Abstract
The mechanism for transcriptional silencing of pericentric heterochromatin is conserved from fission yeast to mammals. Silenced genome regions are marked by epigenetic methylation of histone H3, which serves as a binding site for structural heterochromatin proteins. In the fission yeast Schizosaccharomyces pombe, the major structural heterochromatin protein is Swi6. To gain insight into Swi6 function in vivo, we have studied its dynamics in the nucleus of living yeast. We demonstrate that, in contrast to mammalian cells, yeast heterochromatin domains undergo rapid, large-scale motions within the nucleus. Similar to the situation in mammalian cells, Swi6 does not permanently associate with these chromatin domains but binds only transiently to euchromatin and heterochromatin. Swi6 binding dynamics are dependent on growth status and on the silencing factors Clr4 and Rik1, but not Clr1, Clr2, or Clr3. By comparing the kinetics of mutant Swi6 proteins in swi6− and swi6+ strains, we demonstrate that homotypic protein-protein interactions via the chromoshadow domain stabilize Swi6 binding to chromatin in vivo. Kinetic modeling allowed quantitative estimation of residence times and indicated the existence of at least two kinetically distinct populations of Swi6 in heterochromatin. The observed dynamics of Swi6 binding are consistent with a stochastic model of heterochromatin and indicate evolutionary conservation of heterochromatin protein binding properties from mammals to yeast.
In eukaryotic cells, genomes exist in the form of chromatin. Morphological studies have described two major types of chromatin: euchromatin corresponds to loosely packed chromatin, where most active genes are transcribed, whereas heterochromatin consists of condensed, predominantly transcriptionally repressed chromatin (48). Heterochromatin is molecularly characterized by a high density of nucleosomes containing histone H3 methylated on lysine 9 (H3-K9) (5, 27). In humans, H3-K9 methylation is mediated by methyltransferases Suv39h1 and Suv39h2 (43). The presence of methylated H3-K9 creates a specific binding site for one of the major heterochromatin proteins, HP1 (heterochromatin protein 1) (5, 27, 37, 43). HP1 was originally identified in Drosophila melanogaster as a suppressor of variegation, and it has been shown to modify mammalian position effect variegation in a dose-dependent manner (11, 19). The role of HP1 does not appear to be limited to heterochromatin since HP1 also represses, and in some cases activates, euchromatic genes (24, 28). Consistent with its primary role in the formation and maintenance of heterochromatin, HP1 is predominantly localized in heterochromatin domains (12, 33).
The system of repression involving methylation of H3-K9 as a mark which is recognized by a structural chromatin protein is conserved from mammalian cells to Schizosaccharomyces pombe. The fission yeast homologues of Suv39h and HP1 are Clr4 and Swi6, respectively (25, 30, 47). Swi6 binds to three transcriptionally silent heterochromatic regions, the mating type loci, telomeres, and centromeres (20, 23), all of which have a high concentration of H3-K9 methylation (37, 41). Both Clr4 and Swi6 are essential for maintaining transcriptional silencing and chromatin organization in these regions (2, 16, 17, 30). Similar to mammalian cells, fission yeast cells lacking Clr4 or Swi6 have disrupted heterochromatin and exhibit chromosome segregation defects (16). Swi6 is also involved in centromere function and recruitment of cohesin to heterochromatin regions and is required for proper cohesion of centromeres during mitosis (6, 42). Like all mammalian HP1 homologues, Swi6 is composed of a chromodomain (CD) and a chromoshadow domain (CSD), separated by a hinge region (15, 54). In vitro, the CD interacts directly with the trimethylated H3-K9 histone tail (26, 40), and its function is conserved from S. pombe to mammals (5, 54). The CSD is a protein-protein interaction domain and has been implicated in the formation of HP1 dimers as well as in mediating interactions with HP1 binding proteins, such as LBR, KAP1, CAF1, Suv39, and BRG1 (7, 13, 36, 38, 39, 49, 55, 56).
The structural hallmark of heterochromatin is the high degree of chromatin condensation. The ability of HP1/Swi6 to form oligomers has led to a model in which HP1/Swi6 confers and maintains a condensed heterochromatin state by forming stable interactions between nucleosomes (5, 15). Consequently, chromatin becomes compacted and access of regulatory factors to chromatin sequences is reduced or prevented altogether. Heterochromatic HP1/Swi6 is also instrumental in the propagation of the heterochromatin state by recruiting Suvar39h/Clr4 to existing heterochromatin regions, thus facilitating the methylation of adjacent regions (5). This implicitly static model for heterochromatin has recently been questioned by in vivo studies using mammalian cells (10, 18). Observations in living cells indicate that HP1 is a highly mobile protein and that HP1 binds only transiently to chromatin in vivo despite its role in maintenance of stable heterochromatin domains (10, 18). To ask whether the dynamic aspects of heterochromatin proteins are evolutionarily conserved and to gain further insight into the molecular mechanisms of heterochromatin formation, we have here investigated the dynamics of heterochromatin domains and of Swi6 binding in living yeast cells.
MATERIALS AND METHODS
Yeast strains and growth.
We used yeast strains AL91L (swi6−; h90 swi6::ura4 ura4-D18 leu1-32 ade6-704), SP557 (swi6+; h90 ura4-D18 leu1-32 ade6) (54), EG338 (h90 rik1-304 ura4-D18 leu1-32) (14), PG438 [mat3-M(EcoRV)::ura4] clr1-5 ura4-D18 leu1-32 (ade6-216) (53), KE78 (h90 clr2-E22 ura4 ade6- M216), KE81 (h90 clr3-E36 ade6-M216), and KE108 (h90 clr4-S5 ura4 ade6-M216) (17). All strains were haploid. Fission yeasts were maintained on Edinburgh minimal medium (EMM) with appropriate supplements and transformed according to a standard protocol (35). Plasmids for expression of green fluorescent protein (GFP)-wild-type Swi6 (Swi6-WT; both N- and C-terminal fusions), GFP-Swi6ΔCD, and GFP-Swi6ΔCSD (N-terminal GFP fusion) were constructed as described previously (54) in the pRep1 expression vector (32). Repression of the nmt1 promoter was maintained with 0.4 μg of thiamine/ml. For microscopy yeast strains were grown in liquid EMM at 28°C; cells in exponential phase were observed 20 h after removal of thiamine. Experiments were carried out at an optical density at 600 nm (OD600) of ∼1.5, when most daughter cells were not fully separated and the average length of cells was ∼9 μm. In exponential phase, microscopy was performed only when the two daughter cells were not fully segregated. Cells in stationary phase and spores were observed after 3 days of incubation at an OD600 of over 2, when the shape of cells was more spheroid than in exponential phase and their length was about 6 μm. For microscopy, 2 μl of cell suspension was spread between the coverslide and coverslip. Samples were observed at room temperature (28°C) for no longer than 20 min.
Time-lapse microcopy and FRAP.
Live-cell microscopy and fluorescence recovery after photobleaching (FRAP) were performed as previously described on a Zeiss LSM 510 confocal microscope using the 488-nm line of an argon laser (10). For time-lapse microscopy, stacks of images (256 by 63 pixels) were recorded at maximum speed (64 ms per image, one iteration) with a Z-step of 0.2 to 0.3 μm (20 sections) every minute. Displacements of heterochromatin domains between two stacks were measured with Zeiss LSM 510 software. For high-frequency acquisition, time-lapse microscopy was performed by collecting single two-dimensional images (64 ms, one iteration) every 1.5 s. To calculate the motion of heterochromatin domains over time, we measured the displacement of single heterochromatin domains at several time points along the trajectory.
FRAP on fission yeast was performed by using a 0.5-μm spot bleach with a triple bleach pulse of 64 ms. Bleaching occurred after 15 prebleach images, and 60 images (256 by 63 pixels, one iteration) were collected after the bleach. For heterochromatin recovery measurements were performed only in the bright spots corresponding to heterochromatin. FRAP normalization was performed by measuring the intensity in the bleach area (B), unbleached area (U), and background (bg). Values displayed in recovery curves (R) are double normalized by Rn = (B − bg)/(U − bg), where R = Rn/R0 (R0 is the Rn value corresponding to prebleach). For each condition, at least 30 individual recovery curves were collected from three or more independent experiments. Statistical analyses were performed using a standard t test.
Kinetic modeling.
Several hypotheses were formulated to describe the binding of Swi6 to chromatin as described in Results. Each hypothesis was mathematically described by a system of differential equations constructed using principles of mass action kinetics. As part of the analysis, it was assumed that the nucleus is a well-mixed compartment and that endogenous and GFP-tagged Swi6 molecules behave similarly. In addition, pseudo-first-order kinetics were assumed, where the number of binding sites is in excess of the number of Swi6 molecules. This assumption is supported by the lack of a correlation between the number of Swi6 molecules localized in heterochromatin and the observed mobility (see Fig. S1 in the supplemental material).
To directly compare the solution of a model to the experimental data, the FRAP experiment was simulated in silico. The full description of the model and equations are given in the supplemental material. All models were implemented and solved numerically in Berkeley Madonna. The models were fitted against the data with the curve-fitting algorithm in Berkeley Madonna software using least-squares regression analysis.
RESULTS
Dynamics of heterochromatin domains in vivo.
To study Swi6 in living fission yeast, we used a swi6 null strain expressing GFP-Swi6 under the control of a thiamine-repressible promoter. The fusion protein has previously been shown to be functional and to fully complement the phenotype of a swi6 null strain as assayed by sporulation frequency, formation of normal asci, and ability to maintain linear minichromosomes (54).
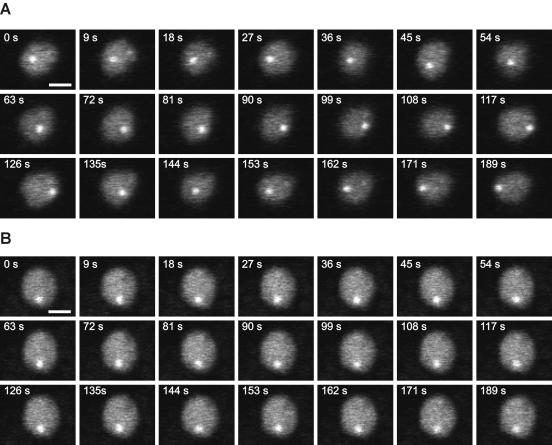
Pericentric heterochromatin regions have previously been shown to be positionally stable within the nuclei of mammalian cells (10). To directly compare the dynamic behavior of heterochromatin domains in yeast and mammals, we performed time-lapse microscopy on exponentially growing yeast cells expressing GFP-Swi6 as a marker for heterochromatin (Fig. 1). GFP-Swi6 accumulates in fission yeast in heterochromatin in the form of two to six bright foci, with most of the cells containing three foci. For the purpose of this study, we defined these bright GFP-Swi6 foci as heterochromatin, whereas euchromatin was defined as the rest of the nucleus. Time-lapse microscopy with high-frequency acquisition was performed to monitor the movement of heterochromatin domains (Fig. 1). We found that heterochromatin regions were highly mobile within the S. pombe nucleus and underwent extensive translational motion as previously shown (Fig. 1A) (46). To quantitatively estimate the motion of heterochromatin domains, we traced the trajectory of single heterochromatin domains over time. During exponential growth, heterochromatin domains moved on average 660 ± 270 nm/min. Single heterochromatin domains were often seen to traverse the entire nuclear volume in an almost uniform movement within less than 1 min (Fig. 1A). As previously shown for budding yeast, the mobility of chromatin loci depends on the growth status of the cell (22). When S. pombe cells in stationary phase were analyzed, it was found that the mobility of heterochromatin domains was significantly decreased and only limited, locally confined motion was observed (Fig. 1B). The reduction in heterochromatin domain mobility was not due to a global increase in nuclear viscosity due to growth arrest since the diffusion of a soluble marker protein (GFP alone) in stationary phase was unaffected (data not shown). Similarly, the reduction of heterochromatin velocity between stationary phase and exponential phase was not due to a reduction of nucleus size since the fission yeast nucleus diameter measures 1.84 ± 0.18 μm in exponentially growing cells and 1.88 ± 0.09 μm in stationary cells (Fig. 1). These results indicate that, although heterochromatin domains are structurally stable in fission yeast, they are, in contrast to those in mammalian cells, positionally highly mobile and their mobility is strongly dependent on the growth status of the cell.
FIG. 1.
Time-lapse microscopy of GFP-Swi6. Optical sections were taken from movies collected at the indicated times from yeast growing in exponential phase (A) and stationary phase (B). Heterochromatin domains are highly mobile in the exponential-growth phase, whereas their mobility is greatly reduced during stationary phase. Bar: 1 μm.
Dynamics of Swi6 binding in vivo.
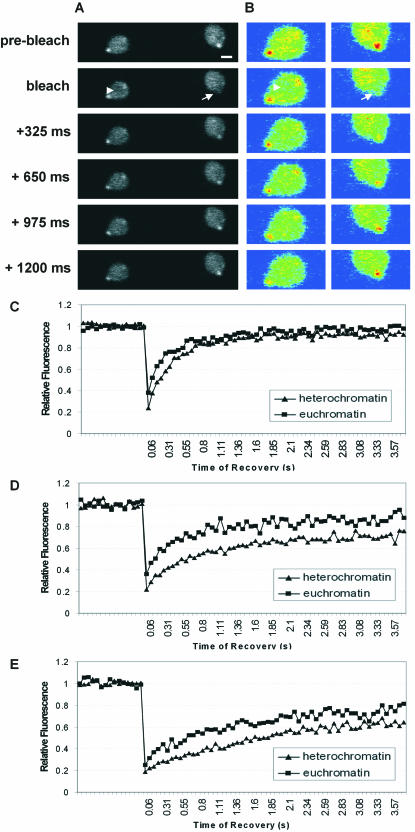
To study the binding of Swi6 to chromatin inside the living fission yeast nucleus, we performed FRAP on yeast expressing GFP-Swi6. In FRAP experiments a region in either heterochromatin or euchromatin was irreversibly bleached by a short laser pulse of appropriate wavelength and the recovery of fluorescence signal as a function of time was measured. For chromatin proteins, the recovery kinetics are a direct indicator of the protein's ability to bind to chromatin (34, 44, 45). To accommodate the small size of the yeast nucleus and the very small size of the yeast heterochromatin domains, we developed optimized experimental conditions which allowed routine FRAP measurements in yeast cells (see Materials and Methods). In these experiments, a small circular region of 300 to 500 nm in diameter was bleached either in euchromatin or heterochromatin and recovery was observed by recording images every 64 ms (Fig. 2A and B). Bleached heterochromatin included mating loci, telomeres, or centric heterochromatin (20, 23). Care was taken to exclude nucleoli from the bleach region. Upon bleaching of either a heterochromatic or euchromatic region, the fluorescence signal for GFP-Swi6 recovered within less than 2 s, demonstrating that GFP-Swi6 is highly mobile and binds only transiently to chromatin in exponentially growing cells (Fig. 2A to C). Similar results were obtained with a C-terminal fusion of GFP to Swi6 (data not shown). Within a cell population, GFP-Swi6 binding is independent of the level of expression of GFP-Swi6 since no correlation between intensity of labeling in heterochromatin and the mobility of Swi6 was observed (see Fig. S1 in the supplemental material). In contrast, GFP-Swi6 mobility correlates with its intensity ratio in heterochromatin versus euchromatin, indicating that Swi6 mobility reflects the relative accumulation of Swi6 in heterochromatin (see Fig. S1 in the supplemental material).
FIG. 2.
Mobility of GFP-Swi6 in S. pombe. (A) FRAP on exponentially growing yeast cells expressing GFP-Swi6 by bleaching an area in heterochromatin (arrow) or in euchromatin (arrowhead). Bar, 1 μm. (B) Pseudocolor images of those in panel A. (C to E) Comparison of GFP-Swi6 recovery in heterochromatin and euchromatin during exponential phase (C), during stationary phase (D), and after sporulation (E). Values represent averages for 35 cells from five experiments.
In comparison to that in exponentially growing cells, GFP-Swi6 mobility was significantly reduced in fission yeast in stationary phase (Fig. 2D) and was even further reduced in spores (Fig. 2E). The signal recovery in heterochromatin 300 ms after the bleach pulse was 60, 38, and 28% in exponential phase, stationary phase, and spores, respectively (Fig. 2C to E). Similarly, in euchromatin the recovery after 300 ms was 75, 64, and 47% in exponential phase, stationary phase, and spores, respectively (Fig. 2C to E). These results demonstrate that Swi6 binding to chromatin is dynamic and depends on the growth status of the cell.
Domain analysis of Swi6 binding in vivo.
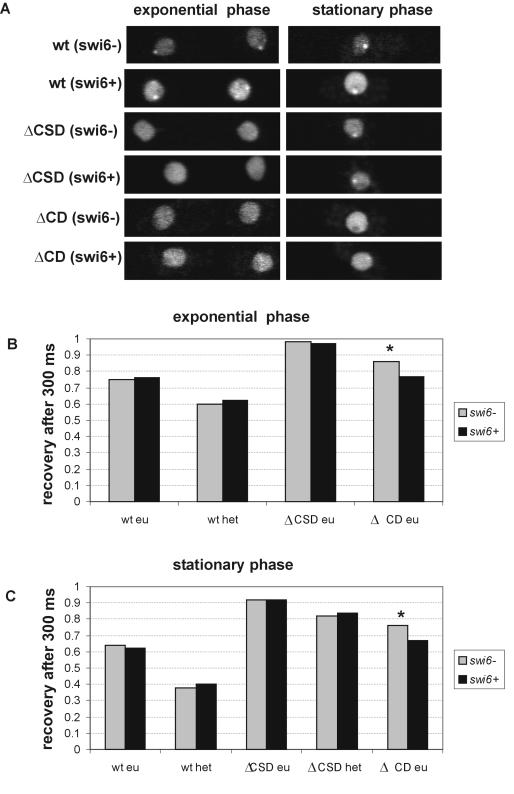
To study the contribution of the various domains of the Swi6 protein to its binding to chromatin in living yeast, we used strains expressing fusions of GFP with Swi6 containing deletions of the CSD (GFP-Swi6ΔCSD) or the CD (GFP-Swi6ΔCD) (54). The mutant fusion proteins were expressed in either a swi6+ or swi6− background (Fig. 3). The localization of these mutants has previously been characterized in detail (54). While GFP-Swi6-WT localizes to heterochromatin in both exponentially growing and stationary cells, GFP-Swi6ΔCSD displays diffuse labeling in most exponentially growing cells and accumulates in heterochromatin in stationary cells (Fig. 3A). GFP-Swi6ΔCD is distributed throughout the nucleoplasm and accumulates to only modest levels in heterochromatin of exponentially growing and stationary-phase swi6+ cells (Fig. 3A).
FIG. 3.
Contribution of the CD and CSD to Swi6 binding. (A) Localization of GFP-Swi6, GFP-ΔCSD, and GFP-ΔCD in swi6− and swi6+ strains growing exponentially or in stationary phase. (B and C) Quantitation of FRAP recovery after 300 ms in swi6− and swi6+ backgrounds for GFP-Swi6-WT, GFP-Swi6ΔCD, and GFP-Swi6ΔCSD during exponential phase (B) and stationary phase (C). Values represent averages for 30 cells from three experiments. *, statistically significant difference in recovery.
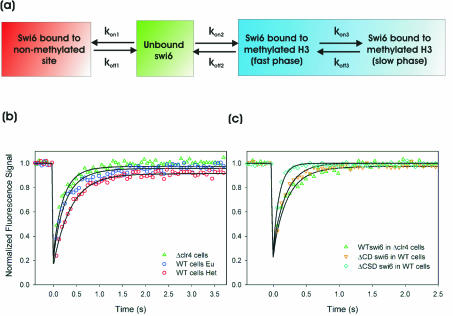
To study the binding dynamics of these constructs, we performed FRAP and used the percentage of recovery reached after 300 ms as an indicator of their binding behavior (Fig. 3). Significant differences in recovery after 300 ms are an indicator of general recovery kinetics (8). During exponential growth, GFP-Swi6-WT reached a recovery of 60% in heterochromatin and 75% in euchromatin after 300 ms during exponential growth (Fig. 3B). This difference is statistically significant (P < 0.001). The mobilities of GFP-Swi6ΔCSD and GFP-Swi6ΔCD were significantly higher than that of GFP-Swi6-WT, indicative of reduced chromatin binding (Fig. 3B) (P < 0.001). These observations show that Swi6 requires both the CSD and the CD to display full binding activity in vivo.
To probe the interactions between the CD and the CSD of Swi6 in vivo, we compared recovery kinetics of the mutant proteins in swi6+ and swi6− strains (Fig. 3B). The mobility of GFP-Swi6-WT in a swi6− strain was indistinguishable from that in a swi6+ background, and no difference was observed for GFP-Swi6ΔCSD between swi6− and swi6+ strains (Fig. 3B). This observation indicates that the endogenous Swi6 is not able to interact with a mutant Swi6 lacking the CSD. In contrast to the CSD mutant protein, the mobility of GFP-Swi6ΔCD was slower in a swi6+ background than in a swi6− strain (Fig. 3B). The recovery after 300 ms of GFP-Swi6ΔCD was 86% in the swi6− strain but only 77% in the swi6+ strain (P < 0.001). This finding indicates that the CSD of the mutant protein interacts with endogenous Swi6-WT, resulting in slowed FRAP recovery of GFP-Swi6ΔCD in WT cells. Similar effects were observed in cells in stationary phase, suggesting that the growth status does not affect the contribution of either domain to chromatin binding (Fig. 3C). We conclude from the ability of Swi6-WT to interact with a CD deletion mutant protein, but not a CSD deletion mutant protein, that homotypic CSD-CSD interactions stabilize binding of Swi6 to chromatin in vivo.
Effect of silencing factors on Swi6 dynamics.
To study the effect of components involved in heterochromatic silencing on Swi6 binding to chromatin in living yeast, we examined GFP-Swi6 dynamics in strains deficient for Clr1, Clr2, Clr3, Clr4, or Rik1. These proteins have previously been identified as critical suppressors of silencing at the mating type locus and centromeres (17, 30, 53). As previously shown, the absence of Clr4 or Rik1 results in loss of heterochromatin localization of Swi6 in exponential- and stationary-phase cells (16) (data not shown). In contrast the absence of Clr1, Clr2, or Clr3 has no effect on the distribution of Swi6 (data not shown).
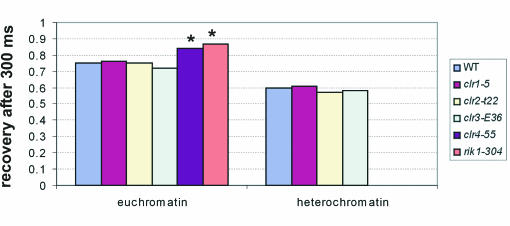
To study the contribution of these proteins to Swi6 binding, we compared the FRAP recovery kinetics of GFP-Swi6 in the WT, clr1-5, clr2-E22, clr3-E36, clr4-S5, or rik1-304 strain (Fig. 4). As expected, mutation of Clr4 or Rik1 significantly affected GFP-Swi6 binding (Fig. 4). In euchromatin, recovery of GFP-Swi6-WT reached 75% after 300 ms but more than 85% in the clr4-S5 and rik1-304 strains (P < 0.001). These results confirm a direct requirement for Rik1 and Clr4 for efficient binding of Swi6 in heterochromatin and euchromatin in vivo. In contrast, the mutations of Clr1, Clr2, and Clr3 had no significant effect on GFP-Swi6 binding in euchromatin or heterochromatin (Fig. 4). Recoveries after 300 ms were statistically indistinguishable from that observed in a WT background (Fig. 4). These results suggest that Clr1, Clr2, and Clr3 do not exert their effect on maintenance of silencing by directly or indirectly affecting Swi6 binding to chromatin and that they do not alter chromatin globally so as to change Swi6 binding.
FIG. 4.
Influence of Clr1, Clr2, Clr3, Clr4, and Rik1 on Swi6 binding. Shown is a quantitation of FRAP recovery after 300 ms in swi6+, clr1-5, clr2-E22, clr3-E36, clr4-S5, and rik1-304 strains during exponential phase. Values represent averages for 30 cells from three experiments. *, statistically significant difference in recovery.
Kinetic binding parameters of Swi6 in vivo.
Although these results clearly demonstrate that GFP-Swi6 binds only transiently to chromatin in vivo, we sought to obtain quantitative information about the properties of Swi6 binding to native chromatin in intact cells. To this end, we analyzed FRAP data using a kinetic modeling approach (44, 45). We generated a kinetic model based on standard principles of chemical kinetics to describe the behavior of Swi6-GFP in living yeast. In this model, Swi6-GFP diffuses rapidly through the nuclear volume and is free to bind to available binding sites in euchromatin and heterochromatin. The model is implemented as a set of differential equations and contains as variables the “on” rates (kon) for potential binding sites, the corresponding “off” rates (koff), and the relative number of binding sites (Fig. 5a; see Fig. S2 in the supplemental material). To first determine the kinetic properties of GFP-Swi6 binding to methylated binding sites, we analyzed the FRAP recovery kinetics of GFP-Swi6-WT in heterochromatin in WT cells. The best-fit values between the experimental data and FRAP simulations were obtained using a model containing two distinct types of methylated binding sites. Least-squares regression analysis yields best-fit parameters indicating that a major fraction, 84%, of Swi6 molecules were bound in methylated heterochromatin with a residence time of about 385 ms, whereas an additional fraction of about 10% had a residence time of 110 s (Table 1). The fraction of Swi6 bound to unmethylated chromatin was less than 1% in heterochromatin, and the percentage of free Swi6 in heterochromatin was about 5% (Table 1).
FIG. 5.
Kinetic model and corresponding fits to experimental data. (a) Minimal kinetic model for simultaneously fitting Swi6 FRAP data in clr4-S5 cells and in euchromatin and heterochromatin of a WT strain. kon and koff are association and dissociation rate constants, respectively (see Fig. S2 in the supplemental material for a full model). (b) Best fits, determined by least-squares regression analysis, to the model for WT Swi6 recovery in clr4-S5 cells and WT cells in euchromatin and heterochromatin. All parameters are listed in Table 1. Note that all rate constants are the same for each curve, the observed differences being attributed to the difference in the number of methylated binding sites. (c) Best fits determined by least-squares regression analysis for the CD and CSD deletion mutant proteins. For Swi6ΔCD, all rate constants describing binding to methylated H3 (kon2, koff2, kon3, and koff3) are set to zero. For Swi6ΔCSD a single set of rate constants (koff2 and kon2) describing binding to methylated H3 was sufficient for an accurate fit. All parameters are listed in Table 1.
TABLE 1.
Kinetic parameters for GFP-Swi6 binding in vivo
| Parameterc | Value ford:
|
||||
|---|---|---|---|---|---|
| clr4-S5, Swi6, eu | WT, Swi6, eu | WT, Swi6, hetero | WT, ΔCD, eu | WT, ΔCSD, eu | |
| kon1 (s−1) | 24.2 | 24.2 | 24.2 | 24.2 | 0.00 |
| koff1 (s−1) | 5.21 | 5.21 | 5.21 | 5.02 | 0.00 |
| kon2 (s−1) | 49.5 | 49.5 | 49.5 | 0.00 | 49.5 |
| koff2 (s−1) | 2.59 | 2.59 | 2.59 | 0.00 | 10.3 |
| kon3 (s−1) | 0.00105 | 0.00105 | 0.00105 | 0.00 | 0.00 |
| koff3 (s−1) | 0.0091 | 0.0091 | 0.0091 | 0.00 | 0.00 |
| kdiff (s−1) | 60 | 60 | 60 | 60 | 60 |
| Pa | 0.06 | 0.18 | 0.99 | 0.06 | 0.99 |
| Swi6 fraction | 0.18 | 0.14 | 0.05 | 0.24 | 0.21 |
| Swi6-H3 fraction | 0.63 | 0.43 | 0.01 | 0.76 | 0.00 |
| Swi6-H3metfast fraction | 0.17 | 0.39 | 0.84 | 0.00 | 0.79 |
| Swi6-H3metslow fraction | 0.02 | 0.04 | 0.10 | 0.00 | 0.00 |
| Residence time for Swi6-H3 (ms) | 192 | 192 | 192 | 199 | NAb |
| Residence time for Swi6-H3metfast (ms) | 385 | 385 | 385 | NA | 97 |
| Residence time for Swi6-H3metslow (s) | 110 | 110 | 110 | NA | NA |
P, relative proportion of methylated binding sites.
NA, not applicable.
Swi6-H3metfast and Swi6-H3metslow, Swi6 bound to methylated H3, fast and slow phases, respectively.
Background, version of Swi6 (Swi6, WT; ΔCD, Swi6ΔCD; ΔCSD, Swi6ΔCSD), type of chromatin (eu, euchromatin; hetero, heterochromatin).
To determine the kinetic properties of GFP-Swi6 binding to nonmethylated binding sites, we analyzed the FRAP recovery kinetics of GFP-Swi6-WT in the clr4-S5 strain, which has reduced in H3 methyltransferase activity. ChIP analysis on two centromeric loci indicated an approximately threefold reduction of methylated H3-K9 (see Fig. S3 in the supplemental material). The recovery curve for GFP-Swi6 in the clr4-S5 strain fits a model containing an unbound pool of GFP-Swi6, the two methylated binding sites identified previously in heterochromatin, and protein bound to a single type of nonmethylated binding site (Fig. 5a, red and green compartments). Kinetic parameters for binding of GFP-Swi6 to nonmethylated sites in the clr4-S5 strain and euchromatin in WT cells were determined for a number of different values of P, representing the fraction of methylated binding sites relative to heterochromatin, ranging from 0.01 to 0.2. In all fits the rate constants for binding to methylated H3 were kept constant as previously defined for heterochromatin. The set of parameters that best matched the experimentally determined ratio of methylated binding sites in WT yeast versus the clr4-S5 strain were determined. These parameters revealed that the relative fractions of methylated binding sites in the clr4-S5 strain and euchromatin in WT cells are 0.06 and 0.18, respectively. This approach yields a residence time of ∼170 ms for GFP-Swi6 bound to unmodified chromatin. In euchromatin about 43% of Swi6 was bound to methylated H3 and another 43% of the protein was bound to nonmethylated sites. Consistent with the reduction of methylated H3 in the clr4-S5 strain 63% of GFP-Swi6 molecules were bound to nonmethylated H3 in clr4-S5 cells while only 19% were bound to methylated sites; this compares to less than 1% and more than 84%, respectively, in heterochromatin of WT cells.
The validity of the kinetic model was confirmed by accurate prediction of the recovery profiles of several mutant Swi6 proteins (Fig. 5C; Table 1). In addition, sensitivity analysis on the data sets showed that the key parameters koff1, koff2, and koff3 were well defined (see Fig. S4 and Table S1 in the supplemental material). An alternative kinetic model assuming direct binding of GFP-Swi6 to two independent types of methylated binding sites provides a reasonable but less-stringent fit (see Fig. S5 in the supplemental material). Regardless, both models imply the presence of at least two kinetically distinct populations of Swi6 bound to methylated sites in intact chromatin.
DISCUSSION
Comparison of mammalian and yeast Swi6 dynamics.
Despite hundreds of millions of years of divergent evolution, the basic principles involved in the silencing of pericentric heterochromatin for mammals and fission yeast are remarkably similar (15, 54). Here we have analyzed the dynamic properties of Swi6 in living cells. In comparing them to our earlier observations on the binding dynamics of mammalian HP1 we find that several dynamic features of the major heterochromatin proteins HP1 and Swi6 are evolutionarily conserved. As in mammalian cells, Swi6 binding to chromatin is transient and overall binding dynamics are affected by the degree of chromatin condensation (10). Similar to our observations on mouse HP1, Swi6 binding dynamics depend on the presence of the histone methyltransferase specific for modification of K9 of histone H3. In addition, the contributions of the two conserved major protein domains, the CD and the CSD, in HP1 and Swi6 were similar. In both cases, the CD was required for targeting to heterochromatin and binding required the CSD for stabilization. However, we also note several differences. First, heterochromatin domains are highly mobile in fission yeast whereas they are almost immobile in the mammalian cell (10, 46). Mammalian heterochromatin domains move ∼140 nm/min (10), whereas they typically move ∼660 nm/min in exponentially growing S. pombe cells. This dramatic difference in mobility of heterochromatin domains might be indicative of fundamental differences in chromatin organization, and possibly nuclear architecture in general, between yeast and mammals. Fission yeast centromeric heterochromatin clusters at the nuclear periphery and interacts with the spindle pole body (SPB). The mobility of both SPB and centromeric heterochromatin is dependant on cytoplasmic microtubules and is thought to be important for nuclear positioning (21, 46). It is not known if these movements have a functional significance with respect to chromatin. Although both fission and budding yeast heterochromatin domains are highly mobile, fission yeast heterochromatin undergoes more-uniform displacements with a reduced velocity, compared to that of budding yeast, where loci undergo more-rapid, diffusive movement (22). A second difference in comparing HP1 and Swi6 dynamics is that the kinetics of binding of Swi6 are at least 1 order of magnitude faster than those of HP1. While we estimated the residence time of the majority of HP1 in heterochromatin to be on the order of 20 to 30 s, kinetic analysis of Swi6 suggests a residence time of the majority of heterochromatic Swi6 on the order of less than 1 s. The overall higher mobility of Swi6 molecules than of HP1 might be indicative of a lower proportion of silenced genes in yeast cells than in mammalian cells and might reflect an overall more open, less heterochromatic nature of yeast chromatin. Consistent with this interpretation we have shown here that Swi6 mobility is increasingly reduced as cells progress from exponential to stationary phase and to spores.
Swi6/HP1 and nucleosome binding.
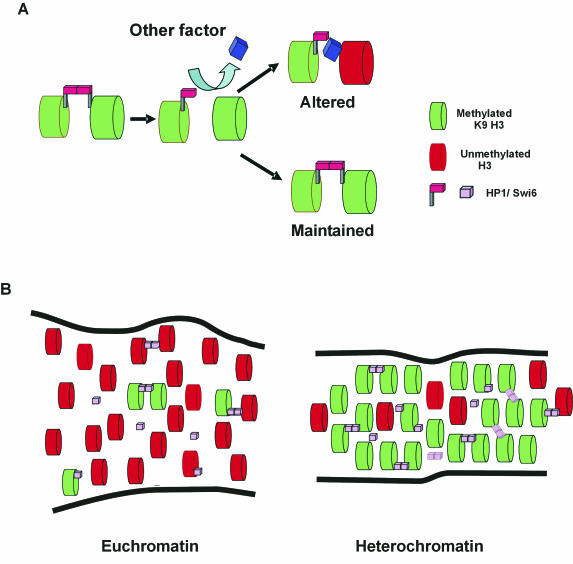
Using domain deletion mutant proteins, we show that both the CD and CSD are required for correct Swi6 binding to chromatin in the context of intact chromatin in a living S. pombe nucleus. These results are in agreement with similar mammalian in vivo and in vitro studies (5, 10, 27, 40). Our observation that recovery of a mutant Swi6 containing the CSD but not the CD is slower in a swi6+ strain than in a swi6− stain indicates that homotypic interactions via the CSD occur among Swi6 molecules in vivo (52, 54, 55). In contrast, the binding of a mutant protein lacking the CSD in a swi6− background was indistinguishable from that in a swi6+ background. These data are consistent with a model where the CD of HP1/Swi6 acts as a recognition site for methylated H3 and the CSD functions as a bridge to stabilize HP1/Swi6 binding (Fig. 6A) (29, 51).
FIG. 6.
Stochastic model for HP1/Swi6 binding to chromatin. (A) HP1/Swi6 dimers cross-link adjacent nucleosomes. The dynamic nature of binding frequently creates vacant binding sites. Competition for the open binding sites determines the fate of the nucleosome. Association of HP1/Swi6 maintains the status quo. Association of a competitor results in an altered nucleosome configuration. (B) Steady-state representation of stochastic HP1/Swi6 binding to euchromatin and heterochromatin domains. HP1/Swi6 has a higher affinity for mK9-H3, and, since this compartment is enriched in mK9-H3, HP1/Swi6 has a higher probability to bind heterochromatin. Since the compaction of the nucleosome is higher in heterochromatin, the probability of establishing a cross-link to an adjacent nucleosome, either on the same fiber or a different chromatin fiber, is also higher in heterochromatin than in euchromatin. Because euchromatin is enriched in nucleosomes bearing histone H3 unmethylated on K9, the fraction of HP1 bound to unmethylated histone H3 on K9 is higher in euchromatin than in heterochromatin. These interactions are less stable than the interactions with methylated K9-H3 and do not result in heterochromatinization. The overall stability of heterochromatin is conferred by the stable methylation of core histones.
Swi6 binding to heterochromatin was not altered in yeast lacking the previously identified silencing factors Clr1, Clr2, and Clr3, suggesting that these factors exert their silencing function independently of Swi6 binding (16). As expected, strains lacking Clr4 or Rik1 showed a strong defect in Swi6 binding in vivo. While it is clear that the Clr4 effect is due to its reduced ability to methylate H3-K9, the function of Rik1 is still enigmatic. The Rik1 protein contains β-propeller-like domains typically found within WD 40 domains, and it has been suggested that these domains function as chaperones that bind chromatin assembly factors (37). The differences observed in Swi6 binding in clr1-5, clr2-E22, and clr3-E36 strains compared to binding in clr4-S5 and rik1-304 strains also reinforce the possible involvement of HP1/Swi6 in chromosome segregation. Mutant Rik1, Clr4, and Swi6 have previously been demonstrated to have defects in chromosome segregation. However, no such defects occur in mutant Clr1, Clr2, or Clr3 (16). Similar phenotypes have been observed in mammalian cells lacking the histone methyltransferase Suv39h (43). Moreover, it was recently shown that Swi6 is required for the recruitment of cohesin to heterochromatin (6). This correlation might be explained if pericentric heterochromatin played an important mechanistic role in chromosome segregation.
A stochastic model for heterochromatin.
Our results demonstrate that Swi6 is involved in maintenance of yeast heterochromatin by making transient interactions with chromatin, as has been shown for HP1 in mammals (10, 18). Despite differences in the rates of binding, the dynamic nature of HP1/Swi6 binding is consistent with the notion that both mammals and fission yeast use stochastic mechanisms to control formation, maintenance, and likely spreading of heterochromatin. We propose that HP1/Swi6 binding to nucleosomes and the formation of internucleosome cross-links are essentially stochastic events. While the long-term stability of heterochromatin domains is determined by the putative irreversible nature of methylation on H3-K9, which acts as a permanent and heritable mark of heterochromatin (4), the short-term stability of heterochromatin is maintained by the steady-state equilibrium of dynamic association and dissociation of HP1/Swi6 from a diffusible nucleoplasmic pool. Since HP1/Swi6 has a higher affinity for binding to histone H3 methylated on K9 than for binding to unmodified sites on chromatin and since modified H3 is enriched in heterochromatin, the probability of the binding of nucleoplasmic HP1/Swi6 to a nucleosome bearing a methylation mark is higher in heterochromatin than in euchromatin (Fig. 6B). We also suggest that the establishment of cross-links between two proximal nucleosomes is similarly stochastic, although the probability is higher in heterochromatin since the density of the nucleosome bearing HP1/Swi6 is higher in heterochromatin than in euchromatin (Fig. 6B). The high number of exchange events within a domain contributes to the overall stability and creates a robust system, whose status is maintained over time but can be altered rapidly.
Advantages of a stochastic model.
A stochastic model for heterochromatin maintenance can account for several observations that are difficult to reconcile with a more static model. First, stronger expression of an activator is required to overcome telomeric repression of a reporter gene than to overcome its repression in other locations. This effect was interpreted to be due to the stochastic nature of silencing mechanisms (3). Second, silenced genes located in heterochromatin can be activated relatively rapidly, indicating that silencing is reversible. For example, expression of gal4 activator reverses the silencing of a variegating transgene containing a gal4 binding site in its promoter (1). Furthermore, a λ5 transgene in mouse pre-B cells undergoes reversible transitions between active and inactive states (31). Third, transcription factors and preinitiation complexes have been shown to be able to bind to silenced chromatin regions (50). In support of this observation, GFP-HP1/Swi6 is readily exchanged from heterochromatin in FRAP experiments; thus, this nuclear compartment is not inaccessible to other proteins, and it is therefore likely that transcriptional regulators can access heterochromatin. Fourth, in yeast, repression is maintained through a continuous requirement of repressors (9). A continuous flux of repressors appears superfluous in a static context, whereas it becomes essential when heterochromatin maintenance is controlled by dynamic equilibria.
Determination of the heterochromatin state by dynamic equilibrium has regulatory consequences. Modulation of dynamic equilibria provides a simple way of propagating or resolving heterochromatin. Dissociation of HP1/Swi6 from chromatin opens a window of opportunity for other chromatin binding proteins to associate and to act on the open binding site (Fig. 6B). Depending on the identity of the newly bound factor, chromatin states may be altered. The dynamic nature of HP1/Swi6 binding may be critical for propagation of heterochromatin states, since more long binding events, as they occur in heterochromatin, favor association of further HP1 and methyltransferase molecules, resulting in the perpetuation of the heterochromatin state, whereas more short binding events, as observed in euchromatin, favor dissociation of HP1/Swi6, resulting in the resolution of the heterochromatic state.
Our results suggest that globally stable heterochromatin domains are generated from dynamic interactions. Dynamic organization of heterochromatin confers the ability of a cell to spatially compartmentalize particular genome regions while at the same time allowing rapid response to changing environmental conditions. It is therefore likely that dynamic maintenance of heterochromatin is a crucial mechanism to ensure stability and responsiveness of genomes in vivo.
Supplementary Material
Acknowledgments
We thank Ken-ichi Noma and Shiv Grewal for the ChIP data in Fig. S3 in the supplemental material.
T.M. is a fellow of the Keith R. Porter Endowment for Cell Biology.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839-847. [DOI] [PubMed] [Google Scholar]
- 2.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 3.Aparicio, O. M., and D. E. Gottschling. 1994. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 8:1133-1146. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 5.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., J. F. Maure, J. F. Partridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 7.Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19:1587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, T. H., and M. R. Gartenberg. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14:452-463. [PMC free article] [PubMed] [Google Scholar]
- 10.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli, T. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 11.Clark, R. F., and S. C. Elgin. 1992. Heterochromatin protein 1, a known suppressor of position-effect variegation, is highly conserved in Drosophila. Nucleic Acids Res. 20:6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowell, I. G., R. Aucott, S. K. Mahadevaiah, P. S. Burgoyne, N. Huskisson, S. Bongiorni, G. Prantera, L. Fanti, S. Pimpinelli, R. Wu, D. M. Gilbert, W. Shi, R. Fundele, H. Morrison, P. Jeppesen, and P. B. Singh, P. B. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111:22-36. [DOI] [PubMed] [Google Scholar]
- 13.Cowieson, N. P., J. F. Partridge, R. C. Allshire, and P. J. McLaughlin. 2000. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10:517-525. [DOI] [PubMed] [Google Scholar]
- 14.Egel, R., M. Willer, and O. Nielsen. 1989. Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant rik1. Curr. Genet. 15:407-410. [Google Scholar]
- 15.Eissenberg, J. C., and S. C. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 16.Ekwall, K., E. R. Nimmo, J. P. Javerzat, B. Borgstrom, R. Egel, G. Cranston, and R. Allshire. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109(Pt. 11):2637-2648. [DOI] [PubMed] [Google Scholar]
- 17.Ekwall, K., and T. Ruusala. 1994. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics 136:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 19.Festenstein, R., S. Sharghi-Namini, M. Fox, K. Roderick, M. Tolaini, T. Norton, A. Saveliev, D. Kioussis, and P. Singh. 1999. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 23:457-461. [DOI] [PubMed] [Google Scholar]
- 20.Grewal, S. 2000. Transcriptional silencing in fission yeast. J. Cell. Physiol. 184:311-318. [DOI] [PubMed] [Google Scholar]
- 21.Hagan, I., and M. Yanagida. 1997. Evidence for cell cycle-specific, spindle pole body-mediated, nuclear positioning in the fission yeast Schizosaccharomyces pombe. J. Cell Sci., 110(Pt. 16):1851-1866. [DOI] [PubMed] [Google Scholar]
- 22.Heun, P., T. Laroche, K. Shimada, P. Furrer, and S. M. Gasser. 2001. Chromosome dynamics in the yeast interphase nucleus. Science 294:2181-2186. [DOI] [PubMed] [Google Scholar]
- 23.Huang, Y. 2002. Transcriptional silencing in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Nucleic Acids Res. 30:1465-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang, K. K., J. C. Eissenberg, and H. J. Worman. 2001. Transcriptional repression of euchromatic genes by Drosophila heterochromatin protein 1 and histone modifiers. Proc. Natl. Acad. Sci. USA. 98:11423-11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanova, A. V., M. L. Bonaduce, S. V. Ivanov, and A. J. Klar. 1998. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19:192-195. [DOI] [PubMed] [Google Scholar]
- 26.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 27.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 28.Li, Y., J. R. Danzer, P. Alvarez, A. S. Belmont, and L. L. Wallrath. 2003. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development 130:1817-1824. [DOI] [PubMed] [Google Scholar]
- 29.Li, Y., D. A. Kirschmann, and L. L. Wallrath. 2002. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16462-16469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorentz, A., L. Heim, and H. Schmidt. 1992. The switching gene swi6 affects recombination and gene expression in the mating-type region of Schizosaccharomyces pombe. Mol. Gen. Genet. 233:436-442. [DOI] [PubMed] [Google Scholar]
- 31.Lundgren, M., C. M. Chow, P. Sabbattini, A. Georgiou, S. Minaee, and N. Dillon. 2000. Transcription factor dosage affects changes in higher order chromatin structure associated with activation of a heterochromatic gene. Cell 103:733-743. [DOI] [PubMed] [Google Scholar]
- 32.Maundrell, K. 1993. Thiamine-repressible expression vectors pREB and pRIB for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 33.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 34.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 35.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 36.Murzina, N., A. Verreault, E. Laue, and B. Stillman. 1999. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell 4:529-540. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, A. L., C. Sanchez, H. Ichinose, M. Cervino, T. Lerouge, P. Chambon, and R. Losson. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. EMBO J. 21:5797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416:103-107. [DOI] [PubMed] [Google Scholar]
- 41.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 42.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 43.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 44.Phair, R. D., S. A. Gorski, and T. Misteli. 2004. Measurement of dynamic protein binding to chromatin in vivo using photobleaching microscopy. Methods Enzymol. 375:393-414. [DOI] [PubMed] [Google Scholar]
- 45.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 46.Pidoux, A. L., S. Uzawa, P. E. Perry, W. Z. Cande, and R. C. Allshire. 2000. Live analysis of lagging chromosomes during anaphase and their effect on spindle elongation rate in fission yeast. J. Cell Sci. 113:4177-4191. [DOI] [PubMed] [Google Scholar]
- 47.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 48.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 49.Ryan, R. F., D. C. Schultz, K. Ayyanathan, P. B. Singh, J. R. Friedman, W. J. Fredericks, and F. J. Rauscher III. 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell Biol. 19:4366-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403-414. [DOI] [PubMed] [Google Scholar]
- 51.Singh, P. B., and S. D. Georgatos. 2002. HP1: facts, open questions, and speculation. J. Struct. Biol. 140:10-16. [DOI] [PubMed] [Google Scholar]
- 52.Smothers, J. F., and S. Henikoff. 2000. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr. Biol. 10:27-30. [DOI] [PubMed] [Google Scholar]
- 53.Thon, G., and A. J. Klar. 1992. The clr1 locus regulates the expression of the cryptic mating-type loci of fission yeast. Genetics 131:287-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, G., A. Ma, C. M. Chow, D. Horsley, N. R. Brown, I. G. Cowell, and P. B. Singh. 2000. Conservation of heterochromatin protein 1 function. Mol. Cell. Biol. 20:6970-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto, K., and M. Sonoda. 2003. Self-interaction of heterochromatin protein 1 is required for direct binding to histone methyltransferase, SUV39H1. Biochem. Biophys. Res. Commun. 301:287-292. [DOI] [PubMed] [Google Scholar]
- 56.Ye, Q., I. Callebaut, A. Pezhman, J. C. Courvalin, and H. J. Worman. 1997. Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272:14983-14989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.