Abstract
Notch signaling plays an essential role in development as well as cancer. We have previously shown that Notch3 is important for lung cancer growth and survival. Notch receptors are activated through the interaction with their ligands, resulting in proteolytic cleavage of the receptors. This interaction is modulated by Fringe, a family of fucose-specific β1,3 N-acetylglucosaminyltransferases that modify the extracellular subunit of Notch receptors. Studies in developmental models showed that Fringe enhances Notch’s response to Delta ligands at the expense of Jagged ligands. We observed that Manic Fringe expression is down-regulated in lung cancer. Since Jagged1, a known ligand for Notch3, is often over-expressed in lung cancer, we hypothesized that Fringe negatively regulates Notch3 activation. In this study, we show that re-expression of Manic Fringe down-regulates Notch3 target genes HES1 and HeyL and reduces tumor phenotype in vitro and in vivo. The mechanism for this phenomenon appears to be related to modulation of Notch3 protein stability. Proteasome inhibition reverses Manic Fringe-induced protein turnover. Taken together, our data provide the first evidence that Manic Fringe functions as a tumor suppressor in the lung and that the mechanism of its anti-tumor activity is mediated by inhibition of Notch3 activation.
Keywords: Jagged1, manic fringe, Notch3, lung cancer
Introduction
Notch signaling plays a highly conserved role in determining cell fate and controlling cell growth in both mammals and lower organisms [1]. In mammals, there are four Notch receptors. They are single transmembrane proteins, containing 29 to 36 epidermal growth factor-like (EGFL) repeats within the extracellular domain [2]. There are two families of Notch ligands, the Delta-like ligands (Delta-like 1, 3 and 4) and the Jagged ligands (Jagged1-2). In the canonical pathway, the interaction between Notch receptors and their ligands results in pathway activation through a series of proteolytic cleavages, releasing the intracellular fragment, which translocates to the nucleus to activate transcription of target genes.
Notch regulates cell fate determination through its effects on differentiation, growth and apoptosis. Unsurprisingly, aberrant activation of Notch proteins has been associated with cancer phenotypes [3-5]. We were first to link Notch3 with lung cancer [6]. We showed that Notch3 is over-expressed in about 40% of non-small cell lung cancers (NSCLC), and suppression of Notch3 results in the loss of the malignant phenotype both in vitro and in vivo models [7]. Furthermore, recent research indicates a role for Notch3 signaling in other cancers such as breast and ovarian cancer [8,9].
Given the important role of Notch signaling in cancer, we seek to better understand its regulation in the context of cancer. Originally described in Drosophila, Fringe was shown to regulate the ligand-sensitivity of Notch signaling, defining the precise signal location at the dorsal/ventral boundary of the wing marginal disc [10]. Mammals have three Fringe homologues, Lunatic Fringe (Lfng), Manic Fringe (Mfng) and Radical Fringe (Rfng). Recent studies showed that Fringe proteins are fucose-specific β1,3-GlcNAc transferases and utilize uridine diphosphate N-acetylglucosamine (UDP-GlcNAc) as a donor substrate. The addition of GlcNAc groups to O-fucose groups within Notch’s extracellular domain modulates the response of Notch to its ligands by potentiating Delta ligand signaling while inhibiting Serrate ligands, such as Jagged1 [11,12].
While the contribution of Fringe to the modulation of Notch activation in development is still being elucidated, Fringe’s role in cancer remains poorly understood. High expression of Jagged1 with reciprocal decrease in expression of Manic Fringe was observed in the progression of human papillomavirus-mediated cervical cancer [13]. Furthermore, the loss of Lunatic Fringe cooperates with Met/Caveolin in the development of basal-like breast cancer [14].
Human Manic Fringe is located on chromosome 22q13.1, a region often found to be deleted in lung cancer using CGH and karyotypic analyses [15-17]. By contrast, the loci for Lunatic Fringe and Radical Fringe, 7p22 and 17q25 appear unaffected in lung cancer, leaving Manic Fringe as the only family member the genetic locus of which is deleted. These observations suggest a role for Manic Fringe in tumor suppression in the context of lung cancer. In the present study, we observed lower expression of Manic Fringe in lung cancer compared to normal tissue. We also noted that re-introduction of Manic Fringe in lung cancer cells decreased Notch3 protein stability and reduced cell proliferation and tumor growth. These findings support hypothesis that Manic Fringe functions as a tumor suppressor in lung cancer.
Materials and methods
Cancer microarray database and data mining platform
Oncomine (Compendia Bioscience, Ann Arbor, MI) was used to retrieve the public cancer database information for the analysis of Fringe expression in lung cancer [18]. Seven datasets containing both normal tissues and cancers were used for analysis [19-25]. The probes for Fringe genes were derived from the following the probe sets: HG-U133_Plus_2 (MFNG 204152_s_at, RFNG 212968_at, and LFNG 215270_at for Hou, Landi, Su and Wachi datasets, HG-U95Av2 (MFNG 41521_at; RFNG 41705_at, and LFNG 431908_at for Bhatta-charjee and Stearman) and IMAGE clone 51817 (Garber). Values for Log2 Mean-Centered Intensity were extracted from Oncomine. Student’s two-sided t-tests were performed to determine the statistical significance of an observation. P-values <0.05 were considered statistically significant. To compare the relative significance of expression among Manic Fringe, Lunatic Fringe and Radical Fringe, we used Oncomine’s gene rank analysis. Each gene rank reflects the rank of that gene in the dataset based on its p-value.
DNA constructs
Plasmid pcDNA4-Notch3 expressing full-length Notch3 with a carboxy-terminal His-tag has been described previously [26]. Human Manic Fringe expressing plasmid with a N-terminal FLAG-tag was generated using PCR amplification followed by directional cloning in pCMV-3tag vectors at EcoRI and HindIII restriction sites.
Cell lines and cell culture
The Notch3-expressing lung cancer cell line HCC2429 was established as previously described [6]. The human embryonic kidney (HEK) 293T cell line and the human non-small cell lung cancer cell line H460 were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) and RPMI supplemented with 10% fetal calf serum, respectively. To establish stable clones expressing Manic Fringe, the pcDNA3.1-mFng (Manic Fringe construct) was transfected into HCC2429 or H460 cells using Lipofectamine (Invitrogen) according to manufacturer’s recommendation. After 48 hours, the cells were selected using G418 for 4 weeks. Incorporation of the Manic Fringe construct was determined using antibody detection of the FLAG epitope tag.
Chemicals and antibodies
Cycloheximide and MG132 were obtained from Sigma and Calbiochem, respectively. Rabbit anti-Notch3 (D11B8) and rat anti-Notch1 antibodies (5B5) were obtained from Cell Signaling Technology Inc. Mouse monoclonal antibodies against α-tubulin, FLAG-tag M2, and GFP were obtained from EMD Millipore Co., Sigma-Aldrich Inc., and Roche Diagnostic GmbH.
RNA extraction and real-time PCR
Total RNA was extracted with PureLink™ RNA Mini Kits by Life Technology from HCC2429 cells or HEK 293 cells after transfection of either Notch3 plasmids or control plasmids. RNA was reverse transcribed with iScript cDNA synthesis kits (Bio-RAD, Hercules, CA). Real-time PCR was performed with the iQ5 multicolor Real-Time PCR detection system (Bio-RAD). For the reactions, 50 μl mixtures were used containing 5 μl cDNA sample (0.5-1 μg/μl), 300 nM of primers for target genes or GAPDH. After incubation at 50°C for 2 min followed by 95°C for 10 min, the reactions were carried out for 40 cycles of the following: 95°C for 15 sec and 55°C for 1 min. The threshold cycle value (Ct) was obtained with iCycler Optical system interface software. The mean Ct of target genes was calculated from triplicate measurements and normalized with the mean Ct of the control gene for GAPDH. The sequences for forward and reverse primers were as followed: HES1, ATG CTC TGA AGA AAG GCT CGC GG and TCC GGA GGT GCT TCA CTG TCA TTT; HEY L, TCA TCT GCA AGA CCT CGG CTT and AGG GCA CTG GCA GTT ATA GGT GTT; NOTCH3, TGT GAA GTG AAC GTG GAC GAC TGT and AGG GCA CTG GCA GTT ATA GGT GTT; MANIC FRINGE, ACG AGG CTG GTA CAG TTC TGG TTT and TCA GGG TGT GCA CTG TGA GAT CAA; and GAPDH, TCT CCT CTG ACT TCA ACA GCG ACA and CCC TGT TGC TGT AGC CAA ATT CGT.
Proliferation and soft agar colony formation assay
Cell proliferation was assessed using alamar blue cell viability reagent (Invitrogen) according to the manufacturer’s protocol. Briefly, HCC2429 cells or H460 cells, stably transfected with or without mFng-expressing plasmid, were plated at the density of 3000 cells/96-well plate and cultured in complete RPMI with 300 μg/ml of G418. On each day of analysis, alamar reagent was added directly to cells in the culture medium and incubated 4 hours at 37°C, after which the absorbance at 570 nm wave length was measured using a synergy HT instrument (BioTek). The experiments were performed in triplicate. For agar colony formation assays, cells expressing mFng were plated into 35 mm dishes at a density of 5000 cells per plate and suspended in 0.4% agar containing 10% FBS RPMI and 300 μg/ml of G418 over 0.8% base agar. The plates were incubated at 37°C and 5% CO2 in a humidified chamber for 14 days. The number and area of the colonies in each dish were determined with NIH ImageJ software.
In vivo tumorigenicity assay
Animal experiments were performed in accordance with the approval by the University of Virginia Institutional Animal Care and Use Committee (IACUC). Briefly, athymic 4-6-week-old female nude mice were used as tumor xenograft model. HCC2429-mFng cells (1 x 106 cells in the volume of 200 μl of PBS) were inoculated subcutaneously (s.c.) into the right posterior legs of the mice. The animals were followed daily and tumors were measured with a caliper after three weeks. Tumor volume (TV) was calculated using the formula: TV = (length) x (Width)/2.
Cycloheximide chase and protein accumulation assays
The HEK cells were transfected with Notch3 and Manic Fringe or vector control plasmids. Twelve hours after transfection, the cells were treated with 40 μg/mL cycloheximide or proteasome inhibitor MG132 at 20 μM for various time intervals and then lysed in 2 x SDS sample buffer. The samples were analyzed with immunoblotting using Notch3 antibodies.
Results
Expression of manic fringe is down-regulated in lung cancer
Fringe modulates Notch signaling via glycosylation of the O-fucose residues that are attached to the epidermal growth factor-like sequence repeats of the Notch receptor. This glycosylation potentiates Notch signaling through Delta-like ligands at the expense of the Jagged ligands [10]. We have previously shown that Jagged1 and Notch3 are over-expressed in lung cancers and that inhibiting Notch3 signaling results in the suppression of tumor phenotype [7,26]. Furthermore, Jagged1 formed a juxtacrine loop with Notch3 to promote cancer growth [27]. Based on these observations, we hypothesized that Fringe may function as a tumor suppressor by inhibiting Jagged1 signaling.
To test our hypothesis, we utilized available lung cancer databases through Oncomine (www.oncomine.org) to query for the expression of the three mammalian Fringes. Seven lung cancer microarray datasets with normal controls were utilized. We found that Manic Fringe expression is statistically decreased in lung tumors compared to controls (Figure 1A). To compare expression of Manic Fringe, Lunatic Fringe and Radical Fringe, we used gene ranking based on the P-value for each analysis (www.oncomine.org) and noted that of the six eligible data sets, Manic Fringe is more likely to be down-regulated compared to Lunatic Fringe or Radical Fringe (Figure 1B). This observation concurs with the finding that Manic Fringe is the only family member whose genetic locus is deleted in lung cancer. We then surveyed the expression of Manic Fringe in 21 lung cancer cell lines. We noted that, compared to the immortalized lung epithelial cell line BEAS2-B, nearly all cell lines exhibited reduced expression of Manic Fringe (Figure 1C). Loss of tumor suppressor expression is a hallmark of many cancers, and the reduced expression of Manic Fringe in lung cancer supports the hypothesis that Manic Fringe is a tumor suppressor.
Figure 1.
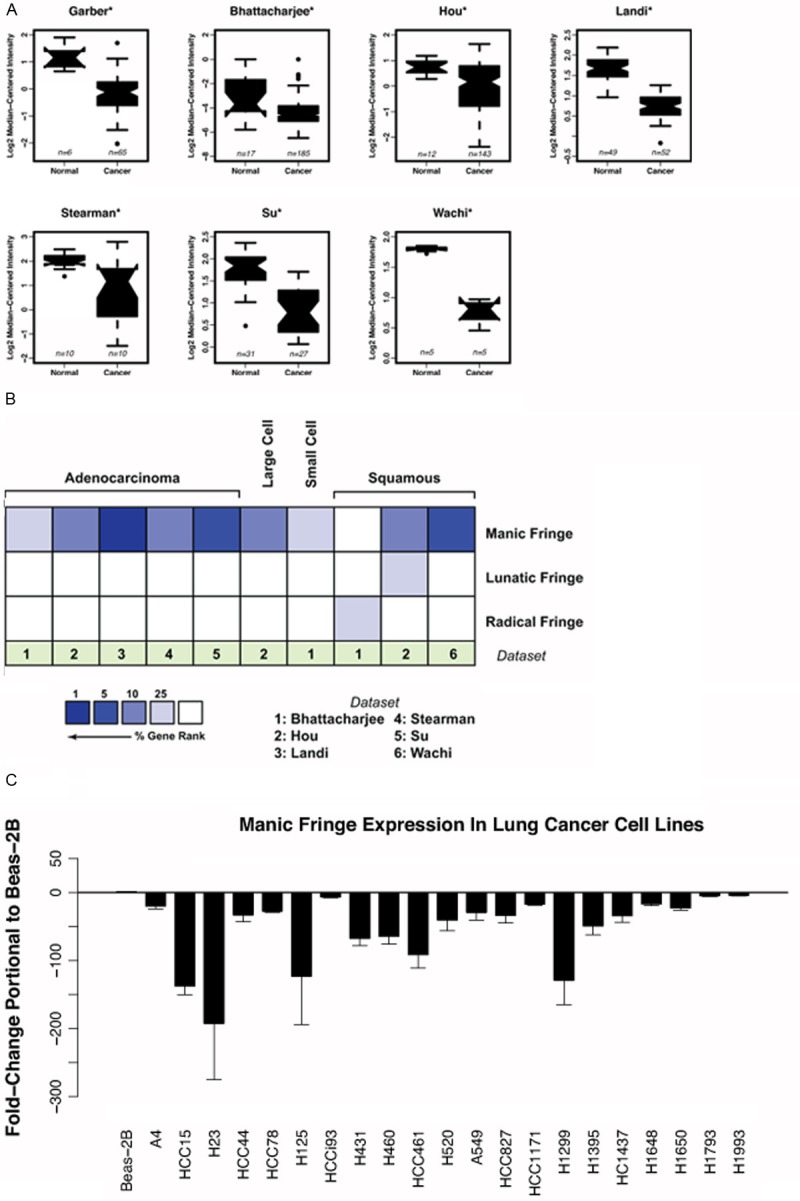
Manic Fringe expression is downregulated in cancer cells. A: Oncomine analysis of deposited expression microarray data shows down-regulation of Manic Fringe transcript in multiple tumor types compared with normal tissue. Reporters: 204152_s_at from Human Genome U133A Array (Hou, Landi, Su and Wachi datasets), cDNA IMAGE:51817 (Garber) and 41521_at from Human Genome U95A-Av2 Array (Bhattacharjee and Stearman). B: Six datasets containing all three Fringes were used for analysis. Using gene rank analysis by Oncomine, the highest reduction in expression was observed for Manic Fringe compared to Lunatic Fringe and Radical Fringe. C: Expression of Manic Fringe in 22 lung cancer cell lines is decreased compared to immortalized epithelial cell line BEAS-2B.
Forced expression of manic fringe reduces tumor growth in vitro and in vivo
To examine the functional role of Manic Fringe in lung cancer, we created stable clones expressing human Manic Fringe in lung cancer cell lines HCC2429 and H460. Using the alamar blue assay, we observed a decrease in the proliferation of H460 cells transfected with Manic Fringe compared with control vector in serum-free condition but not in serum-supplemented media (Figure 2A). We noted a similar effect of Notch3 inhibition on cancer cell proliferation in serum-deprived media in previous studies [26]. One hallmark of cancer cells is anchorage-dependent growth. Manic Fringe diminished either the number or the size of the colonies (Figure 2B). When the clones were injected into the flanks of nude mice, a significant reduction in tumor size was observed. Moreover, whereas all control animals formed tumors after 15 days, approximately half of the experimental animals transfected with clones expressing Manic Fringe formed no appreciable tumors (Figure 2C).
Figure 2.
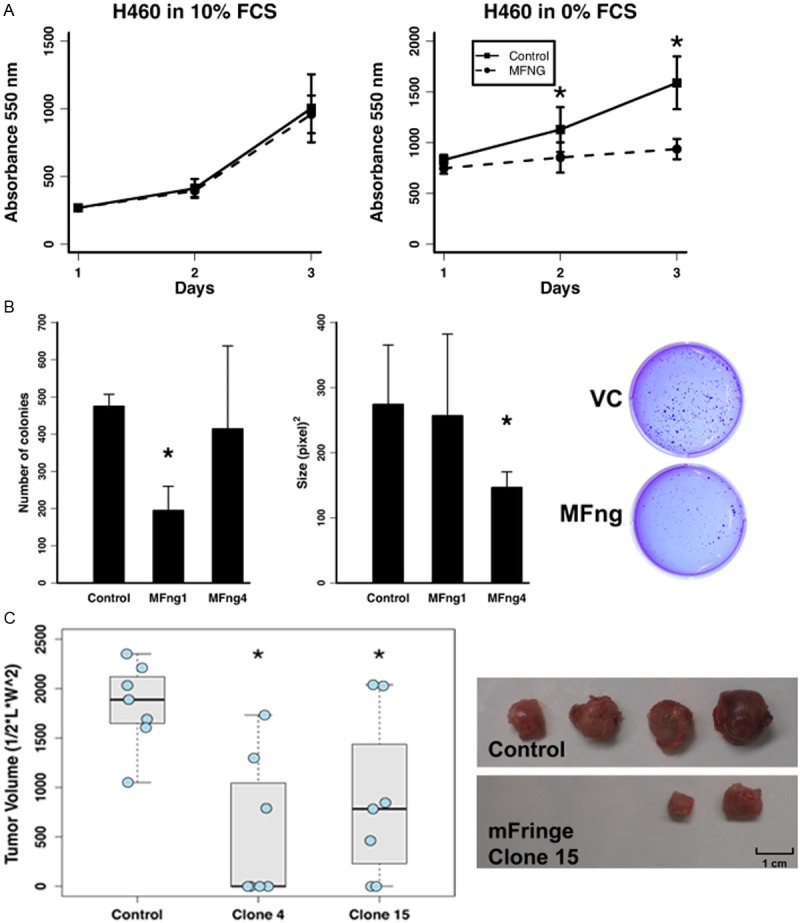
Manic Fringe suppresses the tumor phenotype in lung cancer cells. A: Manic Fringe increases the dependency of H460 cells on exogenous growth factors. Under serum-starved conditions, the proliferative growth of the cells expressing Manic Fringe is severely inhibited compared with that of control. However, with the addition of exogenous growth factors the growth rate is equal to that of VC. B: Manic Fringe (MFng1 and MFng4) markedly reduces the number (First panel) or size (Second panel) of the colonies formed in soft agar, compared with vector control (VC) in HCC2429 cells. A representative soft agar experiment demonstrating a significant decrease in the number of colonies of clones expressing Manic Fringe compared to VC (Third panel). The number and size of colonies formed were measured and calculated using the NIH ImageJ. C: Cells transfected with Manic Fringe formed less tumors in the flanks of nude mice compared to control clones. The sizes of the Manic Fringe tumors were generally smalle. (*) Denotes statistical significance p≤0.05.
Manic fringe suppresses Notch3 levels and expression of Notch-dependent genes in lung cancer cells and 293T cells
Fringe is a key modulator of Notch, by suppressing Jagged1-dependent signaling [27]. We expect that Manic Fringe reduced lung tumor proliferation and growth by inhibiting the interaction between Notch3 and its ligand Jagged1. However, as shown in Figure 3A, the expression levels of activated Notch3 (N3ICD) and full-length Notch3 (N3FL) were decreased with the overexpression of Manic Fringe in both lung cancer cell lines HCC2429 and H460. No change in the level of Jagged1 was noted (Figure 3A, first panel). Furthermore, when HEK 293T cells were transiently transfected with both full-length Notch3 and Manic Fringe, we observed reduced levels of both full-length (N3FL) and activated Notch3 (N3ICD) in the presence of Manic Fringe (Figure 3B). We noted that Manic Fringe also reduced the full-length (N1FL) and activated form (N1ICD) of Notch1, although to a lesser degree (Figure 3C). In addition, Manic Fringe reduced the expression of Notch-dependent genes HES1 and HeyL, suggesting repression of Notch-dependent activation (Figure 3D).
Figure 3.
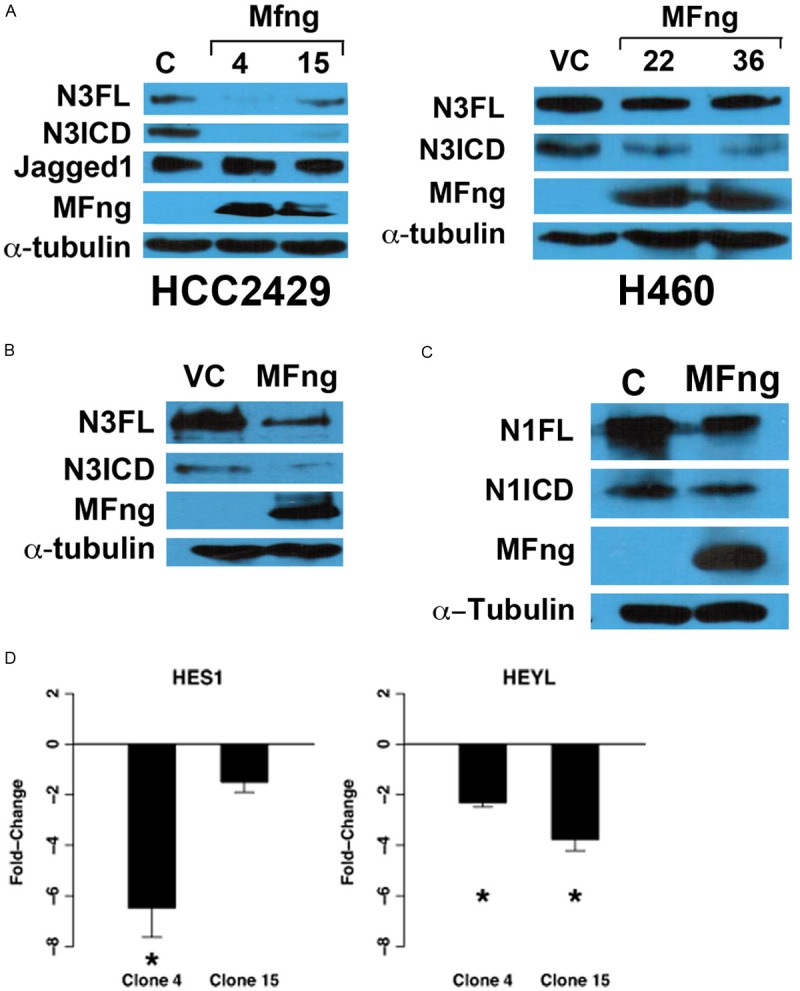
Manic Fringe reduces Notch3 protein levels and down-regulates Notch target genes. A: H460 and HCC2429 cells stably transfected with Manic Fringe showed a decreased in the levels of both full-length and activated forms of Notch3. No change in the level of the Notch ligand Jagged1 was detected. B: Similar results were observed when HEK293 cells were transiently transfected with Manic Fringe. C: Minimal reduction of Notch1 was observed in HEK 293 when Manic Fringe were over-expressed. D: Marked fold-change in transcriptions of Notch-dependent genes HES1 and HeyL was observed with realtime RT-PCR, compared to control clone.
Manic fringe enhances degradation of Notch3
Given the role of Fringe in modulating Notch activation, we also examined the effect of Manic Fringe on Notch3’s interaction with Jagged1. However, we did not observe any difference between the ability of Jagged1 to induce cleavage and activation of Notch3 (data not shown). Furthermore, Manic Fringe did not alter N3ICD’s binding with the Notch transcription factor RBP-Jκ (data not shown). These observations suggest that Manic Fringe does not interfere with Notch3’s activation by Jagged ligands, but rather that it modulates Notch3’s activity by regulating its protein turnover. While the protein level of Notch3 was suppressed by Manic Fringe, the transcriptional expression of Notch3 was not significantly reduced (Figure 4A). In fact, in transient transfections using HEK 293, the transcription of Notch3 in Manic Fringe transfected cells was increased (Figure 4A, second panel). Since we did not detect a reduction in Notch3 transcriptional expression in Manic Fringe transfected cells, we examined the turnover rate of Notch3 protein, using cycloheximide blocking. Immunoblot analysis of Notch3 protein showed that in the presence of Manic Fringe Notch3 protein was cleared more quickly than under control conditions (Figure 4B). The increased turnover rate in cells transfected with Manic Fringe suggests that Manic Fringe enhances the degradation of Notch3.
Figure 4.
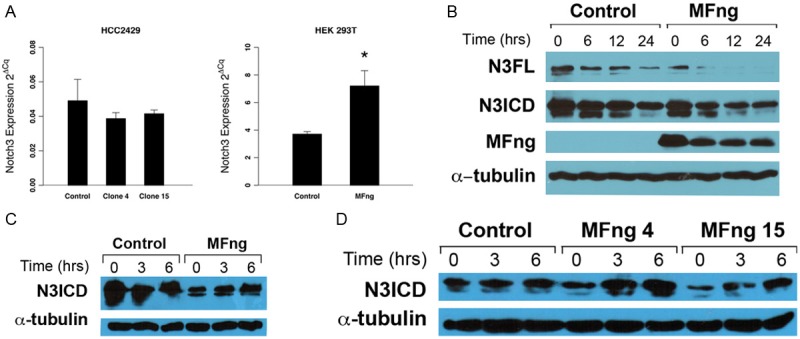
Manic Fringe enhances Notch3 degradation. A: Manic Fringe has no effect on mRNA expression of Notch3 in stably transfected HCC2429 clones. After transient transfection of Manic Fringe in HEK 293 cells, the mRNA of Notch3 was increased compared to control, suggesting that the inhibitory effect on Notch3 is not through transcription. B: Cycloheximide blocking assays showed a noticeable increase in the clearance of both full-length (N3FL) and activated form (N3ICD) of Notch3 in Manic Fringe clones compared to controls, particularly after 12 and 24 hours. C: Extracts from HEK 293 cells were transfected with Notch3 and Manic Fringe or vector control and treated with 20 μM MG132 at different intervals. The activated level of Notch3 increased with treatment in both transiently transfected HEK 293. D: Similar observation was made in stable clones expressing Manic Fringe.
Notch proteins contain a PEST domain at the C-terminus. This domain is a target for ubiquitylation and proteasome-mediated degradation, a common mechanism of inactivation for proteins with regulatory functions. In a time-course experiment, HEK 293 cells transiently transfected with Manic Fringe or vector control were treated with the proteasome inhibitor MG132. Compared with controls, activated Notch3 protein started to accumulate within 3 hours of incubation with MG132 (Figure 4B). Similar observations were made in stable clones expressing Manic Fringe (Figure 4D). The data supports a role for Manic Fringe as a tumor suppressor that diminishes the level of Notch3 by promoting degradation through a proteasome mediated pathway.
Discussion
Three mammalian Fringe paralogs have been described: Manic Fringe, Lunatic Fringe and Radical Fringe. Their collective roles in development are still being explored. Like Notch receptors, the differential expression of the three Fringe proteins in development suggests non-redundancy and context-dependent signaling [28,29]. A recent paper showed that deletion of Lunatic Fringe contributed to basal-like breast cancer through the accumulation of Notch’s intracellular domain [14]. The results of the present study indicate that Manic Fringe, but not Lunatic or Radical Fringe, is a tumor suppressor in lung cancer. While more studies are needed to reach a definitive conclusion, it is likely that, analogous to Notch receptors, the phenotypic outcome of Fringe modulation is context-dependent.
Since Notch’s activated intracellular domain can no longer be modified by extracellular stimuli after receptor signaling, it is not surprising that a major mode of controlling Notch signaling is through the control of protein turnover rates. Indeed, known negative regulators of Notch signaling such as Deltex and Numb promote Notch degradation [30,31]. Since no Notch-independent activity has been described for Fringe, it is likely that the mechanism of Manic Fringe’s anti-tumor effect is modulation of Notch’s activation. Previous studies have shown that Fringe-mediated glycosylation modulates Notch-ligand binding through glycosylation. Our data suggests a novel mechanism for Manic Fringe-mediated Notch3 downregulation, through accelerated proteasome dependent-degradation.
It is not known whether different Fringe enzymes glycosylate different Fucose residues and/or a different number of Fucose residues on Notch receptors. The pattern of glycosylation catalyzed by Manic Fringe may affect the cellular distribution of Notch3, for example by inhibiting its export to the plasma membrane and promoting its recycling into endosomes. Since the turnover of full-length Notch3 was also affected by Manic Fringe, it is possible that Manic Fringe-glycosylated Notch3 is less efficiently exported to the trans-Golgi and/or cleaved by Furin-like convertase. Uncleaved Notch3 would be shunted to an intracellular degradation pathway. Alternatively, glycosylation by Manic Fringe may prevent Jagged1-dependent Notch activation. In the absence of Delta ligands, this may indirectly enhance recycling and turnover of inactive Notch3.
In summary, we have shown that the expression of Manic Fringe is frequently down-regulated in lung cancer and that Manic Fringe inhibits lung cancer cell proliferation and tumorigenesis. These observations support a novel role of Manic Fringe as tumor suppressor, confirm the role for Notch-dependent oncogenesis in lung cancer and reveal a new function of Manic Fringe as a regulator of Notch3 protein turnover.
Acknowledgements
This study was supported by NCI 1R01 CA115707.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi K, Ahn YH, Gibbons DL, Tran HT, Creighton CJ, Girard L, Minna JD, Qin FXF, Kurie JM. Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. J Biol Chem. 2009;284:17766–17774. doi: 10.1074/jbc.M109.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 5.Reedijk M, Odorcic S, Zhang H, Chetty R, Tennert C, Dickson BC, Lockwood G, Gallinger S, Egan SE. Activation of Notch signaling in human colon adenocarcinoma. Int J Oncol. 2008;33:1223–1229. doi: 10.3892/ijo_00000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 7.Konishi J, Kawaguchi KS, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007;67:8051–8057. doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Thiaville MM, Chen L, Stoeck A, Xuan J, Gao M, Shih IM, Wang TL. Defining NOTCH3 target genes in ovarian cancer. Cancer Res. 2012;72:2294–2303. doi: 10.1158/0008-5472.CAN-11-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai JI, Ohwada S, Tatsuta K, Inoue JI, Semba K, Watanabe S. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- 10.Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 11.Kato TM, Kawaguchi A, Kosodo Y, Niwa H, Matsuzaki F. Lunatic fringe potentiates Notch signaling in the developing brain. Mol Cell Neurosci. 2010;45:12–25. doi: 10.1016/j.mcn.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Yuan JS, Tan JB, Visan I, Matei IR, Urbanellis P, Xu K, Danska JS, Egan SE, Guidos CJ. Lunatic Fringe prolongs Delta/Notch-induced self-renewal of committed αβ T-cell progenitors. Blood. 2011;117:1184–1195. doi: 10.1182/blood-2010-07-296616. [DOI] [PubMed] [Google Scholar]
- 13.Veeraraghavalu K, Pett M, Kumar RV, Nair P, Rangarajan A, Stanley MA, Krishna S. Papillomavirus- mediated neoplastic progression is associated with reciprocal changes in JAGGED1 and manic fringe expression linked to notch activation. J Virol. 2004;78:8687–8700. doi: 10.1128/JVI.78.16.8687-8700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu K, Usary J, Kousis PC, Prat A, Wang DY, Adams JR, Wang W, Loch AJ, Deng T, Zhao W, Cardiff RD, Yoon K, Gaiano N, Ling V, Beyene J, Zacksenhaus E, Gridley T, Leong WL, Guidos CJ, Perou CM, Egan SE. Lunatic fringe deficiency cooperates with the Met/Caveolin gene amplicon to induce basal-like breast cancer. Cancer Cell. 2012;21:626–641. doi: 10.1016/j.ccr.2012.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berrieman HK, Ashman JNE, Cowen ME, Greenman J, Lind MJ, Cawkwell L. Chromosomal analysis of non-small-cell lung cancer by multicolour fluorescent in situ hybridisation. Br J Cancer. 2004;90:900–905. doi: 10.1038/sj.bjc.6601569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran JL, Johnston SH, Rauskolb C, Bhalerao J, Bowcock AM, Vogt TF. Genomic structure, mapping, and expression analysis of the mammalian Lunatic, Manic, and Radical fringe genes. Mamm Genome. 1999;10:535–541. doi: 10.1007/s003359901039. [DOI] [PubMed] [Google Scholar]
- 17.Testa JR, Siegfried JM, Liu Z, Hunt JD, Feder MM, Litwin S, Zhou JY, Taguchi T, Keller SM. Cytogenetic analysis of 63 non-small cell lung carcinomas: recurrent chromosome alterations amid frequent and widespread genomic upheaval. Genes Chromosomes Cancer. 1994;11:178–194. doi: 10.1002/gcc.2870110307. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, Murphy SE, Yang P, Pesatori AC, Consonni D, Bertazzi PA, Wacholder S, Shih JH, Caporaso NE, Jen J. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, Baron AE, Keith RL, Nemenoff RA, Malkinson AM, Geraci MW. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CYF. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, Carbone DP, Dang TP. Dominant-negative Notch3 receptor inhibits mitogen-activated protein kinase pathway and the growth of human lung cancers. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 27.Choi JH, Park JT, Davidson B, Morin PJ, Shih IM, Wang TL. Jagged-1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res. 2008;68:5716–5723. doi: 10.1158/0008-5472.CAN-08-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu W, Xu W, Ding T, Guo X. Fringe controls na{ï}ve CD4(+)T cells differentiation through modulating notch signaling in asthmatic rat models. PLoS One. 2012;7:e47288. doi: 10.1371/journal.pone.0047288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thélu J, Viallet JP, Dhouailly D. Differential expression pattern of the three Fringe genes is associated with epidermal differentiation. J Invest Dermatol. 1998;111:903–906. doi: 10.1046/j.1523-1747.1998.00372.x. [DOI] [PubMed] [Google Scholar]
- 30.McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P, Yang Y, Nolo R, Zweidler-McKay PA, Hughes DPM. Regulation of NOTCH signaling by reciprocal inhibition of HES1 and Deltex 1 and its role in osteosarcoma invasiveness. Oncogene. 2010;29:2916–2926. doi: 10.1038/onc.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]


