Abstract
Pancreatic cancer is the fourth leading cause of cancer related death in the US and exhibits aggressive features with short survival rate and high mortality. Therefore, it is important to understand the molecular mechanism(s) involved in the aggressive growth of pancreatic cancers, and further design novel targeted therapies for its treatment with better treatment outcome. In the present study, we found that the expression of miR-221 was significantly up-regulated in pancreatic cancer cell lines and tumor tissues compared to normal pancreatic duct epithelial cells and normal pancreas tissues. Moreover, we found that the pancreatic cancer patients with high miR-221 expression had a relatively shorter survival compared to those with lower expression, suggesting that miR-221 could be an oncogenic miRNA and a prognostic factor for poor survival of patients. Interestingly, transfection of miR-221 inhibitor suppressed the proliferative capacity of pancreatic cancer cells with concomitant up-regulation of PTEN, p27kip1, p57kip2, and PUMA, which are the tumor suppressors and the predicted targets of miR-221. Most importantly, we found that the treatment of pancreatic cancer cells with isoflavone mixture (G2535), formulated 3,3’-diindolylmethane (BR-DIM), or synthetic curcumin analogue (CDF) could down-regulate the expression of miR-221 and consequently up-regulate the expression of PTEN, p27kip1, p57kip2, and PUMA, leading to the inhibition of cell proliferation and migration of MiaPaCa-2 and Panc-1 cells. These results provide experimental evidence in support of the oncogenic role of miR-221 and also demonstrate the role of isoflavone, BR-DIM, and CDF as potential non-toxic agents that are capable of down-regulation of miR-221. Therefore, these agents combined with conventional chemotherapeutics could be useful in designing novel targeted therapeutic strategy for the treatment of pancreatic cancer for which there is no curative therapy.
Keywords: miR-221, proliferation, pancreatic cancer, isoflavone, DIM, CDF
Introduction
Although the incidence and mortality of pancreatic cancers have declined slowly in recent years, pancreatic cancer is still the fourth leading cause of cancer related death in the US with estimated 30,700 new cases and 30,000 deaths expected in 2013 [1]. Pancreatic cancer exhibits aggressive features with shorter 5-year relative survival rate of ~4%. For all stages combined, the 1-year relative survival rate is only 21% [1]. Such high mortality of pancreatic cancer could in part be due to the capacity of pancreatic cancer cells to acquire rapid cell proliferative, invasive, and metastatic characteristics during the development and progression of pancreatic cancer. Only early pancreatic cancers can be removed by surgery. Unfortunately, early pancreatic cancer only accounts for a very small numbers of patients (about 20% of all pancreatic cancer diagnosed). Moreover, chemotherapies for unresectable pancreatic cancer are not effective for most patients. Therefore, it is important to understand the molecular mechanism(s) involved in the aggressive growth characteristics of pancreatic cancer. By knowing the altered molecular signaling in pancreatic cancer, novel targeted and combination therapies could be designed to inhibit the aggressiveness of pancreatic cancer so that the patients with pancreatic cancers could be treated with better outcome.
In recent years, growing evidence demonstrates the importance of microRNAs (miRNAs) in the development and progression of cancers including pancreatic cancer [2-5]. The aberrantly increased or decreased level of specific miRNAs in pancreatic cancer is associated with the aggressiveness of pancreatic cancer [3,4,6]. Experimental studies have also identified some of their molecular targets which are known to regulate the biological behaviors of cancer cells [3-6]. The miR-221 is one of the oncogenic miRNAs which is known to promote the development and progression of various cancers [7-10]. The up-regulation of miR-221 expression has been found in various types of cancers. The miR-221 could inhibit the expression of its targets, HECTD2 and RAB1A, leading to the development of castration resistant prostate cancer [7,10] whereas miR-221 could also promote tumorigenesis in triple negative breast cancer cells through the inhibition of p27kip1 and E-cadherin [8]. In addition, miR-221 has been found to induce cell survival and cisplatin resistance, and reduce apoptosis of osteosarcoma cells through the inhibition of PTEN signaling [9]. In pancreatic cancer, the concentration of plasma miR-221 has been found to be significantly higher compared with benign pancreatic tumors and normal controls while plasma miR-221 concentration was significantly reduced in postoperative samples [11]. A recent study showed that miR-221 could regulate PDGF-mediated EMT phenotype and growth of pancreatic cancer cells [12]. These observations suggest that miR-221 could play important roles in the aggressiveness of pancreatic cancer. Therefore, this miRNA could be a putative oncogenic promoter, and thus strategies to down-regulate its expression may prove to be beneficial in reverting the aggressive phenotype of pancreatic cancer. By investigating the targets and related signaling of miR-221, the targeted therapeutic strategies could be designed for the treatment of pancreatic cancers with better treatment outcome.
We have previously found that the dietary compounds including isoflavone genistein and 3,3’-diindolylmethane (DIM) could enhance the anti-tumor activity of chemotherapeutic agents in various cancers including pancreatic cancers [13,14]. We have also found that isoflavone genistein and DIM could up-regulate the expression of let-7, miR-200, and miR-146a, leading to the reversal of epithelial-to-mesenchymal transition and the suppression of invasive capacity of pancreatic cancer cells [15,16]. In this study, we assessed the expression patterns of miR-221 and its targets in the normal pancreatic duct epithelial cells, pancreatic cancer cell lines, pancreatic cancer tissues, and normal pancreatic tissues. We also investigated whether the treatment of pancreatic cancer cells with either G2535 (a mixture of genistein and other isoflavones), BR-DIM (BioResponse formulated DIM with greater bioavailability [17]), or CDF (a novel difluorinated curcumin analogue) could alter the expression of miR-221 and its targets that are related to the aggressiveness of pancreatic cancer. The effects of miR-221, G2535, BR-DIM, and CDF on relevant molecular regulations and biological behaviors of pancreatic cancer were also investigated in this study.
Materials and methods
Cell lines, reagents, and antibodies
MiaPaCa-2, Panc-1, and BxPC-3 pancreatic cancer cells obtained from ATCC (Manassas, VA) were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 50 U/mL penicillin, and 50 μg/mL streptomycin in a 5% CO2 atmosphere at 37 °C. Human pancreatic duct epithelial (HPDE) cells were obtained from MD Anderson Cancer Center (a generous gift of Dr. Paul J. Chiao), maintained in keratinocyte serum-free medium supplied with 5 ng/mL of epidermal growth factor and 50 μg/mL of bovine pituitary extract (Invitrogen), and cultured in DMED/FBS medium when conducting experiments. The cell lines from ATCC have been tested and authenticated in core facility Applied Genomics Technology Center at Wayne State University. The method used for testing was short tandem repeat (STR) profiling using the PowerPlex® 16 System from Promega (Madison, WI). Isoflavone mixture G2535 (70.54% genistein, 26.34% diadzin, and 0.31% glycitein manufactured by Organic Technologies and obtained from NIH) was dissolved in DMSO to make a stock solution containing 50 mM equivalent to genistein. The concentration of isoflavone we described in this article all refer to the concentration of genistein in the isoflavone mixture. BR-DIM (BioResponse, Boulder, CO) was generously provided by Dr. Michael Zeligs and was dissolved in DMSO to make a 50 mM stock solution. CDF discovered in our institution [18] was dissolved in DMSO to make a 5 mM stock solution. Anti-PTEN, anti-p27kip1, anti-p57kip2, and anti-PUMA antibodies were purchased from Santa Cruz (Santa Cruz, CA), and used for Western Blot analysis.
Tissue collection
All 24 patients in the study were both clinically and pathologically diagnosed as pancreatic cancer. The median age was 65 and gender count was 58.3% female and 41.7% male. Archived formalin-fixed paraffin embedded (FFPE) tumor tissue blocks from these patients with pancreatic adenocarcinoma and morphologically normal appearing pancreas tissue that were anatomically far away from the pancreatic tumor and served as the control were used for the study along with the collection of survival data. The institutional human investigation review board approved the study. The relationship between miR-221 expression and survival was analyzed by Kaplan-Meier survival analysis using GraphPad Prism software (GraphPad Software Inc, San Diego, CA).
Total RNA extraction from tissues and cell lines
Total RNA from FFPE tissue was isolated by miRNeasy FFPE Kit and RNase-free DNase Set (QIAGEN, Valencia, CA) according to the manufacturer’s protocol. Briefly, four freshly cut tissue sections of 10 μm thick and approximately 1 cm in diameter were placed in micro tubes along with 1 ml xylene. RNA was extracted, eluted in a final volume of 25 μl, and quantified using NanoDrop 2000 (Thermo Scientific, Pittsburgh, PA) as described earlier [19]. The ratio of 260/280 varied from 1.8-2.1. Samples with values less or more were considered to be not usable.
Total RNA from cell lines was extracted by using the miRNeasy Mini Kit and RNase-free DNase Set (QIAGEN) following the protocol provided by the manufacturer.
miRNA array and data analysis
Purified RNA pooled separately from normal and tumor tissue samples were analyzed by LC Sciences for miRNA expression profiling using miRBase version 19 (LC Sciences Houston, TX). In LC Sciences, the total RNA samples were enriched for microRNAs and the miRNA arrays were performed on μParaFlo™ microfluidic chips, each of which had a miRNA probe region with multiple repeat regions that detect miRNAs. Multiple control probes were also included on the arrays for assessing various chip and assay qualities. Chips were scanned and the signal intensity data was obtained. Then, the data was analyzed by subtracting the background and normalizing the signals using selected housekeeping genes. The ratio of signals from normal and tumor tissues was calculated.
miRNA and mRNA real-time RT-PCR assay
The expression levels of miR-221 in pancreatic cells and tumors were further quantitated and validated by using TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA) following manufacturer’s protocol. Briefly, total RNA from each sample was subjected to reverse transcription with a specific miR-221 primer (Applied Biosystems). Real-time PCR reactions were then carried out in StepOnePlus (Applied Biosystems). The PCR program was initiated by 10 min at 95 °C before 40 thermal cycles, each of 15 s at 95 °C and 1 min at 60 °C. Data were analyzed according to the comparative Ct method and were normalized by RNU48 expression in each sample. The expression level of miRNA was statistically evaluated by Student’s t-Test using GraphPad StatMate software (GraphPad Software Inc).
The expression level of PTEN, p27kip1, p57kip2, and PUMA mRNAs in pancreatic cancer cells was analyzed by real-time RT-PCR using High Capacity RNA-to-cDNA Kit and SYBR Green Master Mixture (Applied Biosystems). The sequences of primers used were shown in Table 1. The PCR was initiated by 10 min at 95 °C before 40 thermal cycles, each of 15 s at 95 °C and 1 min at 60 °C. Data were analyzed according to the comparative Ct method and were normalized by GAPDH expression in each sample.
Table 1.
The sequences of primers used for real-time PCR
| Primers | Sequences |
|---|---|
| PTEN-2F | TCCAATGTTCAGTGGCGGAA |
| PTEN-2R | CGTGTGGGTCCTGAATTGGA |
| p27-2F | CAGCTTGCCCGAGTTCTACT |
| p27-2R | TGTCCTCAGAGTTAGCCGGA |
| p57v1-F | CTCCGCAGCACATCCACGAT |
| p57v1-R | GGTGCGCACTAGTACTGGGA |
| PUMA-2F | GTTCCAGCTGCAGGGGTG |
| PUMA-2R | CAGAGTGAAGGAGCACCGAG |
| GAPDH-F | TTCTTTTGCGTCGCCAGCCGA |
| GAPDH-R | GTGACCAGGCGCCCAATACGA |
Re-expression and inhibition of miR-221 in pancreatic cancer cells
MiaPaCa-2 (express high levels of miR-221) and Panc-1 (express low levels of miR-221) cells were seeded in 6 well plates. Next day, the MiaPaCa-2 cells were transfected with anti-miR-221 or anti-miR negative control (Applied Biosystems) while Panc-1 cells were transfected with miR-221 mimic or miR mimic negative control (Applied Biosystems) at a final concentration of 30 nM using DharmaFact Transfection Reagent (Dharmacon, Lafayette. CO). After 3 days of transfection, total RNA from each samples were then extracted using the miRNeasy Mini Kit and RNase-free DNase Set (QIAGEN) and subjected for measuring target mRNA expression by real-time RT-PCR. Total proteins from each sample were also extracted and subjected to Western Blot analysis to measure the target protein expression after transfections. The miR-221 mimic or inhibitor transfected cells were also subjected to proliferation assays.
Western Blot analysis
Western Blot analysis was conducted to test the protein expression level of miR-221 targets including PTEN, p27kip1, p57kip2, and PUMA by using our standard protocol. MiaPaCa-2 and Panc-1 cells were transfected with miR-221 mimic or inhibitor, or treated with 25 μM G2535, 25 μM BR-DIM, or 500 nM CDF for 72 hours. The cells were then lysed in RIPA buffer with protease inhibitors and protein concentration was measured using BCA protein assay (PIERCE, Rockford, IL). The proteins were subjected to 10% SDS-PAGE, and electrophoretically transferred to nitrocellulose membrane. The membranes were incubated with specific primary antibodies, and subsequently incubated with secondary antibody conjugated with peroxidase (Bio-rad, Hercules, CA). The signal was detected using the chemiluminescent detection system (PIERCE).
Cell proliferation assay by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
MiaPaCa-2 and Panc-1 cells were seeded in 96 well plates. The cells were then transfected with miR-221 mimic or inhibitor, or treated with 25 μM G2535, 25 μM BR-DIM, or 500 nM CDF for 72 hours. The transfected and treated cells were subjected to cell proliferation assay using MTT assay as described previously [14]. The cell proliferation index of MiaPaCa-2 and Panc-1 cells after transfection and treatment was statistically evaluated by Student’s t-Test using GraphPad StatMate software (GraphPad Software Inc).
Migration assay
The migratory capacity of MiaPaCa-2 cells after treatment with BR-DIM or CDF was accessed using wound-healing assay. The cells were plated into 6-well plates and cultured in the incubator until the cultures were subconfluent. The plates were then scratched linearly in multiple areas with a plastic 200 μl pipette tip. The cells were treated with 0.1% DMSO (vehicle control), 25 μM BR-DIM, or 500 nM CDF. The “wounded” areas were photographed by phase contrast microscopy at 0, 24 or 48 hour time points.
Results
Pancreatic cancer cells and tissues showed significant up-regulation of miR-221 expression
To investigate the difference in miR-221 expression between normal pancreatic duct epithelial cells (HPDE cells) and pancreatic cancer cells, we conducted miRNA RT-PCR assay. We found that the expression level of miR-221 was significantly up-regulated in MiaPaCa-2, Panc-1, and BxPC-3 pancreatic cancer cells compared to HPDE cells (Figure 1A). To reveal whether the up-relation of miR-221 in pancreatic cancer cell lines observed in vitro is also existed in vivo, we conducted miRNA array analysis using pooled total RNA extracted from tissue samples from 24 cases of pancreatic cancer. We found that the expression level of miR-221 was significantly higher in pancreatic cancer tissues than that in adjacent normal pancreatic tissues (Figure 1B). We further conducted real-time RT-PCR analysis of each specimen independently to quantitate and validate array data for assessing the levels of miR-221 expression in paired pancreatic cancer tissues and adjacent normal pancreatic tissues from the 24 cases of pancreatic cancer. The results from miRNA real-time RT-PCR analysis validated the data from miRNA array showing significantly higher expression of miR-221 in pancreatic cancer tissues (Figure 1C). These results clearly demonstrated that miR-221 is aberrantly up-regulated in pancreatic cancer and that miR-221 is an oncogenic miRNA which could promote the development and progression of pancreatic cancer. Because miRNA could regulate cancer development and progression by inhibiting the expression of its targets, we further tested the expression of miR-221 target genes after re-expression or inhibition of miR-221 by transfection studies in pancreatic cancer cells.
Figure 1.
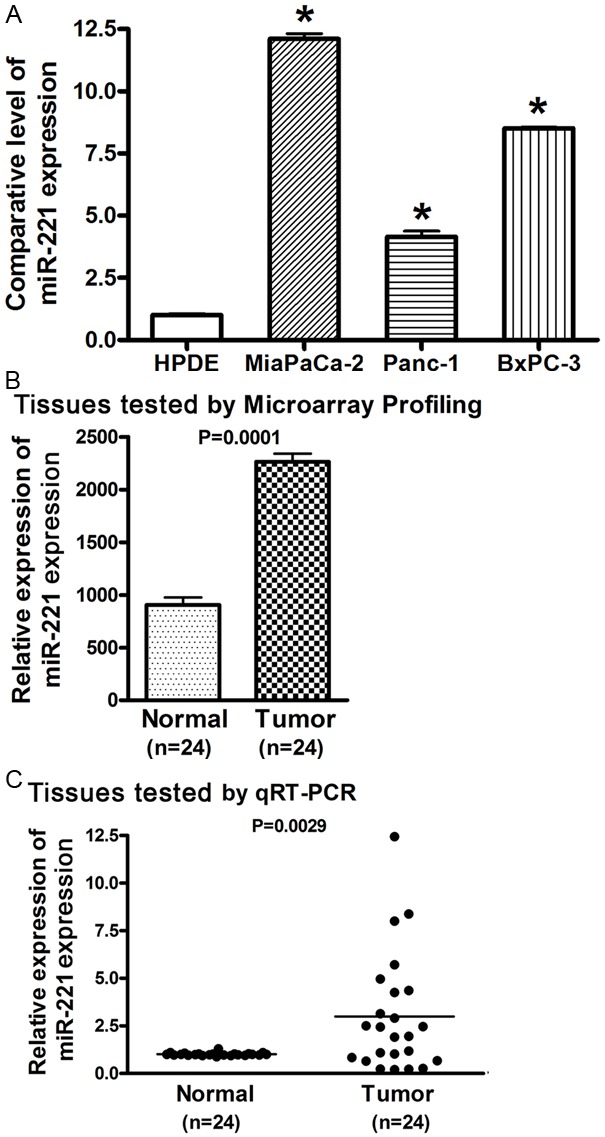
The expression of miR-221 was significantly higher in pancreatic cancer cells (A) and tissues (B, C) compared to normal pancreatic epithelial cells and tissues tested by miRNA real-time RT-PCR (A, C) and miRNA array (B). (*: P< 0.05).
Inhibition of miR-221 led to the up-regulation of its targets in cell proliferation signaling
From miRNA RT-PCR analysis, we observed that MiaPaCa-2 cells expressed significantly higher level of miR-221 while Panc-1 cells had relatively lower expression of miR-221 (Figure 1A). In order to investigate the role of miR-221 in the regulation of its targets and related signaling, we transfected miR-221 inhibitor into MiaPaCa-2 cells and conversely introduced miR-221 mimic into Panc-1 cells. PTEN, p27kip1, p57kip2, and PUMA have been found to be the targets of miR-221 in various cancers [8,9,20-22]. We found that the inhibition of miR-221 by transfection of miR-221 inhibitor caused up-regulation of PTEN, p27kip1, p57kip2, and PUMA in MiaPaCa-2 cells at the mRNA level (Figure 2A). In contrast, over-expression of miR-221 in Panc-1 cells resulted in the down-regulation of PTEN, p27kip1, p57kip2, and PUMA at the mRNA level (Figure 2A). Importantly, further studies showed that transfection of miR-221 inhibitor into MiaPaCa-2 cells induced the expression of PTEN, p27kip1, p57kip2, and PUMA proteins (Figure 2B). We also found that introduction of miR-221 mimic into Panc-1 cells caused the down-regulation of PTEN, p27kip1, p57kip2, and PUMA protein expression (Figure 2B). These results demonstrate that miR-221 could regulate the expression of its targets, PTEN, p27kip1, p57kip2, and PUMA in pancreatic cancer cells. Because PTEN, p27kip1, p57kip2, and PUMA all are the molecules in cell proliferation signaling and critically involved in the control of cancer cell proliferation, we further investigated the role of miR-221 in the progression of pancreatic cancer.
Figure 2.
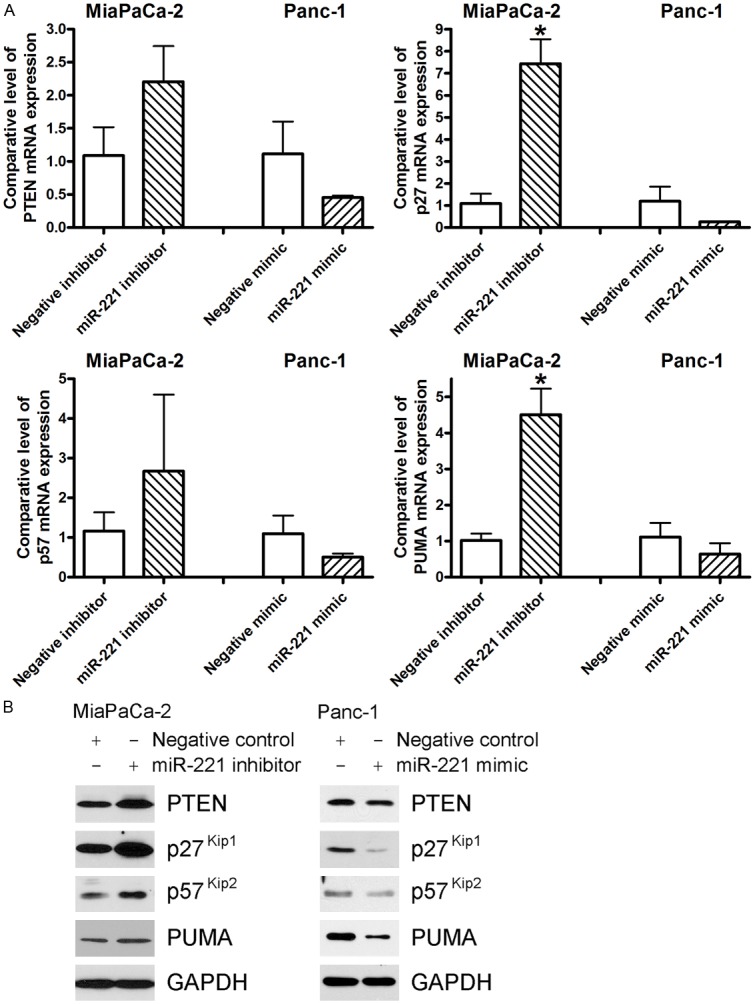
The inhibition of miR-221 by transfection with anti-miR-221 (miR-221 inhibitor) resulted in the up-regulation of PTEN, p27kip1, p57kip2, and PUMA at both mRNA (A) and protein (B) levels in MiaPaCa-2 cells while the introduction of miR-221 by transfection with miR-221 mimic down-regulated the expression of PTEN, p27kip1, p57kip2, and PUMA at mRNA (A) and protein (B) levels in Panc-1 cells. (*: P< 0.05).
High expression of miR-221 led to increased pancreatic cancer cell proliferation and poor survival of pancreatic cancer patients
After we transfected miR-221 inhibitor into MiaPaCa-2 cells, we found that the down-regulation of miR-221 caused inhibition of proliferation of MiaPaCa-2 cells (Figure 3A). Moreover, transfection of miR-221 mimic into Panc-1 cells significantly increased the proliferation of Panc-1 pancreatic cancer cells (Figure 3A). These results demonstrate the role of miR-221 in the promotion of cell proliferation in pancreatic cancer, suggesting that the high expression of miR-221 could be responsible for the aggressive progression of pancreatic cancer in vivo. Indeed, we found that the pancreatic cancer patients with low miR-221 expression had a relatively longer survival compared to the patients with high expression of miR-221 although the difference was not statistically significant (Figure 3B), which appears to be due to relatively low number of patients in this study. Moreover, two patients who are still alive and survive for more than 5 years after diagnosis had low expression of miR-221 (Figure 3B). These results suggest that the high expression of miR-221 could be a prognostic factor for the aggressiveness and poor survival of pancreatic cancer patients. Therefore, targeting miR-221 could be a promising strategy for the inhibition of tumor progression of pancreatic cancer. Thus, we further investigated whether isoflavone mixture G2535, BR-DIM, or CDF could alter the expression of miR-221 and its targets in pancreatic cancer, and thus these non-toxic agents could be novel therapeutics.
Figure 3.
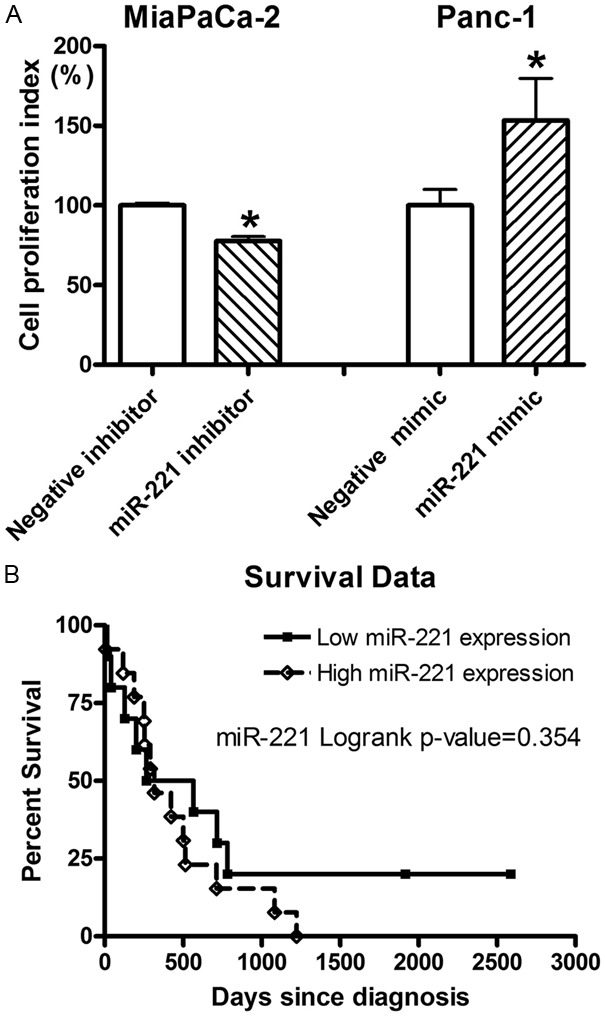
A: Transfection of miR-221 inhibitor into MiaPaCa-2 cells significantly inhibited cell proliferation while the transfection of miR-221 mimic into Panc-1 cell significantly promoted cell proliferation as tested by MTT assay (*: P< 0.05). B: Kaplan-Meier survival analysis showed that the pancreatic cancer patients with lower miR-221 expression had a relatively longer survival compared to the patients with higher expression of miR-221.
G2535, BR-DIM, and CDF inhibited the expression of miR-221
By RT-PCR assay for assessing the expression of miRNA, we found that 25 μM G2535, 25 μM BR-DIM or 500 nM CDF did differentially down-regulate the expression of miR-221 in MiaPaCa-2 and Panc-1 cells (Figure 4). However, the down-regulation of miR-221 expression was different between MiaPaCa-2 and Panc-1 cells. The reason for this difference could in part be due to differences in the basal level expression of miR-221 in these cell lines. MiaPaCa-2 cells had much higher expression of miR-221; therefore, the down-regulation of miR-221 was more obvious in MiaPaCa-2 cells compared with Panc-1 cells (Figure 4). Next, we examined whether G2535, BR-DIM or CDF could alter the expression of PTEN, p27kip1, p57kip2, and PUMA mediated through the regulation of miR-221 expression.
Figure 4.
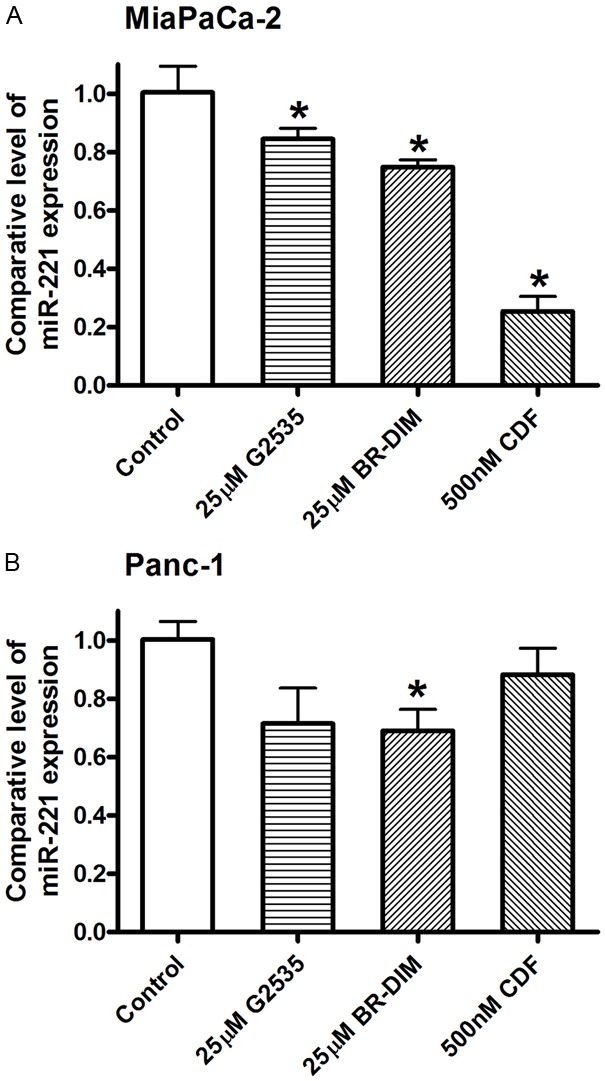
miRNA Real-time RT-PCR analysis showed that 25 μM G2535, 25 μM BR-DIM, and 500 nM CDF inhibited the expression of miR-221 in MiaPaCa-2 (A) and Panc-1 (B) cells (*: P< 0.05).
G2535, BR-DIM, and CDF induced the expression of PTEN, p27kip1, p57kip2, and PUMA
Because PTEN, p27kip1, p57kip2, and PUMA are miR-221 targets, the down-regulation of miR-221 by G2535, BR-DIM, or CDF could consequently up-regulate the expression of PTEN, p27kip1, p57kip2, and PUMA. By real-time PCR analysis, we did find that G2535, BR-DIM, or CDF treatment increased the expression level of PTEN, p27kip1, p57kip2, and PUMA mRNA in MiaPaCa-2 and Panc-1 cells (Figure 5A). Moreover, G2535, BR-DIM or CDF treatment also differentially up-regulated the expressions of PTEN, p27kip1, p57kip2, and PUMA proteins (Figure 5B), which is consistent with mRNA data. These results suggest that the induction of PTEN, p27kip1, p57kip2, and PUMA expression by G2535, BR-DIM or CDF could be due to the suppression of miR-221 expression. We further tested the consequences of regulation of PTEN, p27kip1, p57kip2, and PUMA which control cell proliferation.
Figure 5.
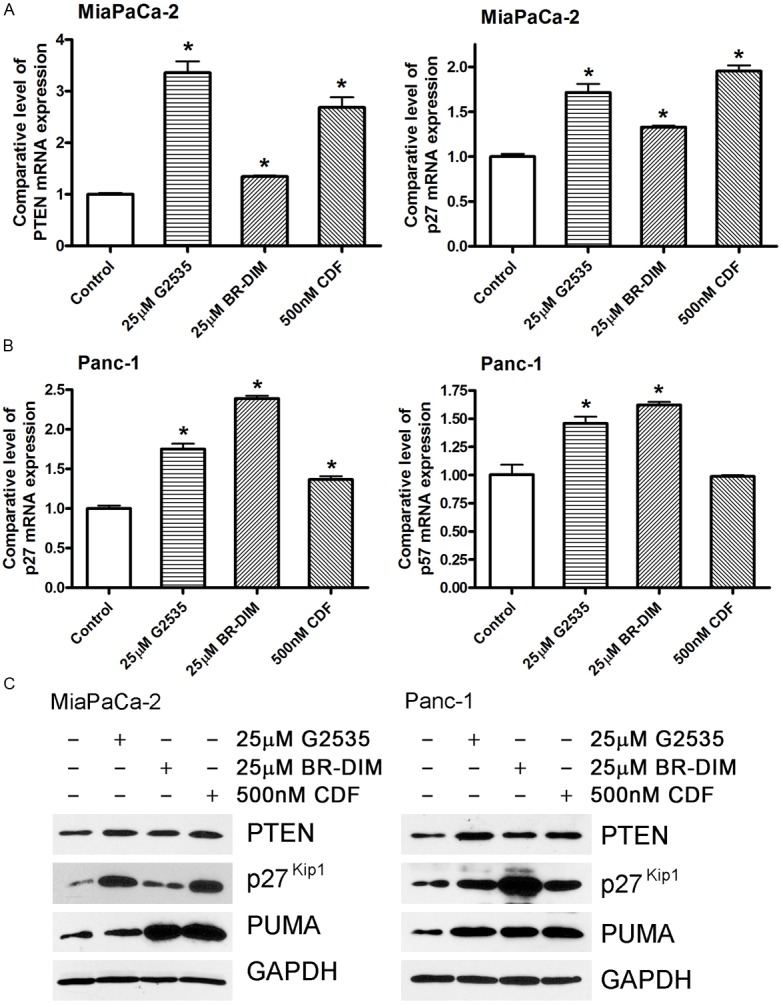
Real-time RT-PCR analysis (A, B) and Western Blot analysis (C) showed that 25 μM G2535, 25 μM BR-DIM, and 500 nM CDF induced the expression of PTEN, p27kip1, p57kip2, and PUMA in MiaPaCa-2 (A, C) and Panc-1 (B, C) cells both at the mRNA and protein levels. (*: P< 0.05).
G2535, BR-DIM, and CDF inhibited cell proliferation and migration of pancreatic cancer cells
Because we observed the inhibited cell proliferation by miR-221 inhibitor transfection (Figure 3A) and the down-regulated miR-221 expression by G2535, BR-DIM, or CDF (Figure 4), we tested whether the treatments by G2535, BR-DIM or CDF could also inhibit cell proliferation through the down-regulation of miR-221. We found that G2535, BR-DIM or CDF at different concentrations significantly inhibited cell proliferation in both MiaPaCa-2 and Panc-1 pancreatic cancer cells (Figure 6A and 6B). We also found that BR-DIM and CDF inhibited cell migration of MiaPaCa-2 cells (Figure 6C). These results suggest that the inhibition of cell proliferation or migration by G2535, BR-DIM or CDF could be partly mediated by the down-regulation of miR-221 expression and subsequent up-regulation of PTEN, p27kip1, p57kip2, and PUMA expression.
Figure 6.
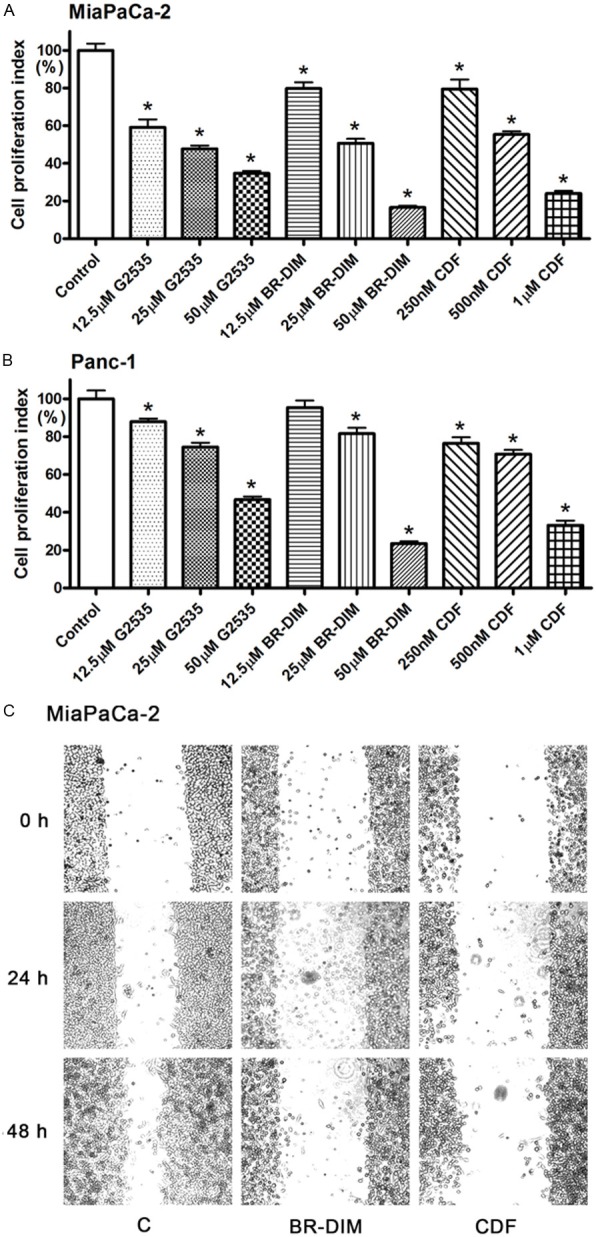
MTT assay showing that G2535, BR-DIM, and CDF at different concentrations significantly inhibited the proliferation of MiaPaCa-2 (A) and Panc-1 (B) cells (*: P< 0.05). (C) 25 μM BR-DIM and 500 nM CDF also inhibited cell migration of MiaPaCa-2 cells.
Discussion
Although aberrant expression of miR-221 has been found to be associated with the development of various cancers and implicated in the accelerated tumor growth, the in vivo prognostic significance of miR-221 in pancreatic cancer is still unclear. The elevated miR-221 levels have been found in most types of cancers including breast [8,23], prostate [24], hepatic [25], gastric [26], colorectal [21], pancreatic [27], and other cancers [28,29] although some controversy exists showing that the expression level of miR-221 was down-regulated in breast cancer tissues [30] and TMPRSS2:ERG fusion-positive prostate cancer [31]. In the present study, we found significantly up-regulated expression of miR-221 in pancreatic cancer cell lines and tumor tissues compared to normal pancreatic duct epithelial cells and normal pancreas tissues, respectively. Our finding is consistent with the report by other investigators [27], suggesting that miR-221 is an oncogenic miRNA in pancreas and is associated with the development of pancreatic cancer. Most importantly, we found that the over-expression level of miR-221 could be an important prognostic factor in predicting the survival of patients with pancreatic cancer. The pancreatic cancer patients with lower expression of miR-221 had a relatively longer survival time compared to those patients with relatively higher expression of miR-221. It is interesting to note that the only two patients found to be alive in this study showed lower expression of miR-221 and these two patients are surviving for more than 5 years after diagnosis. These results collectively suggest that the expression of miR-221 could exert its critical effects in the development and progression of pancreatic cancer and that the down-regulation of miR-221 in pancreatic cancer cells could suppress cell proliferation, and thus could inhibit progression of pancreatic cancer. However, further studies with large number of pancreatic cancer patients are needed to conclude the value of miR-221 as a prognostic factor for poor survival of patients diagnosed with pancreatic cancer.
The molecular mechanism(s) involved in the miR-221 mediated progression of pancreatic cancer are still unclear. From our results, we believe that miR-221 promotes the development and progression of pancreatic cancer partly through the regulation of signaling pathways which controls cell proliferation. It is well known that miRNA regulates physiological and pathophysiological processes through the suppression in the expression of its target genes. The reported targets of miR-221 in other types of cancers include PTEN [9,32,33], p27kip1 [20,34], p57kip2 [20,21], PUMA [35,36], and others [7,37,38]. However, the status and the roles of these miR-221 targets in the development and progression of pancreatic cancer are still unclear. PTEN is an important molecule in the regulation of cell growth and apoptosis. By suppressing PI3K-AKT-mTOR signaling, PTEN controls many cellular processes such as survival, proliferation, energy metabolism and cellular architecture [39]. PTEN is a tumor suppressor gene and shows aberrant expression in cancers due to genetic mutation, epigenetic silencing, transcriptional repression, or miRNA regulation [39]. The progression of cell cycle is driven by cyclins and their associated cyclin-dependent kinases (CDKs). Both p27kip1 and p57kip2 are CDK inhibitors, suggesting their roles in the suppression of cell growth. It is well known that p27kip1 regulates cell proliferation, motility and apoptosis [40]. In human cancers, decreased expression or cytoplasmic mislocalization of p27kip1 causes augmented cell proliferation and migration, leading to the progression of cancer [41]. p57kip2 controls the process of cell cycle exit, cytoskeletal organization, cell migration and differentiation [42]. In cancer cells, its expression is down-regulated through epigenetic changes such as DNA methylation, histone modification, or miRNA regulation [43]. PUMA (p53 upregulated modulator of apoptosis) is a Bcl-2 homology 3 (BH3)-only Bcl-2 family member. PUMA plays a critical role in the regulation of p53-dependent and -independent apoptosis [44]. It has been found that the expression of PUMA is down-regulated in malignant cutaneous melanoma and that the low expression of PUMA was a predictor of poor prognosis in patients [45]. Interestingly, all these molecules (PTEN, p27kip1, p57kip2, and PUMA) are the targets of miR-221 and are critical regulators controlling cell cycle, cell proliferation and apoptosis. In the present study, we found that the transfection of miR-221 mimic could inhibit the expression of PTEN, p27kip1, p57kip2, and PUMA both at the mRNA and protein levels in pancreatic cancer, leading to enhanced cell proliferation of pancreatic cancer cells. These results suggest that the oncogenic effect of miR-221 is mediated through the inhibition of tumor suppressors, PTEN, p27kip1, p57kip2, and PUMA in pancreatic cancer. Moreover, we also found that the transfection with miR-221 inhibitor could induce the expression of PTEN, p27kip1, p57kip2, and PUMA, resulting in the inhibition of cell proliferation in pancreatic cancer. These results suggest that the strategies which down-regulate miR-221 expression could be useful for the suppression of pancreatic cancer growth through the induction in the expression of the tumor suppressor PTEN, p27kip1, p57kip2, and PUMA which are the targets of miR-221.
The inhibition of cancer cell proliferation and cancer progression should be an important strategy for the successful treatment of pancreatic cancer. Therefore, any novel strategies which inhibit the aggressive ability of pancreatic cancer cells by targeting specific molecules should be useful for improving the devastating outcome of patients diagnosed with pancreatic cancer. We have previously reported that isoflavone, DIM or CDF (non-toxic natural agents or analogue) could inhibit the progression of prostate and breast cancer cells [46-48]. However, the molecular mechanisms involved in the inhibition of cancer progression by these agents have not been fully elucidated. In the present study, we found that the non-toxic natural agent such as isoflavone and BR-DIM, and the synthetic compound CDF could inhibit the expression of miR-221 and, in turn, induce the expression of PTEN, p27kip1, p57kip2, and PUMA causing inhibition of cell proliferation and migration of pancreatic cancer cells. These results clearly suggest that instead of in vivo delivery of synthetic miR-221 antisense nucleotide which has several side-effects, one could simply treat pancreatic cancer cells with non-toxic natural agents (isoflavone and BR-DIM) or CDF that will lead to the suppression of miR-221 expression. We believe that such a strategy targeting miR-221 could be useful for the activation of multiple tumor suppressor genes including PTEN, p27kip1, p57kip2, and PUMA which are downstream targets of miR-221 toward pancreatic cancer therapy. However, further in-depth mechanistic studies and in vivo clinical trials are warranted based on our exciting results.
In conclusion, our results clearly demonstrate that the up-regulation of miR-221 and down-regulation of its targets, PTEN, p27kip1, p57kip2, and PUMA, are responsible for the aggressive nature of pancreatic cancer. Our results also exhibited that non-toxic natural agents (isoflavone mixture G2535 and BR-DIM) and CDF could down-regulate miR-221 and inhibit pancreatic cancer cell proliferation and migration partly due to the induction of PTEN, p27kip1, p57kip2, and PUMA, which are miR-221 targets and commonly inactivated in pancreatic cancer. Further in vivo studies and clinical trials are needed for assessment of whether isoflavone mixture G2535, BR-DIM, and CDF could be useful in combination with conventional chemotherapeutics or targeted agents for improving the treatment outcome of pancreatic cancer patients for whom curative therapy is urgently needed.
Acknowledgements
This work was partly funded by the grants from the National Cancer Institute, NIH (5R01CA108535, 5R01CA131151, 5R01CA132794, 5R01CA154321, and 5R01CA164318). We also thank Guido and Puschelberg Foundation for their generous contribution for the completion of this study.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, Gong Q, Wang CY. MicroRNA-141, downregulated in pancreatic cancer, inhibited the cell proliferation and invasion by directly targeting MAP4K4. Mo Cancer Ther. 2013 doi: 10.1158/1535-7163.MCT-13-0296. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Singh S, Chitkara D, Kumar V, Behrman SW, Mahato RI. miRNA profiling in pancreatic cancer and restoration of chemosensitivity. Cancer Lett. 2013;334:211–220. doi: 10.1016/j.canlet.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwagami Y, Eguchi H, Nagano H, Akita H, Hama N, Wada H, Kawamoto K, Kobayashi S, Tomokuni A, Tomimaru Y, Mori M, Doki Y. miR-320c regulates gemcitabine-resistance in pancreatic cancer via SMARCC1. Br J Cancer. 2013;109:502–511. doi: 10.1038/bjc.2013.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Balk S, Lee GS, Kantoff PW. MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene. 2013 doi: 10.1038/onc.2013.230. doi: 10.1038/onc.2013.230. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassirpour R, Mehta PP, Baxi SM, Yin MJ. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8:e62170. doi: 10.1371/journal.pone.0062170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhao G, Cai C, Yang T, Qiu X, Liao B, Li W, Ji Z, Zhao J, Zhao H, Guo M, Ma Q, Xiao C, Fan Q, Ma B. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8:e53906. doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun T, Yang M, Chen S, Balk S, Pomerantz M, Hsieh CL, Brown M, Lee GS, Kantoff PW. The altered expression of MiR-221/-222 and MiR-23b/-27b is associated with the development of human castration resistant prostate cancer. Prostate. 2012;72:1093–1103. doi: 10.1002/pros.22456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361–369. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su A, He S, Tian B, Hu W, Zhang Z. MicroRNA-221 Mediates the Effects of PDGF-BB on Migration, Proliferation, and the Epithelial-Mesenchymal Transition in Pancreatic Cancer Cells. PLoS One. 2013;8:e71309. doi: 10.1371/journal.pone.0071309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali S, Banerjee S, Ahmad A, El-Rayes BF, Philip PA, Sarkar FH. Apoptosis-inducing effect of erlotinib is potentiated by 3,3’-diindolylmethane in vitro and in vivo using an orthotopic model of pancreatic cancer. Mol Cancer Ther. 2008;7:1708–1719. doi: 10.1158/1535-7163.MCT-08-0354. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Li Y, Ahmed F, Ali S, Philip PA, Kucuk O, Sarkar FH. Inactivation of nuclear factor kappaB by soy isoflavone genistein contributes to increased apoptosis induced by chemotherapeutic agents in human cancer cells. Cancer Res. 2005;65:6934–6942. doi: 10.1158/0008-5472.CAN-04-4604. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Vandenboom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Vandenboom TG, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderton MJ, Manson MM, Verschoyle R, Gescher A, Steward WP, Williams ML, Mager DE. Physiological modeling of formulated and crystalline 3,3’-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos. 2004;32:632–638. doi: 10.1124/dmd.32.6.632. [DOI] [PubMed] [Google Scholar]
- 18.Padhye S, Yang H, Jamadar A, Cui QC, Chavan D, Dominiak K, McKinney J, Banerjee S, Dou QP, Sarkar FH. New difluoro Knoevenagel condensates of curcumin, their Schiff bases and copper complexes as proteasome inhibitors and apoptosis inducers in cancer cells. Pharm Res. 2009;26:1874–1880. doi: 10.1007/s11095-009-9900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ali S, Saleh H, Sethi S, Sarkar FH, Philip PA. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer. 2012;107:1354–1360. doi: 10.1038/bjc.2012.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 21.Sun K, Wang W, Zeng JJ, Wu CT, Lei ST, Li GX. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol Sin. 2011;32:375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Martino MT, Gulla A, Cantafio ME, Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F, Cannataro M, Neri A, Giordano A, Tagliaferri P, Tassone P. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget. 2013;4:242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10:507–517. doi: 10.4161/cc.10.3.14754. [DOI] [PubMed] [Google Scholar]
- 24.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–3363. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70–73. doi: 10.1016/j.bbrc.2011.01.111. [DOI] [PubMed] [Google Scholar]
- 26.Liu K, Li G, Fan C, Diao Y, Wu B, Li J. Increased Expression of MicroRNA-221 in gastric cancer and its clinical significance. J Int Med Res. 2012;40:467–474. doi: 10.1177/147323001204000208. [DOI] [PubMed] [Google Scholar]
- 27.Panarelli NC, Chen YT, Zhou XK, Kitabayashi N, Yantiss RK. MicroRNA expression aids the preoperative diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2012;41:685–690. doi: 10.1097/MPA.0b013e318243a905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang CJ, Shen WG, Liu CJ, Chen YW, Lu HH, Tsai MM, Lin SC. miR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells. J Oral Pathol Med. 2011;40:560–566. doi: 10.1111/j.1600-0714.2010.01005.x. [DOI] [PubMed] [Google Scholar]
- 29.Gombos K, Horvath R, Szele E, Juhasz K, Gocze K, Somlai K, Pajkos G, Ember I, Olasz L. miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res. 2013;33:1511–1517. [PubMed] [Google Scholar]
- 30.Hui AB, Shi W, Boutros PC, Miller N, Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, Jurisica I, Penn LZ, Liu FF. Robust global micro-RNA profiling with formalin-fixed paraffin-embedded breast cancer tissues. Lab Invest. 2009;89:597–606. doi: 10.1038/labinvest.2009.12. [DOI] [PubMed] [Google Scholar]
- 31.Gordanpour A, Stanimirovic A, Nam RK, Moreno CS, Sherman C, Sugar L, Seth A. miR-221 Is down-regulated in TMPRSS2:ERG fusion-positive prostate cancer. Anticancer Res. 2011;31:403–410. [PMC free article] [PubMed] [Google Scholar]
- 32.Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer. 2010;10:367. doi: 10.1186/1471-2407-10-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garofalo M, Di LG, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37:1621–1626. doi: 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- 36.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, Pu PY, Cheng JQ, Kang CS. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O’Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, Newman RJ, Yue P, Bourgon R, Modrusan Z, Stern HM, Warming S, de Sauvage FJ, Amler L, Yeh RF, Dornan D. miR-221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:pt5. doi: 10.1126/scisignal.2002258. [DOI] [PubMed] [Google Scholar]
- 38.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 40.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 41.Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin Cancer Res. 2011;17:12–8. doi: 10.1158/1078-0432.CCR-10-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borriello A, Caldarelli I, Bencivenga D, Criscuolo M, Cucciolla V, Tramontano A, Oliva A, Perrotta S, Della RF. p57(Kip2) and cancer: time for a critical appraisal. Mol Cancer Res. 2011;9:1269–1284. doi: 10.1158/1541-7786.MCR-11-0220. [DOI] [PubMed] [Google Scholar]
- 43.Kavanagh E, Joseph B. The hallmarks of CDKN1C (p57, KIP2) in cancer. Biochim Biophys Acta. 2011;1816:50–56. doi: 10.1016/j.bbcan.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–S83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karst AM, Dai DL, Martinka M, Li G. PUMA expression is significantly reduced in human cutaneous melanomas. Oncogene. 2005;24:1111–1116. doi: 10.1038/sj.onc.1208374. [DOI] [PubMed] [Google Scholar]
- 46.Kong D, Heath E, Chen W, Cher M, Powell I, Heilbrun L, Li Y, Ali S, Sethi S, Hassan O, Hwang C, Gupta N, Chitale D, Sakr WA, Menon M, Sarkar FH. Epigenetic silencing of miR-34a in human prostate cancer cells and tumor tissue specimens can be reversed by BR-DIM treatment. Am J Transl Res. 2012;4:14–23. [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Kong D, Wang Z, Ahmad A, Bao B, Padhye S, Sarkar FH. Inactivation of AR/TMPRSS2-ERG/Wnt signaling networks attenuates the aggressive behavior of prostate cancer cells. Cancer Prev Res (Phila) 2011;4:1495–506. doi: 10.1158/1940-6207.CAPR-11-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Upadhyay S, Bhuiyan M, Sarkar FH. Induction of apoptosis in breast cancer cells MDA-MB-231 by genistein. Oncogene. 1999;18:3166–3172. doi: 10.1038/sj.onc.1202650. [DOI] [PubMed] [Google Scholar]


