Abstract
The daily timing of circadian (≅24-h) controlled activity in many animals exhibits seasonal adjustments, responding to changes in photoperiod (day length) and temperature. In Drosophila melanogaster, splicing of an intron in the 3′ untranslated region of the period (per) mRNA is enhanced at cold temperatures, leading to more rapid daily increases in per transcript levels and earlier “evening” activity. Here we show that daily fluctuations in the splicing of this intron (herein referred to as dmpi8) are regulated by the clock in a manner that depends on the photoperiod (day length) and temperature. Shortening the photoperiod enhances dmpi8 splicing and advances its cycle, whereas the amplitude of the clock-regulated daytime decline in splicing increases as temperatures rise. This suggests that at elevated temperatures the clock has a more pronounced role in maintaining low splicing during the day, a mechanism that likely minimizes the deleterious effects of daytime heat on the flies by favoring nocturnal activity during warm days. Light also has acute inhibitory effects, rapidly decreasing the proportion of dmpi8-spliced per transcript, a response that does not require a functional clock. Our results identify a novel nonphotic role for phospholipase C (no-receptor-potential-A [norpA]) in the temperature regulation of dmpi8 splicing.
Circadian rhythms are driven by cellular oscillators known as clocks or pacemakers and are an important aspect of the temporal organization observed in a wide range of organisms from bacteria to humans (11, 12). These clocks exhibit free-running (self-sustaining) periods of ≅24 h in the absence of environmental cues. Nonetheless, an important adaptive feature of circadian oscillators is that they are synchronized (entrained) by daily changes in environmental modalities, most notably visible light and ambient temperature.
Light is almost certainly the predominant entraining agent in nature. Under natural conditions the light-dark (LD) cycle aligns the phases of clocks and evokes daily adjustments in the approximately 24-h endogenous periods of these oscillators such that they precisely match the 24-h solar day. The duration of day length (photoperiod) can modify the temporal alignment between a circadian rhythm and local time (22). A physiologically relevant advantage of this inherent flexibility of clocks is that the daily distributions of physiological and behavioral rhythms are not rigidly locked to local time but can be adjusted for seasonal changes in day length.
Despite the obvious importance of photoperiod, ambient temperature is also a key environmental modality regulating the timing of circadian rhythms (50). This makes intuitive sense, because in temperate latitudes seasonal changes in day length are also accompanied by predictable changes in average daily temperatures. Thus, circadian clocks play an important role in endowing organisms with the ability to anticipate daily and seasonal changes in environmental conditions, resulting in physiological and behavioral rhythms that occur at biologically advantageous times throughout the year. However, it is not clear how these pacemakers integrate environmental cues concerning seasonal changes in photoperiod and temperature.
Work using Drosophila melanogaster has provided numerous insights into our understanding of the time-keeping mechanisms governing circadian rhythms (recently reviewed in references 1, 19, and 47). In D. melanogaster four circadian clock proteins, termed PERIOD (PER), TIMELESS (TIM), dCLOCK (dCLK), and CYCLE (CYC/dBMAL1), function in a transcriptional-translational feedback loop that is a core element of the oscillator mechanism in this species. dCLK and CYC are members of the basic helix-loop-helix (bHLH)/PAS (PER-ARNT-SIM) superfamily of transcription factors that heterodimerize to activate per and tim expression (2, 10, 44). Transcription of per and tim begins in the early to mid-day, resulting in peak mRNA levels being reached in the early night. However, the daytime accumulation in the levels of PER and TIM proteins is slow. This is partly because the DOUBLETIME (DBT) kinase promotes the phosphorylation and degradation of PER (30, 41), and light stimulates the turnover of TIM (26, 38, 55). Eventually, as the levels of per and tim transcripts increase during the late day, TIM and PER reach threshold concentrations that enable them to interact, a process that protects PER against DBT-induced degradation (30, 41) and presumably facilitates the nuclear accumulation of PER and TIM (however, see reference 46). Once in the nucleus PER and TIM interact with the dCLK-CYC transcription factor, blocking its activity (3, 10, 33, 34). Reductions in the concentrations of PER and TIM in the nucleus relieve autoinhibition of dCLK-CYC, beginning another round of per and tim expression and, in an interconnected loop, repression of dClk transcription (9, 16, 17).
We recently used D. melanogaster as a model system to understand the molecular underpinnings governing how changes in daily temperatures modulate activity rhythms. The “evening” activity peak of D. melanogaster exposed to daily LD cycles progressively advances from mainly nocturnal to predominately late day as ambient temperature is decreased (36). We showed that splicing of an intron in the 3′ untranslated region (UTR) of per RNA (dmpi8 [D. melanogaster per intron 8]) is a key aspect of how the Drosophila circadian clock adapts to changes in temperature. At low temperatures, relatively more of the spliced variant (type B′) is present compared to the unspliced variant (type A). The enhanced removal of dmpi8 at low temperatures leads to an advance in the timing of the per mRNA and protein accumulation phases during the day. Because the interaction of PER with TIM is a key event in the progression of the clock, a more rapid increase in the abundance of PER shortens the time necessary to attain threshold concentrations that favor interactions with TIM, contributing to advanced molecular cycles and the preferential daytime activity of flies on cold days. At cold temperatures, mutations that either inhibit splicing (perA mutant) or remove dmpi8 (perB′ mutant) both cause low-amplitude cycles in per mRNA levels and delayed activity rhythms. These results suggested that active splicing of dmpi8 per se, as opposed to the presence or absence of the intron, is the relevant molecular event mediating the cold-enhanced daily increases in the abundance of per mRNA.
How light regulates the clock and the daily distribution of activity in Drosophila is not fully understood and is likely to be complex, involving various ocular and extraocular photoreceptors (15, 25, 43, 51). Early work clearly established an important role for the light-induced degradation of TIM as a key primary clock-specific photoresponse in the entrainment of Drosophila clocks to daily light-dark cycles (26, 35, 38, 49, 54, 55). The putative blue-light photoreceptor CRYPTOCHROME (CRY) enhances the light-induced degradation of TIM in most, if not all, clock cells (18, 25, 27, 28, 48, 51). CRY functions as a deep-brain photoreceptor (15) that apparently directly interacts with TIM (4), presumably leading to the rapid ubiquitination and destruction of TIM by the proteasome (39). Circadian photosensitivity is altered in the cryb mutant, which has severely reduced levels and activity of CRY (4, 13-15, 25, 48). Besides a role in transducing photic signals, CRY also functions as a bona fide clock element in some peripheral circadian pacemakers (27, 32, 48). Although physiologically or anatomically blind, flies manifest robust activity rhythms (e.g., references 24 and 51 to 53), more recent work has supported a role for classic visual signal transduction pathways in the light regulation of daily activity rhythms (e.g., reference 43 and references therein). For example, a mutation in no-receptor-potential-A (norpA), which encodes phospholipase C (PLC/NORPA), is associated with variant patterns in the clock-controlled evening activity (20, 53). As mentioned above temperature (36) and, in addition, mutations in per that shorten or lengthen the periodicities of free-running behavioral rhythms also manifest alterations in the timing of evening activity during entrainment to LD cycles (20).
In this report we show that temperature, light, and the clock interact to regulate dmpi8 splicing. In short photoperiods the daily proportion of the spliced type B′ variant is enhanced and accumulates earlier, contributing to increases in the daily upswing and peak levels of per mRNA. Thus, both short day lengths and low temperatures, which are normally associated under natural conditions, stimulate splicing of dmpi8. Increases in temperature lead to larger amplitude declines in daytime splicing, a response that is regulated by the clock. This more-pronounced clock-dependent inhibition of splicing as temperatures rise likely contributes to the manifestation of mainly nocturnal activity on warm days, enabling flies to avoid the deleterious effects associated with daytime heat. The proportion of the type B′ variant is abnormally high in the norpA mutant and exhibits little daytime decrease even on warm days. Thus, it appears that irrespective of temperature the splicing behavior of dmpi8 in norpA flies exhibits characteristics that are normally observed only at low temperatures, consistent with the mutants advanced evening activity even on warm days. The findings suggest a novel nonphotic role for PLC in transducing the effects of temperature on the splicing behavior of the dmpi8 intron. Our findings suggest that the dual thermal and photoperiodic regulation of dmpi8 splicing acts as a seasonal sensor encoding calendar information.
MATERIALS AND METHODS
Fly strains and collections.
The wild-type (Canton-S [CS]), per01, tim0, cryb, and norpA flies used in this study were previously described (2, 31, 44, 45, 48). All the flies used in this study were wild type for eye and body pigmentation. All flies were reared at room temperature (22 to 25°C) and maintained in vials containing standard agar-cornmeal-sugar-yeast-tegosept medium. Vials containing ∼100 young (2- to 6-day-old) adult flies were placed in controlled environmental chambers (Percival) at the indicated temperature (18 or 29°C) and exposed to at least four 24-h photoperiods of alternating LD cycles (where zeitgeber time zero [ZT0] is defined as lights-on), and in some cases subsequently maintained in constant darkness (DD). Cool white fluorescent light (∼2,000 lx) was used during LD, and the temperature did not vary by more than 0.5°C between the light and dark periods. At selected times during LD and DD, flies were collected by freezing.
Locomotor activity.
Locomotor activity was continuously monitored and recorded in 30-min bins by placing individual adult flies (3- to 7-day-old males) in glass tubes and using a Trikinetics (Waltham, Mass.) system interfaced with an Apple computer as previously described (20). Fly activity data were analyzed using the Periodogram and Phase analysis software available from the web-based Brandeis Rhythm Package (http://hawk.bcm.tmc.edu/brp/brp.cgi). The Phase program was used to calculate the time in a day (in hours relative to ZT0) for the onset (50% of peak) and peak of the evening activity for each individual fly (see Table 1). Values for individual flies were pooled to obtain an average value for each genotype and temperature. Pair-wise comparisons were performed using Student's t test.
TABLE 1.
Locomotor activity rhythms of CS, norpAP41, and cryb flies at different temperaturesa
| Genotype | Temp (°C) | nb | Time (h) relative to ZT0 (mean ± SEM)d
|
|
|---|---|---|---|---|
| 50% onsetc | Peak | |||
| CS | 18 | 32 | 8.9 ± 0.2 | 10.4 ± 0.1 |
| norpAP41 | 18 | 30 | 7.2 ± 0.3* | 9.1 ± 0.2* |
| cryb | 18 | 28 | 7.4 ± 0.2* | 9.2 ± 0.2* |
| CS | 29 | 31 | 10.6 ± 0.2 | 12.1 ± 0.2 |
| norpAP41 | 29 | 30 | 8.5 ± 0.2* | 11.6 ± 0.2** |
| cryb | 29 | 27 | 9.3 ± 0.4* | 11.5 ± 0.4 |
Flies were kept at either 18 or 29°C and exposed to 4 days of 12:12LD. Average data from two independent experiments are shown.
Total number of flies that survived until the end of the testing period.
Onset is the time at which the upswing in evening activity reached 50% of the peak value and is given in hours from the last lights-on transition, where ZT0 is lights-on.
*, significant difference between CS and norpA or cryb at P < 0.001 using Student's t test. **, significant difference between CS and norpA at P < 0.05. No significant differences were observed between norpA and cryb for either onset or peak at either temperature.
RNase protection assay.
For each time point, total RNA was extracted from ∼30 μl of fly heads using Tri reagent (Sigma) and following the manufacturer's recommended procedure. The total (type A and type B′) levels of per transcripts were determined by RNase protection assays using the per 2/3 probe (21). In addition, we also used RNase protection assays to measure the relative levels of type A and type B′ per RNA variants using a novel probe called per3′UTR. per3′UTR was generated by using PCR to amplify a genomic region of per that contains nucleotides (nt) 7009 to 7308 (numbering according to reference 7). The oligonucleotides used in the PCR were P7009 (5′ ACCGAGCACCAGCCAGT 3′) and P7308 (5′ TTAGGGCTGAGAAGGGTGC 3′). The PCR product was subcloned into the pGEM-T easy vector (Promega) and linearized with SpeI, and antisense probe was produced in vitro by using T7 RNA polymerase in the presence of [α-32P]UTP as previously described (36). The antisense probe protects a per RNA region containing the 3′-most 62 nt of dmpi8, followed at the 3′ end of dmpi8 by the next 238 nt of flanking exon. Thus, in the presence and absence of dmpi8, the protected radiolabeled band is 300 and 238 nt long, respectively. As a control for RNA loading in each lane, a ribosomal protein probe (RP49) was included in each protection assay (21). Protected bands were quantified using a PhosphorImager from Molecular Dynamics, and values were normalized relative to those of RP49 and for uridine content.
RT-PCR splicing assay.
The relative levels of the type B′ and type A per RNA variants were measured using a reverse transcriptase PCR (RT-PCR) assay based on a method previously described (36). For each time point, total RNA was extracted from ∼30 μl of fly heads using Tri reagent (Sigma) and following the manufacturer's recommended procedure. Approximately 2 μg of total RNA was incubated in a final volume of 20 μl, and reverse transcription was performed using oligo(dT)20 as a primer and the ThermoScript RT-PCR kit from Invitrogen, according to the manufacturer's recommended procedure. A 2-μl aliquot of the reaction mixture was further processed by PCR in a final volume of 50 μl using the per-specific primers P7197 (5′ TCTACATTATCCTCGGCTTGC 3′) and P6869 (5′ TAGTAGCCACACCCGCAGT 3′). This amplified a region of the 3′ UTR of per from bp 6869 to 7197 (numbering according to reference 7). To control for sample-to-sample differences in total RNA, we also included primers for the noncycling mRNA encoding Cap binding protein 20 (CBP20). Prior work using DNA microarrays has shown that the mRNA levels for CBP20 are constant throughout a daily LD cycle (5; also, see http://expression.gnf.org/cgi-bin/circadian/index.cgi) (Fig. 1A). We further chose cbp20 RNA as an internal control because the levels of per and cbp20 mRNAs are in a similar range in total head extracts, as inferred by the staining intensities following RT-PCR done in the exponential phase (Fig. 1A). The primers used in the RT-PCR for amplification of cbp20 sequences were CBP495R (5′ CAACAGTTTGCCATAACCCC 3′) and CBP362F (5′ GTCTGATTCGTGTGGACTGG 3′). This amplified a region of cbp20 from bp 362 to 495 (numbering according to GenBank accession no. NM079672). PCR products were separated and visualized by electrophoresis on 2% agarose gels followed by staining with Gelstar (Cambrex Co.), and the bands were quantified using a Typhoon 9400 Imager. The two per band intensities were normalized for product length differences and relative to cbp20 levels. Two per-specific bands of the expected sizes for PCR products that contained either dmpi8 (329 bp) or where the dmpi8 intron was removed (240 bp) were detected in total RNA following RT-PCR (Fig. 1A). In addition, one cbp20-specific band of the expected size (134 bp) that had relatively constant intensity irrespective of time of day was detected (Fig. 1A).
FIG. 1.
Photoperiod affects the waveform of the per mRNA abundance profile. Wild-type (CS) flies were kept at 18°C, exposed to 4 days of different photoperiods (6:18LD or 12:12LD as indicated), and collected during the last day of LD. RT-PCR (A to D) or RNase protection assays (E) were used to measure the relative levels of total (i.e., type A and type B′ per variants together) (B and E), dmpi8-spliced type B′ (C), and dmpi8-unspliced type A (D) per transcripts in head extracts. For each time point, values shown are from at least two independent experiments. (A) Representative agarose gel showing the per (dmpi8 unspliced and spliced) and cbp20 RT-PCR products from flies collected at different times during 12:12LD (top of panel, lanes 1 to 6); lane 7, control sample in the absence of RT; lane 8, DNA size markers (M). (B to E) White horizontal bar, lights on; black horizontal bar, lights off.
Numerous control experiments were performed to ensure the accuracy of the results, including the following: (i) to verify that our assays were performed in the exponential phase of PCR for per amplicons, we collected wild-type flies at different times during a daily LD cycle and subjected aliquots from the reverse transcription reaction to PCRs ranging in cycle length from 24 to 28. Under the conditions used, all these cycle lengths resulted in curves for per RNA levels that had indistinguishable amplitudes, peak times, and overall shapes as a function of time in a daily cycle (data not shown). Importantly, the results we obtained using RT-PCR were similar to those using RNase protection assays (e.g., compare Fig. 1B and E). Furthermore, as expected from the linear dose-response for per RNA staining intensities using these different PCR cycle lengths, the different reactions yielded splicing ratios (i.e., relative molar ratio of type B′ to type A) that were very similar and were also similar to those using RNase protection assays. Control experiments using wild-type flies collected throughout a daily cycle and various PCR cycle lengths were performed at least three independent times with very similar results (data not shown). In addition, for each independent experiment, several of the first-strand synthesis reactions (usually expected trough and peak values for wild-type flies) were subjected to at least two different PCR cycle lengths (usually 24 and 26) to verify that per amplicon levels were in the linear range. We also routinely included as part of our experiments separate reactions with serial dilutions ranging from 10- to 50-fold differences in per RNA concentration to ensure linear band intensities (2). We included cbp20 mRNA as an internal noncycling RNA control during the RT-PCR (see above). Between 24 and 28 PCR cycle lengths, both the per and cbp20 amplicons were in the exponential phase (Fig. 1A and data not shown) (3). Similar ratios of type B′ to type A were obtained when reverse transcription was performed with either oligo(dT)20 or P7308 (see above) as primers (4). No per-specific or cbp20-specific amplicons were detected when the RT-PCR was performed in the absence of RT (Fig. 1A) or gene-specific primers (5). The two per-specific RT-PCR bands were extracted from gels and sequenced to verify that they were produced from type A and type B′ per variants (data not shown) (6). We always included wild-type control flies whenever mutant flies were analyzed, and RNA samples were processed contemporaneously (7). Finally, we optimized conditions by using, for example, several different primers, incubation temperatures, RTs, and amounts of total RNA or cDNA. For example, more-consistent results were obtained using ThermoScript instead of SuperScript RT for cDNA synthesis, possibly because the higher temperature during the incubation (55 versus 42°C) minimizes RNA secondary structure.
RESULTS
Photoperiodic and clock regulation of dmpi8 splicing.
We previously showed that in wild-type flies entrained by standard 12-h light-12-h dark cycles (12:12LD; where ZT0 is lights-on), cold temperatures stimulate the relative splicing of dmpi8 (i.e., relative molar ratio of type B′ to type A per variants), which underlies the earlier daily upswing in total per mRNA abundance and higher peak values (36) (see also Fig. 7A and B). Importantly, it is the more-rapid accumulation phase of per mRNA that contributes to the advanced evening activity of Drosophila on cold days (36). In temperate regions, low daily temperatures are normally associated with short day lengths.
FIG. 7.
Higher overall levels of per mRNA were found in norpA flies. Wild-type (CS) and norpAP41 flies were exposed to 4 days of 12:12LD at either 18 or 29°C. RT-PCR was used to measure the relative levels of total (i.e., type A and type B′ per variants together) (A and B), unspliced type A (C), and spliced type B′ (D) per transcripts in head extracts. The RNA samples were from the fly collections shown in Fig. 6. Higher overall levels of total per mRNA in norpAP41 flies were also obtained using RNase protection assays (data not shown).
To determine whether changes in photoperiod also regulate dmpi8 splicing, wild-type CS flies were kept at 18°C and exposed for several days at two different photoperiods (6:18LD and 12:12LD) (Fig. 1). First, we assayed the effects of variations in photoperiod on total per mRNA levels. RNA levels were mainly determined by semiquantitative RT-PCR (Fig. 1A), but similar results were obtained by RNase protection assays (e.g., compare Fig. 1B and E) (see Materials and Methods). To control for sample-to-sample variations in the RT-PCR, we coamplified the noncycling cbp20 mRNA and normalized per values (Fig. 1A) (see Materials and Methods). Consistent with our previous findings (36), total per mRNA levels accumulated earlier during the day and reached higher peak values as a function of shortening the duration of the photoperiod (Fig. 1B and E). An earlier accumulation phase for per mRNA levels under shorter photoperiods was also previously observed at the standard temperature of 25°C (42).
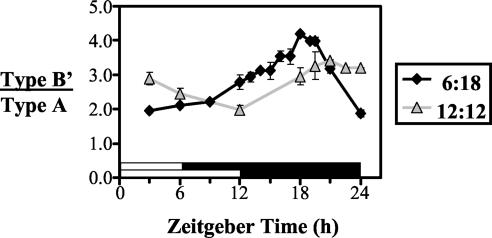
We next analyzed the spliced type B′ (Fig. 1C) and unspliced type A (Fig. 1D) per RNA variants individually. Irrespective of day length, at 18°C the majority of per mRNA transcripts were type B′ (Fig. 2) (36). The relative abundance of type B′ to type A (type B′/type A ratio) at 18°C ranged from ∼2- to 4.5-fold higher at the different photoperiods analyzed (Fig. 2 to 6). In more than 10 independent experiments, we observed that in wild-type flies the proportion of type B′ transcripts changed as a function of time in LD cycles (e.g., Fig. 2 to 6). At all photoperiods analyzed (i.e., 6:18LD, 9:15LD, and 12:12LD), daily trough and peak values in the ratio of type B′ to type A were significantly different (P < 0.0001 by Student's t test; n = 4) (Fig. 2 and data not shown). The most consistent trends in dmpi8 splicing oscillation were that the proportion of the spliced type B′ variant was overall lower during the day, and as photoperiod was shortened it accumulated earlier and reached higher peak values in the night (Fig. 2). These results suggest that the daily mean splicing of dmpi8 is more efficient under short photoperiods, consistent with the earlier accumulation phase and higher levels of total per mRNA (Fig. 1B and E).
FIG. 2.
Effects of photoperiod on the relative splicing of dmpi8. Shown are the relative levels of type B′ to type A per variants as a function of changes in day length. The data are derived from the results shown in Fig. 1B to D. A significant difference was found on comparison of peak to trough values for either 6:18LD or 12:12LD (P < 0.0001 by Student's t test). White horizontal bar, lights on; black horizontal bar, lights off.
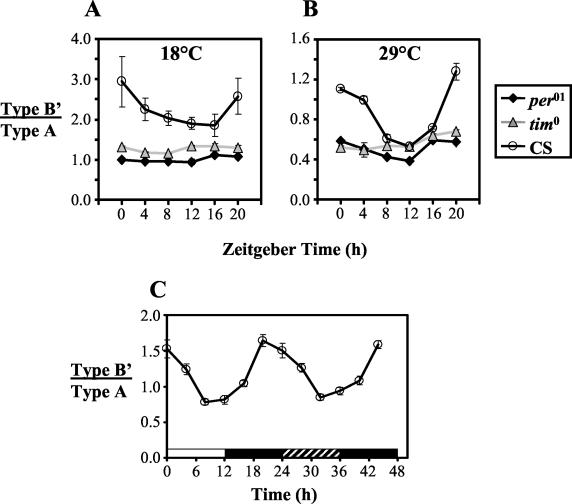
FIG. 6.
Effects of cryb and norpA mutants on splicing of dmpi8. (A and B) Wild-type (CS) and mutant flies (cryb and norpAP41) were exposed to 4 days of 12:12LD at either 18°C (A) or 29°C (B). The RNA samples were from fly collections done at the same time as those shown in Fig. 4. Comparison of peak to trough values in norpA flies at both temperatures did not show a significant difference (P > 0.3 by Student's t test). Similar results were obtained in at least four other experiments (data not shown). (C) In separate experiments, CS and norpA flies were kept at 29°C and collected on the last day of 12:12LD and the first day of DD (the CS splicing profile is the same as that shown in Fig. 4B). In addition, on the first day of DD, groups of CS and norpA flies were exposed to light beginning at 40 h (time from the last dark-to-light transition at ZT0); other groups served as nontreated controls. For each genotype, comparison of light-treated (open symbols) and control samples (closed symbols) showed significant differences (P < 0.05; Student's t test). LP, light pulse-treated samples.
At low temperatures, oscillations in the daily proportion of type B′ to type A per transcripts were observed under DD conditions, albeit with dampened amplitudes (Fig. 3A and C). The dampening in this molecular cycle was mainly attributable to a lowering of peak values in the type B′/type A ratio during DD conditions, whereas trough values remained quite constant (Fig. 3C). Furthermore, trough values remained lower in flies entrained to longer photoperiods even after 2 days in DD (Fig. 3C) (P < 0.0001 by Student's t test for comparison of average trough values during the last day of LD and the first 2 days of DD between 12:12D- and 6:18LD-entrained flies), suggesting that day length has long-range indirect effects on setting the basal splicing efficiency of dmpi8. A likely mechanism for how the duration of day (or night) is “remembered” during DD conditions is via stable changes in the dynamics of the clock as a function of changes in photoperiod length. In any event, the results demonstrated circadian regulation of the daily changes in the splicing of dmpi8. Indeed, irrespective of temperature and photoperiod the overall daily proportion of the spliced type B′ variant in several arrhythmic circadian clock mutants tested (i.e., per01 and tim0) was at or below trough values observed for wild-type control flies (Fig. 4A and B and data not shown). It is possible that the decreased splicing of dmpi8 in the clock-impaired flies contributed to the low to intermediate levels of per mRNA previously observed in these mutants (2, 21, 44, 45) (data not shown). The results also suggested that the clock (or clock components) has a significant influence on the mean splicing efficiency of dmpi8 throughout a daily cycle.
FIG. 3.
Dampening of cycles in dmpi8 splicing levels under DD conditions at cold temperatures. Wild-type (CS) flies were exposed to 4 days of either 6:18LD or 12:12LD at 18°C followed by DD conditions. Flies were collected at different times during the last LD cycle and DD. (A and C) RT-PCR was used to determine the relative levels of spliced (type B′) and unspliced (type A) per RNA. (B) Total per mRNA levels (type A and type B′) were measured using RNase protection assays. The data in panels A and B are representative of results obtained in at least two independent experiments. (C) Peak levels in the type B′/type A ratio for 6:18LD were set to 1, and the rest of the values were normalized to show relative peak and trough amounts during either 6:18LD or 12:12LD (indicated at bottom of the panel) followed by the next 2 days of DD (DD1 and DD2). The data are an average of two independent experiments. A P value of <0.0001 was determined by Student's t test on comparison of average trough values during the last day of LD and first 2 days of DD between 12:12LD- and 6:18LD-entrained flies.
FIG. 4.
Lower splicing of dmpi8 in arrhythmic clock mutants. (A and B) Wild-type (CS) and clock mutant flies (per0 and tim0) were exposed to 4 days of 12:12LD at either 18°C (A) or 29°C (B). Comparisons of peak to trough values resulted in the following P values: per01 at 18 and 29°C, P < 0.01; tim0 at 18 and 29°C, P < 0.03. (C) In separate experiments, CS flies were kept at 29°C and collected on the last day of 12:12LD and the first day of DD. The data are derived from an average of two independent RT-PCR experiments.
In wild-type flies, higher-amplitude rhythms in the proportion of type B′ spliced transcripts occurred at warmer temperatures, despite the overall lower splicing levels (compare Fig. 4A and B with C). At 29°C the dmpi8 daily splicing profile was characterized by a relatively steep daytime decline followed by a nighttime upswing that peaked toward the end of the dark phase. There were two notable differences in the daily splicing rhythm of dmpi8 when comparing cold and warm days. First, there was a larger magnitude decline in daytime splicing at warm temperatures. Second, at the higher temperature the splicing rhythm continued with a robust amplitude in the first day of DD, in sharp contrast to the rapid dampening observed at the colder temperature (compare Fig. 3A and 4C). The results suggest a more pronounced role for the clock in repressing splicing during the daytime on warm days.
Despite the low daily mean splicing of dmpi8 in the per01 and tim0 mutants, overall daytime levels in the proportion of type B′ transcripts were lower compared to nighttime values during LD cycles, especially at 29°C (Fig. 4A and B; see the figure legend for P values showing significant peak-to-trough differences). We did not observe any significant changes in the type B′/type A ratio when the clock mutants were analyzed under DD conditions (data not shown), suggesting that light decreases the splicing efficiency of dmpi8 in a manner that does not require a functional clock and that might be enhanced by increases in temperature (see Fig. 5).
FIG. 5.
Light acutely inhibits the splicing of dmpi8 in wild-type and clock mutant flies. (A and B) Wild-type (CS) flies were kept at either 18°C (A and B) or 29°C (B only) and exposed to 4 days of either 6:18LD, with light beginning at ZT12 or ZT16 (A) or 12:12LD, with light beginning at either ZT15 or ZT21 as indicated (B). On the dark phase of the last LD, groups of flies were exposed to light beginning at different times; other groups served as nontreated controls. (C) per01 flies were exposed to 12:12LD cycles at 29°C, and on the first day of DD groups of flies were exposed to light beginning at 40 h (time from the last dark-to-light transition at ZT0); other groups served as nontreated controls. The data shown are derived from an average of three independent RT-PCR experiments. Symbols: *, P < 0.01; **, < 0.05 using Student's t test for comparison between light-pulsed and control values. Similar photoinhibition of dmpi8 splicing was also obtained in wild-type flies exposed to 12:12LD at 25°C (data not shown).
Light acutely inhibits the splicing of dmpi8.
To further examine whether light has acute effects on the splicing efficiency of dmpi8, wild-type and clock mutant flies were entrained to LD cycles and exposed to light pulses administered at different times during the dark phase (Fig. 5). We noted small but reproducible decreases in the proportion of the spliced type B′ variant by nocturnal light pulses, a response that was observed at all temperatures (i.e., 18, 25, and 29°C) and photoperiods (6:18LD and 12:12LD) (Fig. 5 [see figure legend for P values] and data not shown). Brief light pulses did not reduce dmpi8 splicing below the daytime trough levels observed during the preceding LD cycle (Fig. 5B and data not shown). Thus, it appears that the prevailing conditions of photoperiod and temperature set a basal amount of dmpi8 splicing efficiency that is refractory to acute inhibition by light. This likely explains why we observed reproducible time-of-day differences in the acute photoinhibition of dmpi8 splicing, whereby light pulses given during the first half of the night when the proportion of type B′ is increasing generally evoked larger-magnitude decreases (Fig. 5B). During the late night, the proportion of dmpi8-spliced transcripts was already decreasing in wild-type flies and, therefore, during this phase differences between untreated and light-treated samples were minimized. As a result, the clock modulates the magnitude of the light inhibition in a time-of-day-specific manner. The ability of light to evoke decreases in the proportion of type B′ per transcripts was in agreement with the overall lower dmpi8 splicing during the day and higher peak amounts during short photoperiods (Fig. 2 and 4). Importantly, nocturnal light pulses evoked rapid decreases in dmpi8 splicing in per0 and tim0 flies where possible influences from circadian control were eliminated (Fig. 5C and data not shown).
In summary, the roles of the clock and light in regulating dmpi8 splicing are complex and vary as a function of photoperiod and temperature. A short photoperiod advances the daily upswing in the proportion of the spliced type B′ variant and higher peak values are obtained, responses that are likely largely mediated via the effects of day length on the dynamics (phase and amplitude) of the clock. Light also has acute effects, inhibiting the splicing efficiency of dmpi8. The shape of the daily dmpi8 splicing rhythm is strongly modulated by temperature. The cold splicing phenotype is characterized by high overall splicing with relatively low-amplitude cycling that dampens quickly under DD conditions. Conversely, the warm splicing phenotype is characterized by low overall splicing and a very pronounced clock-mediated daytime decline, a pattern that can persist in the absence of LD cycles.
Distinct roles for PLC and CRY in the regulation of dmpi8 splicing.
To investigate a possible role for CRY in the photoregulation of dmpi8 splicing, we used the cryb mutant, which is thought to either severely inhibit or completely inactivate CRY activity (13, 15, 48). The low daily splicing efficiency of dmpi8 in cryb flies (Fig. 6A and B) implies that a different photoreceptor(s) is mediating the inhibitory effects of light. Indeed, daytime decreases in the relative proportion of type B′ transcripts still occur in the cryb mutant (P < 0.03 by Student's t test for comparison of peak to trough values at either 18 or 29°C). However, completely ruling out any photoreceptor function for CRY in the regulation of dmpi8 is complicated by virtue of the fact that the cryb mutation also inactivates circadian regulation in some peripheral clocks (27, 32, 48). Also, because the majority of per-expressing clock cells in total head extracts are derived from ocular photoreceptors, this would mask a possible photoreceptor role for CRY in the small number of key brain pacemaker neurons that control activity rhythms. Although at present we cannot attribute a causal relationship, the levels of per mRNA are constantly low in head extracts from cryb flies (48) (data not shown), consistent with the lower relative splicing efficiency of dmpi8.
The norpAP41 mutant has been instrumental in revealing novel roles for visual phototransduction pathways in different aspects of how light regulates daily activity patterns in Drosophila (e.g., references 15, 25, 27, 28, and 48). In this regard, it is interesting that the splicing ratio of dmpi8 is essentially constantly high in the norpAP41 mutant (Fig. 6A and B). Although maximal dmpi8 splicing is higher in norpA flies than in wild-type flies, the most dramatic alteration is that daytime values remain high (P > 0.3 for peak to trough comparisons at both temperatures). The overall high splicing with low-amplitude cycling is maintained under DD conditions even at 29°C (Fig. 6C and data not shown), indicating that the norpA mutation plays a role in the regulation of dmpi8 splicing that is independent of the acute effects of light. Indeed, light pulses can still evoke rapid inhibition of dmpi8 splicing in norpA flies (Fig. 6C), revealing that PLC is not a critical or sole component of the photic-transduction pathway leading to the light inhibition of dmpi8 splicing. Rather, the findings strongly suggest that the norpA mutation strongly attenuates the robust clock-regulated daytime decrease in the proportion of the spliced type B′ variant normally observed at high temperatures. Otherwise stated, it appears that inactivation of PLC “locks” the splicing behavior of dmpi8 into a pattern characteristic of that observed on cold days, e.g., high overall splicing with low-amplitude rhythm (see Fig. 9). A greater proportion of the type B′ variant is still observed at 18°C compared to 29°C (Fig. 6A and B), indicating that although the norpA mutation has strong effects on minimizing clock-regulated daytime repression of dmpi8 splicing, it does not completely abolish the splicing sensitivity to temperature.
FIG. 9.
Model for how temperature, photoperiod, clock, and PLC regulate splicing of dmpi8. (A) Schematic representation of how temperature regulates the splicing efficiency of dmpi8. The intensity of a hypothetical heat signal is proportionally transduced via PLC to a splicing repressor that inhibits a more direct pathway, modulating the mean daily splicing efficiency (which has a clock-independent component), and a clock-regulated pathway that decreases daytime splicing. In the norpA mutant, there is less heat signal transduced to the splicing repressor, resulting in a cold splicing phenotype even on warm days. On cold days the cold splicing phenotype is augmented in norpA flies. The broken arrow indicates a possible norpA-independent inhibition of dmpi8 splicing by heat. (B) Schematic representation of how light regulates the splicing efficiency of dmpi8. The photoperiod adjusts the dynamics of the clock, which in turn modulates the timing of increases in the dmpi8 splicing rhythm and peak values (bottom). Light also has acute photoinhibitory effects that are at least partially independent of clock function (top). See the text for more details.
Consistent with the enhanced dmpi8 splicing, the levels of total per mRNA during its accumulation phase were greater in norpA flies at both temperatures compared to the control situations (Fig. 7A and B). Nonetheless, in norpA flies peak levels of total per mRNA were higher and were reached earlier at 18°C than at 29°C (Fig. 7A and B), as previously shown for wild-type flies (36). Thus, as with the daily average proportion of type B′ to type A variants (Fig. 6), the total per mRNA levels in norpA flies still respond to temperature. Intriguingly, the daily levels of the unspliced type A per variant were in a similar range at 18 and 29°C for both wild-type and norpA flies (Fig. 7C). It is mainly the abundance of the spliced type B′ per variant that increases in response to colder temperatures or the norpAP41 mutation (Fig. 7D). These results support our prior findings that cold temperatures lead to advanced and higher-amplitude cycles in total per mRNA levels mainly via a posttranscriptional mechanism that increases the splicing efficiency of dmpi8 (36), and they suggest that the norpA mutation essentially phenocopies cold temperatures.
Advanced evening activity in norpA mutants.
Under standard conditions of 12:12LD at 25°C, wild-type D. melanogaster flies exhibit bimodal daily activity patterns with one peak centered around ZT0 (morning peak) and another around ZT12 (evening peak) (Fig. 8A) (20, 53). The timing of evening activity is clearly regulated by per, whereas the morning peak of activity appears to be comprised of (i) a startle response to lights-on characterized by a rapid and transient increase in activity (20, 53), and (ii) a circadian component that might be independent of per activity (23). Increases in temperature and or photoperiod progressively delay the timing of evening activity and are associated with more pronounced “siesta times” (i.e., mid-day inactivity) (36), likely reflecting adaptive responses that ensure flies avoid desiccation during the hot mid-day hours. We previously showed that the onset and peak of the evening activity was delayed in transgenic lines that produced only type B′ transcripts (perB′) or a variant of type A transcripts having inactivated 5′ and 3′ splice sites (perA) (36). To extend these prior findings, we sought to examine the timing of evening activity in norpA and cryb flies, which have alterations in the overall splicing ratio of dmpi8 (Fig. 6) but still manifest robust circadian activity rhythms that can be entrained by daily LD cycles (15, 48, 53).
FIG. 8.
Daily locomotor activity rhythms of CS, norpAP41, and cryb mutants at different temperatures. Shown are the daily distributions of locomotor activity averaged over the fourth and fifth days of LD for wild-type (CS), norpAP41, and cryb male flies entrained to 12:12LD at either 18°C (left panels) or 29°C (right panels). Vertical bars represent the activity recorded in 30-min bins during times when the lights were on (white bars) or off (black bars). The evening activity occurred around ZT12.
As predicted by its effect on the splicing of dmpi8 (Fig. 6) and the accumulation of per mRNA levels (Fig. 7), the onset of evening activity peak in norpA flies was phase advanced at both temperatures compared to wild-type flies (Fig. 8 and Table 1). This is consistent with earlier findings showing that another allele of norpA (norpAP24) has an advanced evening activity peak under standard conditions of 12:12LD at 25°C (20, 53). The endogenous activity periods of norpA flies are approximately 30 min shorter than those in wild-type flies (20, 53) (data not shown), whereas the perA and perB′ transgenic flies generally have slightly longer periods than control flies (6, 36). While changes in the splicing efficiency of dmpi8 might affect the clock's endogenous period, it is unlikely to fully account for the phase differences in the timing of evening activity. As previously noted for the perA and perB′ transgenics, norpA flies manifest larger differences from wild-type flies in the timing of the onset of evening activity compared to when peak values are attained (Table 1). Although in cryb mutants the low dmpi8 spliced levels are consistent with the decreased amounts of total per transcripts (48) (data not shown), these flies also manifest earlier evening peaks than wild-type controls, although not as early as norpA mutants (Fig. 8 and Table 1). The reason(s) for this apparent discrepancy is not clear but might be related to the fact that in cryb flies TIM is less photosensitive, which could override the effects of decreased per mRNA and protein levels observed in this mutant (see Discussion).
Although we certainly cannot claim that the effects of norpA on the timing of evening activity are solely due to alterations in dmpi8 splicing efficiency and further work will be required to better understand how cryb ultimately alters the timing of the evening activity, the main conclusion with regards to dmpi8 splicing is that a norpA-dependent pathway likely mediates its temperature regulation, whereas neither norpA nor cry appears to play a major role in the acute photoinhibition of dmpi8 splicing.
DISCUSSION
In prior work we showed that cold-induced increases in the splicing of the per dmpi8 intron contributed to the preferential manifestation of daytime evening activity in flies as temperatures drop (36). Findings in the present study expand on this model by revealing that the clock and photoperiod also influence the splicing efficiency of dmpi8. Furthermore, our findings suggest a physiological role for PLC in downregulating the production of type B′ transcripts. The results indicate multipathway integration of photic and temperature signals by the Drosophila clock at the level of dmpi8 intron removal. We propose that, whereas the light-induced degradation of TIM is critical for the synchronization of Drosophila clocks to local time, the dual thermal and photic regulation of dmpi8 splicing acts as a seasonal sensor encoding calendar information. In this model, TIM is a major photoresponsive clock component setting the timing of the evening peak to the LD transition by regulating the posttranslational accumulation rate of PER. The splicing efficiency of dmpi8 responds to changes in temperature and photoperiod, contributing to seasonal adjustments in the timing of evening activity by influencing the per mRNA accumulation phase and, hence, the time window necessary to attain threshold concentrations of PER that favor interactions with TIM.
Regulation of dmpi8 splicing by temperature, light, and the clock.
A network of interacting molecular circuits that respond to the clock, temperature, duration of photoperiod, and acute effects of light controls the daily regulation of dmpi8 splicing efficiency. The effects of temperature and light on dmpi8 splicing are mediated through clock and clock-independent mechanisms. In general, the clock helps maintain lower dmpi8 splicing levels during the day and stimulates it during the night, reaching peak values around midnight to late night, although the shape of the dmpi8 splicing profile is strongly influenced by photoperiod and temperature.
Temperature has a major effect on the overall daily splicing efficiency of dmpi8, probably establishing a dynamic range of upper and lower limits. Under standard 12:12LD conditions the daily proportion of type B′ in wild-type flies ranges from ∼65 to 85% at 18°C and ∼35 to 50% at 29°C. In regards to setting the mean daily splicing efficiency, there is almost certainly a significant clock-independent mechanism that responds to temperature changes. For example, our prior findings demonstrated that in clock-impaired per0 flies, decreases in temperature rapidly led to concomitant increases in the proportion of type B′ to type A transcripts (36). This was also supported by the lower average daily splicing values in per0 and tim0 flies at 29°C compared to 18°C (Fig. 4). How changes in temperature directly modulate the splicing efficiency of dmpi8 is not clear.
Temperature also has a strong influence on the clock regulation of dmpi8 splicing. The amplitude of the splicing rhythm is smaller on cold days and dampens more quickly under DD conditions (Fig. 3 and 4). A notable difference is that there is a relatively more pronounced effect of the clock in driving daytime repression of dmpi8 splicing on warm compared to cold days, a feature that persists under DD conditions. Thus, there is a cold splicing phenotype that is characterized by high overall splicing of dmpi8 and low-amplitude cycling that rapidly dampens in DD. Conversely, the warm splicing phenotype is characterized by low overall splicing of dmpi8 and a relatively strong clock-regulated daytime decline in the proportion of type B′ transcripts. Because the abundance of de novo-synthesized per transcripts began to steadily increase during the day at all temperatures (e.g., Fig. 7), higher daytime values of dmpi8 splicing on cold days accelerates the per mRNA accumulation phase, whereas repression of dmpi8 splicing during the day on warm days delays the upswing in per mRNA levels. It is possible that the nighttime increases in the splicing efficiency of dmpi8 at warm temperatures (Fig. 6) also participate in delaying the per mRNA cycle by slowing down the declining phase in per transcript levels (Fig. 7). As previously shown, the higher splicing efficiency of dmpi8 at low temperatures is a seasonal adaptation that enables flies to exhibit daytime evening activity during short cold (36). In contrast, a more prominent influence of clock regulation on the daily inhibition of dmpi8 splicing during warm days might ensure that flies avoid the deleterious effects of daytime heat by favoring nocturnal evening activity. In summary, the effects of temperature on dmpi8 splicing by regulating the overall daily efficiency and the amplitude of clock-controlled daytime decreases influence the accumulation rates of per mRNA and protein products, which adjust the timing of the evening activity such that it mainly occurs during the day at low temperatures and during the night at high temperatures (36) (Fig. 9A).
The photoperiod adds another control point in the regulation of dmpi8 splicing by further adjusting the daily profile. A major effect of photoperiod is indirectly mediated via its synchronization of the phase of the clock, which regulates the timing of the dmpi8 splicing rhythm. As day length is shortened, the proportion of type B′ transcripts increases earlier and reaches higher peak values. The increased efficiency in the mean splicing of dmpi8 during short photoperiods likely underlies the earlier and more robust rhythm in total per mRNA abundance (Fig. 1). Advances in the accumulation phase of the per mRNA cycle contribute to earlier evening activity (36) (Fig. 9B). Thus, short day lengths and low temperatures, which are normally associated under natural conditions, both increase the overall splicing efficiency of dmpi8. Conversely, long photoperiods likely produce synergistic effects with warm temperatures in decreasing the splicing efficiency of dmpi8, contributing to the manifestation of mainly nocturnal evening activity (36).
Changes in photoperiod also adjust the timing of the nighttime decline in the proportion of type B′ per transcripts such that it occurs earlier under shorter photoperiods (Fig. 2). Thus, the start of clock-regulated nighttime repression essentially anticipates the beginning of the next day, when values of dmpi8 splicing are on average lower than nighttime levels. A similar scenario is also observed for the light-sensitive TIM protein, which begins its clock-controlled declining phase in the late night prior to the onset of morning (38, 55). Because photoperiods adjust the dynamics (phase and amplitude) of the clock, the duration of the light phase is remembered during the night by the timing mechanism, generating different nighttime profiles for dmpi8 splicing as a function of day length. It also appears that the duration of the photoperiod has long-range indirect effects on the basal splicing efficiency of dmpi8 that are maintained for at least 2 days under DD conditions (Fig. 3C). Differential aftereffects on circadian properties measured during DD conditions by variations in the length of the photoperiod are a well-established phenomenon in Drosophila (e.g., reference 43). How the clock ultimately regulates the splicing efficiency of dmpi8 is not clear and could be rather indirect.
In addition to a role of photoperiod in the clock regulation of dmpi8 splicing, light acutely inhibits the proportion of the type B′ variant (Fig. 4 and 5). This likely helps maintain lower daytime levels in the efficiency of dmpi8 intron removal, although the relative strength of photoinhibition appears modulated by temperature and the clock (Fig. 4 and 5). Certainly, even though light pulses evoke rapid decreases in dmpi8 splicing (Fig. 5), wild-type and clock mutant flies exposed to LD cycles of 12 h of light still exhibit a significant amount of dmpi8 splicing at low and high temperatures (Fig. 4). Thus, a certain basal level of dmpi8 splicing efficiency, which varies as a function of temperature, photoperiod, and clock regulation, appears refractory to light-mediated inhibition. The acute photoinhibition in the proportion of the type B′ variant might enable the dmpi8 splicing efficiency to rapidly respond to seasonal changes in day length. For example, as the sun sets later, the prolonged acute inhibition by light could delay the rise in dmpi8 splicing efficiency (Fig. 5), whose timing would reflect the internal clock dynamics set by the previous day's slightly shorter photoperiod. This might also explain why the acute photoinhibition of dmpi8 splicing is at least partially clock independent.
Given the highly integrated molecular circuitry underlying the Drosophila clock, it is certain that temperature and photoperiod regulate a wide range of diverse steps in the oscillatory mechanism, likely ranging from transcriptional rates to clock protein turnover. Our data identify the temperature and photoperiodic control of dmpi8 splicing as one important control center that regulates the dynamics of the per RNA cycle and ultimately seasonal adjustments in the duration of time necessary to reach levels of PER that favor formation of the PER-TIM complex, a critical event in the progression of the clock and the timing of evening activity.
A novel nonphotic role for PLC in the temperature regulation of dmpi8 splicing.
CRY does not appear to be an important photoreceptor in mediating the photoinhibition of dmpi8 splicing, but it does play a role in generating daily cycles in the proportion of the type B′ per variant. This was suggested by the observations that in cryb mutants the mean daily levels of dmpi8 splicing were low and daytime values were lower than during the night (Fig. 6). Because arrhythmic clock mutants (such as per0 and tim0) also resulted in low daily average levels of dmpi8 splicing (Fig. 4), the effect of CRY is more likely to be indirectly related to its core clock function (27, 32, 48) and not in its capacity as a circadian-relevant blue-light photoreceptor. Why do cryb flies have advanced evening activity peaks if they have low dmpi8 splicing and per mRNA levels? Here the answer might lie with the photoreceptor function mediating the rapid light-induced degradation of TIM (4, 13, 14, 48). This raises the possibility that a more robust accumulation of TIM in certain key pacemaker cells of cryb flies might override the lower levels of per mRNA contributing to the observed advanced evening phase. However, the situation is complex, because the cryb mutation has differential effects on PER-TIM dynamics in different groups of brain pacemaker neurons (25, 28, 48, 51). In any case, because the photosensitivity of TIM is regulated by CRY in at least some pacemaker cells and TIM influences the stability of PER, it is unclear at present how the low splicing of dmpi8 in the cryb flies might contribute to the altered activity rhythms observed in this mutant. This also raises a potential limitation of our study, as we used whole-head extracts to analyze the splicing levels of the dmpi8 intron. Because most of the clocks in the head are present in ocular photoreceptors, we cannot rule out the possibility that the splicing behavior of dmpi8 is different in the small number of key brain pacemaker neurons required for locomotor activity rhythms. In any event, while future studies will be required to address how cryb alters the timing of evening activity, our data strongly suggest that the circadian-relevant CRY photoreceptor is not a major factor in mediating the light-induced inhibition of dmpi8 splicing.
In contrast, the constitutively high splicing efficiency in norpA flies suggests that PLC has a role in downregulating dmpi8 splicing (similar results have been obtained by others [8; C. Kyriacou, personal communication]). However, the norpA mutation does not appear to have a major role in blocking the ability of light pulses to inhibit the proportion of type B′ transcripts (Fig. 6C). Rather, the findings suggest a novel nonphotic function in mediating increases in splicing repression as temperatures rise. The most dramatic difference between norpA and wild-type flies is that splicing does not decline during the daytime in norpA flies, especially at higher temperatures, where in wild-type flies the daytime decline is relatively greater than in cold temperatures (compare Fig. 3A and 6C). Thus, even at warm temperatures, norpA flies exhibit the cold splicing phenotype characterized by high overall dmpi8 splicing with relatively little clock-controlled daytime decline, a pattern that persists into the first day of constant darkness. These results suggest that in the absence of PLC, the default splicing behavior is that normally observed on cold days.
While there are different models that can be formulated to explain the effects of the norpA mutation on the daily splicing behavior of dmpi8, we favor a simple model whereby PLC/NORPA participates in the temperature-sensing pathway (Fig. 9) (a similar model was also recently proposed [8]). We posit that there is a heat signal whose strength increases as temperatures rise. PLC is required to transduce the intensity of this heat signal into a proportional splicing repression response. More heat signal leads to more PLC-dependent splicing repression. The splicing inhibition mechanism regulates the overall dmpi8 splicing efficiency and the relative strength of the clock-regulated daytime decline (Fig. 9A). Such a model can explain why overall splicing is high and there is little clock-induced daytime inhibition on cold days (little heat signal, low splicing repression) and how inactivation of norpA results in the cold splicing phenotype and early activity even on warm days (strong heat signal but no transduction to splicing repressor). Because there is still less splicing in norpA flies at warm temperatures compared to cold temperatures (Fig. 6), we also suggest that either the norpA mutation is not totally inactive with regards to dmpi8 splicing and/or there is at least some norpA-independent temperature sensing for dmpi8 splicing regulation. Another possibility is that the temperature sensor is intact in the norpA mutant but PLC is required for correct temperature calibration. High-amplitude cycling of other clock-controlled events in norpA flies, such as per mRNA rhythms (Fig. 7), strongly argues that norpA does not directly affect core clock dynamics but plays a role between the daytime temporal signal and splicing inhibition (Fig. 9A). The data strongly suggest that the main role of PLC in regulating dmpi8 splicing is related to sensing temperature and not light. It will be of interest to determine whether PLC affects the splicing efficiency of other introns.
At present it is not known how PLC might downregulate the production of type B′ transcripts in a temperature-dependent manner. The loss-of-function mutation in norpAP41 flies causes the compound eyes and ocelli to be completely unresponsive to light (40). While some of the effects of norpA on fly activity are likely due to its role in ocular photic signal transduction (e.g., startle response) (53), as noted above this is not the case for dmpi8 splicing efficiency. Further support for this contention is based on findings showing that several physiologically blind mutants that affect visual photic signal transduction (e.g., ninaA1, ninaE5, ninaE7, and ninaE8) do not exhibit an increase in the splicing efficiency of dmpi8 (data not shown). Moreover, in contrast to norpA mutants, several eyeless mutants manifest longer periods and/or delayed evening activity peaks compared to wild-type controls (24, 43, 52). In this context, it is noteworthy that norpA products are expressed in the eyes and brain, possibly including brain pacemaker neurons critical for rhythmic activity (28, 37, 56). Also, different isoforms of norpA gene products arising from alternative splicing are differentially expressed in the eyes and brain, although the function of extraretinal PLC is not known (29). Future work will be required to determine the extent to which the advanced evening peak in norpA flies is due to its effects on dmpi8 splicing, to identify the putative pacemaker cells where norpA-dependent changes in dmpi8 splicing efficiency presumably influence the timing of evening activity, and to understand how temperature might modulate the ability of PLC to inhibit this splicing reaction. Furthermore, the identity of the photic transduction pathway mediating the acute light-mediated inhibition of dmpi8 splicing remains elusive but does not seem to involve major participation from either PLC or CRY.
In summary, our findings suggest that the thermal and photoperiodic regulation of dmpi8 splicing acts as a seasonal sensor conveying calendar information to Drosophila, endowing the fly with the ability to optimize its daily distribution of activity in accordance with the prevailing environmental conditions. The results also identify a novel nonphotic role for norpA in mediating direct and clock-mediated effects of temperature on the splicing efficiency of dmpi8, and they further illustrate the complexity of how day length, temperature, and clocks interface to regulate seasonal adaptation in animals and plants.
Acknowledgments
We are grateful to Ben Collins and Bambos Kyriacou for sharing unpublished data. We thank the members of the Edery lab for comments.
This work was supported by Public Health Service grant NS42088 from the National Institute of Neurological Disorders and Stroke to I.E.
REFERENCES
- 1.Allada, R. 2003. Circadian clocks: a tale of two feedback loops. Cell 112:284-286. [DOI] [PubMed] [Google Scholar]
- 2.Allada, R., N. E. White, W. V. So, J. C. Hall, and M. Rosbash. 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93:791-804. [DOI] [PubMed] [Google Scholar]
- 3.Bae, K., C. Lee, P. E. Hardin, and I. Edery. 2000. dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex. J. Neurosci. 20:1746-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceriani, M. F., T. K. Darlington, D. Staknis, P. Mas, A. A. Petti, C. J. Weitz, and S. A. Kay. 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285:553-556. [DOI] [PubMed] [Google Scholar]
- 5.Ceriani, M. F., J. B. Hogenesch, M. Yanovsky, S. Panda, M. Straume, and S. A. Kay. 2002. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 22:9305-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Y., B. Gvakharia, and P. E. Hardin. 1998. Two alternatively spliced transcripts from the Drosophila period gene rescue rhythms having different molecular and behavioral characteristics. Mol. Cell. Biol. 18:6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Citri, Y., H. V. Colot, A. C. Jacquier, Q. Yu, J. C. Hall, D. Baltimore, and M. Rosbash. 1987. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature 326:42-47. [DOI] [PubMed] [Google Scholar]
- 8.Collins, B. H., E. Rosato, and C. P. Kyriacou. 2004. Seasonal behavior in Drosophila melanogaster requires the photoreceptors, circadian clock, and phospholipase C. Proc. Natl. Acad. Sci. USA 10.1073/pnas.0308240100. [DOI] [PMC free article] [PubMed]
- 9.Cyran, S. A., A. M. Buchsbaum, K. L. Reddy, M. C. Lin, N. R. Glossop, P. E. Hardin, M. W. Young, R. V. Storti, and J. Blau. 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112:329-341. [DOI] [PubMed] [Google Scholar]
- 10.Darlington, T. K., K. Wager-Smith, M. F. Ceriani, D. Staknis, N. Gekakis, T. D. L. Steeves, C. J. Weitz, J. S. Takahashi, and S. A. Kay. 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280:1599-1603. [DOI] [PubMed] [Google Scholar]
- 11.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96:271-290. [DOI] [PubMed] [Google Scholar]
- 12.Edery, I. 2000. Circadian rhythms in a nutshell. Physiol. Genomics 3:59-74. [DOI] [PubMed] [Google Scholar]
- 13.Emery, P., W. V. So, M. Kaneko, J. C. Hall, and M. Rosbash. 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95:669-679. [DOI] [PubMed] [Google Scholar]
- 14.Emery, P., R. Stanewsky, J. C. Hall, and M. Rosbash. 2000. A unique circadian-rhythm photoreceptor. Nature 404:456-457. [DOI] [PubMed] [Google Scholar]
- 15.Emery, P., R. Stanewsky, C. Helfrich-Forster, M. Emery-Le, J. C. Hall, and M. Rosbash. 2000. Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26:493-504. [DOI] [PubMed] [Google Scholar]
- 16.Glossop, N. R., J. H. Houl, H. Zheng, F. S. Ng, S. M. Dudek, and P. E. Hardin. 2003. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 37:249-261. [DOI] [PubMed] [Google Scholar]
- 17.Glossop, N. R., L. C. Lyons, and P. E. Hardin. 1999. Interlocked feedback loops within the Drosophila circadian oscillator. Science 286:766-768. [DOI] [PubMed] [Google Scholar]
- 18.Hall, J. C. 2000. Cryptochromes: sensory reception, transduction, and clock functions subserving circadian systems. Curr. Opin. Neurobiol. 10:456-466. [DOI] [PubMed] [Google Scholar]
- 19.Hall, J. C. 2003. Genetics and molecular biology of rhythms in Drosophila and other insects. Adv. Genet. 48:1-280. [DOI] [PubMed] [Google Scholar]
- 20.Hamblen-Coyle, M. J., D. A. Wheeler, J. E. Rutila, M. Rosbash, and J. C. Hall. 1992. Behavior of period-altered circadian rhythm mutants of Drosophila in light:dark cycles (Diptera: Drosophilidae). J. Insect Behav. 5:417-446. [Google Scholar]
- 21.Hardin, P. E., J. C. Hall, and M. Rosbash. 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343:536-540. [DOI] [PubMed] [Google Scholar]
- 22.Hastings, J. W., B. Rusak, and Z. Boulos. 1991. Circadian rhythms: the physiology of biological timing, p. 435-546. In C. L. Prosser (ed.), Neural and integrative animal physiology. Wiley-Liss Inc., New York, N.Y.
- 23.Helfrich-Forster, C. 2001. The locomotor activity rhythm of Drosophila melanogaster is controlled by a dual oscillator system. J. Insect Physiol. 47:877-887. [Google Scholar]
- 24.Helfrich-Forster, C., T. Edwards, K. Yasuyama, B. Wisotzki, S. Schneuwly, R. Stanewsky, I. A. Meinertzhagen, and A. Hofbauer. 2002. The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J. Neurosci. 22:9255-9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helfrich-Forster, C., C. Winter, A. Hofbauer, J. C. Hall, and R. Stanewsky. 2001. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30:249-261. [DOI] [PubMed] [Google Scholar]
- 26.Hunter-Ensor, M., A. Ousley, and A. Sehgal. 1996. Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 84:677-685. [DOI] [PubMed] [Google Scholar]
- 27.Ivanchenko, M., R. Stanewsky, and J. M. Giebultowicz. 2001. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms 16:205-215. [DOI] [PubMed] [Google Scholar]
- 28.Kaneko, M., M. J. Hamblen, and J. C. Hall. 2000. Involvement of the period gene in developmental time-memory: effect of the per Short mutation on phase shifts induced by light pulses delivered to Drosophila larvae. J. Biol. Rhythms 15:13-30. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S., R. R. McKay, K. Miller, and R. D. Shortridge. 1995. Multiple subtypes of phospholipase C are encoded by the norpA gene of Drosophila melanogaster. J. Biol. Chem. 270:14376-14382. [DOI] [PubMed] [Google Scholar]
- 30.Kloss, B., J. L. Price, L. Saez, J. Blau, A. Rothenfluh, C. S. Wesley, and M. W. Young. 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iɛ. Cell 94:97-107. [DOI] [PubMed] [Google Scholar]
- 31.Konopka, R. J., and S. Benzer. 1971. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 68:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnan, B., J. D. Levine, M. K. Lynch, H. B. Dowse, P. Funes, J. C. Hall, P. E. Hardin, and S. E. Dryer. 2001. A new role for cryptochrome in a Drosophila circadian oscillator. Nature 411:313-317. [DOI] [PubMed] [Google Scholar]
- 33.Lee, C., K. Bae, and I. Edery. 1998. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron 21:857-867. [DOI] [PubMed] [Google Scholar]
- 34.Lee, C., K. Bae, and I. Edery. 1999. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol. 19:5316-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, C., V. Parikh, T. Itsukaichi, K. Bae, and I. Edery. 1996. Resetting the Drosophila clock by photic regulation of PER and a PER-TIM complex. Science 271:1740-1744. [DOI] [PubMed] [Google Scholar]
- 36.Majercak, J., D. Sidote, P. E. Hardin, and I. Edery. 1999. How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24:219-230. [DOI] [PubMed] [Google Scholar]
- 37.Malpel, S., A. Klarsfeld, and F. Rouyer. 2002. Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development 129:1443-1453. [DOI] [PubMed] [Google Scholar]
- 38.Myers, M. P., K. Wager-Smith, A. Rothenfluh-Hilfiker, and M. W. Young. 1996. Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271:1736-1740. [DOI] [PubMed] [Google Scholar]
- 39.Naidoo, N., W. Song, M. Hunter-Ensor, and A. Sehgal. 1999. A role for the proteasome in the light response of the timeless clock protein. Science 285:1737-1741. [DOI] [PubMed] [Google Scholar]
- 40.Pearn, M. T., L. L. Randall, R. D. Shortridge, M. G. Burg, and W. L. Pak. 1996. Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J. Biol. Chem. 271:4937-4945. [DOI] [PubMed] [Google Scholar]
- 41.Price, J. L., J. Blau, A. Rothenfluh, M. Abodeely, B. Kloss, and M. W. Young. 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94:83-95. [DOI] [PubMed] [Google Scholar]
- 42.Qiu, J., and P. E. Hardin. 1996. per mRNA cycling is locked to lights-off under photoperiodic conditions that support circadian feedback loop function. Mol. Cell. Biol. 16:4182-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rieger, D., R. Stanewsky, and C. Helfrich-Forster. 2003. Cryptochrome, compound eyes, Hofbauer-Buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila melanogaster. J. Biol. Rhythms 18:377-391. [DOI] [PubMed] [Google Scholar]
- 44.Rutila, J. E., V. Suri, M. Le, W. V. So, M. Rosbash, and J. C. Hall. 1998. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 93:805-814. [DOI] [PubMed] [Google Scholar]
- 45.Sehgal, A., J. L. Price, B. Man, and M. W. Young. 1994. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263:1603-1606. [DOI] [PubMed] [Google Scholar]
- 46.Shafer, O. T., M. Rosbash, and J. W. Truman. 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J. Neurosci. 22:5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanewsky, R. 2003. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J. Neurobiol. 54:111-147. [DOI] [PubMed] [Google Scholar]
- 48.Stanewsky, R., M. Kaneko, P. Emery, B. Beretta, K. Wager-Smith, S. A. Kay, M. Rosbash, and J. C. Hall. 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95:681-692. [DOI] [PubMed] [Google Scholar]
- 49.Suri, V., Z. Qian, J. C. Hall, and M. Rosbash. 1998. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 21:225-234. [DOI] [PubMed] [Google Scholar]
- 50.Sweeney, B. M., and J. W. Hastings. 1960. Effects of temperature upon diurnal rhythms. Cold Spring Harb. Symp. Quant. Biol. 25:87-104. [DOI] [PubMed] [Google Scholar]
- 51.Veleri, S., C. Brandes, C. Helfrich-Forster, J. C. Hall, and R. Stanewsky. 2003. A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr. Biol. 13:1758-1767. [DOI] [PubMed] [Google Scholar]
- 52.Vosshall, L. B., and M. W. Young. 1995. Circadian rhythms in Drosophila can be driven by period expression in a restricted group of central brain cells. Neuron 15:345-360. [DOI] [PubMed] [Google Scholar]
- 53.Wheeler, D. A., M. J. Hamblen-Coyle, M. S. Dushay, and J. C. Hall. 1993. Behavior in light-dark cycles of Drosophila mutants that are arrhythmic, blind, or both. J. Biol. Rhythms 8:67-94. [DOI] [PubMed] [Google Scholar]
- 54.Yang, Z., M. Emerson, H. S. Su, and A. Sehgal. 1998. Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron 21:215-223. [DOI] [PubMed] [Google Scholar]
- 55.Zeng, H., Z. Qian, M. P. Myers, and M. Rosbash. 1996. A light-entrainment mechanism for the Drosophila circadian clock. Nature 380:129-135. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, L., R. R. McKay, and R. D. Shortridge. 1993. Tissue-specific expression of phospholipase C encoded by the norpA gene of Drosophila melanogaster. J. Biol. Chem. 268:15994-16001. [PubMed] [Google Scholar]