Abstract
The biochemical mechanisms underlying epigenetic control of gene expression are increasingly well known. In contrast, the contributions of individual modifications toward activation of lineage-specific genes during vertebrate development are poorly understood. Class II histone deacetylases (HDACs), which show restricted tissue distribution, regulate muscle-specific gene expression, in part through interactions with myogenic transcription factors. We have combined gene expression profiling with manipulation of fetal mouse intestinal tissue to define roles for other regulatory factors. We found that in the developing mouse intestine class I HDACs are confined to the prospective epithelium and that their levels decline coincidently with activation of differentiation genes, suggesting a functional relationship between these events. Overexpression of wild-type but not of mutant HDACs 1 and 2 in fetal intestine explants reverses expression of certain maturation markers. HDAC inhibitors, including the selective class I antagonist valproic acid, activate the same genes prematurely and accelerate cytodifferentiation. Chromatin immunoprecipitation of freshly isolated organs reveals early HDAC2 occupancy at differentiation gene promoters and corresponding histone hypoacetylation that reverses as HDAC levels fall. Thus, modulation of endogenous class I HDAC levels represents a previously unappreciated mechanism to enable onset of tissue-restricted gene expression in a developing mammalian organ.
Reversible acetylation of selected lysine residues in the conserved NH2-terminal tails of core histone proteins combines with DNA methylation and other modifications to generate an epigenetic code of altered chromatin structure and function (41, 44). The acetylation state of histones and other proteins is dynamically regulated by the competing actions of acetyltransferases and deacetylases (HDACs). Hypoacetylated histones promote chromatin condensation and are associated with transcriptionally silent loci, wherein access to transcription factors or the transcriptional apparatus is limited (5, 23). By determining, in some measure, the complement of genes expressed within individual cell types, such alterations may play a seminal role in tissue differentiation. Establishing lineage-specific patterns of gene expression is especially relevant in development, when sequential epigenetic modifications help distinguish individual cell types. However, the manner in which chromatin is modified locally to allow expression of genes for the first time in a developing embryo is not well understood. Histone acetylation plays a part in this process, as implied originally by studies with Xenopus species embryos (6) and revealed in recent in vitro investigation of muscle differentiation (29).
The four known mammalian class I HDACs (HDAC1 through 3 and 8) are related to yeast Rpd3, share a common domain structure, largely show nuclear localization, and are widely expressed (reviewed in reference 18). HDACs 1 and 2, which are especially closely related in sequence, copurify in multiprotein complexes that contain Sin3 and other transcriptional corepressors (1, 13, 31, 47), consistent with their demonstrated role in inhibiting transcription (12). Recruitment of this complex to the promoters of genes targeted for silencing results in modification of histone proteins and nonhistone transcriptional regulators (19, 22, 25, 34). Class II HDACs (HDACs 4 through 7) also mediate transcriptional repression but are distinguished from the class I enzymes on the basis of larger protein size, closer homology to yeast Hda1 than to Rpd3, exclusion from canonical Sin3 complexes, restricted tissue distribution, and nucleocytoplasmic shuttling (14, 18). Class II HDACs influence muscle gene expression by interacting with basic helix-loop-helix transcription factors like MEF2 through N-terminal domains that are absent in the class I enzymes (24, 29). Nonacetylatible mutants of MyoD are also impaired in in vitro myogenic activity (37), where MyoD may rely additionally on regulatory interactions with HDAC1 (25, 34).
The contribution that individual HDACs might make in the timing of tissue-specific gene expression is sometimes assumed but is unproven. Although the varied roles of HDACs in vertebrate muscle differentiation are revealing, their functions in a broader developmental context remain unknown, in part because investigation of HDACs has focused mainly on biochemical mechanisms. Mutants with mutations of the Rpd3 homolog in Drosophila melanogaster and Caenorhabditis elegans show embryonic lethality with different degrees of severity (26, 39), and among them HDACs are implicated in surprisingly limited aspects of invertebrate embryogenesis (3, 7). The present understanding of mammalian HDACs relies heavily on studies with cultured cells, and developmental epigenetic mechanisms require further elucidation. HDAC1 accounts for much of the total HDAC activity in mouse embryonic stem cells, where it supports cell proliferation. However, HDAC1-null mouse embryos die at embryonic day 10 (E10), before they can be informative about its functions in the differentiation of many tissues (21). Here we explore the role of mammalian class I HDACs in controlling differentiation of the intestinal epithelium during organogenesis.
MATERIALS AND METHODS
Antibodies and plasmids.
Antibodies used to detect HDAC1 and HDAC2, or acetylated forms of histones H3 and H4, were purchased from Abcam (Cambridge, United Kingdom) and Upstate Biotechnology (Lake Placid, N.Y.), respectively. The HDAC2 antibody used for chromatin immunoprecipitation (ChIP; Santa Cruz Biotechnology, Santa Cruz, Calif.) and HDAC3, HDAC4 (Upstate Biotechnology), HDAC6 (Abcam), and HDAC7 (Cell Signaling Technology, www.cellsignal.com) antibodies for immunoblotting were also from commercial sources. Full-length inserts and deacetylase-domain deletion mutants (42) for HDAC1 and HDAC2 were amplified by PCR; subcloned in pCIG (a gift from A. McMahon, Harvard University), which drives simultaneous expression of green fluorescent protein (GFP) and inserted genes by virtue of an internal ribosome entry site, or pK7 (a gift from A. Everett, University of Virginia), which generates GFP-tagged fusion proteins; and verified by DNA sequencing.
SAGE.
mRNA expression levels in the small intestine of the developing mouse at postcoital days E12, E13, and E15 were determined by serial analysis of gene expression (SAGE) according to the published protocol (46). RNA was isolated from unfractionated bowel segments extending from the duodenum to the ileum, and we obtained sequences for 65,585, 69,040, and 67,522 SAGE tags from the respective developmental stages. Data were extracted, filtered, and tabulated by using SAGE 300/2000 software, kindly provided by K. Kinzler (Johns Hopkins University, Baltimore, Md.). Gene annotations corresponding to ∼400 SAGE tags were checked manually, with >90% confirmation, and temporal variation in expression levels of selected genes was further verified by reverse transcription (RT)-PCR on independently isolated tissue samples.
Organ explant culture, injection, and electroporation.
Fetal mouse intestines from E12 to E14 were isolated under a dissecting microscope, and 3 to 8 explants were placed on 0.8-μm filter disks (Millipore) supported over a stainless plastic mesh (McMaster-CARB) in 700 μl of medium in 6-well plates. The medium consisted of Fitton-Jackson modified BGJb (Life Technologies) supplemented with 0.1 mg of ascorbic acid (Sigma)/ml and 40 μg of gentamicin/ml. Explants were maintained in a humidified atmosphere of 95% air and 5% CO2, and the medium was changed every 2 days. Solutions of sodium butyrate (SB; Sigma), valproic acid (VPA; Sigma), or 2 μg of plasmid DNA/μl were flushed into the intestinal lumen by using a capillary pipette and Nanoject injection device (Drummond Scientific). To introduce DNA into epithelial cells, injected explants bathed in Dulbecco's modified Eagle medium (DMEM; Life Technologies) were placed between platinum electrodes (BTX, San Diego, Calif.), exposed to three 10-msec pulses of 80 V each by using the BTX830 square-wave pulse electroporator, washed in DMEM, and cultured.
Histone isolation.
Nuclear histones were isolated as described previously (43). Fetal intestines were washed in phosphate-buffered saline (PBS), incubated in ice-cold lysis buffer (10 mM Tris-Cl [pH 6.5], 50 mM sodium sulfite, 10 mM MgCl2, 10 mM sodium butyrate, 8.6% sucrose, 1% Triton X-100, 0.5 mM phenylmethylsulfonylfluoride, and 1 μg each of leupeptin, aprotinin, and pepstatin/ml), ground with a tissue homogenizer (Corning), and centrifuged at 1,000 × g for 10 min. The nuclear pellet was washed twice with lysis buffer, washed once in 10 mM Tris-Cl (pH 7.4)-13 mM EDTA, resuspended in 0.2 M H2SO4, incubated on ice for 1 h, and then centrifuged at 20,000 × g for 5 min. The histone-enriched supernatant was mixed with 10 vol of acetone, and proteins were precipitated overnight at −20°C, collected, and air dried.
RT-PCR analysis.
RNA was extracted with Trizol (Gibco) and reverse-transcribed with oligo(dT) priming. RT-PCR was performed in the presence of 0.1 μCi of [α-32P]dCTP, the number of PCR cycles was varied to ensure that amplification was in the linear range, and products were resolved in 4 or 5% native polyacrylamide gels. The PCR primers (5′ to 3′) and the sizes of the amplified fragments are Apo1a (499 bp), CAGAGACTATGTGTCCAGTTTGA and GGTGTGGTACTCGTTCAAGGTAG; Fabpl (370 bp), TCTCCGGCAAGTACCAATTGCA and TCTCTTGCTGACTCTCTTGTAGA; Fabpi (501 bp), GTAGACCGGAACGAGAACTATG and TAGCTTTGACAAGGCTGGAGAC; Upa (527 bp), GAGATCTACAGCTTCGCCATTC and AAGGAGTGGAAGAGTGGTTAGG; Mt2 (318 bp), ATGCAAATGTACTTCCTGCAAGA and AAGGCTAGGCTTCTACATGGTCTA; Villin (164 bp), TTCTCTGGCACCGTCACTC and CGTAGCAAACCCATGTTCCT; HDAC1 (569 bp), CTGTCCGGTATTTGATGGCT and CACGAACTCCACACACTTGG; HDGF (525 bp), CTCCCTTCCTATACACCCTGTG and AAGTAGATGAAGGCAGCAGGTCT; and Gapdh (341 bp), CTGCACCACCAACTGCTTAG and CCTGCTTCACCACCTTCTTG.
Histology, in situ hybridization, and immunohistochemistry.
Cultured fetal gut explants were fixed for 6 h in 4% paraformaldehyde and immersed in 15% sucrose for 4 h at 4°C. The samples were embedded in Tissue-Tek OCT compound (Sakura), and 12-μm frozen tissue sections were stained with hematoxylin and eosin. Digoxigenin (DIG)-labeled sense and antisense probes, transcribed from linearized plasmid templates using a DIG-labeling kit (Boehringer), were hybridized overnight at 60°C. Sections were washed three times in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.15 sodium citrate) at 60°C and twice in PBT (PBS containing 2 mg of bovine serum albumin/ml and 0.1% Triton X-100) at room temperature, blocked with 10% goat serum in PBT for 1 h, incubated overnight with alkaline phosphatase-conjugated DIG antibody (1:2,000; Boehringer), and washed three times in PBT. Signals were visualized after exposure to NBT/BCIP solution (Boehringer) for 24 to 48 h. For immunohistochemistry, paraffin sections were cleared with xylenes and rehydrated with PBS, and endogenous peroxidases were inactivated by a 30-min incubation in 0.3% H2O2 in methanol followed by three 5-min washes in PBS. After a 30-min blocking step with 5% milk in PBST (PBS, 0.05% Tween-20), slides were incubated overnight with HDAC1 or HDAC2 antibodies (1:2,000 in blocking solution) at 4°C, washed three times in PBST, incubated with goat anti-rabbit horseradish peroxidase (1:4,000; Santa Cruz) for 60 min at room temperature, and washed. Peroxidase staining was visualized by using DAB substrate (Vector).
ChIP.
ChIP analysis was done according to a published protocol (4), with modifications. DNA-protein cross-linking was achieved by injection of 1% formaldehyde into the fetal gut lumen, incubation for 15 min at room temperature with gentle agitation, and treatment with 0.125 M glycine for 5 min to quench the reaction. Six to 10 untreated or cultured E13 intestines or 1 to 2 freshly isolated E16 guts were then washed twice in PBS, resuspended in 1 ml of cell lysis buffer (10 mM Tris-Cl [pH 8.0], 10 mM NaCl, 0.2% NP-40, and protease inhibitors), and ground with a tissue homogenizer (Corning Inc., Corning, N.Y.). Cell nuclei were lysed by incubation in 600 μl of nuclear lysis buffer (25 mM Tris-Cl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, and protease inhibitors) for 10 min, sonicated on ice to shear cross-linked chromatin to an average size of 0.5 to 1 kb, and centrifuged at 12,000 × g for 10 min. The supernatants were diluted in 20 mM Tris-Cl [pH 8.1]-150 mM NaCl-2 mM EDTA-1% Triton X-100 and precleared with a 50% slurry of protein A-Sepharose (Amersham Pharmacia) and 2 μg of herring sperm DNA for 2 h. Samples were incubated overnight with specific antibodies and then for 1 h with 50 μl of protein A-Sepharose and 2 μg of herring-sperm DNA. Immunoprecipitates were recovered by centrifugation and washed successively in RIPA buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1.0% Nonidet P-40), high-salt solution (50 mM Tris-Cl [pH 8.0], 500 mM NaCl, 0.1% SDS, 1.0% Nonidet P-40), LiCl (250 mM LiCl, 50 mM Tris-Cl [pH 8.0], 1 mM EDTA, 0.5% sodium deoxycholate, 1.0% Nonidet P-40), and 10 mM Tris-Cl [pH 8.0]-1 mM EDTA. Beads were extracted with 1% SDS-0.1 M NaHCO3, and eluates were reverse cross-linked over a 6-h period at 65°C. DNA fragments were isolated by using a DNA purification kit (QIAGEN) and amplified by PCR using the following primers (5′ to 3′) corresponding to promoter regions: Apo1a, GTGGGCTCCATGAGACTATCTT and CAGCTCTTCTTCCCTGGTCTAT; Fabpl, GTGCATTGCTGGAGATGTGATTCA and AGGTCACCCACTGTTGCCTATTTT; Fabpi, CGGAGAGCAGCGATTAAAAG and GTCCTGTCCACTAGAGAGAA; Mt2, AGGTGTCCTGGAACCGGTTC and CACGCGGAACGCGACCTTTA; histone1H4a, CATGGTTGATGGGAGGGATTTG and AATGGAGTCAGAGCTGAGAGTC; and Gapdh, CCAATGTGTCCGTCGTGGATCT and GTTGAAGTCGCAGGAGACAACC.
RESULTS AND DISCUSSION
Regulated expression of class I HDACs in mammalian fetal gut development.
In mRNA expression profiles of discrete stages in mouse intestine development, we observed that the abundance of transcripts for both HDAC1 and HDAC2 declines significantly between E13 and E15, whereas other HDAC mRNA levels remain stable (Fig. 1A). The period in question coincides with conversion of the gut endoderm from a pseudostratified squamous epithelium into a rudimentary villous structure and with activation of intestine-specific genes (40). Consistent with the changes in mRNA abundance, HDAC1 and HDAC2 protein levels decrease significantly after E15, with an expected time lag in relation to mRNA levels, and this decrease persists after birth (Fig. 1A); these trends parallel the embryo-wide reduction in HDAC1 mRNA reported previously (21). We detected no regional variation along the rostro-caudal axis of the developing alimentary tract and little to no protein expression by three different class II HDACs (HDAC4, -6, and -7) in the fetal gut (data not shown).
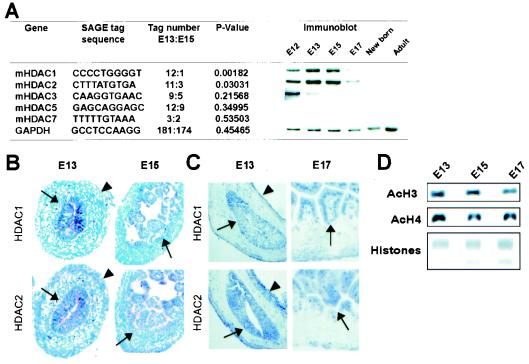
FIG. 1.
Expression of HDAC1 and HDAC2 in the developing mouse intestine. (A) Frequency of tags specific for various HDAC mRNAs in SAGE (46) in the midgestation small bowel of the mouse at E13 and E15, showing selective and significant down-regulation of HDAC1 and HDAC2 mRNAs in this interval. Over 65,000 tags were sequenced from each library, and the temporal variation in expression was confirmed and extended by immunoblot analysis. (B and C) In situ hybridization (B) and immunohistochemistry (C) reveal localized expression of the mammalian class I HDACs in the periluminal epithelium (arrows) and weak expression in outer serosal cells (arrowheads), with conspicuous exclusion from the subepithelial mesenchyme. This spatial restriction persists at later developmental stages, when longer staining permits detection of weaker signals. (D) Immunoblot analysis showing modest reduction in global acetylation of histones H3 and H4 in mouse intestine development. Loading was controlled with total histones, detected by Coomassie blue staining.
Although the functions attributed to class I HDACs are relevant to all cell types, within the developing intestine HDAC1 and HDAC2 mRNA and protein localize principally in the prospective epithelium. Both genes are expressed at lower levels in peripheral serosal cells but are excluded from the subepithelial mesenchyme (Fig. 1B and C). Importantly, the developmentally regulated decline in expression is not associated with a change in total tissue histone acetylation (Fig. 1D). Together, these observations suggest the possibility of a function for class I HDACs in regulating gut epithelial differentiation. We hypothesized that this role is to enable silencing of selected genes associated with tissue maturation until their activation is contextually appropriate in differentiating cells.
Development of a reliable ex vivo model for intestine differentiation.
To investigate the functions of class I HDACs and other factors in mammalian gut development, we first developed an ex vivo model for tissue differentiation in fetal mouse intestine organ explants. Because epithelial differentiation relies on signals that originate in the underlying mesenchyme (2, 16), we built on published studies (8, 10, 17, 35) and harvested mouse fetal intestines en bloc (Fig. 2A). Culture of gut explants in a chemically defined medium over 3 to 5 days results in organ growth and peristalsis, preserves cell viability and replication (Fig. 2B), and culminates in typical cytodifferentiation, signified by villous morphogenesis (Fig. 2C). Two features of the experimental model merit attention. First, molecular markers of epithelial differentiation appear in a manner that mimics their induction in vivo, as assessed by RT-PCR (Fig. 2D). Concomitantly, genes that are down-regulated in the course of normal gut development, including HDAC1, are similarly extinguished in the explants, although histologic and molecular maturation both occur more slowly than in vivo. Second, the intestinal lumen provides a conduit to deliver materials that might influence cell differentiation, including chemical compounds and DNA. Introducing plasmids by luminal injection and electroporation leads to robust, epithelium-restricted expression of exogenous genes, as illustrated here for GFP (Fig. 2E). Significant numbers of epithelial cells begin to express transduced GFP or GFP-tagged fusion proteins 6 to 12 h after transfection and maintain expression for 1 to 2 days (Fig. 2F and data not shown). These features enhance the value of fetal organ explant cultures to probe tissue differentiation.
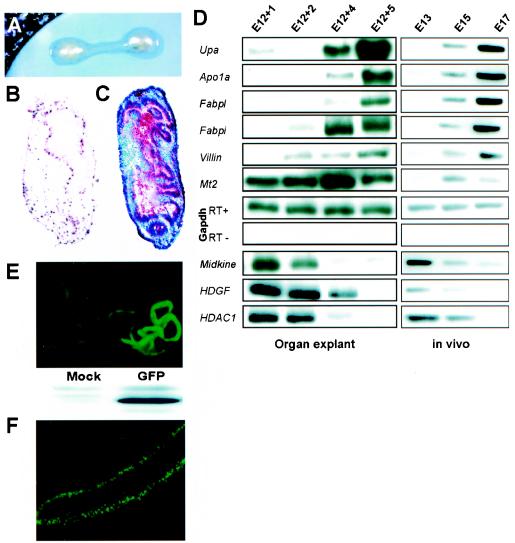
FIG. 2.
Differentiation of mouse fetal gut explants in vitro. (A) Intestines harvested from mouse embryos between E12 and E14 were placed on filter disks and cultured at the air fluid-interface in chemically defined BGJb medium. (B and C) Bromodeoxyuridine (B) and histologic (C) staining after 6 days of continuous culture of E12 intestines reveals tissue viability, persistent cell replication, and the appearance of villiform structures in the epithelium; histology at the onset of organ culture resembled the E13 squamous epithelium shown in Fig. 1B and C. (D) Expression of molecular markers of gut epithelial differentiation in fetal organ explants (left) or freshly isolated intestine (right), as assessed by RT-PCR. Apo1a, apolipoprotein 1A; Fabpi and Fabpl, intestinal and liver fatty acid-binding proteins 1 and 2, respectively; Upa, uterine-specific proline-rich acidic protein; Mt2, metallothionein 2; HDGF, hepatoma-derived growth factor; Gapdh, glyceraldehyde 3-phosphate dehydrogenase (loading control). (E) Fluorescent and immunoblot detection of GFP expressed in transfected organ explants. (F) UV micrograph showing expression of GFP-tagged HDACs in a representative E14 explant 20 h after plasmid electroporation.
Persistent HDAC expression delays expression of gut epithelial differentiation genes.
If the decline in endogenous levels of class I HDACs during intestine development is partially responsible for activating maturation-associated genes, then their forced expression may be expected to reverse or delay the process. Because HDAC1 and HDAC2 are believed to function within a common protein complex (12), we expressed both proteins simultaneously in intestines explanted from E14 mouse fetuses, when endogenous mRNA levels are declining (Fig. 1A) and before expression of differentiation markers is activated (Fig. 2D). We used plasmids that drive expression of both GFP and HDAC1 or HDAC2 by virtue of an internal ribosome entry site and observed robust epithelial GFP expression (Fig. 2F). Over 2 days of subsequent culture, mRNA levels of several intestine-specific markers were significantly reduced: Apo1a, the liver (Fabpl)- and intestine (Fabpi)-specific fatty acid-binding protein genes, Upa, and metallothionein 2 (Mt2) (Fig. 3A). Levels of the intestine- and kidney-specific villin transcript (28) are unresponsive to HDAC overexpression, indicating that class I HDACs influence differentiation genes with some selectivity. Together with the preserved expression of glyceraldehyde 3-phosphate dehydrogenase (Gapdh) mRNA, the latter result also attests to the viability of HDAC-overexpressing explants. Moreover, levels of other markers were greater after 2 days of culture than on the first day, suggesting recovery of differentiation as transgene expression is gradually extinguished.
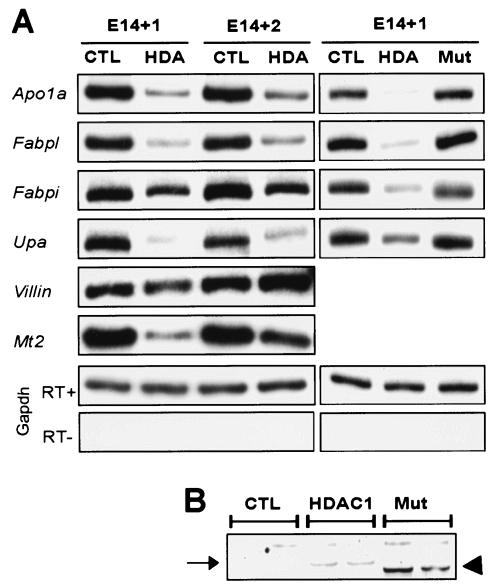
FIG. 3.
Overexpression of HDAC1 and HDAC2 cDNAs in mouse fetal gut explants delays activation of some epithelial markers. (A) RT-PCR analysis of molecular markers of gut epithelial differentiation in response to forced expression of either GFP (CTL) alone, GFP-tagged HDAC1 and HDAC2 (HDA), or mutant HDAC forms with internal deletions (Mut). Explant RNA was isolated 24 and 48 h after transfection. The deletion mutants reveal specificity of HDAC effects. (B) Immunoblot confirming that mutated HDAC1 (arrowhead) was expressed at levels comparable to those of the full-length protein (arrow).
To control for the specificity of these effects, we tested constructs in which a portion of the deacetylase domain of each HDAC is deleted. Unlike the full-length HDACs, these constructs have no effect on expression of differentiation-related genes (Fig. 3A), even though levels of the truncated protein are equivalent or greater than that of the full-length protein (Fig. 3B). Thus, overexpression of class I HDACs during gut development is sufficient to impair expression of cell differentiation markers; the high HDAC levels we observed in the fetal intestine may serve in this role prior to the endoderm-epithelial transition.
HDAC inhibitors accelerate gut epithelial differentiation.
These results suggest that pharmacologic inhibition of HDAC would induce premature differentiation. To examine this possibility, we treated E13 gut explants with HDAC antagonists. To reduce the risks of nonspecific effects, we applied two chemically distinct inhibitors, SB and VPA, in separate experiments; these compounds target all or only class I HDACs, respectively (11, 33, 38). Both agents caused the expected increase in acetylation of histone substrates (Fig. 4A and data not shown), and VPA specifically reduced endogenous HDAC2 levels (Fig. 4A), as predicted (20). Both compounds accelerate induction of several intestine-specific markers that are inhibited upon HDAC overexpression (Fig. 4B). Apo1a, Fabpl, and Fabpi mRNA levels, especially, were increased within 2 days of culture compared to the levels seen in mock-treated explants after 2 or 3 days. Increases in mRNA concentrations of two other markers, Upa and Mt2, were less dramatic and more variable, whereas villin levels were consistently unaffected by either HDAC inhibitor. Besides inducing expression of certain molecular markers, VPA also accelerates cytodifferentiation; a villiform epithelium is evident earlier with compound exposure than in mock-treated explants (Fig. 4C). The effects of both HDAC antagonists on Apo1a and other mRNA levels varies in proportion to drug concentrations (Fig. 4D and data not shown).
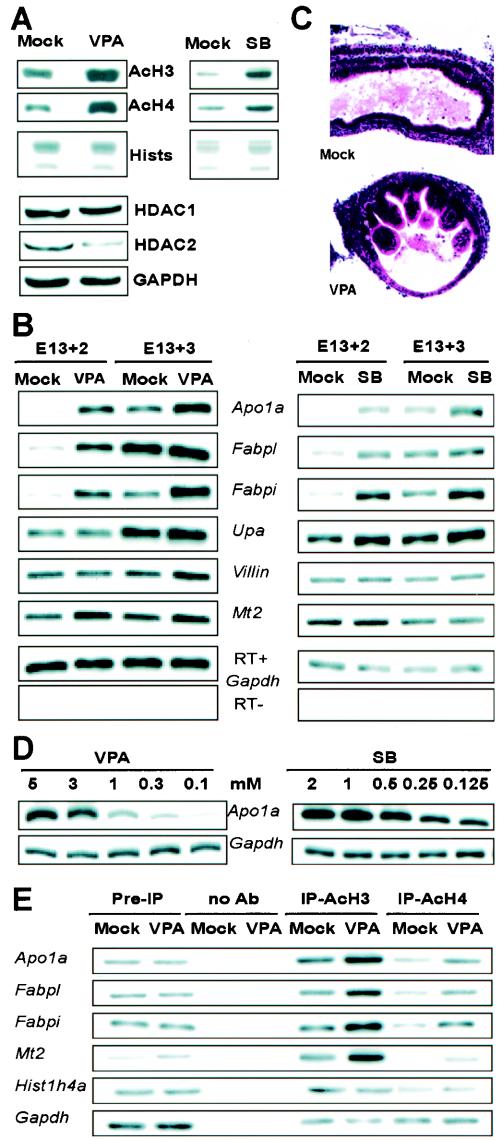
FIG. 4.
Chemical HDAC inhibitors induce epithelial differentiation in mouse fetal gut explants. (A) Immunoblots showing hyperacetylation of histones H3 and H4 in E13 explants treated with 3 mM VPA or 2 mM SB for 24 h and reduced HDAC2 levels in response to VPA. (B) Effects of VPA and SB on expression of molecular markers of epithelial differentiation, as analyzed by RT-PCR, in E13 gut explants cultured for 48 or 72 h after chemical exposure. Apo1a, apolipoprotein 1A; Fabpi and Fabpl, intestinal and liver fatty acid-binding proteins 1 and 2, respectively; Upa, uterine-specific proline-rich acidic protein; Mt2, metallothionein 2; HDGF, hepatoma-derived growth factor;Gapdh, glyceraldehyde 3-phosphate dehydrogenase (loading control). Each lane includes two pooled explants, and the results are representative of five independent experiments. (C) Accelerated appearance of villiform epithelium in E13 gut explants treated with VPA and cultured for 4 days. (D) Dose-dependent induction of the representative differentiation marker Apo1a by VPA and SB. (E) ChIP analysis shows increased association of acetylated histones H3 and H4 with promoters of the Apo1a, Fabpl, Fabpi, and Mt2 genes following injection of 3 mM VPA into the E13 gut lumen and 24-h culture. PCR was performed with a radiotracer, and input DNA for PCR (Pre-IP) was 1/50 the amount used for immunoprecipitation; chromatin subjected to the same treatment without antibody served as the negative control. Each lane contains samples pooled from six to seven gut explants, and the results are representative of three separate experiments with identical results.
Whereas HDAC inhibitors can induce differentiation of various cell types (11, 30), our results in the developing gut are notable for two reasons. First, SB and VPA induce maturation markers whose native activation coincides with declining endogenous HDAC levels, thus supporting the idea of a physiologic function for this decline. Second, our panel of maturation markers reveals different degrees of sensitivity to the activity of the target enzymes, just as HDAC inactivation in another context altered expression of a remarkably small fraction of cellular genes (45).
Besides core histone proteins, HDACs regulate other substrates, including transcription factors and cytoskeletal components (15, 24, 25, 27, 32, 34). Accordingly, there are several mechanisms by which HDACs could influence gene expression in the developing intestine. In consideration of the most direct mechanism, we performed ChIP of E13 fetal gut explants 24 h after exposure to VPA. After cross-linking chromatin complexes, we precipitated with antibodies specific for acetylated forms of histones H3 and H4 and conducted PCR analysis for the 5′-flanking regions of four genes among the induced differentiation markers. All four putative promoters show greater representation in VPA-treated than in mock-treated organ explants (Fig. 4E), whereas control reactions for the housekeeping histone1H4a and Gapdh gene promoters reveal no such difference. These results demonstrate a correlation between the activation of intestinal differentiation-related genes and acetylation of core histone proteins at their promoters.
Direct observation of HDAC effects at differentiation-related gene promoters.
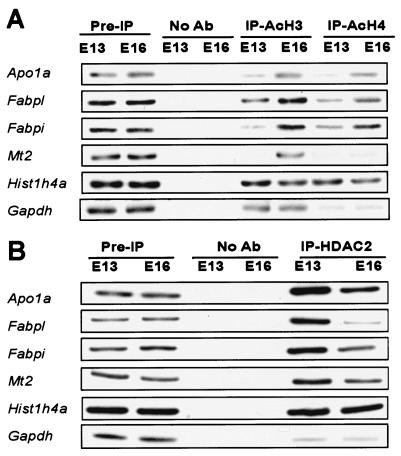
The foregoing observations imply that endogenous levels of class I HDACs are functionally relevant to development of the mammalian intestine. If these enzymes operate directly at differentiation-related gene loci, then promoters targeted by HDAC regulation in gut development should exhibit two features: (i) preferential association with class I HDACs prior to the endoderm-epithelial transition and (ii) histone hyperacetylation after the villous transformation. To test these predictions, we performed ChIP on freshly isolated mouse fetal intestines. The 5′-flanking regions from the differentiation-related genes Apo1a, Fabpl, Fabpi, and Mt2 each reveals greater acetylation of histones H3 and H4 at E16 compared to that for E13 (Fig. 5A). Although we could not amplify villin promoter sequences from the same immunoprecipitated samples, the histone1H4a and Gapdh promoters provided essential controls. Furthermore, a specific antibody revealed physical association of HDAC2 with the promoters of the same genes at E13 (Fig. 5B). Consistent with the greatly reduced levels of HDAC2 after E15, a similar chromatin complex is found at comparably lower levels in the E16 intestine. Thus, proximity with an Rpd3-related HDAC complex early in mammalian gut development is associated with histone hypoacetylation at differentiation loci. Our data suggest that this modification contributes toward a repressive transcriptional state that is reversed when endogenous HDAC levels subsequently decline (Fig. 6).
FIG. 5.
Association of acetylated (Ac) histones H3 and H4 with promoters of the Apo1a, Fabpl, Fabpi, and Mt2 genes increases during gut development. (A) E13 and E16 intestines were cross-linked with 1% formaldehyde and chromatin immunoprecipitates were analyzed by PCR with primers flanking the promoter region of each differentiation-related or control (Histone1h4a and Gapdh) gene. Other controls are similar to those used for Fig. 4. (B) Conversely, HDAC2 is found in physical association with the same gene promoters at higher levels in the gut at E13 than at E16. Ab, antibody; IP, immunoprecipitate.
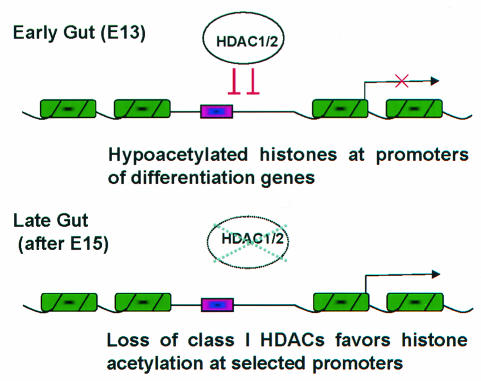
FIG. 6.
Model for regulation of differentiation gene loci in the mammalian gut. Chromatin is represented in green and promoters are represented in pink; the arrow denotes active transcription. Early in intestine development, the abundance of class I HDACs limits histone acetylation at selected differentiation loci. Subsequently, reduced cellular levels of these enzymes favor local chromatin modifications that promote gene transcription. Thus, modification of endogenous class I HDAC levels is likely one of several mechanisms that impose tight temporal control over developmental gene activation.
Implications for mechanisms of tissue differentiation.
Our studies highlight a mechanism that developing tissues may deploy broadly to impose temporal control over gene expression. Early in the genesis of the alimentary tract and possibly of other organs, the abundance of class I HDACs limits histone acetylation at selected loci, whereas subsequent reductions in cellular levels of these enzymes favor local chromatin modifications that promote transcription. Endogenous enzyme levels thus appear to be limiting for gene regulation during development. This model is analogous to one in which rhythmic histone acetylation is a component of the mammalian circadian clock (9), although the latter occurs cyclically over a time scale of hours and developmental regulation occurs noncyclically over a longer period. The relative contributions of acetyltransferases and HDACs in both cases remain unclear, although the inverse correlation between class I HDAC levels and histone acetylation at intestinal differentiation loci hints at the importance of this group of enzymes.
Our results do not, however, exclude the possibility of additional, nonhistone targets for protein deacetylation among the many potential substrates, including transcription factors (24, 25, 27, 32, 34). Because only a few transcriptional regulators of intestinal genes are known, we have not asked whether attenuation of endogenous class I HDACs affects tissue-specific gene expression through multiple, distinct substrates. Histone hyperacetylation is also associated with increased cellular levels of p21WAF1/Cip1 and G1 cell cycle arrest (36), which suggests yet other mechanisms to facilitate differentiation. Our data do, however, indicate that direct modification of promoter-associated histones at differentiation gene loci is one facet of the likely complex process whereby lineage-specific gene expression is initiated during development.
In this study, tissue-restricted genes exhibited differential sensitivity toward HDAC activity, much as remarkably few (≤2%) transcripts respond to the treatment of cultured cells with HDAC inhibitors (45). Furthermore, developmentally regulated decay of class I HDAC expression in the intestine has minor consequences on global histone acetylation. These findings agree with the idea that the major portion of mammalian genomes contains chromatin that is constitutively restrictive for transcription through several possible mechanisms. In contrast, some differentiation markers, represented in the developing mouse intestine by Fabpi, Fabpl, and Apo1a, are particular targets for developmental modulation of class I HDAC levels. This mode of regulation likely acts in concert with ATP-dependent chromatin remodeling, other epigenetic changes, and the presence of key transcription factors to specify the developmental stage and cellular compartment in which tissue-restricted genes are activated.
Acknowledgments
We thank Saverio Minucci, Scott Heller, Jacob Hecksher-Sorensen, Maina Lepourcelet, and Sanjay Tiwari for technical advice and helpful discussions; Ken Kinzler for permission to use SAGE and analytic software; Tao Wang and David Rowitch for assistance with tissue electroporation; Andy McMahon and Allen Everett for plasmids; and Myles Brown, Jeremy Green, and Yoshihiro Nakatani for comments on the manuscript.
This study was supported by a fellowship from the Robert Black Charitable Foundation and grant R01DK61139 from the National Institutes of Health. R.A.S. is a Scholar of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 2.Birchmeier, C., and W. Birchmeier. 1993. Molecular aspects of mesenchymal-epithelial interactions. Annu. Rev. Cell Biol. 9:511-540. [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y. L., Y. H. Peng, I. C. Pan, D. S. Sun, B. King, and D. H. Huang. 2001. Essential role of Drosophila Hdac1 in homeotic gene silencing. Proc. Natl. Acad. Sci. USA 98:9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79-82. [DOI] [PubMed] [Google Scholar]
- 5.Deckert, J., and K. Struhl. 2002. Targeted recruitment of Rpd3 histone deacetylase represses transcription by inhibiting recruitment of Swi/Snf, SAGA, and TATA binding protein. Mol. Cell. Biol. 22:6458-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrov, S., G. Almouzni, M. Dasso, and A. P. Wolffe. 1993. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev. Biol. 160:214-227. [DOI] [PubMed] [Google Scholar]
- 7.Dufourcq, P., M. Victor, F. Gay, D. Calvo, J. Hodgkin, and Y. Shi. 2002. Functional requirement for histone deacetylase 1 in Caenorhabditis elegans gonadogenesis. Mol. Cell. Biol. 22:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duh, G., N. Mouri, D. Warburton, and D. W. Thomas. 2000. EGF regulates early embryonic mouse gut development in chemically defined organ culture. Pediatr. Res. 48:794-802. [DOI] [PubMed] [Google Scholar]
- 9.Etchegaray, J. P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421:177-182. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda, K., N. Sakamoto, T. Narita, K. Saitoh, T. Kameda, H. Iba, and S. Yasugi. 2000. Application of efficient and specific gene transfer systems and organ culture techniques for the elucidation of mechanisms of epithelial-mesenchymal interaction in the developing gut. Dev. Growth Differ. 42:207-211. [DOI] [PubMed] [Google Scholar]
- 11.Gottlicher, M., S. Minucci, P. Zhu, O. H. Kramer, A. Schimpf, S. Giavara, J. P. Sleeman, F. Lo Coco, C. Nervi, P. G. Pelicci, and T. Heinzel. 2001. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20:6969-6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassig, C. A., J. K. Tong, T. C. Fleischer, T. Owa, P. G. Grable, D. E. Ayer, and S. L. Schreiber. 1998. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proc. Natl. Acad. Sci. USA 95:3519-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 14.Huang, E. Y., J. Zhang, E. A. Miska, M. G. Guenther, T. Kouzarides, and M. A. Lazar. 2000. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 14:45-54. [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbert, C., A. Guardiola, R. Shao, Y. Kawaguchi, A. Ito, A. Nixon, M. Yoshida, X. F. Wang, and T. P. Yao. 2002. HDAC6 is a microtubule-associated deacetylase. Natur. 417:455-458. [DOI] [PubMed] [Google Scholar]
- 16.Kedinger, M., I. Duluc, C. Fritsch, O. Lorentz, M. Plateroti, and J. N. Freund. 1998. Intestinal epithelial-mesenchymal cell interactions. Ann. N. Y. Acad. Sci. 859:1-17. [DOI] [PubMed] [Google Scholar]
- 17.Kedinger, M., P. M. Simon-Assmann, B. Lacroix, A. Marxer, H. P. Hauri, and K. Haffen. 1986. Fetal gut mesenchyme induces differentiation of cultured intestinal endodermal and crypt cells. Dev. Biol. 113:474-483. [DOI] [PubMed] [Google Scholar]
- 18.Khochbin, S., A. Verdel, C. Lemercier, and D. Seigneurin-Berny. 2001. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides, T. 1999. Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev. 9:40-48. [DOI] [PubMed] [Google Scholar]
- 20.Kramer, O. H., P. Zhu, H. P. Ostendorff, M. Golebiewski, J. Tiefenbach, M. A. Peters, B. Brill, B. Groner, I. Bach, T. Heinzel, and M. Gottlicher. 2003. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22:3411-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagger, G., D. O'Carroll, M. Rembold, H. Khier, J. Tischler, G. Weitzer, B. Schuettengruber, C. Hauser, R. Brunmeir, T. Jenuwein, and C. Seiser. 2002. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 21:2672-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 23.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73-84. [DOI] [PubMed] [Google Scholar]
- 24.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 25.Mal, A., M. Sturniolo, R. L. Schiltz, M. K. Ghosh, and M. L. Harter. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mannervik, M., and M. Levine. 1999. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc. Natl. Acad. Sci. USA 96:6797-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maunoury, R., S. Robine, E. Pringault, C. Huet, J. L. Guenet, J. A. Gaillard, and D. Louvard. 1988. Villin expression in the visceral endoderm and in the gut anlage during early mouse embryogenesis. EMBO J. 7:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 30.Munster, P. N., T. Troso-Sandoval, N. Rosen, R. Rifkind, P. A. Marks, and V. M. Richon. 2001. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 61:8492-8497. [PubMed] [Google Scholar]
- 31.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 32.Ozawa, Y., M. Towatari, S. Tsuzuki, F. Hayakawa, T. Maeda, Y. Miyata, M. Tanimoto, and H. Saito. 2001. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 98:2116-2123. [DOI] [PubMed] [Google Scholar]
- 33.Phiel, C. J., F. Zhang, E. Y. Huang, M. G. Guenther, M. A. Lazar, and P. S. Klein. 2001. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276:36734-36741. [DOI] [PubMed] [Google Scholar]
- 34.Puri, P. L., S. Iezzi, P. Stiegler, T. T. Chen, R. L. Schiltz, G. E. Muscat, A. Giordano, L. Kedes, J. Y. Wang, and V. Sartorelli. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8:885-897. [DOI] [PubMed] [Google Scholar]
- 35.Quaroni, A. 1985. Development of fetal rat intestine in organ and monolayer culture. J. Cell Biol. 100:1611-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richon, V. M., T. W. Sandhoff, R. A. Rifkind, and P. A. Marks. 2000. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 97:10014-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 38.Sealy, L., and R. Chalkley. 1978. The effect of sodium butyrate on histone modification. Cel. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., and C. Mello. 1998. A CBP/p300 homolog specifies multiple differentiation pathways in Caenorhabditis elegans. Genes Dev. 12:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, T. C., and J. I. Gordon. 1995. Intestinal epithelial cell differentiation: new insights from mice, flies and nematodes. Curr. Opin. Genet. Dev. 5:577-586. [DOI] [PubMed] [Google Scholar]
- 41.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 42.Taplick, J., V. Kurtev, K. Kroboth, M. Posch, T. Lechner, and C. Seiser. 2001. Homo-oligomerisation and nuclear localisation of mouse histone deacetylase 1. J. Mol. Biol. 308:27-38. [DOI] [PubMed] [Google Scholar]
- 43.Taplick, J., V. Kurtev, G. Lagger, and C. Seiser. 1998. Histone H4 acetylation during interleukin-2 stimulation of mouse T cells. FEBS Lett. 436:349-352. [DOI] [PubMed] [Google Scholar]
- 44.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 45.Van Lint, C., S. Emilaiani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-254. [PMC free article] [PubMed] [Google Scholar]
- 46.Velculescu, V. E., L. Zhang, B. Vogelstein, and K. W. Kinzler. 1995. Serial analysis of gene expression. Science 270:484-487. [DOI] [PubMed] [Google Scholar]
- 47.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]