Abstract
O -glycopeptides are often acidic owing to the frequent occurrence of acidic saccharides in the glycan, rendering traditional proteomic workflows that rely on positive mode tandem mass spectrometry (MS/MS) less effective. In this report, we demonstrate the utility of negative mode ultraviolet photodissociation (UVPD) MS for the characterization of acidic O-linked glycopeptide anions. This method was evaluated for a series of singly- and multiply-deprotonated glycopeptides from the model glycoprotein kappa casein, resulting in production of both peptide and glycan product ions that afforded 100% sequence coverage of the peptide and glycan moieties from a single MS/MS event. The most abundant and frequent peptide sequence ions were a/x-type products, which, importantly, were found to retain the labile glycan modifications. The glycan-specific ions mainly arose from glycosidic bond cleavages (B, Y, C, and Z ions) in addition to some less common cross-ring cleavages. Based on the UVPD fragmentation patterns, an automated database searching strategy (based on the MassMatrix algorithm) was designed that is specific for the analysis of glycopeptide anions by UVPD. This algorithm was used to identify glycopeptides from mixtures of glycosylated and non-glycosylated peptides, sequence both glycan and peptide moieties simultaneously, and pinpoint the correct site(s) of glycosylation. This methodology was applied to uncover novel site-specificity of the O-linked glycosylated OmpA/MotB from the “superbug” A. baumannii to help aid in the elucidation of the functional role that protein glycosylation plays in pathogenesis.
INTRODUCTION
Glycosylation is one of the most common protein post-translational modifications, regulating an array of structural and functional processes in cellular systems.1–5 The site-specificity of glycan attachment is categorized in two general ways: (1) N-linked glycosylations modify asparagine at a specific sequon of Asn-Xxx-Ser or Asn-Xxx-Thr (for which Xxx can be any amino acid except proline) and (2) O-linked glycosylation modifies serine or threonine with no characterized specific sequence motif. Prokaryotic cells were long thought not to contain glycoproteins,3 until the mid-1970s when surface layer (S-layer) proteins were first identified to be glycosylated.6, 7 More recently, non-S-layer glycoproteins have been increasingly identified in a variety of pathogenic Gram negative bacteria3–5, 8 and have been found to play a significant role in biological function.3–5 For example, glycosylation via an N-linked heptasaccharide has been well characterized in Campylobacter jejuni3, 9–11 and has been identified in over 50 unique proteins (a total of 150 proteins are predicted to be modified by this glycan).10 O-linked glycosylation of multiple proteins, including flagellar and/or pilin proteins, have also been identified in several pathogenic bacteria including Campylobacter jejuni, Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa, and Helicobacter pylori (among others).3–5 The aerobic Gram negative bacteria Acinetobacter baumannii was recently discovered to possess a novel O-linked glycosylation system, which was found to affect biofilm formation and virulence in three infection models.12 Due to the increasing appearance of multi- and panresistant strains, this bacterium is considered a “superbug”, and thus understanding the mechanisms of its pathogenesis may undercover new strategies for intervention against A. baumannii infections. The O-glycosylated proteins found in A. baumannii represent novel targets important for the development of next-generation antibiotics.12
Mass spectrometry (MS) has become an indispensable tool in the characterization of glycoproteins.13 Tandem MS (MS/MS) has been particularly useful as it can identify both glycan construction and sequence sites of glycosylation in glycopeptides (depending on the MS/MS method employed) by activation and dissociation of precursor ions into interpretable glycan and/or peptide fragments. Collision-induced dissociation (CID) is the most routinely used MS/MS technique and has been particularly effective for sequencing the glycan portion of glycoproteins via predictable cleavage at glycosidic bonds.13–16 N-linked glycans are routinely released from glycoproteins by the selective enzyme PNGase F,17 and can be enriched by a number of methods based on their significantly higher hydrophilicity compared to proteins. The combination of PNGase F-induced glycan release and CID is a powerful duet for the elucidation of N-glycan structures.13 Unfortunately, release of O-glycans via enzymes or chemicals is not selective, and thus no universal method has been established for this task.18–20 O-glycan structures therefore are most accurately deduced from protease-digested glycopeptides.14, 18, 19, 21–31 This point and the fact that O-glycan core structure are very heterogeneous (significantly more so compared to N-glycans) makes structure elucidation a substantial analytical challenge.14, 20
Key hurdles in defining functional roles of protein glycosylation are not only the elucidation of the glycan structures, but also the determination of their sites of attachment to the protein. For N-linked glycosylation, the specific sequon of N-X-S/T makes site determination fairly predictable by employing conventional bottom-up proteomic approaches.13, 14, 32 Moreover, N-linked sequons as specific as D/E-X-N-X-S/T have been observed, such as in Campylobacter jejuni.10, 33 CID in general cleaves only the glycan portion of glycopeptides leaving the peptide portion intact (and consequently the peptide sequence unidentified).13–16 While peptide fragments are sometimes observed (usually with higher-energy, beam-type and TOF-TOF collision activation),10, 34, 35 these products are typically devoid of glycans, thus rendering site determination difficult. Glycan sites can still often be deduced, however, due to the specific N-linked sequon.10, 34–38 MS/MS/MS of [peptide – glycan] product ions has proven useful for increasing peptide sequence coverage and determining N-linked sites (given the specificity of the sequon)39–41 but is difficult to perform in an automated fashion due to the heterogeneity of the attached glycans. Conversely, O-linked glycosylation can occur at both serine and threonine residues, and no known sequence motif exists. These glycan types are often observed in areas rich in serine and threonines, making site-specific determination far more challenging.18, 19, 25, 42 For example, given a typical proteolyzed glycopeptide containing several S and T residues, almost complete peptide sequence coverage would be needed (with no detachment of glycan) in order to confidently pinpoint the exact site(s) of modification - a demanding task for any MS/MS method.
To increase confidence in site-specific glycosylation analysis, alternative methods such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) have recently been employed to increase peptide sequence information of both N- and O-glycopeptides.10, 15, 16, 18, 23, 26–28, 30, 31, 42,44 The combination of CID to sequence the glycan portion and ECD/ETD to sequence the peptide portion has shown promise in the total sequencing of glycopeptides.15, 16 More recently, glycan-specific oxonium ions produced by higher energy collision dissociation (HCD) were used to trigger ETD scans on-the-fly, facilitating high-throughput characterization of N-linked glycopeptides by both dissociation techniques.43 Ye et al. combined the use of HCD, ETD, and CID in a complementary manner to afford complete structural elucidation of glycopeptides.44 In another approach, Zhang and Reilly reported the use of high-energy 157 nm UV photodissociation for analysis of protonated N-glycopeptides to simultaneously extract both glycan and peptide sequence information.45
MS analysis of glycopeptides has been performed predominantly in the positive mode (i.e., the detection of cations).13, 22 This widespread use of the positive mode is a result of standard “bottom-up” protocols involving the use of trypsin as a protease, which yields peptides with at least two basic protonation sites (i.e. N-terminus and R or K at C-terminus). However, glycan attachment sites are often located in regions devoid of lysines and arginines,18, 19, 25, 42 and can moreover block the activity of trypsin.13 Furthermore, acidic saccharides such as sialic acid, glucuronic acid, and pseudaminic acid make glycopeptides substantially less basic. Several laboratories have used sample preparation techniques that cleave off acidic monosaccharides (e.g., sialic acid) prior to positive mode MS, but these methods often impede the elucidation of native glycan conformations.14, 19 Other groups have sought to derivatize the glycans (such as with permethylation) to increase glycan and/or glycopeptide basicity and thus improve positive mode detection,13, 19 at the expense of longer sample preparation time.13, 19 Recently, Nwoso, Lebrilla, and coworkers highlighted the benefits of negative mode analysis over the positive mode owing to detection of substantially more glycopeptides as anions.22 It was also shown in an inter-laboratory assessment of methods used for O-glycosylation characterization that native glycopeptides analysis by negative mode MS yielded superior results compared to other common techniques (including ones utilizing positive mode MS).19
A key reason negative mode analysis is not as widespread as its positive mode counterpart is the lack of MS/MS techniques that can yield predictable and extensive peptide and glycan sequence information for glycopeptides (i.e., CID generally yields less predictable fragmentation patterns for peptide anions46–48 and ECD/ETD are not applicable to anion analysis). Recently, strides have been made in peptide anion dissociation from the development of MS/MS methods such as electron detachment dissociation (EDD),49, 50 negative electron transfer dissociation (NETD),51–53 and activated – electron photodetachment dissociation (a-EPD).54, 55 These methods have shown merits for the production of predictable a/x-type peptide sequence ions and high overall sequence coverage (although not yet widely applied to glycopeptides).
Recently, we used 193 nm photodissociation (UVPD) in the negative mode to characterize a mixture of mitogen-activated pathway kinases and demonstrated the ability to produce high peptide sequence coverage for a range of charge states (from singly up to triply deprotonated) in a high-throughput LCMS workflow.56 We also applied this technique to the characterization of deprotonated acidic oligosaccharides (i.e., ones containing sialic acid), and illustrated how UVPD generates increased amounts of diagnostic product ion relative to CID.57 The analytical metrics of UVPD for identification of negatively charged peptides were assessed more recently by characterizing complex proteomics samples with a novel database searching strategy, facilitating the identification of significantly more proteins compared to CID, HCD, and ETD alone.58 Furthermore, negative mode UVPD was employed to uncover the first tyrosine sulfation in prokaryotes, after traditional positive mode CID failed to identify this important post-translational modification.59 Based on our promising results for deprotonated peptides and oligosaccharides individually,55,56 we extended our UVPD capabilities to the characterization of O-linked glycopeptide anions as described in this report. This method is applied to singly and multiply deprotonated glycopeptides, and the resulting fragmentation behavior is described herein. We also designed an automated database searching strategy (based on the MassMatrix algorithm60–63) which is specific for the analysis of glycopeptide anions by UVPD. The O-linked glycosylated protein bovine kappa casein was digested with GluC to evaluate the performance of UVPD and MassMatrix for identifying glycopeptides from mixtures of glycosylated and non-glycosylated species, sequencing both the glycan and peptide simultaneously, and pinpointing the correct site of modification. We then utilized this methodology for site-specific analysis of O-linked acidic glycopeptide species from A. baumannii OmpA/MotB.
EXPERIMENTAL
Materials, Reagents, and Protein Preparation
Zwitterionic hydrophilic interaction chromatography solid phase extraction (ZIC-HILIC SPE) cartridges with 25 mg of packing material were obtained from SeQuant (Southboro, MA). GluC, Trypsin Gold, and ProtaseMax were purchased from Promega (Madison, WI). All solvents were obtained from Fisher Scientific (Fair Lawn, NJ). Bovine kappa casein and all other reagents were purchased from Sigma Aldrich (St. Louis, MO). OmpA/MotB was cloned as described in Supplemental Information and purified from the soluble membrane protein extract via Ni2+-NTA chromatography using the ATKA explorer.
Protein processing
Proteins were reduced, alkylated, and digested using GluC or a cocktail of GluC and trypsin as described in Supplemental Information. Digested glycoproteins were dried, reconstituted in 1 mL of 80% acetonitrile/15% water/5% formic acid, and enriched using a ZIC-HILIC SPE cartridge.
Liquid Chromatography and Mass Spectrometry
Separation of enriched glycopeptides was undertaken using a Dionex Acclaim PepMap RSLC C18 analytical column (Santa Clara, CA) (75 μm × 15 cm, 2 μm particle size). Separation was performed with eluent A consisting of 0.05% acetic acid in water and eluent B consisting of 0.05% acetic acid in acetonitrile with a 120 min linear gradient from 5% to 30% eluent B at a flow rate of 300 nL/min.
Mass spectrometric analysis was performed in a data-dependent mode on a Thermo Fisher Scientific LTQ Velos dual linear ion trap mass spectrometer (San Jose, CA) equipped with a Coherent (Santa Clara, CA) ExciStar XS excimer laser operated at 193 nm. The coupling of the laser to a linear ion trap mass spectrometer was described in detail previously.64, 65 A series of four 2 mJ UV pulses (applied during an 8 ms activation period) was used per MS/MS scan with a q-value set to 0.1. A normalized collision energy of 35%, a q-value of 0.25, and an activation period of 10 ms was used for CID. The maximum ion injection time for all experiments was 100 ms for both survey scan and MS/MS events.
MassMatrix Automated Searching of Glycopeptides
UVPD generates product ions from both the peptide and glycan portions of glycopeptide anions in a single spectrum. A modified version of the MassMatrix60–63 database searching algorithm was developed in this study for the simultaneous identification of both types of ions, thus allowing the complete sequencing of glycopeptides. Based on fragmentation behavior previously characterized,56, 58 MassMatrix was first modified to search peptide-specific product ions (ones generated from cleavage of the peptide backbone) associated with UVPD of peptide anions. The algorithm was then adapted further to search glycan-specific product ions (ones generated from cleavages throughout the glycan) and neutral-loss ions from the fragmentation trends observed in this study. The peptide and glycan-specific ions incorporated into MassMatrix are summarized in Supplemental Table 1 and Supplemental Table 2, respectively. The main scoring models used for MassMatrix are the pp and pp2 scores, which are a statistical measure of the number of matched product ions and the total abundance of matched product ions, respectively.60 A set of pp and pp2 scores are generated for both the peptide and glycan products for each spectrum collected from LC-MS/UVPD runs; these scores can then be combined to create a “glycopeptide pp score” (peptide pp2 x glycan pp). Additional details about the MassMatrix database search algorithm and parameters are provided in Supplemental Information.
RESULTS AND DISCUSSION
The overall experimental strategy developed and utilized in this report is illustrated in Supplemental Scheme 1. First, glycoproteins were digested with GluC or a cocktail of specific proteases (e.g., GluC + trypsin). The resulting glycopeptides were then enriched via ZIC-HILIC solid phase extraction, separated using reversed phase nanoHPLC, transferred as anions into the mass spectrometer via nanoESI in the negative mode, and activated and dissociated by UVPD. Lastly, the database searching algorithm, MassMatrix, was adapted for glycopeptide identification by means of simultaneous automated sequencing of both the glycan and peptide moieties and for pinpointing the correct site(s) of amino acid modification.
Evaluation of the fragmentation behavior of glycopeptide anions by UVPD
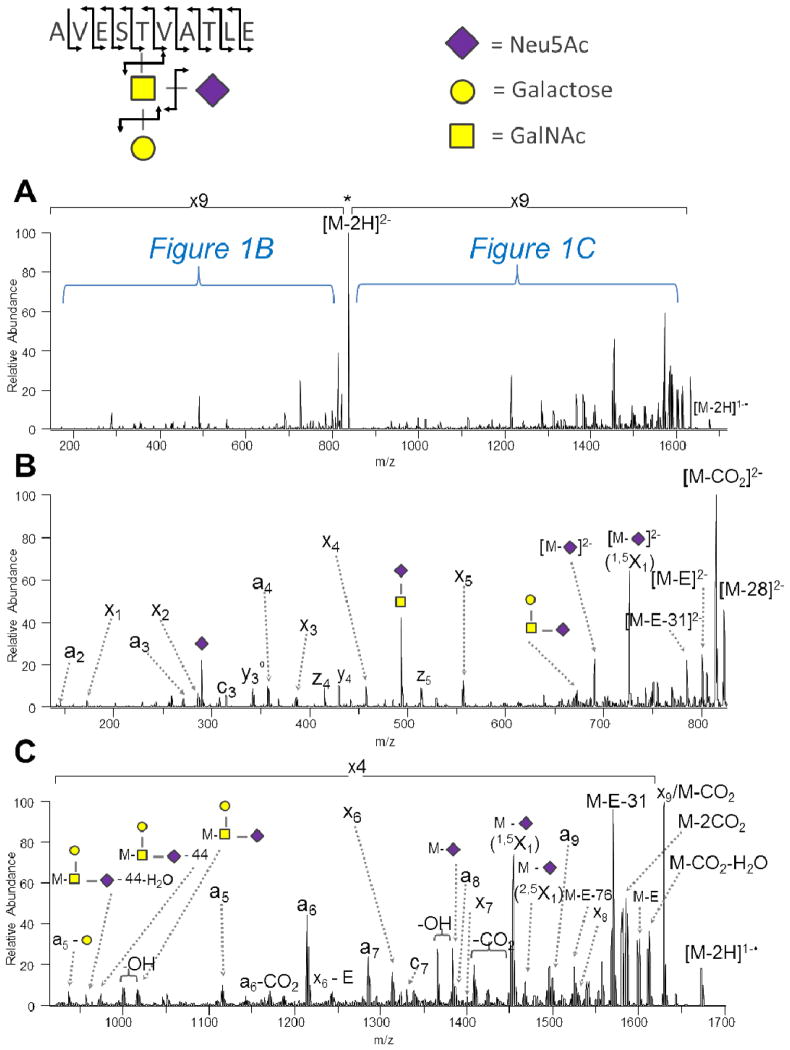
To establish the utility of UVPD for sequencing acidic glycopeptides, protease-digested glycoproteins with known glycans and glycosylation sites were used to develop the workflow, optimize the methodology, and evaluate the general fragmentation trends associated with peptide and glycan bond cleavages. The UVPD fragmentation behavior of glycopeptide anions was mapped using GluC-digested kappa casein, a glycoprotein consisting of a total of six known O-glycosylation sites located in serine- and threonine-rich regions and a wide variety of glycan types, many of which include sialic acid (Neu5Ac). GluC was used to generate glycopeptides with at least one acidic amino acid (glutamic acid or aspartic acid) at the C-terminus, and therefore facilitate ionization in the negative mode and generate higher charge states. The great majority of kappa casein glycopeptide anions observed after ZIC-HILIC enrichment, RP-HPLC separation, and negative ESI-MS detection were doubly deprotonated. The dominance of [M – 2H]2− peptide ions upon negative mode ESI has been observed previously53, 56 and is well suited for UVPD analysis.56 As one example, Figure 1 illustrates the fragmentation pattern for the doubly deprotonated glycopeptide AVEST(HexNAc + Hex + Neu5Ac)VATLE from a GluC digest of kappa casein. This glycopeptide has a very low pI due to the presence of two acidic amino acids (both glutamic acid), no basic residues, and the sialic acid saccharide in the glycan, thus favoring efficient ionization in the negative mode. To better illustrate the spectral detail of the UVPD spectrum, the range from m/z 150 to 800 is expanded in Figure 1B and the m/z range beyond m/z 800 is shown in Figure 1C. Similar to what was previously reported by Zhang and Reilly for vacuum UV fragmentation of positively charged N-glycopeptides,45 glycan fragments attached to an intact peptide and peptide fragments attached to an intact glycan are also observed upon UVPD of negatively charged O-glycopeptides, a characteristic that facilitates the complete characterization of glycopeptides. The most abundant and frequent peptide sequence ions observed in the anion UVPD spectrum are a/x-type products, which are also the main diagnostic ions observed previously for unmodified, phosphorylated, and sulfated peptide anions upon UVPD.56, 58, 59 Importantly, this series of ions not only yields 100% sequence coverage (as illustrated by the dotted lines denoting cleavage sites at the upper left corner of Figure 1), but all the product ions that contain the modified T also retain the glycan moiety. For example, in Figure 1C the product ions a5, a6, a7, a8, a9 and x6, x7, x8, x9 all contain the trisaccharide. This information can readily be used to pinpoint the correct site of glycosylation (the fifth threonine from the N-terminus) even with several other possible serine and threonine residues present in the peptide (one of which is directly adjacent to the modified residue). Glycan-specific ions are also present in the spectrum. These products are most commonly of two varieties: low m/z glycan-only product anions (i.e., B and C ions) and neutral losses of glycan segments from the intact peptide (i.e., [M – glycan moiety]−, Y and Z ions). Due to mass differences between these two varieties of ions, the glycan-only product ions are dominant in the lower m/z range (Figure 1B), and the [M – glycan moiety]− ions types are more prominent in Figure 1C (higher m/z range). The culmination of these glycan-specific ions allows the complete sequencing of the glycan section of the glycopeptide. Glycan analysis in ion traps has often relied on multiple stages of MS (MSn) as pioneered by the Reinhold group;66–68 however, the work herein does not rely on higher order MSn to elucidate glycan structures. In addition, the disaccharide product ion of m/z 493 consisting of a HexNAc bound to an Neu5Ac allows for facile differentiation of structural isomers. That is, the sialic acid is confidently determined to be bound to HexNAc, not hexose. A few cross-ring cleavage products are also observed in Figure 1 (e.g., 1,5X1 and 2,5X1) that aid in deciphering the saccharide linkage positions, but these ions are not so common that the spectrum becomes cluttered and difficult to interpret. Neutral losses of carbon dioxide (44 Da), methoxyl radical (31 Da), hydroxyl radical (17 Da), water (18 Da), the glutamic acid (E) side-chain (72 Da) and combinations thereof are also observed – most commonly from the glycan moiety or the radical, charge-reduced photoelectron detachment products.
Figure 1.
UVPD of the doubly deprotonated kappa casein glycopeptide AVEST(glycan)VATLE (tr = 28.6 min): (A) UVPD spectrum, (B) expansion of the low m/z region, and (C) expansion of the high m/z region. The neutral loss of water from sequence ions is denoted by °.
The doubly deprotonated ST(HexNAc + Hex + Neu5Ac2)VATLE glycopeptide anion was also analyzed by UVPD to evaluate differences in fragmentation behavior for glycopeptides with shorter peptide sequences and correspondingly larger glycan portions (i.e., the glycan moiety is more massive than the peptide portion). Supplemental Figure 1 illustrates the resulting fragmentation pattern of this kappa casein glycopeptide, with the lower and upper m/z ranges again expanded for easier inspection. More glycan-specific ions are observed compared to the array in Figure 1 due to the significantly increased glycan:peptide mass ratio as well as the additional sialic acid bound to the hexose. The presence of two product ions of m/z 493 (HexNAc bound to Neu5Ac) and 468 (Hex bound to Neu5Ac) confirm the identity of the tetrasaccharide. Peptide sequence ions remained prominent as well for ST(HexNAc + Hex + Neu5Ac2)VATLE, again resulting in 100% sequence coverage via a/x-type ions. These latter ions also retained the entire glycan moiety and therefore may be used to pinpoint the correct site of glycosylation (in this case the second threonine from the N-terminus). While significantly more glycan-specific ions are observed in Supplemental Figure 1 compared to Figure 1, the site of glycosylation even in the peptide containing multiple and adjacent unmodified serine and threonine residues can still be elucidated despite the fact that the glycan moiety (947 Da) was significantly more massive compared to the peptide (719 Da), thus highlighting the versatility of the UVPD method.
The fragmentation patterns of peptides by UVPD are known to be charge-state dependent due to differences in proton mobility (just like in CID), coulombic repulsion and electron photodetachment behavior;56, 58 therefore, the UVPD patterns of glycopeptides in charge states other than z = 2 were investigated. Besides doubly deprotonated species, singly and triply charged glycopeptides were also commonly produced upon negative mode ESI. Supplemental Figures 2 and 3 illustrate the UVPD spectra of singly charged AVEST(HexNAc + Hex + Neu5Ac)VATLE with structural isomer “A” (HexNAc bound to Neu5Ac) and the triply charged AVEST(HexNAc + Hex + Neu5Ac)VATLE with structural isomer “B” (Hex bound to Neu5Ac), respectively. The singly charged glycopeptide anion yielded a slightly simpler fragmentation pattern compared to that seen for the doubly species in Figure 1, and the triply charged species resulted in several multiply charged product ions. Importantly, both charge states as well as the doubly charged species all exhibit similar fragmentation behavior in terms of the production of both a/x peptide sequence ions and glycan-specific ions, thus facilitating the development of algorithms for stream-lined spectral interpretation.
MassMatrix automated searching of glycopeptides
Using the fragmentation trends established above, we developed a modified version of MassMatrix for the automated spectral analysis of glycopeptide anions by UVPD. The algorithm matches both the peptide and glycan information from each individual spectrum with theoretical glycopeptides generated from protein databases (the specifics of the searching method are outlined in the methods and supplemental sections). Therefore, given a known protein sequence, MassMatrix was designed to identify unknown glycopeptides from a mixture of glycosylated and non-glycosylated peptides, to sequence both the glycan and peptide portions simultaneously, and to pinpoint the correct site of modification all from a single spectrum. To test the automated search performance and to develop a scoring system, the modified MassMatrix algorithm was applied to the UVPD spectra generated from the ZIC-HILIC-enriched GluC-digested kappa casein. The pp (a statistical measure of number of matched product ions) and pp2 (a statistical measure of total abundance of matched product ions) scores60 for both peptide and glycan moieties were plotted individually against the site-specific identification false discovery rate for identified glycopeptides (false positive site ID/(true positive site ID + false positive site ID)) and are illustrated in Supplemental Figure 4 A – D. The site-specific false discovery rate is analogous to false discovery rate calculations common to shotgun proteomics but also considers the accuracy of the glycan identification and its corresponding site on the peptide (for which lower values represent greater confidence). Spectra were labeled as “true positive site ID” if the peptide sequence, glycan composition, and amino acid site(s) of modification were all identified correctly; in contrast, spectra labeled as “false positive site ID” had at least one incorrect identification. Owing to the fact that ZIC-HILIC usually does not result in 100% glycopeptide enrichment, several unmodified peptides were also identified. These peptides and other hits with a glycan pp score of “0” or “NA” were not pertinent to this study and were therefore removed. The hits with a glycan pp of “0” are likely false IDs since no glycan ions were observed in the spectra. The ones with glycan pp of “N/A” are non-glycosylated peptides. However, identifications that exhibited high site-specific ID false discovery rates (lower confidence) and low pp and pp2 scores are included in the figure in order to better visualize the distribution of high confidence and low confidence results. Based on the figures, the pp2 score for the peptide portion (a measure of the abundance of peptide sequence ions) and the pp score for the glycan moiety (a measure of the number of matches of glycan-specific ions) yielded the lowest site-specific ID false discovery rates (higher confidence) at the highest pp or pp2 scores, respectively, and therefore represent the two best scoring regimes. The peptide pp2 scoring system likely outperformed the peptide pp scoring mode due to the large search space and rich spectra which resulted in many false positives with high pp scores, whereas the pp2 score more accurately differentiated the abundant sequence ions (e.g., a/x-type products) from the noise. The site-specific ID false discovery rates associated with the glycan pp and pp2 scoring modes were fairly similar, likely due to the lower search space for glycan-related fragment ions. To take advantage of both the peptide and glycan information from each spectrum, the peptide pp2 was combined with the glycan pp score by simple multiplication to form a net glycopeptide pp score. Using this combined score, significantly lower site-specific ID false discovery rates were observed (see Supplemental Figure 4E) compared to any other single scoring regime and therefore represents the best score for the accurate identification of glycopeptides by UVPD. Importantly, the highest glycopeptide pp scores are only obtained when both the peptide pp2 and glycan pp exhibit high scores, resulting in greater confidence in the overall scoring.
The upper portion of Table 1 shows the unique true positives identified for kappa casein glycopeptides (listed from highest to lowest glycopeptide pp) by coupling negative mode LC-MS/UVPD with the MassMatrix algorithm. Only the highest score for each peptide spectral match for each unique glycopeptide is listed. The true positive hits with low scores were derived from spectra of poorer quality arising from low abundance precursor ions. In comparison, kappa casein digested by GluC and enriched by ZIC-HILIC yielded no confident glycopeptide hits by traditional positive mode LC-MS/CID and automated database searching (b and y ions). A few glycosylation sites were poorly represented compared to previous studies,21 an outcome that is attributed due to bias in the HILIC enrichment step. Furthermore, accurately identifying glycosylation sites with two or more sites poses an additional challenge for the MassMatrix algorithm, especially when search specificity is limited due to relatively low mass accuracy of the ion trap data. We are currently implementing negative mode UVPD analysis of O-glycopeptides on a high mass accuracy Orbitrap mass spectrometer to improve algorithm performance; however, this is beyond the scope of this current study.
Table 1.
Unique glycopeptide species identified by LC-MS/UVPD and MassMatrix automated spectral interpretation. The sites of glycosylation are denoted in the farthest left column by (X) where X can be either serine or threonine.
| Glycopeptide | Charge | glycopeptide pp | Glycan |
|---|---|---|---|
| bovine kappa casein (GluC) | |||
| S(T)VATLE | 2- | 150.38 | HexNAc + Hex + Neu5Ac2 |
| S(T)VATLE | 1- | 115.64 | HexNAc + Hex + Neu5Ac2 |
| AVES(T)VATLE | 2- | 86.84 | HexNAc + Hex + Neu5Ac2 |
| S(T)VATLE | 2- | 60.72 | HexNAc + Hex + Neu5Ac-B |
| S(T)VATLE | 1- | 48.28 | HexNAc + Hex + Neu5Ac-B |
| AVES(T)VATLE | 1- | 44.46 | HexNAc + Hex + Neu5Ac-A |
| AVES(T)VATLE | 3- | 38.34 | HexNAc + Hex + Neu5Ac-B |
| AVES(T)VATLE | 2- | 33.37 | HexNAc + Hex + Neu5Ac-B |
| AVES(T)VATLE | 2- | 32.64 | HexNAc + Hex + Neu5Ac-A |
| S(T)VATLE | 1- | 29.25 | HexNAc + Hex |
| S(T)VATLE | 2- | 28.06 | HexNAc + Hex |
| AVES(T)VATLE | 3- | 24.64 | HexNAc + Hex + Neu5Ac2 |
| S(T)VATLE | 1- | 21.60 | HexNAc |
| S(T)VATLE | 2- | 16.24 | HexNAc + Hex + Neu5Ac-A |
| AVES(T)VATLE | 1- | 15.05 | HexNAc + Hex + Neu5Ac2 |
| AVES(T)VATLE | 2- | 12.48 | HexNAc + Hex |
| P(T)S(T)PTTE | 2- | 10.88 | HexNAc + Neu5Ac & HexNAc + Hex |
| KTEIP(T)INTIASGE | 2- | 1.69 | HexNAc |
| INTVQV(T)STAV* | 2- | 48.18 | HexNAc |
| INTVQV(T)STAV* | 2- | 36.48 | HexNAc + Hex |
| INTVQV(T)STAV* | 2- | 30.00 | HexNAc + Hex + Neu5Ac-B |
| A. baumannii OmpA/MotB (GluC + Trypsin) | |||
| AA(S)GVE | 2- | 116.16 | HexNAc2 + Hex2 + OAcGlcA |
| AA(S)GVE | 1- | 80.04 | HexNAc2 + Hex2 + OAcGlcA |
extra glycopeptides identified from a separate run using less specific HILIC enrichment
To facilitate the identification of more hydrophobic glycopeptides such as INTVQVT(glycan)STAV by this LC-MS/UVPD/MassMatrix automated strategy, a run using ZIC-HILIC enrichment without any ion-pairing reagent was performed (thus reducing the specificity of HILIC for the hydrophilic species).69 In addition to identification of other more hydrophilic glycopeptides (as seen for other HILIC runs with an ion-pairing reagent), this run resulted in identification of several glycopeptide forms of INTVQVT(glycan)STAV as seen in Table 1. Importantly, the glycan site was accurately pinpointed to the seventh threonine (from the N-terminus) in the presence of three adjacent serine and threonine residues, representing a total of four possible modification sites.
Site-specific glycosylation analysis of OmpA/MotB from A. baumannii
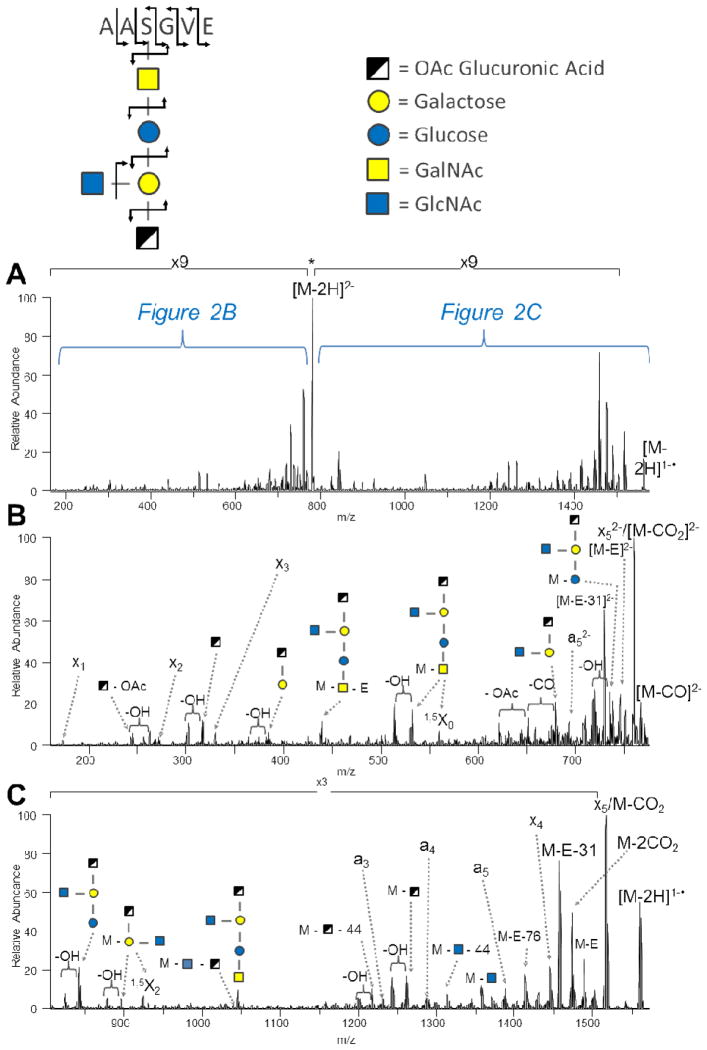
Recently, a general O-linked protein glycosylation system was discovered for the pathogenic bacteria Acinetobacter baumannii, and the glycan found in the strain ATCC 17978 was identified as a pentasaccharide with the formula β-GlcNAc3NAcA4OAc-4-(β-GlcNAc-6-)α-Gal-6-β-Glc-3-β-GalNAc-S/T.12 Interestingly, the glycan contains a novel negatively charged O-acetylated glucuronic acid derivative. The pentasaccharide was identified on the membrane protein Ompa/MotB using traditional trypsin digestion and positive mode collision activation on an Orbitrap Velos mass spectrometer; however, the site of modification could not be localized to any of several possible serine and threonine residues.12 Therefore, given the acidity of the A. baumannii glycan and the low pI (approximately 5.4) of the Ompa/MotB protein sequence, we applied our UVPD method as described herein in an attempt to more fully characterize the Ompa/MotB glycosylation system. The protein processing method was revised to utilize a GluC/trypsin cocktail instead of GluC alone in order to generate smaller, more acidic peptides while maintaining a degree of enzymatic cleavage specificity. The HILIC-enriched sample was analyzed by LC-MS/UVPD followed by MassMatrix spectral interpretation first targeting the peptide portion. The UVPD spectrum of the doubly deprotonated glycopeptide is displayed in Figure 2. As seen for other glycopeptides previously, UVPD generated a/x ions that retained the full glycan as well as many products from cleavage along the pentasaccharide. The abundant glycan-specific ions seen in the spectrum were then incorporated into MassMatrix as per Supplemental Table 2. An acidic glycopeptide with the sequence AAS(glycan)GVE containing the pentasaccharide glycan was identified via MassMatrix for both the doubly and singly charged species with confident peptide pp and pp2 scores of 7.6 and 9.6 and 10.9 and 8.7, respectively. The UVPD spectral data was subsequently re-analyzed by MassMatrix utilizing the glycan-specific ions in order to calculate the glycopeptide pp and to identify the glycopeptide as AAS(pentasaccharide)GVE as shown in the upper section of Figure 2. As listed in the bottom of Table 1, the resulting glycopeptide pp scores were 116.16 and 80.04 for the doubly and singly charged species (respectively), illustrating the confidence and specificity achievable for the total characterization of A. baumannii acidic glycopeptides. Importantly, the same sample run by traditional positive mode LC-MS/CID analysis and database searching yielded no confident hits, presumably due to the acidity of the glycopeptide (i.e., lower positive ion signals) and the lack of b and y product ions.
Figure 2.
UVPD of the doubly deprotonated Acinetobacter baumannii Ompa/MotB glycopeptide AAS(glycan)GVE (tr = 12.9 min): (A) UVPD spectrum, (B) expansion of the low m/z region, and (C) expansion of the high m/z region.
CONCLUSION
The combination of enzymatic glycoprotein digestion, ZIC-HILIC glycopeptide enrichment, reversed phase LC-MS/UVPD, and automated data interpretation was shown to be an effective strategy for identifying acidic glycopeptide anions, sequencing both the glycan and peptide moieties concurrently, and pinpointing the correct site(s) of amino acid modification. Using this methodology, novel site-specificity of O-linked acidic glycopeptide species from A. baumannii OmpA/MotB was identified, an outcome which had proven elusive via traditional positive MS/MS methods such as ion trap CID as well as orbitrap HCD.12 Work performed previously (without confirmation of the exact site of glycosylation) indicated that by knocking out the O-OTase gene responsible for O-glycosylation in A. baumannii, both biofilm formation and virulence were significantly affected.12 Considering that conjugate vaccines for pathogenic bacteria are often comprised of surface glycoproteins, the outer membrane OmpA/MotB may become a key player in future therapeutics against the “superbug” A. baumannii, and therefore its total characterization has great importance. The ability to sequence both the glycan and peptide portions as well as pinpoint the site of glycosylation in a single spectrum by negative mode UVPD represents a compelling advance in the development of tandem mass spectrometry strategies for characterizing O-linked glycoproteins.
Supplementary Material
Acknowledgments
Funding from NIH (R21GM099028) and the Robert A. Welch Foundation (F1155) is gratefully acknowledged. M.F.F acknowledges funding from the Alberta Glycomics Centre. MFF is a CIHR New Investigator and AHFMR Scholar.
References
- 1.Olden K, Parent JB, White SL. Biochim Biophys Acta. 1982;650:209–232. doi: 10.1016/0304-4157(82)90017-x. [DOI] [PubMed] [Google Scholar]
- 2.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nothaft H, Szymanski CM. Nat Rev Microbiol. 2010;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- 4.Benz I, Schmidt MA. Mol Microbiol. 2002;45:267–276. doi: 10.1046/j.1365-2958.2002.03030.x. [DOI] [PubMed] [Google Scholar]
- 5.Logan SM. Microbiology. 2006;152:1249–1262. doi: 10.1099/mic.0.28735-0. [DOI] [PubMed] [Google Scholar]
- 6.Sleytr UB. Nature. 1975;257:400–402. doi: 10.1038/257400a0. [DOI] [PubMed] [Google Scholar]
- 7.Mescher MF, Strominger JL. J Biol Chem. 1976;251:2005–2014. [PubMed] [Google Scholar]
- 8.Sandercock LE, MacLeod AM, Ong E, Warren RAJ. FEMS Microbiol Lett. 1994;118:1–8. doi: 10.1111/j.1574-6968.1994.tb06795.x. [DOI] [PubMed] [Google Scholar]
- 9.Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, Aebi M. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 10.Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AVG, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P, Packer NH, Larsen MR, Cordwell SJ. Mol Cell Proteomics. 2011;10:1–18. 18. doi: 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young NM, Brisson J-R, Kelly J, Watson DC, Tessier L, Lanthier PH, Jarrell HC, Cadotte N, StMichael F, Aberg E, Szymanski CM. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 12.Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Wittal R, Scild S, Feldman MF. PLoS Pathog. 2012;8:002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An HJ, Froehlich JW, Lebrilla CB. Curr Opin Chem Biol. 2009;13:421–426. doi: 10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seipert RR, Dodds ED, Lebrilla CB. J Proteome Res. 2009;8:493–501. doi: 10.1021/pr8007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hakansson K, Cooper HJ, Emmett MR, Costello CE, Marshall AG, Nilsson CL. Anal Chem. 2001;73:4530–4536. doi: 10.1021/ac0103470. [DOI] [PubMed] [Google Scholar]
- 16.Hogan JM, Pitteri SJ, Chrisman PA, McLuckey SA. J Proteome Res. 2005;4:628–632. doi: 10.1021/pr049770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarentino AL, Plummer TH., Jr Methods Enzymol. 1994;230:44–57. doi: 10.1016/0076-6879(94)30006-2. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi K, Wall SB, Suzuki H, Smith ADIV, Hall S, Poulsen K, Kilian M, Mobley JA, Julian BA, Mestecky J, Novak J, Renfrow MB. Mol Cell Proteomics. 2010;9:2545–2557. doi: 10.1074/mcp.M110.001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, Backstrom M, Costello CE, Hansson GC, Hiki Y, Ishihara M, Ito H, Kakehi K, Karlsson N, Hayes CE, Kato K, Kawasaki N, Khoo KH, Kobayashi K, Kolarich D, Kondo A, Lebrilla C, Nakano M, Narimatsu H, Novak J, Novotny MV, Ohno E, Packer NH, Palaima E, Renfrow MB, Tajiri M, Thomsson KA, Yagi H, Yu SY, Taniguchi N. Mol Cell Proteomics. 2010;9:719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christiansen MN, Kolarich D, Nevalainen H, Packer NH, Jensen PH. Anal Chem (Washington, DC, U S) 2010;82:3500–3509. doi: 10.1021/ac901717n. [DOI] [PubMed] [Google Scholar]
- 21.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. J Proteome Res. 2011;10:2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nwosu CC, Strum JS, An HJ, Lebrilla CB. Anal Chem (Washington, DC, U S) 2010;82:9654–9662. doi: 10.1021/ac101856r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Proc Natl Acad Sci U S A. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, Hiki Y, Toma K, Emmett MR, Mestecky J, Marshall AG, Novak J. J Biol Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 25.Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, Poulsen K, Emmett MR, Marshall AG, Novak J. Anal Bioanal Chem. 2007;389:1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 26.Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 27.Chalkley RJ, Thalhammer A, Schoepfer R, Burlingame AL. Proc Natl Acad Sci U S A. 2009;106:8894–8899. doi: 10.1073/pnas.0900288106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirgorodskaya E, Roepstorff P, Zubarev RA. Anal Chem. 1999;71:4431–4436. doi: 10.1021/ac990578v. [DOI] [PubMed] [Google Scholar]
- 29.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. Mol Cell Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Darula Z, Medzihradszky KF. Mol Cell Proteomics. 2009;8:2515–2526. doi: 10.1074/mcp.M900211-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Udeshi ND, O’Malley M, Shabanowitz J, Hunt DF, Hart GW. Mol Cell Proteomics. 2010;9:153–160. doi: 10.1074/mcp.M900268-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apweiler R, Hermjakob H, Sharon N. Biochim Biophys Acta, Gen Subj. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 33.Kowarik M, Young NM, Numao S, Schulz BL, Hug I, Callewaert N, Mills DC, Watson DC, Hernandez M, Kelly JF, Wacker M, Aebi M. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irungu J, Go EP, Zhang Y, Dalpathado DS, Liao HX, Haynes BF, Desaire H. J Am Soc Mass Spectrom. 2008;19:1209–1220. doi: 10.1016/j.jasms.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bykova NV, Rampitsch C, Krokhin O, Standing KG, Ens W. Anal Chem. 2006;78:1093–1103. doi: 10.1021/ac0512711. [DOI] [PubMed] [Google Scholar]
- 36.Conboy JJ, Henion JD. J Am Soc Mass Spectrom. 1992;3:804–814. doi: 10.1016/1044-0305(92)80003-4. [DOI] [PubMed] [Google Scholar]
- 37.Huddleston MJ, Bean MF, Carr SA. Anal Chem. 1993;65:877–884. doi: 10.1021/ac00055a009. [DOI] [PubMed] [Google Scholar]
- 38.Wuhrer M, Hokke CH, Deelder AM. Rapid Commun Mass Spectrom. 2004;18:1741–1748. doi: 10.1002/rcm.1546. [DOI] [PubMed] [Google Scholar]
- 39.Demelbauer UM, Zehl M, Plematl A, Allmaier G, Rizzi A. Rapid Commun Mass Spectrom. 2004;18:1575–1582. doi: 10.1002/rcm.1521. [DOI] [PubMed] [Google Scholar]
- 40.Wada Y, Tajiri M, Yoshida S. Anal Chem. 2004;76:6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 41.Olivova P, Chen W, Chakraborty AB, Gebler JC. Rapid Commun Mass Spectrom. 2008;22:29–40. doi: 10.1002/rcm.3330. [DOI] [PubMed] [Google Scholar]
- 42.Sihlbom C, van Dijk Haerd I, Lidell ME, Noll T, Hansson GC, Baeckstroem M. Glycobiology. 2009;19:375–381. doi: 10.1093/glycob/cwn144. [DOI] [PubMed] [Google Scholar]
- 43.Singh C, Zampronio CG, Creese AJ, Cooper HJ. J Proteome Res. 2012;11:4517–4525. doi: 10.1021/pr300257c. [DOI] [PubMed] [Google Scholar]
- 44.Ye H, Boyne MT, Buhse LF, Hill J. Anal Chem. 2013;85:1531–1539. doi: 10.1021/ac3026465. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, Reilly JP. Journal of Proteome Research. 2009;8:734–742. doi: 10.1021/pr800766f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boontheung P, Alewood PF, Brinkworth CS, Bowie JH, Wabnitz PA, Tyler MJ. Rap Comm Mass Spectrom. 2002;16:281–286. doi: 10.1002/rcm.577. [DOI] [PubMed] [Google Scholar]
- 47.Ewing NP, Cassady CJ. J Am Soc Mass Spectrom. 2001;12:105–116. doi: 10.1016/S1044-0305(00)00195-1. [DOI] [PubMed] [Google Scholar]
- 48.Bowie JH, Brinkworth CS, Dua S. Mass Spectrometry Reviews. 2002;21:87–107. doi: 10.1002/mas.10022. [DOI] [PubMed] [Google Scholar]
- 49.Kjeldsen F, Silivra OA, Ivonin IA, Haselmann KF, Gorshkov M, Zubarev RA. Chem--Eur J. 2005;11:1803–1812. doi: 10.1002/chem.200400806. [DOI] [PubMed] [Google Scholar]
- 50.Kjeldsen F, Horning OB, Jensen SS, Giessing AMB, Jensen ON. J Am Soc Mass Spectrom. 2008;19:1156–1162. doi: 10.1016/j.jasms.2008.04.031. [DOI] [PubMed] [Google Scholar]
- 51.Coon JJ, Shabanowitz J, Hunt DF, Syka JEP. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 52.Huzarska M, Ugalde I, Kaplan DA, Hartmer R, Easterling ML, Polfer NC. Anal Chem. 2010;82:2873–2878. doi: 10.1021/ac9028592. [DOI] [PubMed] [Google Scholar]
- 53.McAlister GC, Russell JD, Rumachik NG, Hebert AS, Syka JEP, Geer LY, Westphall MS, Pagliarini DJ, Coon JJ. Anal Chem (Washington, DC, U S) 2012;84:2875–2882. doi: 10.1021/ac203430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antoine R, Joly L, Tabarin T, Broyer M, Dugourd P, Lemoine J. Rap Comm Mass Spectrom. 2007;21:265–268. doi: 10.1002/rcm.2810. [DOI] [PubMed] [Google Scholar]
- 55.Larraillet V, Vorobyev A, Brunet C, Lemoine J, Tsybin YO, Antoine R, Dugourd P. J Am Soc Mass Spectrom. 2010;21:670–680. doi: 10.1016/j.jasms.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 56.Madsen JA, Kaoud TS, Dalby KN, Brodbelt JS. Proteomics. 2011;11:1329–1334. doi: 10.1002/pmic.201000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ko BJ, Brodbelt JS. Anal Chem. 2011;83:8192–8200. doi: 10.1021/ac201751u. [DOI] [PubMed] [Google Scholar]
- 58.Madsen JA, Xu H, Robinson MR, Horton AP, Shaw JB, Giles DK, Kaoud TS, Dalby KN, Trent MS, Brodbelt JS. Mol Cell Proteomics. 2013 doi: 10.1074/mcp.O113.028258. ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han SW, Lee SW, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC. Nat Commun. 2012;3:1153. doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Freitas Michael A. BMC Bioinformatics. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu H, Yang L, Freitas Michael A. BMC Bioinformatics. 2008;9:347. doi: 10.1186/1471-2105-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu H, Freitas MA. J Proteome Res. 2008;7:2605–2615. doi: 10.1021/pr800002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu H, Freitas MA. Proteomics. 2009;9:1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Anal Chem. 2008;80:4807–4819. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- 65.Madsen JA, Boutz DR, Brodbelt JS. J Proteome Res. 2010;9:4205–4214. doi: 10.1021/pr100515x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ashline D, Singh S, Hanneman A, Reinhold V. Anal Chem. 2005;77:6250–6262. doi: 10.1021/ac050724z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashline DJ, Lapadula AJ, Liu YH, Lin M, Grace M, Pramanik B, Reinhold VN. Anal Chem. 2007;79:3830–3842. doi: 10.1021/ac062383a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reinhold V, Zhang H, Hanneman A, Ashline D. Mol Cell Proteomics. 2013;4:866–873. doi: 10.1074/mcp.R112.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M. Anal Chem. 2010;82:5598–5609. doi: 10.1021/ac100530w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.