Summary
Dynamic changes in 5-methylcytosine (5mC) have been implicated in the regulation of gene expression critical for consolidation of memory. However, little is known about how these changes in 5mC are regulated in the adult brain. The enzyme Tet methylcytosine dioxygenase 1 (TET1) has been shown to promote active DNA demethylation in the nervous system. Therefore, we took a viral-mediated approach to overexpress the enzyme in the hippocampus and test its potential involvement in memory formation. We found that Tet1 is a neuronal-activity regulated gene and that its overexpression leads to changes in global modified cytosine levels. Furthermore, expression of TET1 or a catalytically inactive mutant (TET1m) resulted in the up-regulation of several neuronal memory-associated genes and impaired contextual fear memory. In summary, we show that neuronal Tet1 regulates DNA methylation levels and that its expression, independent of its catalytic activity, regulates the expression of CNS activity- dependent genes and memory formation.
Introduction
In recent years, epigenetic modifications of DNA and chromatin have been identified as essential mediators of memory formation through their regulation of gene expression (Sultan and Day, 2011), with methylation of cytosine bases in DNA (5mC) playing a critical role in both memory consolidation and storage (Feng et al., 2010a; Lubin et al., 2008; Miller et al., 2010; Miller and Sweatt, 2007; Monsey et al., 2011). Although early studies identified 5mC as a stable transcriptional silencer (Bonasio et al., 2010; Feng et al., 2010b), new evidence of rapid and reversible changes in DNA methylation at memory-associated genes implies the presence of an active DNA demethylation mechanism in response to neuronal activity (Guo et al., 2011b; Lubin et al., 2008; Ma et al., 2009; Miller and Sweatt, 2007).
The near-simultaneous discoveries of a hydroxylated form of 5mC (5hmC) (Kriaucionis and Heintz, 2009) and the Ten-eleven translocation (Tet) family of enzymes required for its conversion (Tahiliani et al., 2009) has now offered insight into how these observations of rapid changes in DNA methylation might occur. Specifically, all three Tets (TET1-3) have been shown to catalyze the conversion of 5mC to 5-hydroxymethyl cytosine (5hmC) as well as the further oxidation of 5hmC into 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), respectively (He et al., 2011; Ito et al., 2010; Ito et al., 2011). These modified bases can then function as DNA demethylation intermediates subject to deamination, glycosylase-dependent excision and repair culminating in a reversion back to unmodified cytosine (Guo et al., 2011b; Shen et al., 2013; Zhang et al., 2012; for review; Branco et al., 2012; Tan and Shi, 2012). However, it has now become apparent that 5hmC is not merely a DNA demethylation intermediate, but also functions as a stable epigenetic mark that is enriched within gene bodies, promoters and transcription factor binding sites, where it may influence gene expression (Hahn et al., 2013; Mellen et al., 2012; Szulwach et al., 2011).
In the adult brain, alterations in global DNA methylation patterns in response to neuronal activity (Guo et al., 2011a; Miller-Delaney et al., 2012) are at least partially mediated by TET1, which is both necessary and sufficient for demethylation of the fibroblast growth factor 1 (Fgf1) and the brain-derived neurotrophic factor (Bdnf) promoters in response to electroconvulsive shock (Guo et al., 2011b). Complementary studies have shown that BDNF is critical for memory formation (Bekinschtein et al., 2008; Mizuno et al., 2000), and its promoter region undergoes rapid demethylation following associative learning in a fear conditioning paradigm in rodents (Lubin et al., 2008), suggesting the possibility that TET1 may contribute to memory formation. However, at present, the role of Tet-mediated regulation of 5hmC and subsequent active demethylation in relation to the expression of neuronal plasticity genes and memory has not been extensively explored, although Zhang et al. recently reported that Tet1 deletion in a knockout mouse model resulted in altered neurogenesis and a deficit in spatial memory in the Morris water maze (Zhang et al., 2013).
In this study, we sought to investigate the role of TET1 enzymatic activity in memory formation, through its ability to regulate 5hmC levels and therefore, gene expression. We found that endogenous TET1 is expressed in neurons throughout the hippocampus and that its transcript levels are regulated by neuronal activity. In addition, we used an AAV-mediated approach to overexpress the catalytic domain of TET1 (TET1) or a catalytically-inactive mutant version (TET1m) in the hippocampus and found that active TET1 drove hydroxylation of 5mC and resulted in active demethylation in vivo. Surprisingly, we observed that overexpression of either TET1 or a catalytically inactive TET1m increased expression of many immediate early genes (IEGs) implicated in memory, and induced a selective deficit in long term contextual fear memory.
Results
TET1 is primarily expressed in neurons and its transcript levels are regulated by neuronal activity
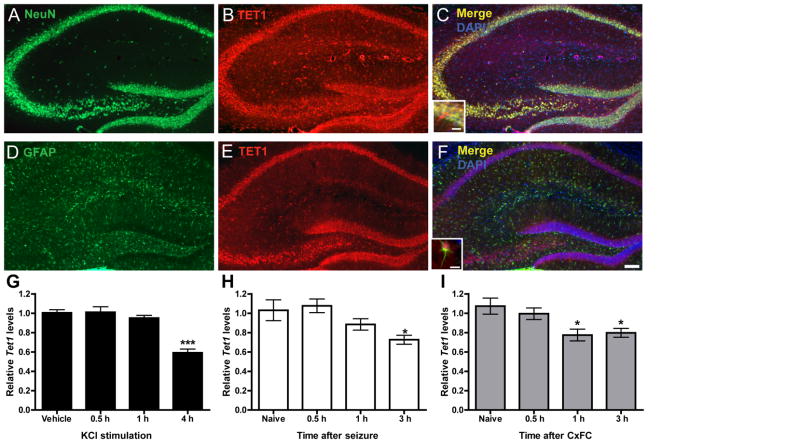
Although TET1 has recently been shown to regulate the expression of genes in the DG following neuronal activation (Guo et al., 2011b), little is known about TET1 localization within the hippocampus. To address this, we double-labeled hippocampal tissue sections with the neuronal marker NeuN and an antibody against TET1. Immunohistohemical analysis revealed strong co-localization of TET1 and NeuN signals in neurons throughout the hippocampus (Figures 1A–C). Within neurons, the 5-methylcytosine dioxygenase was found to be present in both the nucleus and soma (Figure 1C, inset). In addition, we asked if TET1 was also expressed in non-neuronal cells in the CNS by double-labeling sections with the astrocytic marker GFAP and TET1. At lower magnification we did not observe obvious co-localization (Figure 1D–F) but under higher magnification, we did detect low levels of TET1 staining in the soma of several astrocytes (Figure 1F, inset).
Figure 1. TET1 is expressed in neurons and its transcript levels are altered by neuronal activity.
(A, B) NeuN labeled neurons and TET1 labeled cells in the hippocampus. (C) Merged image of NeuN and TET1 double labeling, counterstained with DAPI. Inset, higher magnification of the CA1 pyramidal cell layer showing merged signal present in the soma of neurons. (D, E) GFAP labeled astrocytes and TET1 labeled cells in the hippocampus. (E) Merged image of GFAP and TET1 double labeling, counterstained with DAPI. Inset, higher magnification of a GFAP positive cell with TET1 labeling in the soma. Scale bar, 200μm. Inset scale bar 20μm.(G) Quantitative reverse-transcription PCR (qRT-PCR) analysis of Tet1 expression in primary hippocampal neuron cultures depolarized with 25 mM KCl for 0.5, 1 and 4 h compared to vehicle controls. Data represent the combined results of two independent experiments (F3, 22 = 23.91; n = 5–6 total/group). Vehicle vs. 4 h KCl treatment. ***p < 0.001, one-way ANOVA followed by Bonferroni post hoc test. (H) qRT-PCR analysis of Tet1 expression in dorsal CA1 subregion 0.5, 1 and 3 h after flurothyl-induced seizures, compared to controls. Data represent the combined results of three independent experiments (F3, 25 = 4.443; n = 6–7 total/group). Naive vs. 3 h. *p < 0.05, one-way ANOVA followed by Bonferroni post hoc test. (I) qRT-PCR analysis of Tet1 expression in dorsal CA1 0.5, 1 and 3 h after fear conditioning compared to naïve controls. Data represent the combined results of three independent experiments (F3, 35 = 5.352; n = 9 total/group). Naive vs. 1 and 3 h. *p < 0.05, one-way ANOVA followed by Bonferroni post hoc test. All data are presented as mean ± s.e.m. See also Figures S1 and S2.
Next we sought to determine if the transcript levels of Tet1, like those of other epigenetic regulators necessary for memory formation, may be modified following neuronal stimulation, fear conditioning, or both (Miller and Sweatt, 2007; Monsey et al., 2011; Oliveira et al., 2012). To determine if Tet1 expression levels were regulated by neuronal activity, we utilized a primary hippocampal neuronal culture system and examined the effect of KCl-induced cell depolarization on its transcription. We found that prolonged KCl incubation of hippocampal neurons consistently resulted in a significant reduction in Tet1 transcript levels compared to vehicle controls (Figure 1G). Next, using a flurothyl-induced epileptic seizure paradigm we sought to establish whether or not Tet1 message could also be transcriptionally regulated by neuronal activity in vivo. Again, we observed a significant reduction in Tet1 levels several hours post-episode (Figure 1H). Finally, we trained animals using a context plus cued fear conditioning paradigm to ascertain whether the expression of Tet1was also modulated during memory formation. Like the two experiments before, a consistent downregulation of Tet1 was observed following fear learning (Figure 1I). The transcript levels of the other two Tet-family members, Tet2 and Tet3, did not consistently respond to stimulation using any of our activity-inducing paradigms (Figure S1B, C). In all experiments, we monitored the expression of the gene activity-regulated cytoskeleton-associated protein (Arc) as a positive control to ensure that neuronal activation had indeed occurred (Figure S1A).
Considering the role of TET1 in active DNA methylation, we asked whether other genes whose products act downstream of TET1 to convert 5hmC back to an unmodified cytosine were also regulated by neuronal activity. We focused our attention on four genes previously implicated in the active DNA demethylation pathway which included the cytidine deaminase activation-induced cytidine deaminase (AID)/apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (Apobec1) (Guo et al., 2011b; Popp et al., 2010; Zhu, 2009) and three glycosylases, thymine-DNA glycosylase (Tdg) (Cortellino et al., 2011), strand selective monofunctional uracil-DNA glycosylase 1 (Smug1) (Kemmerich et al., 2012) and methyl-CpG-binding domain protein 4 (Mbd4) (Rai et al., 2008). qRT-PCR for these genes revealed a general trend towards downregulation several hours after neuronal activation both in vitro and in vivo, similar to that observed for Tet1 (Figure S2). However, unlike Tet1, these trends were not observed consistently across all our paradigms. Together, these data reveal that TET1 is broadly expressed in neurons throughout the hippocampus and exhibits activity-dependent changes in its mRNA levels, both in vitro and in vivo. In addition, other active DNA demethylation genes also appear to be transcriptionally regulated following neuronal activity. Furthermore, the alterations in expression of active DNA demethylation machinery observed here temporally overlaps with previously reported changes in DNA methylation following fear conditioning (Lubin et al. 2008; Miller and Sweatt, 2007).
Global alteration of modified cytosines following neuronal activity
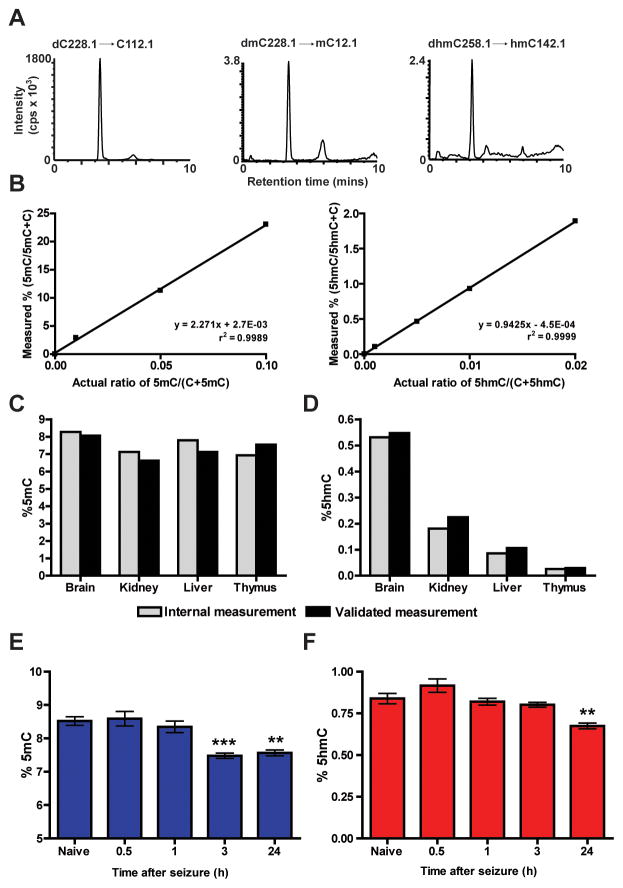
Using an approach similar to those previously reported (Globisch et al., 2010; Le et al., 2011) we next developed an HPLC/MS system for the accurate, precise, and simultaneous measurement of 5mC and 5hmC levels in biological samples (Figures 3A, B). Our rationale for the development of this quantitative analytical chemistry approach was to directly test whether TET1 oxidase is capable of actively regulating 5mC oxidation to 5hmC in vivo. To confirm that our system was accurate and sensitive, we measured the global 5mC and 5hmC levels from commercially available genomic DNA samples validated previously using a similar HPLC/MS methodology. The amount of 5mC and 5hmC determined from our system was similar to the commercial samples, suggesting our system was able to accurately measure modified cytosines (Figures 3C, D). In agreement with the results of earlier studies (Globisch et al., 2010; Khare et al., 2012; Nestor et al., 2012), 5hmC levels varied by tissue type, with nervous system tissue containing the highest amounts of the epigenetic mark, whereas the amount of 5mC was similar across all tissues tested (Figures 3E, F).
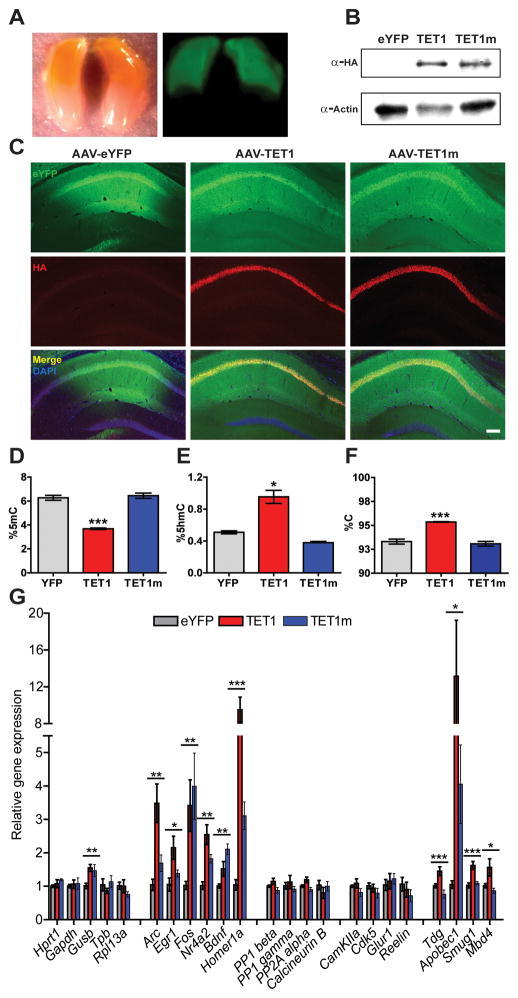
Figure 3. Functional characterization of AAV-mediated expression of TET1 and TET1m in the dorsal hippocampus.
(A) Representative images of YFP expression 14 d after AAV injection along the anterior-posterior axis of the hippocampus under white and UV light, respectively. (B) Protein samples from area CA1 tissue expressing YFP, HA-TET1 or HA-TET1m analyzed by western blot to confirm expression of both peptides. Actin was used as a loading control. (C) Representative images of dorsal hippocampal sections 14 d after AAV-mediated expression of YFP, TET1 and TET1m. Sections were double labeled with anti-GFP, anti-HA and conterstained with DAPI. Robust viral expression was restricted to area CA1. Scale bar, 200 μm. (D) Percent 5mC in microdissected area CA1 (F2, 12 = 66.68; n = 4–5/group). YFP vs. TET1. ***p < 0.001, one-way ANOVA followed by Bonferroni post hoc test. (E) Percent 5hmC in microdissected area CA1 14 d after AAV injection (F2, 11 = 37.34; n = 4/group). YFP vs. TET1. ***p < 0.001, one-way ANOVA followed by Bonferroni post hoc test. (F) Percent unmodified cytosines in microdissected area CA1 (F2, 12 = 31.04). YFP vs. TET1. ***p < 0.001, one-way ANOVA followed by Bonferroni post hoc test (n = 4–5/group). Data are presented as mean ± s.e.m. (G) qRT-PCR analysis of genes involved in synaptic plasticity and memory formation 14 d after viral injection (Gusb, F2,11 = 4.97; Arc, F2,11 = 11.42; Egr1, F2,11 = 5.57, Fos, F2,11 = 4.66; Bdnf, F2,11 = 11.96; Nr4a2, F2,11 = 14.92; Homer1a, F2,11 = 27.23; Tdg, F2,24 = 10.17; Apobec1, F2,24 = 5.37; Smug1, F2,24 = 13.92; Mbd4, F2,24 = 5.52). (n =4/group from one representative experiment). For Tdg, Apobec1, Smug1 and Mbd4 (n = 8–9 combined from two independent experiments).*p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA. All data are presented as mean ± s.e.m.
Based on our expression analysis of Tet1 and other genes implicated in active DNA demethylation (Figure 1 and S2), we examined whether changes in 5mC and 5hmC could be detected on a global scale following neuronal activity. To explore this possibility we used our flurothyl-seizure inducing paradigm to facilitate generalized seizures in mice and subsequently collected dorsal CA1 tissue from animals at varying time points upon recovery. Surprisingly, we observed a significant reduction in the relative percentage of 5mC at both 3 and 24 h after seizure when compared to our naive animals (Figure 3G). In addition, the levels of 5hmC were also reduced at the 24 h time point (Figure 3H). Thus, using our HPLC/MS system, we discovered that neuronal activation alters the global levels of both 5mC and 5hmC in vivo. Overall, these studies serve to validate this HPLC/MS method as an accurate analytical technique to quantitatively measure the levels of 5mC and 5hmC, the proposed substrate and product of TET1 in the CNS.
Viral-mediated overexpression of TET1 catalytic domain results in global changes in modified cytosines
To assess whether TET1 is capable of catalyzing 5mC hydroxylation and triggering a decrease in 5mC levels via active demethylation, we stereotaxically injected AAVs overexpressing a HA-tagged catalytic domain of human TET1, or a catalytically inactive version (TET1m), into the dorsal hippocampus (Guo et al., 2011b). At two weeks post-infection, AAV-mediated expression was consistently observed throughout the entire dorsal half of the hippocampus (Figure 3A). Immunostaining of coronal sections and western blots confirmed consistent expression of both peptides in areas CA1 and portions of CA3, respectively (Figure 3B, C). We next assessed the functional consequences of TET1 and TET1m overexpression by measuring the global levels of 5hmC, 5mC and cytosine in microdissected CA1 tissue using the HPLC/MS analysis system previously optimized for accuracy and sensitivity (Figure 2A–F). We found that after 14 d, 5hmC levels in CA1 increased from 0.49% in controls to 0.95% of all cytosines in tissue overexpressing TET1 (Figure 3D). Likewise, the amount of 5mC in TET1 samples was reduced by 41%, as would be expected by conversion of 5mC into 5hmC (Figure 3E). Finally, in AAV-TET1 injected samples, we observed a significant increase in the global levels of unmodified cytosines compared to both controls (Figure 3F). No statistically significant alterations in the levels of 5hmC, 5mC or unmodified cytosine were observed from tissue infected with the catalytically inactive TET1m. Our analyses of global modified cytosines provide the first direct evidence that overexpression of TET1 in vivo in the CNS leads to increased conversion of 5mCs to 5hmCs followed by active DNA demethylation, which results in an increased percentage of unmodified cytosines in the genome.
Figure 2. Measurement of global 5mC and 5hmC levels in the hippocampus following neuronal activation.
(A) LC-MS/MS-MRM chromatograms of nucleosides using three commercial 948-bp standard DNA fragments (dmC .01, dhmC .001, and dC 1.0) showing peaks corresponding to the response obtained from gas phase transitions of dC to C, dmC to mC, and dhmC to hmC. cps, counts per second. (B) Standard curves for 5mC and 5hmC. The percentages of 5mC and 5hmC are plotted against the known ratios of methylated and hydroxymethylated DNA to the total amount of cytosine in the standard samples. (C, D) Validation of HPLC/MS system for 5mC and 5hmC detection accuracy was performed using a set of previously characterized genomic DNA samples (Zymo Research). (E, F) Percentages of 5mC and 5hmC in genomic DNA from several different tissue sources (n = 3/group). (G, H) 5mC and 5hmC levels in area CA1 of adult mice at several time points following flurothyl induced seizures compared to controls (F4, 29 = 13.41; each sample represents the average of 3 technical replicates, n = 6/group). Naive vs. 3 or 24 h. **p < 0.01, ***p < 0.001; one-way ANOVA followed by Bonferroni post hoc test. In figures E- H data are presented as mean ± s.e.m.
Overexpression of Tet1 catalytic domains dysregulates genes known to be induced by neuronal activity and memory formation
Previous studies suggest that overexpression of the TET1 catalytic domain in the dentate gyrus (DG) results in the increased expression levels of both brain-derived neurotrophic factor (Bdnf) and the brain-specific isoform of the gene fibroblast growth factor 1 (Fgf1B). Therefore, we reexamined the effects of TET1 on the expression of Bdnf and several other candidate genes formerly reported to either positively and negatively impact memory formation (Figure 3G). As a control, we examined a number of genes normally used for qRT-PCR normalization due to their constitutive activity as it is related to their roles in the maintenance of basic cellular functions and thus, not generally influenced by epigenetic mechanisms. With the exception of Glucuronidase beta (GusB), expression of either TET1 or TET1m had no effect on the expression levels of these “housekeeping” genes. In addition, the expression levels of phosphatase-encoding genes such as calcineurin, protein phosphatase 1 isozymes beta and gamma (PP1) and protein phosphatase 2A (PP2A), which are thought to negatively influence memory formation, remained unaffected. Similarly, the transcripts of genes involved in synaptic plasticity, like Ca2+/calmodulin-dependent kinase 2A (CamKIIa), Cyclin-dependent kinase 5 (Cdk5), Glutamate receptor 1 (Glur1) and Reelin (Rln), were also unchanged. However, in contrast, we found that overexpression of TET1 as well as the catalytically inactive TET1m significantly increased the mRNA levels of not only Bdnf, but other activity-dependent, immediate early genes (IEGs) including Fos, Arc, Early growth response 1 (Egr1), Homer1a and Nuclear receptor subfamily 4, group A, member 2 (Nr4a2), Finally, based on our earlier findings of changes in the expression of genes thought to act downstream of TET1 5mC hydroxylation (Figure S2), we reexamined the transcript levels of Tdg, Apobec1, Smug1 and Mbd4, to investigate whether they too were affected by TET1 or TET1m overexpression. Indeed, the mRNA levels of all four were significantly increased following TET1 infection. However, we found that only the transcript levels of Apobec1 were elevated following the expression of both peptides (Figure 3G). Overall, our mRNA expression analysis of memory-related genes indicates that loci whose transcriptional regulation are tightly coupled to and rapidly induced by neuronal activation as well as genes encoding enzymes acting downstream of TET-mediated 5mC hydroxylation are sensitive to increases in TET1 enzyme levels. Lastly, the upregulation of memory-associated IEGs and the deaminase Apobec1, do not appear to be directly dependent on increased levels of 5hmC as the catalytically inactive TET1m elicited a similar effect.
Long-term memory formation is impaired by expression of TET1, independent of its catalytic activity
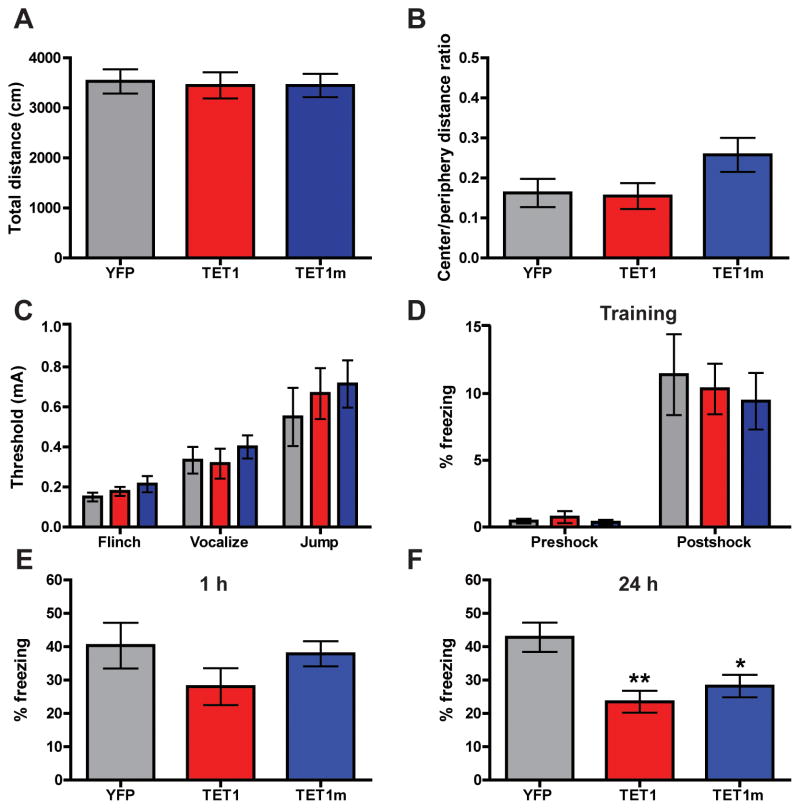
Having observed that AAV-mediated overexpression of TET1 in the dorsal hippocampus regulates the transcript levels of a number of genes involved in synaptic plasticity and memory formation (Figure 3G), and that TET1 is capable of driving the production of 5hmC in the hippocampus (Figure 3D–F), we next sought to investigate potential cognitive effects of TET1 overexpression. Two weeks after viral injection of TET1 and TET1m constructs, animals were subjected to several behavioral paradigms to evaluate locomotion, anxiety and memory formation. We found open field activity levels of all groups tested to be similar; demonstrating that exploratory behavior in a novel context was unaffected by elevated TET1 levels (Figure 4A). To measure levels of basal anxiety, we calculated the ratio of time spent in the center of the open field in relation to time spent on the periphery. No differences in anxiety-like behavior were observed (Figure 4B). In addition, all groups tested exhibited similar responses during the shock threshold test, which is critical for the proper interpretation of fear conditioning results (Figure 4C). Next, mice were fear conditioned using a background (novel context plus auditory cue) training paradigm consisting of a single presentation of a mild footshock. Time spent freezing during the training session --- either before or after the presentation of the footshock --- was similar between groups (Figure 4D). Contextual fear memory was assessed both 1 hr and 24 hrs after the training session. At 1 h following training, all groups exhibited similar levels of freezing behavior, indicating that overexpression of the TET1 catalytic domains did not have a significant effect on short term memory formation (Figure 4E). However, animals injected with AAV-TET1 or AAV-TET1m displayed an impairment of long term memory compared to AAV-YFP controls 24 h after training (Figure 4F). Taken together, these behavioral data suggest that overexpression of TET1 and TET1m in the dorsal hippocampus specifically impairs long term memory formation, while leaving general baseline behaviors and learning intact. Furthermore, it appears that the catalytic activity of TET1 is not necessary for this inhibition, as the TET1m blocks memory to a similar degree as observed with the catalytically active TET1; however, it is certainly possible that the two constructs inhibit memory consolidation by parallel and partially overlapping mechanisms (Figure S3).
Figure 4. Behavioral characterization of mice overexpressing TET1 and TET1m in the dorsal hippocampus.
(A) Total distance traveled during 15 min in the open field. (B) The ratio of time spent in the center versus time spent in the periphery of the open field, a measure of anxiety. (C) Shock threshold test. (D) Percent of time freezing before and after presentation of the foot shock during the 3 min training session. (E) Percent of time freezing during a 5 min context test, 1 h after training. For experiments A–C, E n = 9 for all groups. (F) Percent of time freezing during a 5 min context test, 24 h after training (F2, 58 = 7.185). YFP vs. TET1 and TET1m. **p < 0.01, *p < 0.05; one-way ANOVA followed by Bonferroni post hoc test. For experiments D and F; AAV-YFP (n = 17), AAV-TET1 (n = 21), AAV-TET1m (n = 21). All data are presented as mean ± s.e.m.
Discussion
Epigenetic regulation of gene expression through chromatin remodeling and DNA methylation are two important mechanisms required for long term information storage within the brain. Until recently, the mechanisms underlying active DNA demethylation during memory formation have remained mysterious and contentious (Day and Sweatt, 2010; Dulac, 2010). However, the discovery of 5hmC and its generation by the Tet family of proteins has lead to the identification of an active DNA demethylation pathway involved in many biological processes, including those pertaining to nervous system function. In the present study, we took a viral-mediated approach to genetically manipulate the enzymatic activity of TET1 in an attempt to determine whether this 5-methylcytosine dioxygenase might regulate learning and memory. We found endogenous TET1 to be strongly expressed in neurons throughout the hippocampus and that its transcript levels (Figure 1), as well as genes involved in active DNA demethylation (Figure S2), were reduced in response to neuronal activation under physiological conditions. Importantly, we observed similar reductions following fear conditioning, implicating Tet1 in the epigenetic regulation of gene expression necessary for memory formation.
Development of our HPLC/MS system (Figure 2) allowed for the sensitive, simultaneous measurement of 5mC, 5hmC and unmodified cytosines in CNS tissue. Using this system, we detected a small, but statistically significant reduction in both 5mC and 5hmC levels in area CA1 24 h after induction of a generalized-seizure episode, indicative of active DNA demethylation. In agreement with our results, genome-wide methylation analysis found evidence of promoter region hypomethylation at >90% of genes that were differentially expressed following status epilepticus (Miller-Delaney et al., 2012). Our findings add further support to the growing number of studies implicating changes in DNA methylation in response to neuronal activation across diverse experimental paradigms (Feng et al., 2010; Guo et al., 2011a; Guo et al., 2011b; Lubin et al., 2008; Ma et al., 2009; Miller et al., 2010; Miller and Sweatt, 2007; Monsey et al., 2011).
We observed that injection of an AAV virus expressing the TET1 catalytic domain resulted in a dramatic increase in global levels of 5hmC, as was also shown previously (Guo et al., 2011b). Moreover, using an accurate and sensitive HPLC/MS method we also observed a decrease in global 5mC and a significant increase in the fraction of unmodified cytosines compared to either control or TET1m infected samples (Figures 3D–F). Together these data provide evidence for an active DNA demethylation process at the global level, driven by TET1 oxidase activity and utilizing 5hmC as an intermediate. In agreement with this general model, we also observed a significant increase in the expression levels of several genes involved in TET-oxidase mediated DNA demethylation, including Tdg, Apobec1, Smug1 and Mbd4, after TET1 manipulation (Figure 3G). These findings suggest the transcription of these genes maybe coupled to changes in 5hmC as part of a transcriptionally coordinated system in neurons.
TET1 expression has been shown to induce increases in the expression of Bdnf and the brain-specific Fgf1B while providing no effect on the developmentally expressed Fgf1G, indicating target specificity (Guo et al, 2011b). Similarly, gene expression analysis of our survey of memory-related genes in our study not only confirmed that Bdnf is positively regulated by TET1, but also revealed significant regulation of many other IEGs, including Arc, Egr1, Fos, Homer1a and Nr4a2 (Figure 3G). Interestingly, TET1 did not have any significant effect on the expression of other genes we surveyed including reference genes, genes involved in synaptic plasticity and genes generally thought to negatively regulate memory. Unexpectedly, we found that the same set of genes whose expression was promoted by TET1 were also significantly elevated in response to the catalytically inactive TET1m, suggesting TET1 regulates the expression of these genes, at least in part, independently of 5mC to 5hmC conversion. These findings are contradictory to those previously reported by Guo et al.; where TET1m had no effect on the expression of Bdnf or Fgf1B in the DG (Guo et al., 2011b). One distinct possibility for this difference may include our targeting of pyramidal cells in area CA1 in comparison to the previous study’s focus on granule cells of the DG, which exhibit different gene expression profiles and thus, differences in the regulation of their transcriptomes (Datson et al., 2004).
Interestingly, data generated in an earlier study investigating TET1 and its role in embryonic stem (ES) cells lends support for our findings that TET1m regulates gene expression independent of its catalytic activity. Specifically, it was reported that shRNA-mediated knockdown (KD) of Tet1 in Dnmt triple knockout ES cells led to similar changes in gene expression as those observed in Tet1-depleted wild type cells (Williams et al., 2011). These findings suggest that in the absence of its 5mC substrate, TET1 retains the ability to both positively and negatively influence the expression of its gene targets. The mechanism through which the TET1m peptide, encompassing only 718 amino acids of the C-terminal end of TET1, positively regulates the expression of the genes examined in our study remains an open question. Presumably it is through an allosteric, as opposed to catalytic, mechanism.
In line with our finding that both TET1 and TET1m dysregulate the expression of the same group of memory-related genes, they similarly disrupted the formation of long-term memory formation following context fear conditioning (Figure 4F). The impairment of this process could be the result of several possibilities that are not mutually exclusive (see Figure S4). Our preferred hypothesis is that the constitutive increases observed for IEG mRNAs in mice selectively expressing TET1 and TET1m could result in memory dysfunction. Specifically, the increased expression of the transcription factors Fos (both constructs) and Egr1 (TET1 catalytic domain) and the subsequent activation of their downstream gene targets in the absence of the appropriate neuronal stimulus context may impair their ability to facilitate the correct response (James et al., 2005). Likewise, Bdnf (mutant construct) and Arc (catalytic domain) could lead to inappropriate signaling cascades and structural changes. Most importantly, it has been shown that the selective overexpression of Homer1a in the dorsal hippocampus of disrupts both LTP and spatial working memory (Celikel et al., 2007) offering direct evidence for how memory could be disrupted by expression of either construct.
In conclusion, this study revealed for the first time that the 5-methylcytosine dioxygenase Tet1 is regulated by neuronal activity, that TET1 oxidase activity drives active demethylation in the CNS and positively regulates several genes implicated in learning and memory, and that its overexpression impairs hippocampus-dependent long term associative memory. Surprisingly, expression of both the TET1 catalytic domain and a catalytically inactive mutant affected gene expression and memory formation similarly, prompting future studies into the roles of both oxidase-dependent and oxidase-independent functions of TET1 in transcription and memory.
Experimental Procedures
Detailed experimental procedures can be found in Supplemental Experimental Procedures online.
Supplementary Material
Acknowledgments
The authors thank Stephen Moore, Alison Margolies, and Faraz Sultan for help with experiments, and Adam Petterson at Zymo Research for technical assistance regarding measurement of 5mC and 5hmC. This work was supported by NIH grants MH091122, MH57014, and NR012686 to J.D.S, and the McKnight Brain Research Foundation. Further support was provided by NIH grants NS07344, ES021957 and SFARI to H.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celikel T, Marx V, Freudenberg F, Zivkovic A, Resnik E, Hasan MT, Licznerski P, Osten P, Rozov A, Seeburg PH, et al. Select overexpression of homer1a in dorsal hippocampus impairs spatial working memory. Front Neurosci. 2007;1:97–110. doi: 10.3389/neuro.01.1.1.007.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nat Neurosci. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010a;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011a;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011b;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmerich K, Dingler FA, Rada C, Neuberger MS. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res. 2012;40:6016–6025. doi: 10.1093/nar/gks259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Delaney SF, Das S, Sano T, Jimenez-Mateos EM, Bryan K, Buckley PG, Stallings RL, Henshall DC. Differential DNA methylation patterns define status epilepticus and epileptic tolerance. J Neurosci. 2012;32:1577–1588. doi: 10.1523/JNEUROSCI.5180-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan FA, Day JJ. Epigenetic mechanisms in memory and synaptic function. Epigenomics. 2011;3:157–181. doi: 10.2217/epi.11.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, et al. Tet1 Regulates Adult Hippocampal Neurogenesis and Cognition. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.