Abstract
MBD1 is a vertebrate methyl-CpG binding domain protein (MBD) that can bring about repression of methylated promoter DNA sequences. Like other MBD proteins, MBD1 localizes to nuclear foci that in mice are rich in methyl-CpG. In methyl-CpG-deficient mouse cells, however, Mbd1 remains localized to heterochromatic foci whereas other MBD proteins become dispersed in the nucleus. We find that Mbd1a, a major mouse isoform, contains a CXXC domain (CXXC-3) that binds specifically to nonmethylated CpG, suggesting an explanation for methylation-independent localization. Transfection studies demonstrate that the CXXC-3 domain indeed targets nonmethylated CpG sites in vivo. Repression of nonmethylated reporter genes depends on the CXXC-3 domain, whereas repression of methylated reporters requires the MBD. Our findings indicate that MBD1 can interpret the CpG dinucleotide as a repressive signal in vivo regardless of its methylation status.
Cytosine methylation is the major DNA modification in eukaryotes. In vertebrates it is found almost exclusively in the 5′ CpG context, where it adds epigenetic information to the genomic DNA sequence and can function to maintain stable gene silencing through mitotic cell divisions. In genetic approaches, DNA methylation has been shown to be essential for normal development in both mice and frogs (21, 29, 33). Most CpGs are methylated in mammals, and (due to the mutagenic nature of 5-methylcytosine) the dinucleotide is much underrepresented in mammalian genomes (3). Exceptions to this rule are the CpG islands, short CG-rich regions that are found at the promoter of 60% of human genes, including housekeeping genes and some genes that show a tissue-specific expression profile (2). Most CpG island promoters remain nonmethylated irrespective of expression state, but in cases in which methylation occurs somatically (e.g., at imprinted genes and on the inactive X chromosome), the associated gene is silenced. Aberrant gene silencing in tumors via CpG island methylation is also well documented (1).
The effect of DNA methylation is due in part to structural alteration of DNA, which prevents some transcription factors from binding to their cognate sequences. In addition, DNA methylation affects chromatin structure (9) due to recruitment of corepressors and chromatin-modifying activities by proteins that bind specifically to methylated DNA (24, 27, 30, 37). Most of the methyl-DNA binding proteins described so far belong to the MBD family, defined by the methyl-CpG binding domain (MBD; pfam01429). Proteins containing regions with homology to the MBD have been identified in plants and animals, but not every domain has the ability to bind methylated DNA (17). MBD1 is a member of the subfamily of MBD proteins that do bind methylated CpG, which in mammals also includes MeCP2, MBD2, and MBD4. MBD1 orthologues have been identified in a range of vertebrates (Fugu rubripes, Xenopus laevis, and various mammals) but are apparently absent from invertebrates, including the chordate Ciona intestinalis (17). The Mbd1 protein is not essential for mouse development, but Mbd1 null animals have defects in adult neurogenesis and show elevated aneuploidy in neurons (41).
In keeping with the silencing effect of DNA methylation, the three MBD proteins MBD1, MBD2, and MeCP2 are all transcriptional repressors (36). A transcriptional repression domain (TRD) that represses transcription when fused to the Gal4 DNA-binding domain has been identified at the C terminus of mammalian MBD1 (26). Unlike MBD2 and MeCP2, MBD1 does not appear to interact with histone deacetylase 1 (HDAC1) or HDAC2 (26). Also, repression by the TRD is variably sensitive to trichostatin A, indicating that histone deacetylase activity is not consistently involved (12, 26). Recently, yeast two-hybrid screening has revealed several protein-binding partners for MBD1. The histone methyltransferase enzyme SUV39h1 and p150, a subunit of CAF-1 (chromatin assembly factor 1), both interact with the MBD (13, 31). MBP, a methylpurine-DNA glycosylase (39), and MCAF (MBD1-containing chromatin-associated factor; also named mAM) (12) both bind to the TRD of MBD1 and appear to act as corepressors of transcription. Interestingly, MCAF/mAM is a cofactor for the histone methyltransferase ESET, which causes transcriptional repression by trimethylating dimethylated lysine 9 of histone H3 (38).
Uniquely among MBD proteins, MBD1 has three zinc-coordinating CXXC domains (zf-CXXC; pfam02008) (8, 11). Previous work established that the third CXXC domain of MBD1 (CXXC-3) is differentially spliced in both humans and mice and that splice variants that contain CXXC-3 show DNA methylation-independent repression (11). Moreover, MBD1 was independently cloned as a protein that binds using the CXXC-3 domain to a promoter element of the fibroblast growth factor 2 (FGF2) promoter (34). CXXC domains are also found in other chromatin-associated proteins, including DNA methyltransferase 1 (DNMT1), mixed lineage leukemia (MLL), and CpG binding protein (CGBP). In the cases of CGBP and MLL, DNA sequence-specific binding has been shown in vitro for the CXXC domains, both of which require nonmethylated CpG sites (4, 20, 35). Here we establish that the CXXC-3 in the methyl-CG binding protein Mbd1 also binds specifically to nonmethylated CpGs in vitro. We use an in vivo assay to show that this CXXC domain can target nonmethylated CpGs in living cells and can direct repression of nonmethylated reporter genes by Mbd1.
MATERIALS AND METHODS
Cell lines and transfection.
DNA methylation-deficient Dnmt1n/n (21), p53−/−, and control p53−/− Dnmt1+/+ mouse embryonic fibroblast lines were derived from embryonic day 9.5 embryos (I. Ben-Porath and H. Cedar, unpublished data) and maintained in Dulbecco's modified Eagle's medium (Gibco) supplemented with 15% bovine calf serum, nonessential amino acids, sodium pyruvate, and antibiotics (Gibco). The presence of a homozygous p53 mutation allows survival of Dnmt1-deficient cells (18). In all mouse cell experiments described here, both control Dnmt1+/+ and DNA methylation-deficient Dnmt1n/n cells were p53−/−. HeLa cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% bovine calf serum and antibiotics (Gibco). Cells were transfected using JetPEI (QBiogene) according to the manufacturer's instructions. For reporter assays, cells were transfected in quadruplicate. Three wells were analyzed for luciferase activity with luciferase (firefly or Renilla) reporter assays (Promega), and one well was analyzed for expression with Western blotting using standard procedures.
Plasmids and recombinant protein.
Murine Mbd1a cDNA was cloned into pCMV-Flag-2 (Sigma) to produce pFlag-mMbd1a. pFlag-mMbd1c was constructed by replacing the 3′ end of the Mbd1a cDNA with the coding sequence of the alternative 3′ end found by 3′ rapid amplification of cDNA ends (RACE). pFlag-mMbd1b and pFlag-mMbd1d were produced by removing exon 10 from pFlag-mMbd1a and pFlag-mMbd1c, respectively. pcDNA3 constructs containing untagged Mbd1a to Mbd1d cDNAs were in vitro transcribed and translated using a TNT kit (Promega) according to the manufacturer's instructions. The Mbd1a sequence corresponding to amino acids 1 to 75, 171 to 297, or 349 to 439 was cloned into pET30b+ (Novagen) to produce bacterial expression constructs for His-tagged MBD, CXXC-1/2, or CXXC-3 peptides, respectively. Point mutations corresponding to R22A (in Mbd1a-R22A and the MBD-R22A peptide) and C356A (in the CXXC-C356A peptide) (see Fig. 3) were introduced using QuikChange protocol (Stratagene). Recombinant peptides were purified from 250 ml of induced BL21(DE3) cultures on Ni-nitrilotriacetic acid agarose (Qiagen) using the manufacturer's instructions. Gal4-DBD-MBD1 expression constructs were described previously (26). pGL-1.2FGF2 was kindly provided by F. Gage (University of California, Los Angeles). pRL-TK (which expresses the luciferase gene under a thymidine kinase [TK] gene promoter) and pGL2 (which expresses the firefly luciferase gene under control of the simian virus 40 promoter and enhancer) were from Promega.
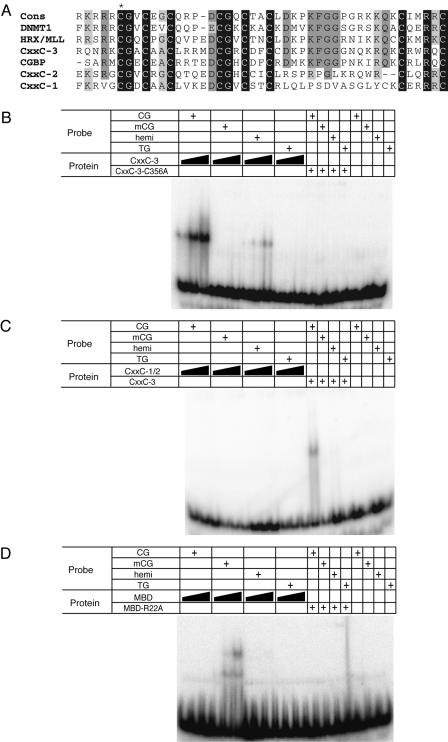
FIG. 3.
The CXXC-3 domain of Mbd1 recognizes nonmethylated CpG. (A) Alignment of the three CXXC domains (CXXC-1, CXXC-2, and CXXC-3) from Mbd1 with the CXXC domains from DNMT1, MLL, and CGBP. The consensus sequence of the domain is given above. An asterisk marks cysteine C356 that is mutated in the CXXC-3-C356A peptide. (B and C) Band shifts with probes containing nonmethylated (CG), methylated (mCG), or hemimethylated CG or a probe lacking CG entirely (TG). (B) Increasing amounts (50, 250, or 750 ng) of recombinant wild-type CXXC-3 (CxxC-3) or mutant CXXC-3 (CxxC-3-C356A; 750 ng) peptide were added. (C) Increasing amounts (100, 500, or 1,000 ng) of a peptide spanning CXXC-1 and CXXC-2 (CxxC-1/2) or CXXC-3 (250 ng) were added. (D) Increasing amounts (50, 200, or 500 ng) of recombinant wild-type (MBD) or mutant (MBD-R22A; 500 ng) MBD peptide were added.
Band shifts.
Binding reaction mixtures including 0 to 750 ng of purified His-CXXC-3, 0 to 1,000 ng of purified His-CXXC-1/2, or 0 to 500 ng of purified His-MBD in 6 mM Tris-Cl (pH 7.5)-6 mM MgCl2-3% glycerol-1 mM dithiothreitol-150 mM KCl-0.05 μg of poly(dAdT)/μl (Sigma) were preincubated for 10 min at room temperature before the addition of 25 fmol of end-labeled double-stranded oligonucleotides. After a further 25 min of incubation at room temperature, the reaction mixtures were loaded onto 6% polyacrylamide-0.5× Tris-borate-EDTA gels and run 2 h at 240 V at 4°C. Gels were dried onto 3-mm Whatman paper, and radioactivity was detected using phosphor screening and a Storm 840 apparatus. The oligonucleotides used for band shifting were as follows: m0-f (GTAGGCGGTGCTACACGGTTCCTGAAGTG) and m0-r (CACTTCAGGAACCGTGTAGCACCGCCTAC), m2-f (GTAGGMGGTGCTACAMGGTTCCTGAAGTG) and m2-r (CACTTCAGGAACMGTGTAGCACMGCCTAC), and TG-f (GTAGGTGGTGCTACATGGTTCCTGAAGTG) and TG-r (CACTTCAGGAACCATGTAGCACCACCTAC). Oligonucleotides were annealed in 10 mM Tris-Cl (pH 8)-1 mM EDTA-50 mM NaCl as follows: for CG, m0-r and m0-f were annealed; for mCG, m2-r and m2-f were annealed; for TG, TG-f and TG-r were annealed; and for hemimethylated (hemi) CG, m0-r and m2-f were annealed.
Immunostaining and Western blotting.
Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature followed by permeabilization in 0.2% Triton X-100-PBS. Slides were blocked in 3% bovine serum albumin-PBS before incubation with primary antibody (anti-Flag M2 [Stratagene], anti-Gal4 [Santa Cruz], or anti-Mbd1 [M245] [Santa Cruz] [all at 1:1,000]) for 60 min. Following washing and incubation with secondary antibody (anti-mouse 594 or anti-rabbit 594; Molecular Probes) (1:1,000), slides were washed and mounted in Vector shield with DAPI (4′,6′diamidino-2-phenylindole) (Vector). Images were obtained using a Zeiss microscope fitted with a charge-coupled device camera and processed using Adobe PhotoShop. For Western blotting, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a PROTRAN membrane (Schleicher and Schuell). Blots were blocked in a 5% milk-Tris-buffered saline-T solution, incubated with primary antibody followed by horseradish peroxidase-conjugated secondary antibody (anti-rabbit or anti-mouse; Amersham), and visualized using enhanced chemiluminescence.
RACE and RT-PCR.
3′ RACE was performed using Marathon-Ready cDNA from mouse brain (Clontech) and the Mbd1-specific primer (5′-CCACGCTGCAGTCTGGCTTCCCTAGC) according to the manufacturer's description. For reverse transcription-PCR (RT-PCR), TRI reagent (Sigma) was used to purify total RNA from mouse embryonic fibroblasts. RNA (1 μg) was DNase treated (RQ-DNase; Promega) and reverse transcribed in a 25-μl reaction mixture containing 200 ng of random hexamer primers and 1 U of Moloney murine leukemia virus reverse transcriptase (Promega). Aliquots (1 μl) of the RT reaction mixture were used for PCR.
Southern blotting.
Genomic DNA (5 μg) from mouse embryonic fibroblasts (p53−/− Dnmt1+/+, p53−/−, and Dnmt1n/n) was digested with TaiI (Helena Biosciences), resolved on a 1.2% agarose gel, and blotted onto Hybond-N membranes. The blot was hybridized with a major satellite probe produced by PCR using the primers mouse sat-for (CTGTAGGACGTGGAATATGGC) and mouse sat-rev (CCGTGATTTTCAGTTTTCTCGC) and washed using standard procedures. The membrane was exposed to a phosphor screen and analyzed on a Storm 840 apparatus.
RESULTS
Mbd1 splice variants in mouse cells.
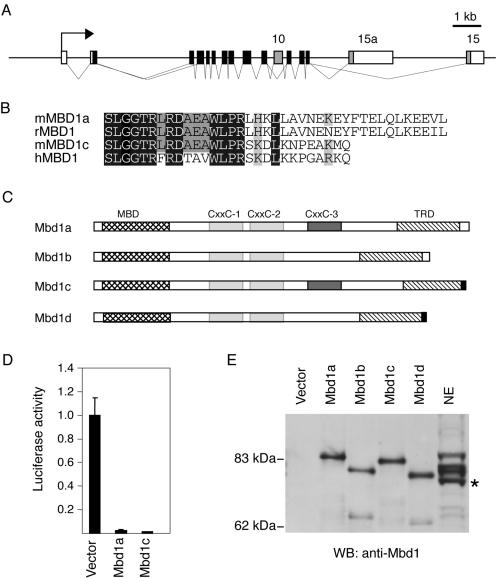
Several different human MBD1 splice variants have been described previously (7, 11), whereas at present only two Mbd1-encoding transcripts (corresponding to Mbd1a and Mbd1b) (Fig. 1C) have been identified in the mouse (16). These two mouse variants differ only in the presence or absence of exon 10, which encodes CXXC-3. A transcript lacking exon 3 has been described previously (16), but the presence of this transcript gives rise to a frameshift; hence, no Mbd1 protein is produced. Anti-Mbd1 antibodies directed to the C terminus of the murine protein specifically recognize three polypeptides in mouse nuclear extracts that have different mobilities. All bands were unaffected by phosphatase treatment (data not shown), indicating that they do not represent phosphorylated forms. To search for additional isoforms in the mouse, we performed 3′ RACE with cDNA from murine brain where Mbd1 is highly expressed (41). This identified a novel Mbd1 transcript with an alternative 3′ end (exon 15a) (Fig. 1A) encoding the C-terminal sequence KNPEAKMQ, which is very similar to the C terminus of human MBD1v1 (11) (Fig. 1B). We named splice isoforms carrying this 3′ end Mbd1c and Mbd1d (Fig. 1C). Transient expression of Mbd1c caused repression of a methylated reporter, indicating that the function of the TRD is not disrupted by the presence of the alternative C terminus (Fig. 1D). The presence of the alternative C terminus in brain and cultured murine fibroblasts was verified by RT-PCR (data not shown). In combination with the differential use of exon 10, there are four possible Mbd1 transcripts (Fig. 1C). To assign the bands present on an anti-Mbd1 Western blot, we in vitro translated the untagged splice variants and ran them alongside nuclear extracts (Fig. 1E). The three Mbd1-specific bands comigrate with Mbd1a, Mbd1b, and Mbd1d, suggesting that these are the three Mbd1 species found in mouse cells. Transcripts that contained exon 15a in combination with exon 10 (Mbd1c) were not detected by either Western blotting or RT-PCR (data not shown). Although both Mbd1a/b and Mbd1d type C termini are found in bovine expressed sequence tags (GenBank AW479704 and BE750602), there are no reports of the Mbd1a/b C-terminal isoform among more than 16 human expressed sequence tags, suggesting that it might be absent or a minor form in Homo sapiens.
FIG. 1.
Mbd1 isoforms in mouse. (A) Schematic of the murine Mbd1 locus showing the differentially spliced exons (exons 10, 15, and 15a) shaded in gray. (B) Alignment of the C termini of the predicted MBD1 proteins from the human MBD1v1 (hMBD1), rat MBD1 (rMBD1), and murine Mbd1a/b (mMBD1a) and Mbd1c/d (mMBD1c) cDNA. (C) Schematic representation of the predicted Mbd1 proteins in mouse. CXXC domains 1 and 2 are shown shaded in light grey; the differentially spliced CXXC-3 domain is shaded in dark gray. The MBD (cross-hatched) and TRD (hatched) are indicated, and the novel alternative C terminus is shaded in black. (D) Mbd1a and Mbd1c isoforms both repress a methylated reporter gene. HeLa cells were transfected with the methylated pGL2 reporter and expression vectors for Mbd1a or Mbd1c. Means and standard deviations of the luciferase activity of triplicate transfections from a representative experiment are shown. The value for the activity without an Mbd1 expression vector is arbitrarily set to 1. (E) A Western blot (WB) of in vitro transcription and translation reactions of the possible splice variants alongside a sample of nuclear extract (NE) from murine fibroblasts. Bands in the NE lane are somewhat bowed due to the high protein concentration and are therefore aligned at their leading edges. The asterisk marks a cross-reacting band that is not Mbd1.
Localization of Mbd1 in cells lacking DNA methylation.
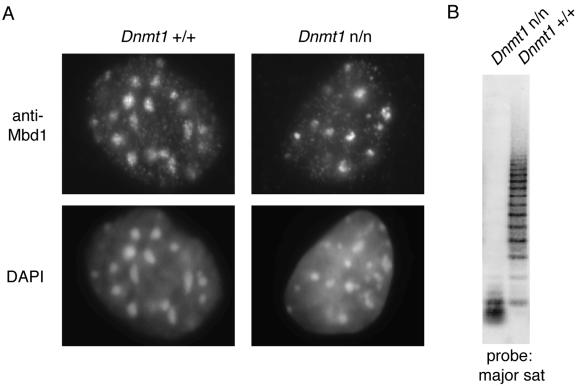
A functional MBD directs MBD proteins to the heterochromatic regions of mouse cells that are brightly stained with DAPI (16, 25). These foci contain the pericentromeric major satellite repeat that carries about half of all methylated CpGs in the mouse genome. Cells deficient in the DNA methyltransferase Dnmt1 lack methylation at these CpG-rich foci, and Mbd2, Mecp2, and Mbd4 fail to localize in these cells. It was noted previously, however, that Mbd1 still localizes to the DAPI bright foci even in mouse Dnmt1n/n embryonic stem cells (16). We revisited this finding with Dnmt1n/n mouse embryonic fibroblasts that have less than 5% of the normal level of DNA methylation (Ben-Porath and Cedar, unpublished). These cells have (as expected) very low levels of DNA methylation in the mouse major satellite, as demonstrated by Southern blotting results (Fig. 2B), but retain heterochromatin. Previously, it was shown that the speckled localization of Mecp2 and Mbd2 is lost in DNA methylation-deficient cells (16, 25). Mbd1, on the other hand, localizes to the heterochromatic foci in both the Dnmt1 wild-type and mutant cells (Fig. 2A), confirming that the protein is targeted to heterochromatic foci in spite of their unmethylated status.
FIG. 2.
Localization of endogenous Mbd1 in DNA methylation-deficient cells. (A) Immunocytochemistry of Dnmt1+/+ (left panels) or Dnmt1n/n (right panels) cells as determined using anti-Mbd1 antibodies (top panels). DNA was counter stained using DAPI (bottom panels). (B) Southern blot of DNA from Dnmt1+/+ and Dnmt1n/n cells. TaiI-digested genomic DNA was blotted and probed with a major satellite (sat) sequence. TaiI cleaves at the sequence ACGT and is blocked by methylation of CpG.
Mbd1 has a CpG binding CXXC domain.
Mbd1 localization to heterochromatin in cells that lack DNA methylation could be due to protein-protein interactions between Mbd1 and another heterochromatic protein. Alternatively, Mbd1 might bind the satellite regions in these cells by virtue of another DNA-binding activity in the protein. Recently, two unrelated proteins, CGBP and MLL, were shown to bind to nonmethylated DNA through the CXXC domain (4, 20, 35). We aligned the three CXXC domains of Mbd1 with these CpG binding domains (Fig. 3A). All three Mbd1 regions have the eight signature cysteines conserved, but (unlike CXXC-1 and CXXC-2) the third domain (CXXC-3) showed a striking match throughout the region. We tested the Mbd1 CXXC-3 region for DNA binding activity in vitro using band shift assays. A recombinant peptide spanning this region bound to the nonmethylated CG11 probe (23) but did not bind to methylated CpG (data not shown). The specificity for nonmethylated sequences was confirmed using four synthetic 29-mer duplex probes that contain two CpGs with various methylation states (Fig. 3B). Again, the CXXC-3 domain complexed with the nonmethylated substrate but did not bind to the methylated probe. Weak binding to the hemimethylated probe was observed. Significantly, the Mbd1-CXXC-3 domain did not bind to a substrate entirely lacking CpGs (TG), demonstrating that nonmethylated CpGs are necessary and sufficient for binding. The CXXC-1 and CXXC-2 domains did not show this CpG-dependent DNA binding activity (Fig. 3C). As the MBD of Mbd1 binds preferentially to methylated CpGs (Fig. 3D), it follows that the Mbd1 protein has two DNA-binding domains. One domain is specific for methylated CpG, and the other domain is specific for nonmethylated CpG.
Mbd1 localization in methyl-CpG-deficient cells is dependent on the CXXC-3 domain.
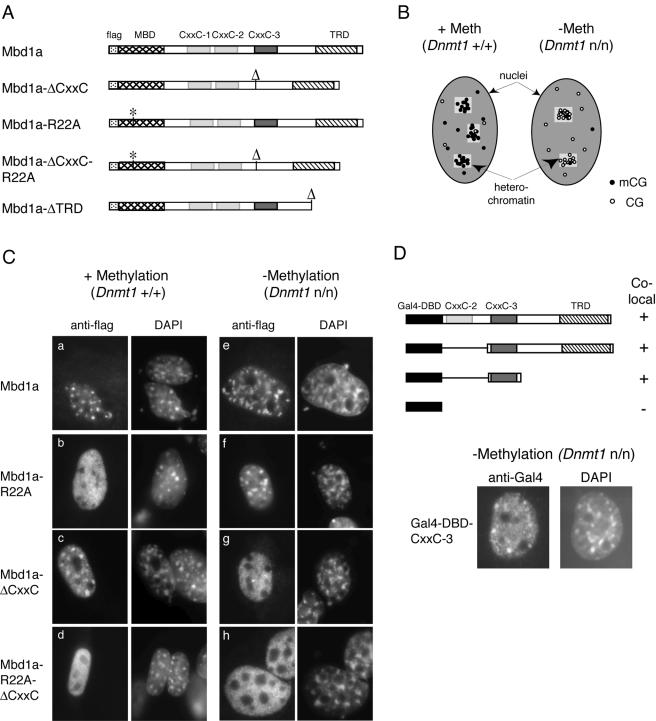
We next asked whether the CXXC-3 domain is required for localization of Mbd1 to CpG sites in vivo. We took advantage of the CpG richness of mouse satellite DNA, which is found concentrated in multiple heterochromatic foci in mouse cell nuclei. In normal cells, the foci are methyl-CpG dense, but in Dnmt1n/n cells, they contain high concentrations of nonmethylated CpG (Fig. 4B). By comparing the localization characteristics of exogenous Mbd1 forms in each cell type, we were able to ask which domains of the protein were required for binding to methylated and nonmethylated DNA. Flag-tagged full-length Mbd1a and various mutants were expressed, and their localization was assayed by immunostaining with anti-Flag antibodies. Like the endogenous protein, recombinant full-length Mbd1a protein localized to the CpG-rich heterochromatic foci in both the wild-type and the methylation-deficient cells (Fig. 4C, panels a and e). In wild-type cells, heterochromatic localization was lost in Mbd1 mutants that lack the ability to bind methylated DNA either by introduction of the R22A point mutation, which abolishes DNA binding of the MBD (28), or by deletion of the MBD (Fig. 4C, panel b, and data not shown). Deletion of the CXXC-3 domain had no effect on localization in these cells. The picture was strikingly reversed in the methylation-deficient cells, in which a functional MBD is dispensable for focal localization of Mbd1 (Fig. 4C, panel f, and data not shown). Deletion of the CXXC-3 domain resulted in a failure of localization to DAPI-bright regions in these cells and led to diffusely distributed Mbd1 (Fig. 4C, panel g). These results demonstrate that the Mbd1 protein can bind in vivo to nonmethylated DNA and that this feature depends on the presence of the CXXC-3 domain. The CXXC-1 and CXXC-2 domains were by themselves unable to target MBD1 to either methylated or nonmethylated DNA in vivo (Fig. 4C, panel g) or in bandshift assays in vitro (Fig. 3C). We further found that the CXXC-1 and CXXC-2 domains are not required to assist CXXC-3 targeting, as the CXXC-3 domain alone efficiently targeted nonmethylated DNA in vivo (Fig. 4D).
FIG. 4.
Localization of Mbd1 in DNA methylation-deficient cells requires the CXXC-3 domain. (A) Schematic representation of the Flag-Mbd1a expression constructs employed. The positions of the MBD (cross-hatched), CXXC-3 (dark gray), and the TRD (hatched) are shown. The CXXC-1 and CXXC-2 positions are shown shaded in light gray, and the Flag tag position is dotted. An asterisk marks the point mutations in the MBD (R22A), and “Δ” marks the sites of deletion of CXXC-3 or the TRD. (B) Diagram illustrating the enrichment for methyl CpG or nonmethyl CpG in the heterochromatic regions of wild-type (Dnmt1+/+) or methylation-deficient (Dnmt1n/n) cells, respectively. Methyl-CpG is shown with filled circles and nonmethyl-CpGs with open circles. The heterochromatic regions are represented by lightly shaded boxes. (C) Dnmt1+/+ (panels a to d) or Dnmt1n/n (panels e to h) mouse embryonic fibroblasts were transfected with constructs expressing Flag-tagged Mbd1a (either full length or the indicated mutants). At 48 h after transfection, the cells were fixed and stained using an anti-Flag antibody (left panels). DNA was counterstained with DAPI (right panels). (D) Schematic of Gal4-DBD-MBD1 fusion constructs analyzed for colocalization with heterochromatic foci in Dnmt1+/+ or Dnmt1n/n mouse embryonic fibroblasts. A plus sign indicates colocalization with heterochromatic foci in Dnmt1n/n cells and diffuse staining in Dnmt1+/+ cells, whereas a minus sign indicates diffuse staining in cells of either genotype. The fusion constructs encode amino acids 81 to 556, 261 to 556, and 261 to 346 of the MBD1 isoform PCM1. The Gal4-DBD is shown in black. CXXC-2 is shown shaded in light gray, CXXC-3 is shown shaded in dark gray, and the TRD is hatched. (Lower panels) Colocalization with heterochromatic foci of the Gal4-DBD-MBD1 (amino acids 261 to 346) (Gal4-DBD-CXXC-3) in Dnmt1n/n cells is shown.
Mbd1 represses transcription from nonmethylated as well as methylated templates.
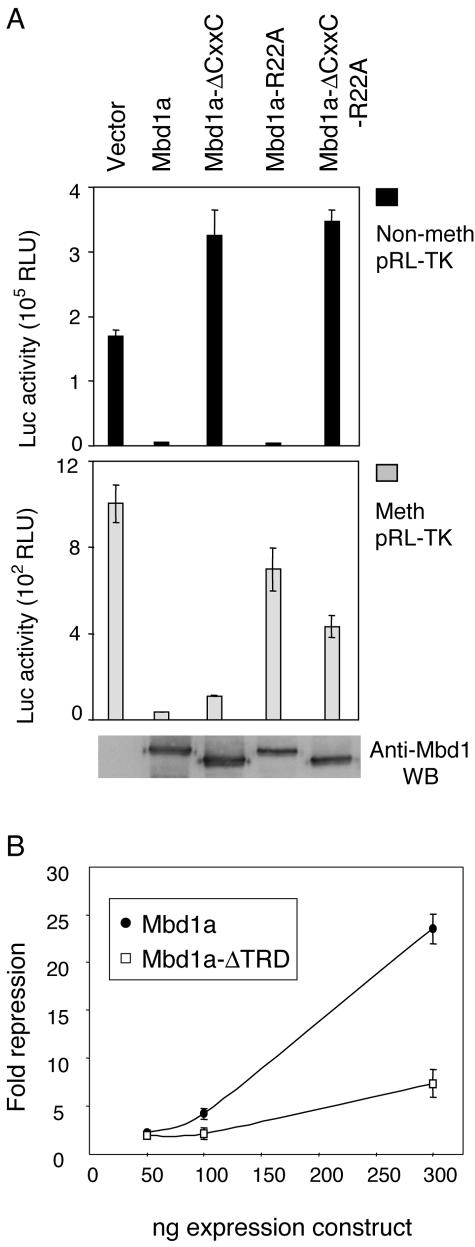
DNA methylation correlates with and can cause transcriptional repression, whereas nonmethylated promoters are usually in a transcriptionally competent or active state. Does binding of Mbd1 to nonmethylated CpGs bring about transcriptional repression or activation? To answer this question, we cotransfected reporter plasmids with the various Mbd1 expression constructs into HeLa cells. We did not include an internal standard in the experiments, as this too would be affected by a CpG-binding transcription factor. To ensure uniform transfection efficiency, the levels of Flag-Mbd1 expression were monitored by Western blotting and the experiments were performed in triplicate and repeated three to six times. Variation between experiments was found to be small. We established that full-length Mbd1a efficiently repressed both methylated and nonmethylated constructs (Fig. 5A). Repression of the nonmethylated reporter depended on an intact CXXC-3 domain, whereas a functional MBD was dispensable. The somewhat increased luciferase activity in cells transfected with constructs lacking CXXC-3 is most likely due to sequestration of a limiting repressing cofactor by the overexpressed repressor (“squelching”). Conversely, repression of methylated constructs required an intact MBD but was indifferent to the presence or absence of the CXXC-3 domain. Equivalent results were observed with reporter constructs encoding either Renilla or firefly luciferase and driven by a variety of promoters (TK, 3-phosphoglycerate kinase, simian virus 40, FGF2, and cytomegalovirus [data not shown]).
FIG. 5.
CXXC-3 is required for transcriptional repression of a nonmethylated reporter construct by Mbd1. (A and B) HeLa cells were transfected with constructs expressing Flag-Mbd1a or mutant derivatives plus a TK promoter-driven reporter construct. (A) Luciferase activities of methylated (Meth) or nonmethylated (Non-meth) reporter constructs are shown. Means and standard deviations of triplicate transfections from a representative experiment are shown. The result of an anti-MBD1 Western blot of whole-cell lysates of cells transfected in parallel (nonmethylated reporter) is shown in the bottom panel. (B) Repression of a nonmethylated reporter by Mbd1a is enhanced by the TRD. The graph plots severalfold repression of luciferase activity from a nonmethylated pRL-TK construct by the indicated amounts of a cotransfected construct expressing Mbd1a or Mbd1a lacking the TRD (Mbd1-ΔTRD) (Fig. 4A). Error bars show standard deviations of three data sets.
The TRD has previously been shown to be necessary for repression when MBD1 is tethered upstream of a promoter (26). To ask whether repression of nonmethylated reporters depends on the TRD, we coexpressed Mbd1a lacking the TRD with a nonmethylated reporter. The results showed significantly reduced repression in the absence of the TRD (Fig. 5B). Residual repression by the truncated protein may be due to the presence of other repression domains within Mbd1 or to steric interference upon binding of the CXXC-3 domain to CpGs within the reporter transcription unit.
DISCUSSION
MBD1 (initially called PCM1) was identified as a protein that binds specifically to methylated CpG dinucleotides in DNA. We show here that a major Mbd1 isoform also contains the CXXC-3 domain that binds specifically to the nonmethylated CpG dinucleotide. In vitro binding to nonmethylated CpG has been established previously for CXXC domains from CGBP and MLL (4, 20). The binding specificity of these proteins in vivo has been unclear, however. CGBP accumulates in nuclear sites of transcriptional activity, and this localization is not disrupted by mutation of the DNA-binding CXXC domain (19). By analyzing intranuclear localization of wild-type and mutant versions of Mbd1, we were able to establish that the CXXC-3 domain specifically localizes Mbd1 to nonmethylated CpGs in vivo. Localization to CpG-rich foci was disrupted either by methylation of the target CpG-rich foci or by mutation of the CXXC-3 domain. The MBD1 protein therefore makes use of two distinct DNA binding domains to target CpGs, the MBD requiring methylated CpG and the CXXC-3 domain requiring nonmethylated CpG. Although only some MBD1 isoforms carry the CXXC-3 DNA binding domain, this new finding affects the previous view of MBD1 as a protein that exclusively interprets the DNA methylation signal.
The biological significance of the dual DNA binding capability of MBD1 is currently unknown, but several possible scenarios can be considered. (i) The MBD and CXXC-3 domains may confer indifference to the methylation state of CpG sites, allowing MBD1 to function at certain CpGs regardless of their methylation status. (ii) The CXXC-3-containing isoform might be targeted to nonmethylated CpGs, where it could repress expression of CpG island promoters at tissue-specific genes in nonexpressing tissues—e.g., human alpha globin, neurofilament, and erythropoietin genes. (iii) MBD1 might utilize the CXXC-3 and MBD regions at the same time to bind a contiguous DNA sequence containing both a CpG and a methyl-CpG motif. Simultaneous usage of both DNA binding domains may increase the affinity of MBD1 for DNA, as both the MBD and the CXXC domain recognize a very short sequence and each domain alone has a moderate binding constant (4, 10). This latter scenario would imply a more gene-specific role of MBD1, in line with recent findings concerning involvement of MeCP2 at the murine Bdnf and Xenopus Hairy2A gene promoters (6, 22, 32).
Concerning the possible role of MBD1 as a repressor of nonmethylated CpG island promoters (see ii above), it is of interest that MBD1 was previously found to bind a repeat element in the human FGF2 CpG island promoter via the CXXC-3 domain (34). In that study, overexpression of MBD1 down-regulated endogenous FGF2 expression in cultured cells, suggesting that FGF2 might be an MBD1 target gene. The MBD1 binding element contains three CpG sites, but we have found that point mutations that change all three CpGs within this repeat in an FGF2-luciferase reporter did not abolish repression by MBD1 (data not shown). This suggests that CpGs elsewhere in the gene might attract repression by MBD1. Further work is needed to identify bona fide target genes for MBD1.
The close proximity of the MBD1 and CGBP genes in humans, mice, rats, and pufferfish suggests an evolutionary ancestry for the CXXC domains. Each CXXC is encoded by a single exon, making exon duplication a likely mechanism for domain dispersal. Unlike MBD1, CGBP is also found in the invertebrate C. intestinalis and is therefore the likely ancestor of the CXXC domains in MBD1. Intriguingly, MBD2, the only obvious MBD protein found in invertebrate animals, is also closely linked to MBD1 in both humans and mice (15). It is therefore possible that both DNA binding domains in MBD1 were donated by neighboring ancestral genes.
Our data indicate that MBD1 has additional TRDs outside the C-terminal TRD. Previous studies have established interactions between the MBD and potential corepressors SUV39h1 and the p150 subunit of CAF-1 (13, 31). Similar involvement of an MBD in corepressor recruitment has been described for MBD2 (5). There is also evidence that the CXXC-3 domain itself can bring about transcriptional repression, as a Gal4 fusion of the MBD1 CXXC-3 region alone was found to repress transcription in a DNA methylation-independent manner (26). The CXXC domains of MLL and DNMT1 both also repress transcription (14, 40). Hence, the CXXC-3 domain and the MBD of MBD1 may each have dual functions: to bind DNA and to act as docking sites for transcriptional corepressors.
Acknowledgments
We are grateful to Howard Cedar for the Dnmt1-deficient cell line and Karen Wilson for excellent technical assistance. We thank Skirmantas Kriaucionis, Eilidh MacDougall, and Jennifer Berger for comments on the manuscript.
This work was funded by a grant from The Wellcome Trust to A.P.B.
REFERENCES
- 1.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A. P. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8:1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birke, M., S. Schreiner, M. P. Garcia-Cuellar, K. Mahr, F. Titgemeyer, and R. K. Slany. 2002. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Res. 30:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke, J., O. Ammerpohl, S. Kegel, U. Moehren, and R. Renkawitz. 2000. The minimal repression domain of MBD2b overlaps with the methyl-CpG-binding domain and binds directly to Sin3A. J. Biol. Chem. 275:34963-34967. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W. G., Q. Chang, Y. Lin, A. Meissner, A. E. West, E. C. Griffith, R. Jaenisch, and M. E. Greenberg. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885-889. [DOI] [PubMed] [Google Scholar]
- 7.Cross, S. H., V. H. Clark, and A. P. Bird. 1999. Isolation of CpG islands from large genomic clones. Nucleic Acids Res. 27:2099-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross, S. H., R. R. Meehan, X. Nan, and A. Bird. 1997. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat. Genet. 16:256-259. [DOI] [PubMed] [Google Scholar]
- 9.Fournier, C., Y. Goto, E. Ballestar, K. Delaval, A. M. Hever, M. Esteller, and R. Feil. 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J. 21:6560-6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraga, M. F., E. Ballestar, G. Montoya, P. Taysavang, P. A. Wade, and M. Esteller. 2003. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31:1765-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita, N., S. Takebayashi, K. Okumura, S. Kudo, T. Chiba, H. Saya, and M. Nakao. 1999. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol. Cell. Biol. 19:6415-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, N., S. Watanabe, T. Ichimura, Y. Ohkuma, T. Chiba, H. Saya, and M. Nakao. 2003. MCAF mediates MBD1-dependent transcriptional repression. Mol. Cell. Biol. 23:2834-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita, N., S. Watanabe, T. Ichimura, S. Tsuruzoe, Y. Shinkai, M. Tachibana, T. Chiba, and M. Nakao. 2003. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 278:24132-24138. [DOI] [PubMed] [Google Scholar]
- 14.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Hendrich, B., C. Abbott, H. McQueen, D. Chambers, S. Cross, and A. Bird. 1999. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm Genome. 10:906-912. [DOI] [PubMed] [Google Scholar]
- 16.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hendrich, B., and S. Tweedie. 2003. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19:269-277. [DOI] [PubMed] [Google Scholar]
- 18.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. H., and D. G. Skalnik. 2002. CpG-binding protein is a nuclear matrix- and euchromatin-associated protein localized to nuclear speckles containing human trithorax. Identification of nuclear matrix targeting signals. J. Biol. Chem. 277:42259-42267. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. H., K. S. Voo, and D. G. Skalnik. 2001. Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 276:44669-44676. [DOI] [PubMed] [Google Scholar]
- 21.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 22.Martinowich, K., D. Hattori, H. Wu, S. Fouse, F. He, Y. Hu, G. Fan, and Y. E. Sun. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890-893. [DOI] [PubMed] [Google Scholar]
- 23.Meehan, R. R., J. D. Lewis, S. McKay, E. L. Kleiner, and A. P. Bird. 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58:499-507. [DOI] [PubMed] [Google Scholar]
- 24.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 25.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, H.-H., P. Jeppesen, and A. Bird. 2000. Active repression of methylated genes by the chromosomal protein MBD1. Mol. Cell. Biol. 20:1394-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 28.Ohki, I., N. Shimotake, N. Fujita, M. Nakao, and M. Shirakawa. 1999. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 18:6653-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 30.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reese, B. E., K. E. Bachman, S. B. Baylin, and M. R. Rountree. 2003. The methyl-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol. Cell. Biol. 23:3226-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stancheva, I., A. L. Collins, I. B. Van den Veyver, H. Zoghbi, and R. R. Meehan. 2003. A mutant form of MeCP2 protein associated with human Rett syndrome cannot be displaced from methylated DNA by notch in Xenopus embryos. Mol. Cell 12:425-435. [DOI] [PubMed] [Google Scholar]
- 33.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ueba, T., B. Kaspar, X. Zhao, and F. H. Gage. 1999. Repression of human fibroblast growth factor 2 by a novel transcription factor. J. Biol. Chem. 274:10382-10387. [DOI] [PubMed] [Google Scholar]
- 35.Voo, K. S., D. L. Carlone, B. M. Jacobsen, A. Flodin, and D. G. Skalnik. 2000. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol. Cell. Biol. 20:2108-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wade, P. A. 2001. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene 20:3166-3173. [DOI] [PubMed] [Google Scholar]
- 37.Wade, P. A., P. L. Jones, D. Vermaak, G. J. Veenstra, A. Imhof, T. Sera, C. Tse, H. Ge, Y. B. Shi, J. C. Hansen, and A. P. Wolffe. 1998. Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase. Cold Spring Harbor Symp. Quant. Biol. 63:435-445. [DOI] [PubMed] [Google Scholar]
- 38.Wang, H., W. An, R. Cao, L. Xia, H. Erdjument-Bromage, B. Chatton, P. Tempst, R. G. Roeder, and Y. Zhang. 2003. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12:475-487. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe, S., T. Ichimura, N. Fujita, S. Tsuruzoe, I. Ohki, M. Shirakawa, M. Kawasuji, and M. Nakao. 2003. Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin. Proc. Natl. Acad. Sci. USA 100:12859-12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeleznik-Le, N. J., A. M. Harden, and J. D. Rowley. 1994. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc. Natl. Acad. Sci. USA 91:10610-10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao, X., T. Ueba, B. R. Christie, B. Barkho, M. J. McConnell, K. Nakashima, E. S. Lein, B. D. Eadie, A. R. Willhoite, A. R. Muotri, R. G. Summers, J. Chun, K. F. Lee, and F. H. Gage. 2003. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA 100:6777-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]