Abstract
Microarray analysis has been used to understand how gene regulation plays a critical role in neuronal injury, survival and repair following ischemic stroke. To identify the transcriptional regulatory elements responsible for ischemia-induced gene expression, we examined gene expression profiles of rat brains following focal ischemia and performed computational analysis of consensus transcription factor binding sites (TFBS) in the genes of the dataset. In this study, rats were sacrificed 24 h after middle cerebral artery occlusion (MCAO) stroke and gene transcription in brain tissues following ischemia/reperfusion was examined using Affymetrix GeneChip technology. The CONserved transcription FACtor binding site (CONFAC) software package was used to identify over-represented TFBS in the upstream promoter regions of ischemia-induced genes compared to control datasets. CONFAC identified 12 TFBS that were statistically over-represented from our dataset of ischemia-induced genes, including three members of the Ets-1 family of transcription factors (TFs). Microarray results showed that mRNA for Ets-1 was increased following tMCAO but not pMCAO. Immunohistochemical analysis of Ets-1 protein in rat brains following MCAO showed that Ets-1 was highly expressed in neurons in the brain of sham control animals. Ets-1 protein expression was virtually abolished in injured neurons of the ischemic brain but was unchanged in peri-infarct brain areas. These data indicate that TFs, including Ets-1, may influence neuronal injury following ischemia. These findings could provide important insights into the mechanisms that lead to brain injury and could provide avenues for the development of novel therapies.
Keywords: Ischemia, Microarray, Reperfusion, Stroke, Transcription Factors, Rat
1. Introduction
Ischemic stroke occurs when the blood supply to the brain is obstructed. The neuronal death that ensues results from the induction of genes associated with a number of cellular functions including apoptosis, inflammation and oxidative stress. Currently tissue plasminogen activator (t-PA) is the only approved treatment for ischemic stroke. Unfortunately, t-PA has a limited time window for therapeutic use, and only 3–5% of stroke patients arriving at the hospital will qualify for treatment (Fisher et al., 2009). Thus, there is a strong need to understand the molecular mechanisms associated with ischemic stroke so that more effective modes of treatment can be investigated.
It is well established that the transcription of new genes plays a major role in the delayed neuronal injury that occurs in the ischemic penumbra following stroke. A number of laboratories, including ours, have used high throughput microarray analysis to understand how the genome transcriptionally responds to ischemic challenge (Lu et al., 2003; Kury et al., 2004; Xu et al., 2005; Ford et al., 2006; Sarabi et al., 2008; Ridder et al., 2009; Vangilder et al., 2012). Previously, we published microarray studies demonstrating that distinct patterns of gene expression were seen in tissues from rat and non-human primate middle cerebral artery occlusion (MCAO) ischemic stroke models (Xu et al., 2005; Ford et al., 2006; Rodriguez-Mercado et al., 2012). Our microarray analysis indicated that many classes of genes were activated following ischemia, but inflammation and cell death were primary gene categories that appear to be associated with ischemic stroke models.
Genes can be regulated by transcription factors (TFs) that bind to regulatory elements in their promoters to induce or suppress gene expression. The analysis of transcriptional regulators can be useful to understand how large sets of genes can be controlled by a small set of upstream signaling molecules. In this study, we performed a computational analysis to predict the transcriptional regulators of genes induced ischemia/reperfusion using the CONserved transcription FACtor binding site finder (CONFAC) program (Karanam and Moreno, 2004). CONFAC identifies transcription factor binding sites within conserved promoters from a subset of genes. In addition, CONFAC enables the comparison of baseline gene expression from non-treated samples with gene expression that occurs following a given treatment. Our data predicted multiple transcription factors that could be important in mechanisms that regulate ischemia/reperfusion-induced genes. In addition, our data identified a potential novel role for the Ets-1 family of TFs following ischemia.
2. Results
2.1 Microarray and CONFAC Analysis
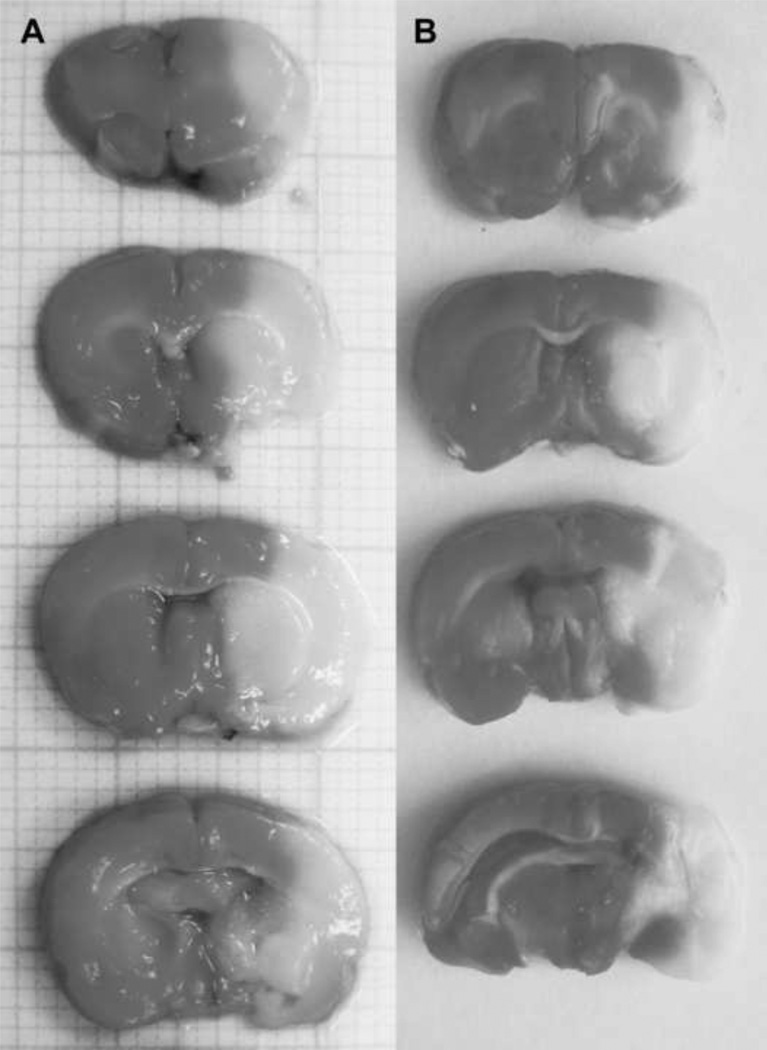
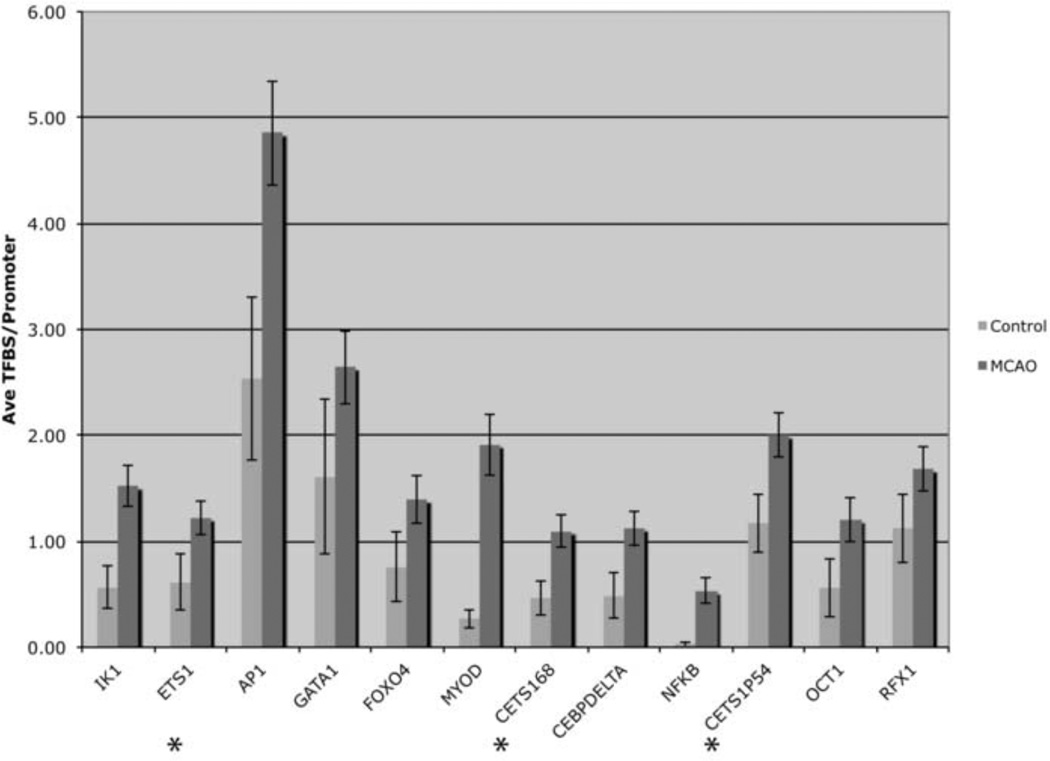
We examined gene expression profiles of animals subjected to permanent MCAO (pMCAO) and transient MCAO (tMCAO; ischemia reperfusion) and sham controls using Affymetrix rat genome U34 microarray genechips containing ~9,000 probe sets (known genes and ESTs). We performed TTC labeling of brain sections to measure neuronal injury that occurred in our MCAO stroke models. Representative TTC staining figures reveal large infarct regions following both pMCAO (Figure 1A) and tMCAO (Figure 1B). Infarcts seen following pMCAO are typically larger than in animals subjected to tMCAO, as we previously showed (Ford et al., 2006; Li et al., 2007). We examined genes that were induced by 2-fold or more only following tMCAO, but not in the pMCAO model when compared to sham controls. We used this dataset to further examine the transcriptional regulation of genes induced by tMCAO. The Affymetrix microarray results showed that there were 233 genes increased 2-fold or more following tMCAO compared to sham control animals that were not increased 2-fold or more following pMCAO. The CONFAC program discriminated 105 of the 233 genes that were annotated and contained at least one TFBS in the promoter that could be used for further TFBS analysis. Computational analysis of our microarray data using CONFAC produced lists of TFBS that were over-represented based on three separate random control datasets. A Mann-Whitney U-test was used to indicate TFBS that were statistically over-represented. We selected TFBS that were over-represented in two of the three analyses and showed that 12 TFBS were over-represented in tMCAO induced genes compared to a random set of genes (Table 1). AP1 showed the highest total number of TFBS in the dataset while IK1 was the most significantly over-represented. The over-represented TFBS included several previously associated with ischemia including AP1, C/EBP delta, FOXO4 and NFκB (Schneider et al., 1999; Hillion et al., 2006; Kapadia et al., 2006; Ridder and Schwaninger, 2009). Other TFBS were identified that bind TFs not previously associated with ischemia, such as GATA, OCT-1 and ETS-1. Ets-1 family member TFBS were represented three times in the analysis as ETS1, cETS1-68 and cETS1-P54 (Figure 2).
Figure 1. Brain injury associated with tMCAO model.
Representative triphenyltetrazolium chloride (TTC) staining of normal (dark areas) and ischemia-injured brain regions (white, indicate infarct) following pMCAO (A) and tMCAO (B).
Table 1. Predicted Transcription Factor Binding Site Activity of Genes Following tMCAO.
A prediction of transcription factor binding site activity for 105 genes was identified using CONFAC. Displayed are the number of genes analyzed, the number of binding sites, the number of genes with binding sites, the percentage of genes with site and the respective p value. A Mann-Whitney test, of predicted transcription factor binding sites (TFBS) revealed TFBS which are significantly associated with tMCAO when compared to a random set of control genes. These TFBS were identified as ETS family members (ETS1, CETS168 and CETS1P54), AP1, GATA FOXO4, MYOD, IK1, CEBPDELTA, NFKB, OCT1 and RFX1. All p-values are represented as (p<0.05).
| Transcription Factor | # Sites | # Genes w/Site(s) | % Genes w/Site(s) | P-value |
|---|---|---|---|---|
| IK1 | 155.00 | 62.00 | 59.05 | 0.00018 |
| ETS1 | 125.00 | 60.00 | 57.14 | 0.00056 |
| AP1 | 502.00 | 81.00 | 77.14 | 0.00080 |
| GATA | 97.00 | 53.00 | 50.48 | 0.00095 |
| FOXO4 | 145.00 | 58.00 | 55.24 | 0.00127 |
| MYOD | 199.00 | 46.00 | 43.81 | 0.00140 |
| CETS168 | 110.00 | 58.00 | 55.24 | 0.00152 |
| CEBPDELTA | 117.00 | 53.00 | 50.48 | 0.00234 |
| NFKB | 53.00 | 23.00 | 21.90 | 0.00384 |
| CETS1P54 | 203.00 | 77.00 | 73.33 | 0.00605 |
| OCT1 | 125.00 | 45.00 | 42.86 | 0.01968 |
| RFX1 | 174.00 | 63.00 | 60.00 | 0.02627 |
Figure 2. Predicted Transcription Factor Binding Site Activity of Gene Promoters Following tMCAO.
A prediction of transcription factor binding site (TFBS) activity for gene promoters were identified using CONFAC. These TFBS significantly over-represented were IK1, ETS family members (ETS1, CETS168 and CETS1P54), AP1, GATA1, FOXO4, MYOD, IK1, CEBPDELTA, NFKB, OCT1 and RFX1. The graph indicates the number of TFBS in control and tMCAO datasets for each TF listed. Asterisks indicate ETS-1 family members.
2.2 Ets-1 regulated genes induced following tMCAO
After the identification of Ets-1 as a novel, potential regulator of ischemia-induced gene expression, we determined the genes in our dataset with Ets-1 binding sites in their promoters (Table 2). The gene with the most Ets-1 promoter binding sites was guanine nucleotide binding protein (G protein alpha z polypeptide; GNAZ; 10 TFBS). The promoters of Inhibitor of DNA binding (ID3) and LIM domain kinase 2 (LIMK2) both contain 5 Ets-1 promoter binding sites. Many of the genes with Ets-1 promoter binding sites were associated with inflammation including interleukin-1 beta (IL1B), matrix metallopeptidase-9 (MMP9), signal transducer and activator of transcription 3 (STAT3), transforming growth factor, beta 1 (TGFB1), TIMP metallopeptidase inhibitor 1 (TIMP1), complement factor B (BF) and an FC receptor (FCGR2B) (Iadecola and Anrather, 2011).
Table 2.
Genes exhibiting a 2-fold or greater increase following tMCAO, which contain ETS-1 transcription factor binding sites (TFBS).
| Gene Name | Gene Symbol |
# of ETS-1 TFBS |
|---|---|---|
| Aquaporin 1 | AQP1 | 2 |
| Complement factor B | BF | 3 |
| Ciliary neurotrophic factor receptor | CNTF | 2 |
| Doublecortin-like kinase 1 | DCAMKL1 | 3 |
| Fc receptor, IgG, low affinity IIb | FCGR2B | 2 |
| Guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 | GNAI2 | 10 |
| Guanine nucleotide binding protein (G protein), alpha z polypeptide | GNAZ | 5 |
| Inhibitor of DNA binding 3 | ID3 | 3 |
| Interleukin 1 beta | IL1B | 1 |
| Jun B proto-oncogene | JUNB | 4 |
| Matrix metallopeptidase 9 | MMP9 | 2 |
| Signal transducer and activator of transcription 3 | STAT3 | 3 |
| Transforming growth factor, beta 1 | TGFB1 | 4 |
| TIMP metallopeptidase inhibitor 1 | TIMP1 | 4 |
| Thioredoxin reductase 1 | TXNRD1 | 1 |
| Upstream transcription factor 1 | USF1 | 1 |
| Potassium channel, subfamily K, member 3 | KCNK3 | 4 |
| LIM domain kinase 2 | LIMK2 | 5 |
| SMAD family member 1 | MADH1 | 2 |
| Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | 4 |
| Nth (endonuclease III)-like 1 (E.coli) | NTHL1 | 2 |
| Phospholipase C, delta 1 | PLCD1 | 2 |
| Syndecan 1 | SDC1 | 2 |
| Syndecan 4 | SDC4 | 2 |
2.3 Ets-1 expression following tMCAO
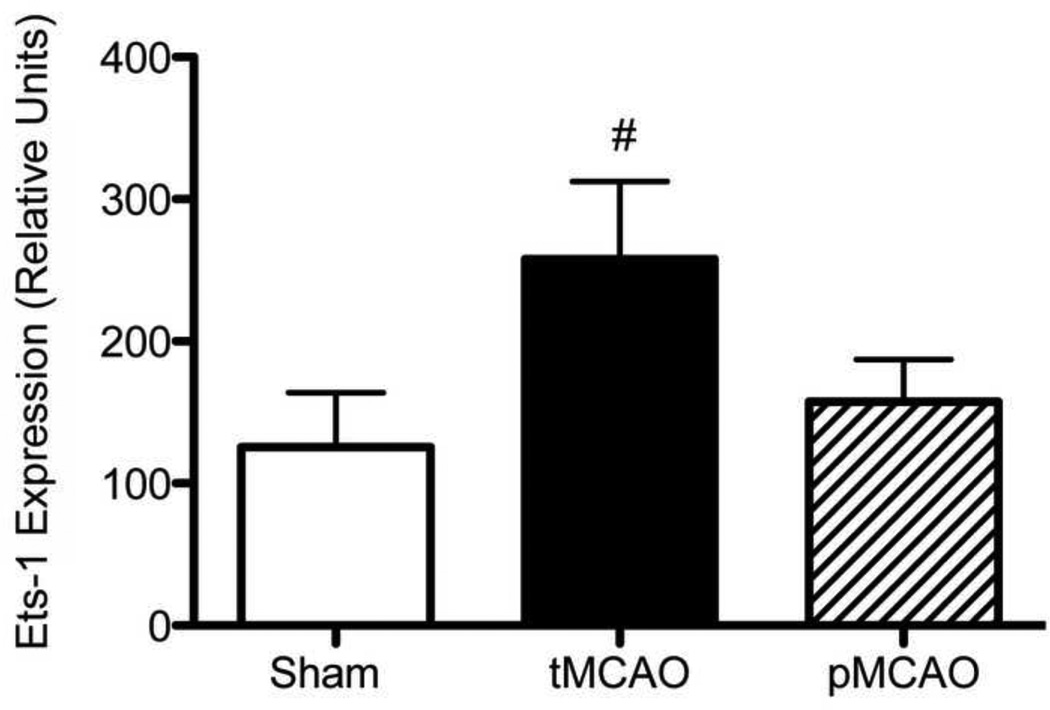
We examined the microarray dataset to determine whether Ets-1 mRNA changed in response to MCAO (Figure 3). Ets-1 mRNA was determined to be absent in all three control brain samples based on Affymetrix detection calls at the 24 h time point examined. Ets-1 was present or marginal in all three tMCAO samples and increased 2-fold following tMCAO based on signal intensity. Ets-1 detection calls were extremely variable following pMCAO (one each absent, marginal and present) and the signal intensity was close to control values.
Figure 3. Ets-1 mRNA following tMCAO.
Ets-1 mRNA levels in sham controls and animals following tMCAO and pMCAO was determined by microarray. Relative Ets-1 mRNA levels from the microarray analysis are shown in the graph. In the comparison between ets-1 expression between sham and MCAO animals, ets-1 was statistically absent in all three sham samples and statistically present or marginal in all stroke samples (indicated by #). However, due to the small sample size, statistical significance was not achieved when comparing the absolute signal values of all sham samples to that of all the controls (p=0.11).
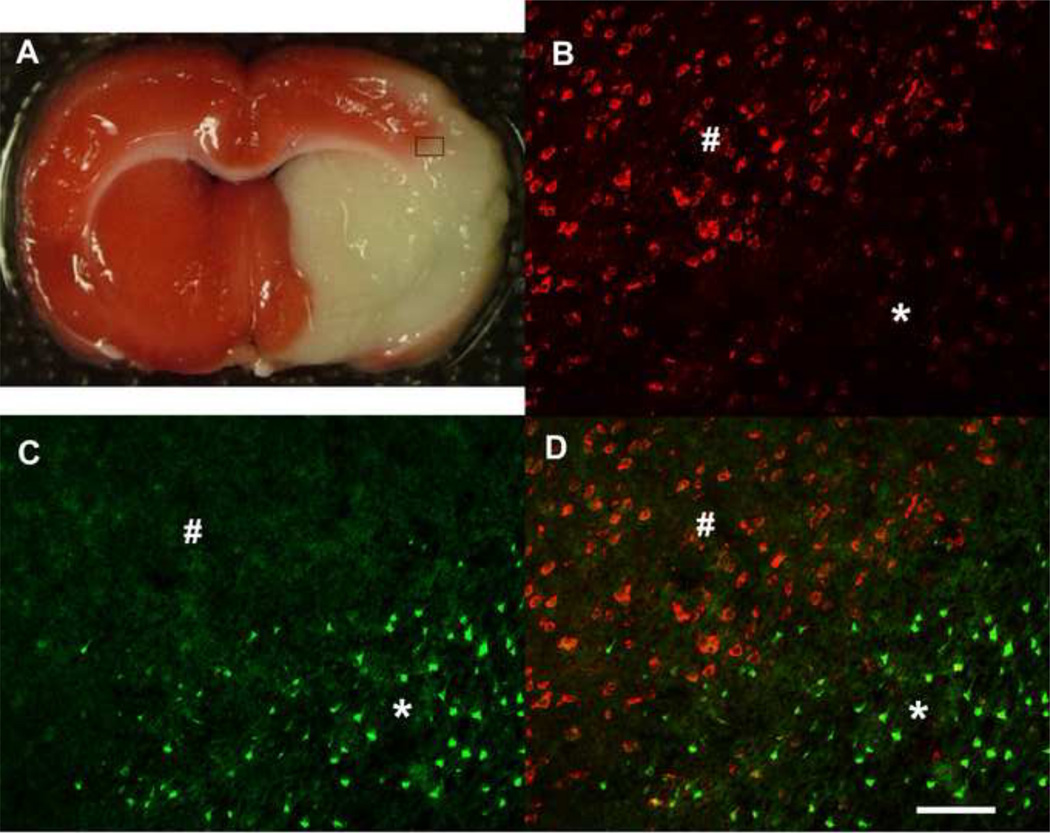
To validate the microarray results, we investigated the expression of Ets-1 protein in rat brains following tMCAO by immunohistochemistry (Figure 4). Figure 4A is a TTC labeled brain slice and the inset box indicates the infarct border where we examined Ets-1 immunolabeling. Ets-1 immunolabeling was uniformly present in non-ischemic brain regions (Figure 4B; indicated by #) and the contralateral side of the brain (not shown). The expression of Ets-1 decreased in the ischemic cortex following tMCAO (Figure 4B; indicated by asterisks) as shown in the representative photomicrograph taken at the infarct border. To determine the expression of Ets-1 in injured neurons, we double labeled brain tissues for FJB and Ets-1. Numerous FJB-positive cells were seen in the ischemic area (Figure 4C; indicated by asterisks) but not in the non-ischemic, peri-infarct area (Figure 4C; indicated by #). The expression of Ets-1 was dramatically reduced in the area containing FJB-positive cells while expression remained constant in the peri-infarct area (Figure 4D). Ets-1 labeling co-localized with NeuN immunostaining indicating that the TF was primarily expressed in neurons (Figure 5). To quantify the levels of Ets-1 protein, we counted the number of Ets-1-positive cells in the peri-infarct area, comparable areas in the contralateral side of the brain and sham controls. While the number of Ets-1-positive cells dramatically decreased on the infarct, the numbers of Ets-1 labeled cells was similar in other brain areas measured (not shown).
Figure 4. Double-labeling Ets-1 with Fluoro-JadeB.
Figure 4A is a TTC labeled brain slice and the inset box indicates the infarct border where we examined Ets-1 immunolabeling. (B) Ets-1 expression (red) in the non-ischemic brain regions (indicated by #) but is reduced in ischemic the cortex (indicated by asterisks). (C) Fluoro jade B (FJB) positive cells (green) are associated with the region of injury following tMCAO. (D) Double-labeled image shows that Ets-1 positive cells demonstrate lack of co-localization with FJB in the ischemia brain cortex. Scale bar is 100 µm.
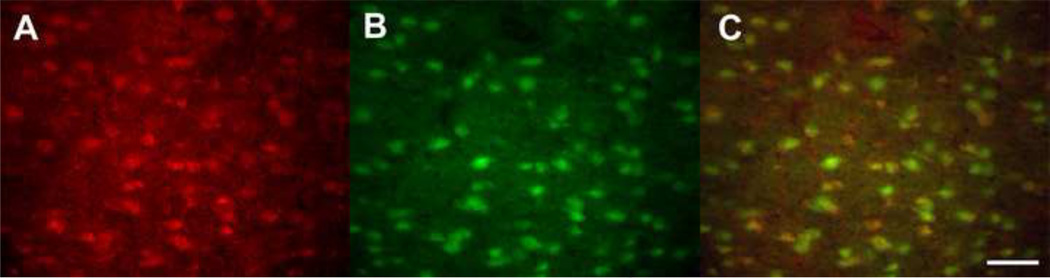
Figure 5. Double-labeling Ets-1 with NeuN.
Ets-1 expression (A; red) and NeuN labeling (B; green) show co-localization (C; yellow) in rat brain. Scale bar is 50 µm.
3. Discussion
The goal of this study was to examine microarray data to computationally predict transcriptional regulators of ischemia-induced genes. We previously showed that hundreds of genes were induced in rat brain following both transient and permanent MCAO (Xu et al., 2005; Ford et al., 2006). We used CONFAC bioinformatics software, to predict conserved TFBS over-represented within a population of genes that were uniquely up-regulated in our tMCAO focal stroke model. The CONFAC analysis utilizes the assumptions that (1) a set of genes are regulated by common transcription factors, (2) the more tightly clustered a gene list is, the more likely the genes will be regulated by a common transcription factor, (3) the regulatory human orthologs will be regulated similarly in transcription and (4) the binding sequence is 3 KB upstream of the start site and the first intron (Karanam and Moreno, 2004). From our analysis, 12 TFBS were identified as being over-represented in the promoters of ischemia-induced genes compared to control gene datasets. A number of TFBS previously associated with ischemia and stroke were shown to be over-represented, including AP1, C/EBP, FOXO4 and NFκB (Schneider et al., 1999; Hillion et al., 2006; Kapadia et al., 2006; Ridder and Schwaninger, 2009). AP1 showed the highest total number of TFBS in the dataset. Studies have shown that the induction and activation of immediate early transcription factors c-fos and c-jun, which bind the AP1 TFBS, regulate the expression of late effector genes following stroke (Salminen et al., 1995). NFκB has a well-described role in promoting neuronal injury cerebral ischemia (Ridder and Schwaninger, 2009). NFκB is activated in the brain following focal ischemia and NFκB knockout mice show reduced ischemia-induced neuronal damage compared to wild-type mice. C/EBP beta was similarly identified in silico as a TF potentially involved in neuronal injury following permanent MCAO (Ridder et al., 2009). C/EBP beta knockout mice displayed significantly smaller infarcts, reduced neurological deficits, decreased TUNEL labeling-positive cells and a reduced inflammatory response compared with their wild-type littermates (Kapadia et al., 2006). FOXO4 was shown to be phosphorylated after ischemia leading to its inactivation in vitro using oxygen-glucose deprivation with PC12 cells (Hillion et al., 2006). The study findings suggested that activation of FOXO4 leads to neuronal death following ischemia, which is blocked by pro-apoptotic factors such as Akt and ischemic preconditioning.
Other TFBS were identified that bind TFs not previously associated with ischemia, such as IK1, GATA, OCT-1 and ETS-1. Although AP1 showed the highest number of TFBS, the Ets-1 family members were present in multiple instances within the dataset as ETS-1, cETS1-68 and cETS1-P54. The Ets-1 family of transcription factors was first identified on the basis of the region of primary sequence homology with protein products from v-ets oncology encoded by E26 avian erythroblast virus (Karim et al., 1990). The Ets-1 family of proteins can act as activators or repressors of transcription and the functional roles of the Ets-1 family have been described in cell proliferation, activation and development (Dittmer, 2003; Oettgen, 2006; Russell and Garrett-Sinha, 2010; Hollenhorst et al., 2011). An essential role for Ets-1 has been identified as Ets-1 knock out animals are embryonic lethal and Ets-1 disruption can lead to immune cell defects such as T cell apoptosis and reduced B cell differentiation (Bories et al., 1995; Muthusamy et al., 1995). Ets-1 has been implicated in the regulation of chemokines associated with the immune response and in caspase-3 associated apoptosis (Liu et al., 2002; Oettgen, 2006; Russell and Garrett-Sinha, 2010). Ets-1 is also involved in angiogenesis as evidenced by its ability to mediate neovasularization following retinal ischemia and to increase capillary density and blood flow after hind limb ischemia (Hashiya et al., 2004; Watanabe et al., 2004). Ets-1 binds with low affinity to DNA and utilizes transcriptional partners to bind to DNA (Crepieux et al., 1994). Interestingly, AP1 is a known interacting partner with Ets-1 transcriptional activity, thus further supporting the notion that transcriptional activity associated with Ets-1 family members is involved in cerebral ischemia (Dittmer, 2003).
CONFAC analysis identified genes in the dataset that contained Ets-1 binding sites and many of these genes have known roles in stroke and inflammation. These included interleukin-1 beta (IL1B), matrix metallopeptidase-9 (MMP9) and signal transducer and activator of transcription 3 (STAT3), transforming growth factor, beta 1 (TGFB1), TIMP metallopeptidase inhibitor 1 (TIMP1), complement factor B (BF) and an FC receptor (FCGR2B) (Iadecola and Anrather, 2011). Ets-1 binding sites were also found in the promoter of aquaporin 1 (AQP1), a water channel shown to contribute to neuronal death following ischemic stroke (Kim et al., 2010). Whether Ets-1 induced or suppressed the genes in the dataset is currently under investigation. To gain insight to the function of Ets-1 in ischemia we examined protein expression of the TF following tMCAO. Ets-1 protein was present in neurons of sham control animals but was dramatically downregulated in the infarct regions of the brain after ischemia. While Ets-1 levels in neurons of the peri-infarct brain areas were comparable to controls, Ets-1 does not co-localize with FJB positive cells in the infarct. While we examined the effect of MCAO on Ets-1 mRNA and protein levels, we should point out that a key feature of CONFAC is to predict whether the activity of specific transcriptional regulators is changed in an experimental condition. It is not necessary that the levels of the TF are altered for it to induce or repress gene expression.
Taken together, these findings could suggest that Ets-1 is required for normal neuronal gene induction or suppression and its absence leads to inflammation and neuronal death. In support of this hypothesis, Ets-1 family member Elk-3 has been shown to suppress nitric oxide synthetase-2 (NOS2; iNOS) and heme oxygenase-1 (HO-1) expression under non-inflammatory conditions (Chen et al., 2003; Chung et al., 2006). In response to endotoxin, Elk-3 mRNA levels are quickly reduced resulting in the upregulation of HO-1 and NOS2. Alternatively, increased activation of Ets-1 could lead to inflammation and neuronal death. Increased Ets-1 expression following ischemia could induce a shift to a more pro-inflammatory/Th1 type response leading to neuronal death. In support of the alternative hypothesis, Ets-1 acts as coregulator along with the Th1 TF T-bet to stimulate interferon gamma production (Barton et al., 1998). In addition, T cells from Ets-1 deficient mice display decreased production of the Th1 cytokine interferon gamma while the levels of the anti-inflammatory cytokine IL-10 increased. The effects of Ets-1 after ischemia likely depend on the activity of co-regulatory factors. For example, cooperative activity of Ets-1 and AP-1 TFs activate the TNFα promoter while Ets-1, AP-1 and GATA-3 cooperatively activate the inhibitory transcription factor interleukin-5 (Kramer et al., 1995; Wang et al., 2006; Russell and Garrett-Sinha, 2010).
The results of this study support the premise that CONFAC can be used as a screening tool for transcriptional regulators associated with ischemia or other neurological conditions. A limitation of this study was the analysis of a single time point following ischemia to identify transcriptional regulators associated with stroke, however studies are underway to incorporate additional time points and provide a more precise view for the role of Ets-1 and other TFs following ischemic brain injury. Future studies will examine the specific roles for Ets-1 in ischemia using Ets-1 knockout mice and in vitro regulation of Ets-1 levels in neuronal cells. Understanding the role Ets-1 and transcriptional regulation in ischemic neuronal injury may reveal new molecular targets and strategies for developing treatment strategies for stroke.
4. Experimental Procedures
4.1 The MCAO Stroke Model
All animals used in these studies were treated humanely and with regard for alleviation of suffering and pain and all protocols involving animals were approved by the IACUC of Morehouse School of Medicine prior to the initiation of experimentation. Adult male Sprague-Dawley rats (250–300g; Charles River, Wilmington, MA, USA) were housed individually in standard plastic cages in a temperature-controlled room (22 ± 2°C) on a 12 h reverse light-dark cycle. Food and water were provided ad libitum. Animals were subjected to transient (tMCAO) or permanent (pMCAO) middle cerebral artery occlusion as previously described (Ford et al., 2006). Briefly, a 4 cm length 4-0 surgical monofilament nylon suture coated with silicon was inserted from the ECA into the ICA and then into the Circle of Willis to occlude the origin of the left middle cerebral artery (MCA). After 1.5 h of ischemia, the nylon suture was withdrawn and the ischemic brain was reperfused for tMCAO. Regional cerebral blood flow (CBF) was measured by continuous laser Doppler flowmetry with a laser doppler probe placed 7mm lateral and 2mm posterior to bregma in a thinned cranial skull window from the beginning of the tMCAO surgery until 15 min after reperfusion. For the pMCAO model, the suture remained in place to block the MCA for the entire 24 h duration. The rats were sacrificed 24h post-MCAO and the brains were removed for RNA isolation or histology.
4.2 RNA Preparation and GeneChip Microarray Analysis
Microarray analysis was performed as previously described (Ford et al., 2006). Animals were sacrificed 24 h after MCAO and the brains were removed and sliced into 2 mm coronal sections (approximately +3.0 to −5.0 from bregma) using a brain matrix. The ipsilateral tissues from the two middle slices (+1 to −3 from bregma) of tMCAO subjected animals (n=3) or sham controls (n=3) were used for subsequent RNA isolation while the outer two slices were used to confirm infarct formation by staining with 2,3,5-triphenyltetrazolium chloride (TTC). Total RNA was extracted with TRIzol Reagent (Life Technologies, Rockville, MD, USA), cleaned (RNAqueous Kit, Ambion, Austin, TX, USA) and converted to double-stranded cDNA (Invitrogen, Superscript Choice System, Carlsbad CA, USA) using a T7-(dT)24 primer. Cleanup of double-stranded cDNA used Phase Lock Gels (Eppendorf, Westbury, NY, USA)-Phenol/Chloroform/Isoamyl Alcohol (Sigma, St. Louis, MO, USA). cRNA was synthesized using a RNA transcript labeling kit (Enzo Diagnostics, Farmingdale, NY, USA). Biotin labeled cRNA was cleaned up using a GeneChip Sample Cleanup Module (Affymetrix Inc., Santa Clara, CA, USA) and then quantified using a spectrophotometer. Twenty micrograms of the in vitro transcription product was fragmented by placing at 94°C for 35 min in fragmentation buffer. Following fragmentation, 15 µg of the biotinylated cRNA was hybridized to either an Affymetrix Rat Genome U34A GeneChip. The chips were hybridized at 45°C for 16 h, and then washed, stained with streptavidin–phycoerythrin and scanned according to manufacturing guidelines.
4.3 Microarray Data Analysis
We used this dataset to further examine the transcriptional regulation of genes induced by tMCAO. Data analysis was performed using Affymetrix Expression Console software (Santa Clara, CA) that supports probe set summarization and CHP file generation of 3’ expression using the MAS5 Statistical algorithm. Affymetrix microarrays contain the hybridization, labeling and housekeeping controls that help determine the success of the hybridizations. Affymetrix Expression Console software analyzed Raw DAT files and generated CEL files. CEL files were analyzed using the Affymetrix MAS5 program. The Affymetrix Expression Analysis algorithm uses the Tukey’s biweight estimator to provide a robust mean signal value for the target-specific intensity differences of the probe pair (perfect match (PM) – mismatch (MM)) relative to its overall hybridization intensity (PM+MM). The MAS5 algorithm output files represented the difference in intensities between the perfect match (PM) and mismatched (MM) probe sets and used the Wilcoxon’s rank test to calculate a significance or p-value and Detection call for each probe set. The Detection call p-values are represented as present (P), p<0.05; marginal (M), p>0.05 and p<0.06; or absent (A), p>0.06.
The data set produced by the Affymetrix MAS5 software contains gene identifiers and corresponding expression values. Data were exported to Microsoft Excel and analyzed for calculation of fold change and whether the genes were confirmed as present in the tissue samples (as determined by the Affymetrix software). Gene Pattern software (http://genepattern.broadinstitute.org/ gp/pages/login.jsf; Broad Institute, MIT) was used to calculate the False Discovery Rate (FDR) and genes that fell within the 95% confidence interval were included in the CONFAC analysis. Three chips were used for each experimental group: sham, pMCAO and tMCAO. Genes in the injured brain that increased in expression by 2-fold or more compared to control and were present in all three tMCAO or pMCAO samples were identified and further analyzed.
4.4 CONFAC Analysis
Using CONFAC, we examined genes that were induced by 2-fold or more only following tMCAO, but not in the pMCAO model when compared to sham control. There were 1577 genes that exhibited a 2-fold or greater increase in mRNA expression following tMCAO and not after pMCAO. Putative transcriptional regulators and genes with associated transcription factor binding sites (TFBS) were identified using the CONserved transcription FACtor (CONFAC) binding site finder program (Karanam and Moreno, 2004). Gene accessions were loaded into the CONFAC program and ENSEMBL was used to convert the rat genes sequences to mouse orthologs. We also used the RESOURCERER program (Tsai et al., 2001) to identify human orthologs. The programs produced similar results following CONFAC analysis. The CONFAC program compared our gene list to a random set of default genes and performed a statistical analysis using a Mann-Whitney test to identify significantly over-represented TFBS. Over-representation describes a class of TFBS that appear more often in a list of interest than would normally be predicted by their distribution among all TFBS assayed. Significantly over-represented transcription factors and genes with TFBS and were identified by using a p-value (p<0.05).
4.5 Histology and Immunocytochemistry
At 24 h post injury, rats were deeply anesthetized with 5% isoflurane and perfused transcardially with saline followed by cold 4% PFA (paraformaldehyde) solution in PBS for 30 min (n=3 animals/group). Brains were quickly removed and cryoprotected in 30% sucrose. The brains were then frozen in OCT compound and stored at −80°C until sectioning. Coronal sections of 20 µm thickness were cryosectioned and mounted on slides which were then stored at −80°C until further processed. Immunocytochemical localization of Ets-1 was determined using a mouse monoclonal antibody (1:200; Millipore, Billerica, MA, USAand a donkey anti-mouse IgG-Cy3 secondary antibody (1:400, Jackson ImmunoResearch Laboratories, Inc., PA, USA). For double labeling, sections were first incubated with a mouse anti-neuN Alexa Fluor 488 conjugated monoclonal antibody (1:200, Millpore) overnight at 4°C, After the sections were washed, they were then incubated in normal mouse serum (5% in PBS) for 6 hours at 4°C. The purpose is to block the open binding sites on the labeled secondary antibody. And then the staining of anti-Ets-1 antibody was performed. For Ets-1 and FJB (Chemicon International, Temecula, CA, USA) double labeling, Ets-1 immunostaining was performed first and then sections were incubated with 0.0015% potassium permanganate for 1 min, washed with distilled water for 2 min and treated with 0.0001% FJB in 0.1% acetic acid for 10 min. Sections were then washed and coverslipped in glycerol/0.3% acetic acid mounting medium. A Zeiss microscope equipped with CCD camera (Carl Zeiss Microimaging Inc, Thornwood, NY) was used to capture all digital images of sections at the same level across rat brains (as determined using a brain atlas) at 200X magnification.
4.6 Quantification of Ets-1 immunopositive cells
Two sections obtained from coronal sectioning of the brain of each rat (n = 3) were labeled with Ets-1. A Zeiss microscope equipped with CCD camera (Carl Zeiss Microimaging Inc, Thornwood, NY) was used to capture digital images of the sections at the same level (as determined using a brain atlas) at 200X magnification. The number of Ets-1-positive cells was determined using Image Pro Plus software (Media Cybernetics, Inc., Bethesda, MD) by an individual who was blinded to the experimental treatments. Only profiles of neuronal somas were counted and Ets-1 -positive fragments were excluded. A mean value of Ets-1-positive cells per unit area within the brain regions was obtained for each individual rat. Data were expressed as mean ± SEM. These mean values from each individual rat were used as the statistical unit of measure for analysis by one-way ANOVA to determine statistically significant treatment effects.
Highlights.
Microarray analysis showed gene induced in brain by ischemia
CONFAC identified over-represented transcription factor binding sites
TFBS for three Ets-1 family members were over-represented in ischemia dataset
Ets-1 levels are regulated by ischemia
Acknowledgements
This work was supported by NIH grants R01 NS34194 (B.F.), U01 NS 057993 (B.F.), U54 NS060659 (B.F.), K12 GM000680 (J.P.) and the W.M. Keck Foundation. The project described was supported by Grants Number U54 RR026137, G12RR003034 and S21MD000101 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. The work described in this article must have been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans; EC Directive 86/609/EEC for animal experiments and Uniform Requirements for manuscripts submitted to Biomedical journals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- Bories JC, Willerford DM, Grevin D, Davidson L, Camus A, Martin P, Stehelin D, Alt FW. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 1995;377:635–638. doi: 10.1038/377635a0. [DOI] [PubMed] [Google Scholar]
- Chen YH, Layne MD, Chung SW, Ejima K, Baron RM, Yet SF, Perrella MA. Elk-3 is a transcriptional repressor of nitric-oxide synthase 2. The Journal of biological chemistry. 2003;278:39572–39577. doi: 10.1074/jbc.M308179200. [DOI] [PubMed] [Google Scholar]
- Chung SW, Chen YH, Yet SF, Layne MD, Perrella MA. Endotoxin-induced downregulation of Elk-3 facilitates heme oxygenase-1 induction in macrophages. J Immunol. 2006;176:2414–2420. doi: 10.4049/jimmunol.176.4.2414. [DOI] [PubMed] [Google Scholar]
- Crepieux P, Coll J, Stehelin D. The Ets family of proteins: weak modulators of gene expression in quest for transcriptional partners. Critical reviews in oncogenesis. 1994;5:615–638. [PubMed] [Google Scholar]
- Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher M, Feuerstein G, Howells DW, Hurn PD, Kent TA, Savitz SI, Lo EH. Update of the stroke therapy academic industry roundtable preclinical recommendations. Stroke; a journal of cerebral circulation. 2009;40:2244–2250. doi: 10.1161/STROKEAHA.108.541128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford G, Xu Z, Gates A, Jiang J, Ford BD. Expression Analysis Systematic Explorer (EASE) analysis reveals differential gene expression in permanent and transient focal stroke rat models. Brain Res. 2006;1071:226–236. doi: 10.1016/j.brainres.2005.11.090. [DOI] [PubMed] [Google Scholar]
- Hashiya N, Jo N, Aoki M, Matsumoto K, Nakamura T, Sato Y, Ogata N, Ogihara T, Kaneda Y, Morishita R. In vivo evidence of angiogenesis induced by transcription factor Ets-1: Ets-1 is located upstream of angiogenesis cascade. Circulation. 2004;109:3035–3041. doi: 10.1161/01.CIR.0000130643.41587.DB. [DOI] [PubMed] [Google Scholar]
- Hillion JA, Li Y, Maric D, Takanohashi A, Klimanis D, Barker JL, Hallenbeck JM. Involvement of Akt in preconditioning-induced tolerance to ischemia in PC12 cells. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2006;26:1323–1331. doi: 10.1038/sj.jcbfm.9600286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C, Anrather J. The immunology of stroke: from mechanisms to translation. Nature medicine. 2011;17:796–808. doi: 10.1038/nm.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapadia R, Tureyen K, Bowen KK, Kalluri H, Johnson PF, Vemuganti R. Decreased brain damage and curtailed inflammation in transcription factor CCAAT/enhancer binding protein beta knockout mice following transient focal cerebral ischemia. J Neurochem. 2006;98:1718–1731. doi: 10.1111/j.1471-4159.2006.04056.x. [DOI] [PubMed] [Google Scholar]
- Karanam S, Moreno CS. CONFAC: automated application of comparative genomic promoter analysis to DNA microarray datasets. Nucleic Acids Res. 2004;32:W475–W484. doi: 10.1093/nar/gkh353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim FD, Urness LD, Thummel CS, Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA, Gunther CV, Nye JA, et al. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee YW, Park KA, Lee WT, Lee JE. Agmatine attenuates brain edema through reducing the expression of aquaporin-1 after cerebral ischemia. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2010;30:943–949. doi: 10.1038/jcbfm.2009.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B, Wiegmann K, Kronke M. Regulation of the human TNF promoter by the transcription factor Ets. The Journal of biological chemistry. 1995;270:6577–6583. doi: 10.1074/jbc.270.12.6577. [DOI] [PubMed] [Google Scholar]
- Kury P, Schroeter M, Jander S. Transcriptional response to circumscribed cortical brain ischemia: spatiotemporal patterns in ischemic vs. remote non-ischemic cortex. Eur J Neurosci. 2004;19:1708–1720. doi: 10.1111/j.1460-9568.2004.03226.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu Z, Ford GD, Croslan DR, Cairobe T, Li Z, Ford BD. Neuroprotection by neuregulin-1 in a rat model of permanent focal cerebral ischemia. Brain Res. 2007;1184:277–283. doi: 10.1016/j.brainres.2007.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lein PJ, Liu C, Bruun DA, Tewolde T, Ford G, Ford BD. Spatiotemporal pattern of neuronal injury induced by DFP in rats: A model for delayed neuronal cell death following acute OP intoxication. Toxicology and applied pharmacology. 2011;253:261–269. doi: 10.1016/j.taap.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Wang G, Yakovlev AG. Identification and functional analysis of the rat caspase-3 gene promoter. The Journal of biological chemistry. 2002;277:8273–8278. doi: 10.1074/jbc.M110768200. [DOI] [PubMed] [Google Scholar]
- Lu A, Tang Y, Ran R, Clark JF, Aronow BJ, Sharp FR. Genomics of the periinfarction cortex after focal cerebral ischemia. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2003;23:786–810. doi: 10.1097/01.WCB.0000062340.80057.06. [DOI] [PubMed] [Google Scholar]
- Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circulation research. 2006;99:1159–1166. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- Ridder DA, Schwaninger M. NF-kappaB signaling in cerebral ischemia. Neuroscience. 2009;158:995–1006. doi: 10.1016/j.neuroscience.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Ridder DA, Bulashevska S, Chaitanya GV, Babu PP, Brors B, Eils R, Schneider A, Schwaninger M. Discovery of transcriptional programs in cerebral ischemia by in silico promoter analysis. Brain Res. 2009;1272:3–13. doi: 10.1016/j.brainres.2009.03.046. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mercado R, Ford GD, Xu Z, Kraiselburd EN, Martinez MI, Eterovic V, Colon E, Rodriguez IV, Portilla P, Ferchmin PA, Gierbolini L, Rodriguez-Carrasquillo M, Powell MD, Pulliam JVK, McCraw CO, Gates A, Ford BD. Acute Neuronal Injury and Blood Genomic Profiles in a Nonhuman Primate Model for Ischemic Stroke. J Comp Med. 2012;62:427–438. [PMC free article] [PubMed] [Google Scholar]
- Russell L, Garrett-Sinha LA. Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine. 2010;51:217–226. doi: 10.1016/j.cyto.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Salminen A, Liu PK, Hsu CY. Alteration of transcription factor binding activities in the ischemic rat brain. Biochem Biophys Res Commun. 1995;212:939–944. doi: 10.1006/bbrc.1995.2060. [DOI] [PubMed] [Google Scholar]
- Sarabi AS, Shen H, Wang Y, Hoffer BJ, Backman CM. Gene expression patterns in mouse cortical penumbra after focal ischemic brain injury and reperfusion. Journal of neuroscience research. 2008;86:2912–2924. doi: 10.1002/jnr.21734. [DOI] [PubMed] [Google Scholar]
- Schneider A, Martin-Villalba A, Weih F, Vogel J, Wirth T, Schwaninger M. NF-kappaB is activated and promotes cell death in focal cerebral ischemia. Nature medicine. 1999;5:554–559. doi: 10.1038/8432. [DOI] [PubMed] [Google Scholar]
- Tsai J, Sultana R, Lee Y, Pertea G, Karamycheva S, Antonescu V, Cho J, Parvizi B, Cheung F, Quackenbush J. RESOURCERER: a database for annotating and linking microarray resources within and across species. Genome biology. 2001;2 doi: 10.1186/gb-2001-2-11-software0002. SOFTWARE0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vangilder RL, Huber JD, Rosen CL, Barr TL. The transcriptome of cerebral ischemia. Brain research bulletin. 2012 doi: 10.1016/j.brainresbull.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Shannon MF, Young IG. A role for Ets1, synergizing with AP-1 and GATA-3 in the regulation of IL-5 transcription in mouse Th2 lymphocytes. International immunology. 2006;18:313–323. doi: 10.1093/intimm/dxh370. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Takagi H, Suzuma K, Suzuma I, Oh H, Ohashi H, Kemmochi S, Uemura A, Ojima T, Suganami E, Miyamoto N, Sato Y, Honda Y. Transcription factor Ets-1 mediates ischemia- and vascular endothelial growth factor-dependent retinal neovascularization. The American journal of pathology. 2004;164:1827–1835. doi: 10.1016/S0002-9440(10)63741-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Ford GD, Croslan DR, Jiang J, Gates A, Allen R, Ford BD. Neuroprotection by neuregulin-1 following focal stroke is associated with the attenuation of ischemiainduced pro-inflammatory and stress gene expression. Neurobiol Dis. 2005;19:461–470. doi: 10.1016/j.nbd.2005.01.027. [DOI] [PubMed] [Google Scholar]