing specificities; leading to the question of how germline encoded receptors could provide the specificity observed in nature. We hypothesize that this is due to an overall avidity integrated from binding of multiple receptors or splice variants to different polymorphic residues on the fuhc protein. In turn, this would require that each individual undergo an epigenetic education process whereby the right combination of receptors and/or splice variants are expressed on the cell surface. This is an equivalent to that hypothesized for vertebrate NK cells [33], and in fact we speculate that this education process is a fundamental characteristic of immunity conserved through evolution [34]. In our case, the apparent individual-specific splice repertoire of fester ectodomain exons may be indicative of this type of an education process. In addition, the intracellular splice variants may create assembly domains which pairs splice variants with different signaling molecules, thus segregating activating and inhibitory function. This has yet to be directly demonstrated, but is an active area of research in our laboratory.
Why does allorecognition exist in Botryllus?
The constant asexual development in B. schlosseri has major implications for allorecognition. Fusion of compatible colonies allows mixing of the progenitor cells responsible for budding, and following fusion new buds in each colony can contain tissues derived from the other individual. Often it is found that somatic or germline tissues are completely usurped by the other, such that one stem cell population is parasitizing the other genotype, and this will remain the case for the life of that individual, even if the two are surgically separated. This process is called stem cell parasitism (SCP) and is likely a major selective pressure on the evolution of allorecognition polymorphism- both the generation of new fuhc alleles, as well as the ability of an effector system to respond appropriately. SCP is a property intrinsic to the stem cells themselves, as the phenotype is retained even with experimental transplantation. In addition, it appears to be genetically controlled, as we have found lineages which produce SCP winners and losers in the lab, and equivalent phenotypes from natural populations [35]. In summary, the interaction of fusibility and SCP are countered by the evolution of polymorphism and effector specificity, essentially limiting the movement of stem cells throughout a population.
Future directions
Allorecognition in B. schlosseri is a tractable model for dissecting allorecognition responses. The recognition events take place on the surface of a single cell-layer epithelium that makes up the ampullae, which are the tips of an extracorporeal vasculature and accessible to external reagents. The ability to combine regeneration and siRNA-mediated knockdown will allow us to functionally dissect allorecognition responses in vivo, using multiple permutations of this assay, for example, pairing colonies with different genes knocked down, or to unmanipulated colonies. Expression studies during embryogenesis, and during regeneration of the ampullae in adults may provide an excellent model for dissecting putative education processes as they occur. Finally, with the proteins in hand, in vivo, ex vivo and in vitro approaches can now be taken to formally demonstrate binding, and provide a model to dissect the molecular basis of specificity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burnet FM. “Self-recognition” in Colonial Marine Forms and Flowering Plants in relation ot the Evolution of Immunity. Nature. 1971;232:230–235. doi: 10.1038/232230a0. [DOI] [PubMed] [Google Scholar]
- 2.Boehm T. Quality control in self/nonself discrimination. Cell. 2006;25:845–858. doi: 10.1016/j.cell.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Pancer Z, Amemlya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte recpetors in the agnathan sea lamprey. Nature. 2004;430:174–180. [PubMed] [Google Scholar]
- 4.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nature Review Immunology. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 5.Bancroft FW. Variation and fusion in compund ascidians. Proc Calif Acad Sci. 1903;3(3):137–186. [Google Scholar]
- 6.Oka H, Watanabe H. Problems of colony specificity in compounds ascidians. Bull Biol Stn Asamushi. 1960;10:153–155. [Google Scholar]
- 7.Mukai H, Watanabe H. On the occurrence of colony specificity in some compund ascidians. Biological Bulletin. 1974;147:411–421. doi: 10.2307/1540458. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe H. Studies on the regulation in fused colonies in Botryllus primigenus (Ascidiae Compositae) Sci Rep Tokyo Bunrika Gaigagu. 1953;7:193–198. [Google Scholar]
- 9.Milkman R. Genetic and Developmental Studies on Botryllus schlosseri. Biological Bulletin. 1967;132(2):229–243. doi: 10.2307/1539891. [DOI] [PubMed] [Google Scholar]
- 10.Manni L, Zaniolo G, Cima F, Burighel P, Ballarin L. Botryllus schlosseri: a model ascidian for the study of asexual reproduction. Developmental Dynamics. 2007;236(2):335–352. doi: 10.1002/dvdy.21037. [DOI] [PubMed] [Google Scholar]
- 11.Scofield VL, Schlumpberger JM, Weissman IL. Protochordate allorecognition is controlled by a MHC-like gene system. Nature. 1982;295:499–502. doi: 10.1038/295499a0. [DOI] [PubMed] [Google Scholar]
- 12.Weissman IL, Saito Y, Rinkevich B. Allorecognition Histocompatibility in a Protochordate Species: Is the Relationship to MHC Semantic or Structural. Immunological Reviews. 1990;113:227–241. doi: 10.1111/j.1600-065x.1990.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 13.De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Luddington WB, Mitchel K, et al. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatvies of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 15.Sabbadin A. The compound ascidian Botryllus schlosseri in the field and in the laboratory. Pubbl Staz Zool Napoli. 1969;37(suppl):62–72. [Google Scholar]
- 16.Ballarin L, Cima F, Sabbadin A. Morula cells and histocompatibility in the colonial ascidian Botryllus schlosseri. Zoological Science. 1995;12:757–764. [Google Scholar]
- 17.De Tomaso AW, Weissman IL. Initial characterization of a protochordate histocompatibility locus. Immunogenetics. 2003;55:480–490. doi: 10.1007/s00251-003-0612-7. [DOI] [PubMed] [Google Scholar]
- 18.De Tomaso AW, Weissman I. Construction and characterization of large-insert genomic libraries (BAC and fosmid) from the Ascidian Botryllus schlosseri and initial physical mapping of a histocompatibility locus. Mar Biotechnol. 2003;5:103–115. doi: 10.1007/s10126-002-0071-4. [DOI] [PubMed] [Google Scholar]
- 19.De Tomaso AW, Weissman IL. Evolution of a Protochordate Allorecognition of Locus. Science. 2004;303:977. doi: 10.1126/science.1094952. [DOI] [PubMed] [Google Scholar]
- 20.Wu CT, Morris JR. Transvection and other homology effets. Current Opinion in Genetics and Development. 1999;9:237–246. doi: 10.1016/S0959-437X(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 21.Karre K, Ljunggren HG, Piontek G, Kiessling R. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 22.Raulet DH. Natural Killer Cells. In: Paul WE, editor. Fundamental Immunology. 5. Philadelphia: Lippincot WIlliams & Wilkins; 2003. pp. 365–391. [Google Scholar]
- 23.Raulet DH, Vance RE, McMahon CW. Regulation of the natural killer cell receptor repertoire. Annual Review of Immunology. 2001;19:291–330. doi: 10.1146/annurev.immunol.19.1.291. [DOI] [PubMed] [Google Scholar]
- 24.Scofield VL, Nagashima LS. Morphology and Genetics of Rejection Reactions between Oozodis from the Tunicate Botryllus schlosseri. Biological Bulletin. 1983;165(3):733–744. doi: 10.2307/1541475. [DOI] [PubMed] [Google Scholar]
- 25.Grave C, Woodbridge H. Botryllus schlosseri (Pallas): The behavior and morphology of the free-swimming larva. J Morph. 1924;39:207–247. [Google Scholar]
- 26.Grave C, Riley G. Development of the sense organs of the larva of Botryllus schlosseri. Journal of Morphology. 1935;57(1):185–211. [Google Scholar]
- 27.Cloney RA. Larval Adhesive Organs and Metamorphosis in Ascidians. Cell Tissue Research. 1977;183:423–444. doi: 10.1007/BF00225658. [DOI] [PubMed] [Google Scholar]
- 28.Nyholm SV, Passegue E, Luddington WB, Voskoboynik A, Mitchel K, Weissman IL, et al. Isolation and characterization of fester, a candidate allorecognition receptor from a primitive chordate. Immunity. 2006;25:163–173. doi: 10.1016/j.immuni.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Tiozzo S, Voskoboynik A, Brown FD, De Tomaso AW. A conserved role of the VEGF pathway in angiogenesis of an ectodermally-derived vasculature. Developmental Biology. 2008;315(1):243–255. doi: 10.1016/j.ydbio.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laird DJ, Chang W, Weissman IL, Lauzon RJ. Identification of a Novel Gene Involved in Asexual Organogenesis in the Budding Ascidian Botryllus schlosseri. Developmental Dynamics. 2005;234:997–1005. doi: 10.1002/dvdy.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirose E, Shirae M, Saito Y. Colony Specificity in the Xenogeneic Combinations among Four Botrylloides Species (Urochordata, Ascidiacea) Zoological Science. 2002;19:747–753. doi: 10.2108/zsj.19.747. [DOI] [PubMed] [Google Scholar]
- 32.Saito Y. Xenogeneic Rejection among Three Botryllids (Compound Ascidians) Zoological Science. 2003;20:581–589. doi: 10.2108/zsj.20.581. [DOI] [PubMed] [Google Scholar]
- 33.Karre K. NK cells, MHC class I molecules and the missing self. Scand J Immunol. 2002;55(3):221–228. doi: 10.1046/j.1365-3083.2002.01053.x. [DOI] [PubMed] [Google Scholar]
- 34.De Tomaso AW. Sea squirts and immune tolerance. Disease Models & Mechanisms. 2009;2:440–445. doi: 10.1242/dmm.001156. [DOI] [PubMed] [Google Scholar]
- 35.Stoner DS, Rinkevich B, Weissman IL. Heritable germ and somatic cell lineage competitions in chimeric colonial protochordates. Proc Natl Acad Sci. 1999;96:9148–9153. doi: 10.1073/pnas.96.16.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]