Abstract
CD8+ T cell responses focus on a small fraction of pathogen- or vaccine-encoded peptides, and for some pathogens, these restricted recognition hierarchies limit the effectiveness of anti-pathogen immunity. We found that simian immunodeficiency virus (SIV) protein-expressing Rhesus Cytomegalovirus (RhCMV) vectors elicit SIV-specific CD8+ T cells that recognize unusual, diverse and highly promiscuous epitopes, including dominant responses to epitopes restricted by class II major histocompatibility complex (MHC) molecules. Induction of canonical SIV epitope-specific CD8+ T cell responses is suppressed by the RhCMV-encoded Rh189 (US11) gene, and the promiscuous MHC class I- and class II-restricted CD8+ T cell responses only occur in the absence of the Rh157.4-.6 (UL128-131) genes. Thus, CMV vectors can be genetically programmed to achieve distinct patterns of CD8+ T cell epitope recognition.
INTRODUCTION
CD8+ T cells detect intracellular pathogens by T cell receptor (TCR)-mediated recognition of short pathogen-derived peptides selected and transported to the cell surface by MHC class I proteins (MHC-I) and an intricate system of intracellular peptide sampling and transport (1). Although pathogens can potentially generate many thousands of different peptides of the appropriate length for CD8+ T cell recognition, requirements for proteolytic processing, peptide transport, binding to available MHC-I allomorphs and TCR repertoire matching, as well as poorly understood immunoregulatory mechanisms, winnow down these candidates to a relative handful of peptide epitopes that actually serve as targets for the CD8+ T cells that comprise anti-pathogen effector and memory responses (2–4). Remarkably, despite the complexity of the process, pathogen-specific CD8+ T cell responses mounted by individuals with shared MHC-I alleles tend to recognize an overlapping set of immunoprevalent epitopes (2, 3, 5). For the vast majority of pathogens, CD8+ T cell responses targeting such immunoprevalent epitopes are able to both recognize pathogen-infected cells and mount effective anti-pathogen effector and memory responses.
This is not the case, however, for agents with efficient immune evasion capabilities such as HIV and its simian counterpart SIV. The massive replication of these viruses, combined with their high rate of mutation and functional plasticity, allows escape from most CD8+ T cell responses (5, 6). Indeed, CD8+ T cell responses in the majority of subjects infected with these viruses fail to target epitopes containing conserved, functionally critical viral sequences, and do not effectively control viral replication (7). Although vaccination against these viruses can greatly augment the magnitude of CD8+ T cell responses after infection, these larger CD8+ T cell responses target many of the same immunoprevalent epitopes as infection of unvaccinated individuals, and therefore are still subject to immune escape (6, 8, 9). Although the AIDS vaccine field has endeavored to develop strategies capable of eliciting HIV/SIV-specific CD8+ T cell responses targeting “vulnerable” epitopes across diverse MHC-I haplotypes (by either increasing recognition breadth or the focusing of responses to conserved sequences), this effort has not, to date, yielded strategies capable of substantially modifying CD8+ T cell immunodominance hierarchies, nor achieved the goal of establishing protective CD8+ T cell responses in the majority of individuals.
We recently reported an HIV/AIDS vaccine strategy that uses SIV protein-encoding RhCMV as a persistent vector to generate and maintain SIV-specific effector memory T cell responses intended to intercept SIV infection prior to the viral amplification needed for efficient immune evasion (6). Although this approach was not designed to prevent acquisition of infection, it proved to be highly successful with about 50% of RhCMV/SIV vector-vaccinated rhesus macaques (RM) challenged with highly pathogenic SIV manifesting immediate, stringent and durable virologic control (10). During the course of these studies, we noticed that RhCMV/SIV vectors did not elicit the canonical CD8+ T cell responses restricted by the well characterized Mamu-A1*001:01 (A*01) MHC-I allele, raising the questions of what CD8+ T cell epitopes were targeted by these effective responses and whether differential targeting might have contributed to efficacy. Here, we show that delivery of SIV antigens to the immune system via strain 68-1-based RhCMV/SIV vectors fundamentally changes CD8+ T cell recognition. The SIVgag-specific CD8+ responses elicited by the RhCMV/gag vector are 3-fold as broad as conventional SIVgag-specific CD8+ T cell responses, and target entirely different epitopes, including an abundance of highly promiscuous epitopes (“supertopes”) and dominant class II MHC (MHC-II)-restricted CD8+ T cell responses that are rarely, if ever, observed in CD8+ T cell responses to any other infectious agent. Moreover, we demonstrate that this atypical CD8+ T cell targeting is under the genetic control of CMV, allowing, for the first time, the ability to genetically manipulate a vaccine vector to achieve distinct patterns of CD8+ T cell epitope recognition.
RESULTS
Distinct CD8+ T cell epitope targeting with RhCMV/SIV vectors
We have previously demonstrated that in contrast to other CD8+ T cell response-inducing SIV vaccines (6), the protection associated with RhCMV/SIV vector vaccination does not correlate with expression of protective MHC-I alleles (10, 11). Moreover, among a group of 8 Mamu-A*01+ RM given RhCMV/gag and rev/tat/nef (rtn) vectors in these efficacy studies, none developed measureable frequencies of CD8+ T cells recognizing the normally immunodominant Gag181-189 (CM9) and Tat28-35 (SL8) epitopes, as measured by MHC-peptide tetramer analysis, although 4/4 of these RM that were subsequently administered Adenovirus5 (Ad5) vectors expressing SIVgag and SIVtat inserts did manifest robust responses to these canonical epitopes (10). To confirm this effect in Mamu-A*01+ RM and to determine whether a lack of response to Mamu-A*01-restricted canonical epitopes in RhCMV/SIV vector-vaccinated RM extends to canonical epitopes restricted by other MHC-I alleles (table S1), we used a sensitive, flow cytometric intracellular cytokine (ICS) assay (10, 12) to analyze such canonical responses in RM expressing one or more of the Mamu-A*01 (n =8), -A1*002:01 (A*02; n = 4), -B*008:01 (B*08; n = 2), and -B*017:01 (B*17; n = 4) alleles and vaccinated with our standard strain 68-1 RhCMV/gag,/rtn,/env and/pol vectors (Fig. 1A; table S2). In all cases, RhCMV/SIV vector-vaccinated RM manifested robust CD8+ T cell responses to the SIV protein containing the canonical epitopes, but these CD8+ T cell responses did not include a detectable response to any of the usually dominant or sub-dominant canonical epitopes restricted by any of these alleles.
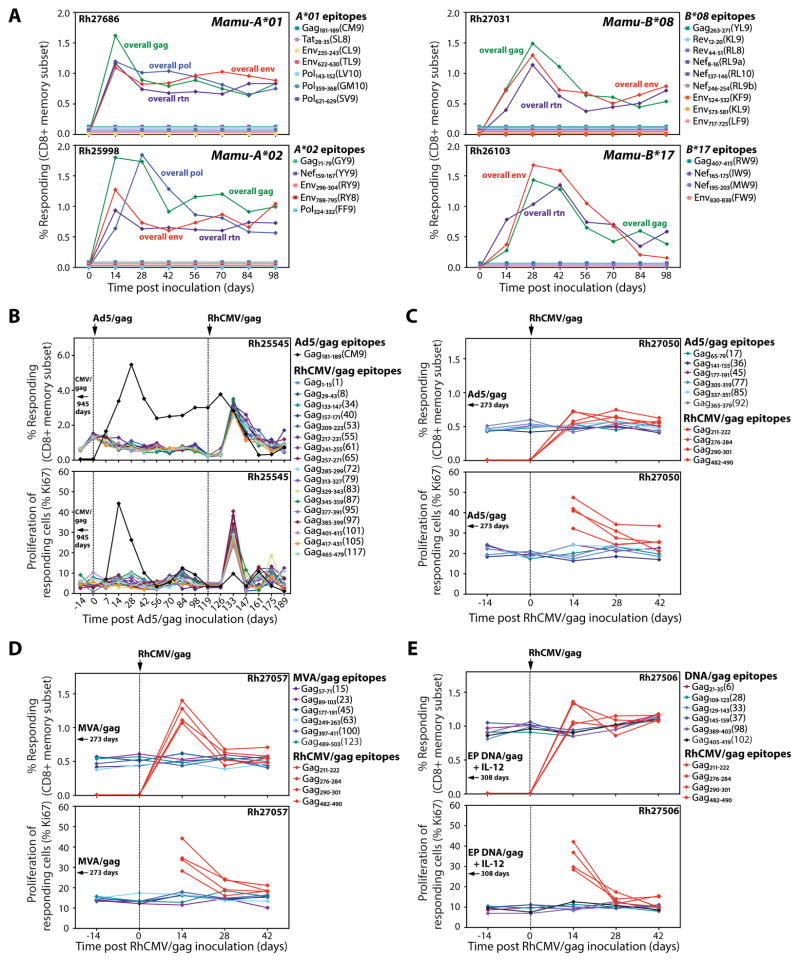
Fig. 1.
Lack of epitope overlap between RhCMV vector-elicited and conventional SIV-specific CD8+ T cell responses. (A) Flow cytometric ICS was used to follow the development of overall SIV insert- and canonical epitope-specific CD8+ T cell responses (responder = TNF-α + and/or IFN-γ +) in the blood of 4 RM, each with a different well characterized MHC-I allele (Mamu-A*01, -A*02, -B*08 and -B*17), after vaccination with strain 68-1 RhCMV/SIVgag, rev/tat/nef (rtn), env, and pol vectors. No response above background was observed for any canonical epitope. (B) The CD8+ T cell response to individual SIVgag 15mer peptides and the canonical Gag181-189 (CM9) epitope was determined in a long-term strain 68-1 RhCMV/gag-vaccinated, Mamu-A*01+ RM (Rh25545) using flow cytometric ICS. The frequency of these responses in blood and the proliferative status of the responding cells (as measured by Ki-67 expression on TNF-α + and/or IFN-γ + events) were followed after boosting with Ad5/gag, and then 119 days later, after re-vaccination with strain 68-1 RhCMV/gag. Gag181-189 (CM9)-specific responses were not detected prior to administration of Ad5/gag. (C–E) The CD8+ T cell response to individual SIVgag 15mer peptides was determined in RM vaccinated with Ad5/gag (C, Rh27050), MVA/gag (D, Rh27057), and electroporated DNA/gag + IL-12 (E, Rh27506), and these responses were followed after vaccination with strain 68-1 RhCMV/gag as described in B. These figures also show induction of 4 SIVgag 15mer-specific CD8+ T cell responses after RhCMV/gag vaccination that were not detectable in the pre-existing SIVgag-specific responses elicited by the conventional vaccines.
These data indicate that the epitope hierarchies of CD8+ T cell responses elicited by RhCMV/SIV vectors are quite different from those elicited by SIV itself or by conventional vaccine vectors, but do not rule out low level sensitization (below detection threshold) to canonical epitopes by RhCMV/SIV vectors or partial overlap between the epitopes targeted by RhCMV/SIV vector- vs. conventional vector-elicited responses. To explore these possibilities in outbred RM, we used a heterologous prime/boost approach designed to determine the extent to which individual epitope-specific CD8+ T cell responses initially elicited by priming with RhCMV/SIV or conventional vectors can be subsequently boosted by the other vector type. We first demonstrated the feasibility of this approach for RhCMV/gag by showing that SIVgag epitope-specific CD8+ T cell responses elicited by this vector are consistently boosted (as measured by a post-boost increase in the response frequency and proliferation of the epitope-specific CD8+ T cells) by a second administration of the same vector (fig. S1). We then determined the SIVgag epitope recognition profile of a Mamu-A*01+ RM (Rh25545) primed with RhCMV/gag and the effect of a subsequent Ad5/gag boost on these individual epitope CD8+ T cell responses (Fig. 1B). We identified 17 distinct CD8+ T cell responses to individual SIVgag 15mer peptides after RhCMV/gag vaccination, but no response to the canonical Mamu-A*01-restricted Gag181-189 (CM9) epitope. None of these 17 RhCMV/gag-elicited SIVgag epitope-specific CD8+ T cell responses were boosted by Ad5/gag, although all were boosted by subsequent administration of the RhCMV/gag vector. CD8+ T cells targeting the canonical Mamu-A*01-restricted Gag181-189 (CM9) epitope were generated de novo by the Ad5/gag vaccine, but were not boosted following the second administration of RhCMV/gag (indeed, these responses fell in frequency). Similar results were observed with additional RM primed with Ad5/gag (Rh27050), Modified Vaccinia Ankara (MVA)/gag (Rh27057) and DNA/gag + IL-12 (the latter administered by electroporation; Rh27506 and Rh27520) and then boosted with RhCMV/gag (Fig. 1C–E; fig. S2), as well as in 2 SIV+ RM (Rh25429 and Rh26557) on fully suppressive anti-retroviral therapy that were administered RhCMV/SIV vectors (fig. S3). None of the SIVgag epitope-specific CD8+ T cell responses elicited by the priming with conventional vectors or by infection with SIV itself were boosted by RhCMV/gag, but administration of the latter vector did, in all cases, generate new CD8+ T cell responses to different (not previously detected) SIVgag epitopes. Taken together, these data indicate that from the perspective of CD8+ T cell responses, the strain 68-1 RhCMV/SIV vectors and conventional vectors expressing the same SIV protein (as well as SIV itself) behave as if they present entirely different peptides to the immune system, manifesting non-overlapping epitope recognition profiles.
Characteristics of unconventional CD8+ T cell epitope targeting
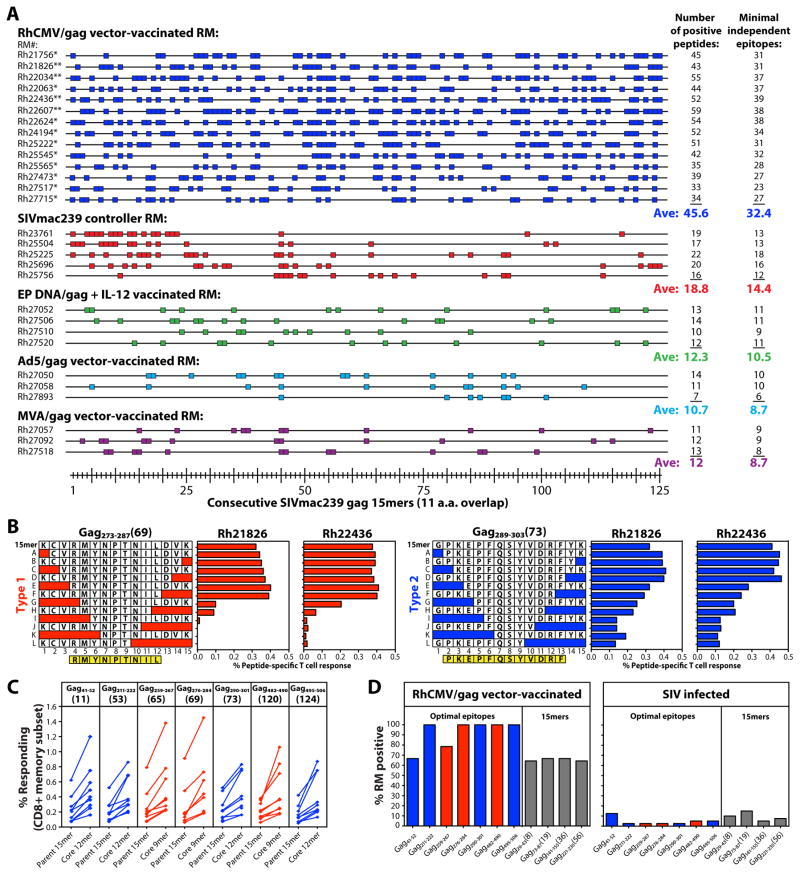
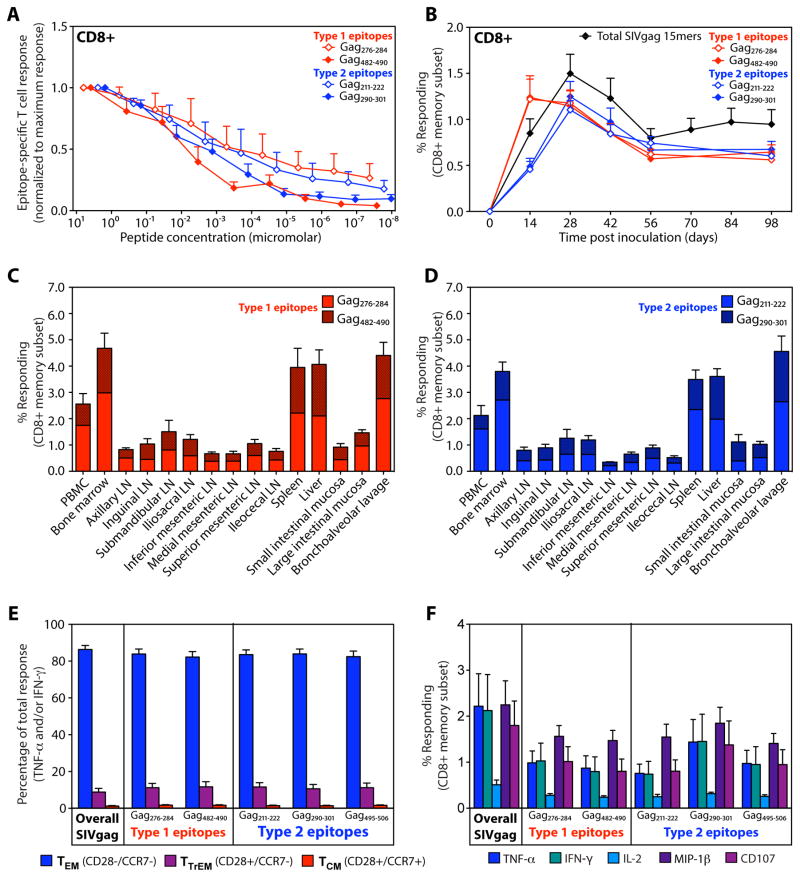
To comprehensively compare the epitope targeting profiles of SIVgag-specific CD8+ T cell responses elicited by strain 68-1 RhCMV/gag vectors vs. conventional vectors and SIV itself, we used flow cytometric ICS to individually quantify CD8+ T cell responses to each of 125 consecutive 15mer peptides (with 11 amino acid overlap) comprising the entire SIVgag protein in 29 RM: 14 vaccinated with RhCMV/gag vectors, 4 with electroporated DNA/gag + IL-12, 3 each with Ad5/gag and MVA/gag and 5 RM with SIV infections controlled by conventional immune responses. Strikingly, peripheral blood CD8+ T cells from RhCMV/gag vector-vaccinated RM responded to an average of 46 of these 125 SIVgag peptides, corresponding to a minimum of ~32 distinct epitopes (Fig. 2A). In contrast, SIV-infected controllers and RM vaccinated with electroporated DNA/gag + IL-12, Ad5/gag and MVA/gag responded to an average of 11-19 peptides, corresponding to an average of ~9–14 distinct epitopes. The breadth of the CD8+ T cell responses in the RhCMV/gag vector-vaccinated RM was so great that many of SIVgag 15mer peptides were targeted by CD8+ T cells in most or even all of the 14 outbred RM studied (Fig. 2A). To determine whether this finding reflects promiscuous recognition of a single common epitope (“supertope”) or simply are the result of CD8+ T cell recognition “hot spots” including multiple overlapping, but different, epitopes, we analyzed the CD8+ T cell response to a series of truncated peptides corresponding to 7 of these commonly recognized 15mers in 3 RM per response, so as to identify core epitopes in each RM (Fig. 2B; fig. S4). Intriguingly, we identified two distinct response patterns with truncated peptides: Type 1, in which response frequencies dropped abruptly with loss of a critical amino acid residue, typically yielding a 9mer core epitope (Gag259-267, Gag276-284, Gag482-490), and Type 2, in which response frequencies only gradually declined as the optimal sequence was truncated, typically leaving a 12mer core epitope (Gag41-52, Gag211-222, Gag290-301, Gag495-506). These truncation response patterns and core peptides were the same in all RM studied for each response, and in all cases, the core peptides manifested superior stimulation (higher response frequencies) than the parent 15mer (Fig. 2C). Taken together, these data strongly suggest that many of the SIVgag epitopes targeted by CD8+ T cells in RhCMV/gag vector-vaccinated RM are specific determinants that are commonly or even universally recognized across disparate MHC haplotypes. Indeed, a detectable CD8+ T cell response to the core (optimal) peptide for 5 of these truncated 15mers (including both Type 1 and 2 truncation patterns) was found in 100% of 42 RhCMV/gag-vaccinated, outbred RM and responses to 6 other peptides (2 optimal peptides and 4 15mers) were found in >60% of RM (Fig. 2D, left panel). Notably, CD8+ T cells in 40 SIV-infected RM rarely recognized these epitopes (Fig. 2D, right panel). Thus, strain 68-1 RhCMV vector-elicited CD8+ T cell responses to SIVgag are ~3-fold as broad as conventional SIVgag-specific CD8+ T cell responses, and are uniquely characterized by frequent targeting of broadly recognized “supertopes”.
Fig. 2.
RhCMV vector-elicited and conventional SIVgag-specific CD8+ T cell responses differ in epitope breadth and promiscuity. (A) CD8+ T cell responses to SIVgag were epitope-mapped using flow cytometric ICS to detect recognition of 125 consecutive 15mer gag peptides (with an 11 amino acid overlap) in RM vaccinated with strain 68-1 RhCMV/gag vectors [*BAC-derived RhCMV/gag; **non-BAC-derived RhCMV/gag(L); n = 14], electroporated DNA/gag + IL-12 vectors (n = 4), Ad5/gag vectors (n = 3), and MVA/gag vectors (n = 3) and in SIV+ RM with controlled infection (n = 5). Peptides resulting in above background CD8+ T cell responses are indicated by a colored box, with the total number of these positive responses and the minimal number of independent epitopes potentially contained within these reactive peptides in each RM designated at right. p < .0001, epitope breadth of RhCMV/gag-vaccinated RM compared to RM pooled over the other groups, using two-tailed Wilcoxon rank sum tests. (B) The core epitopes of selected 15mer peptides targeted by CD8+ T cells from strain 68-1 RhCMV/gag-vaccinated RM were determined by flow cytometric ICS analysis of CD8+ T cell responses to the indicated truncated peptides. The figure shows representative examples of the 2 response patterns observed with truncated peptide sets: Type 1 (red), with abrupt loss of peak responsiveness and a 9mer core epitope, and Type 2 (blue), with gradual loss of peak responsiveness and a 12mer core epitope (see also fig. S4). (C) CD8+ T cell response frequencies to the parent 15mer peptides and the core peptides derived from these 15mers (Type 1, red; Type 2, blue) were compared by flow cytometric ICS in 9 RM for each response. (D) CD8+ T cell responses to selected SIVgag core epitopes (Type 1, red; Type 2, blue), as well as selected additional SIVgag 15mers (gray), were evaluated by flow cytometric ICS in 42 strain 68-1 RhCMV/gag vector-vaccinated RM (left panel) and 40 SIV-infected RM (right panel) with the % of RM in each category with detectable responses to these peptides shown in the figure.
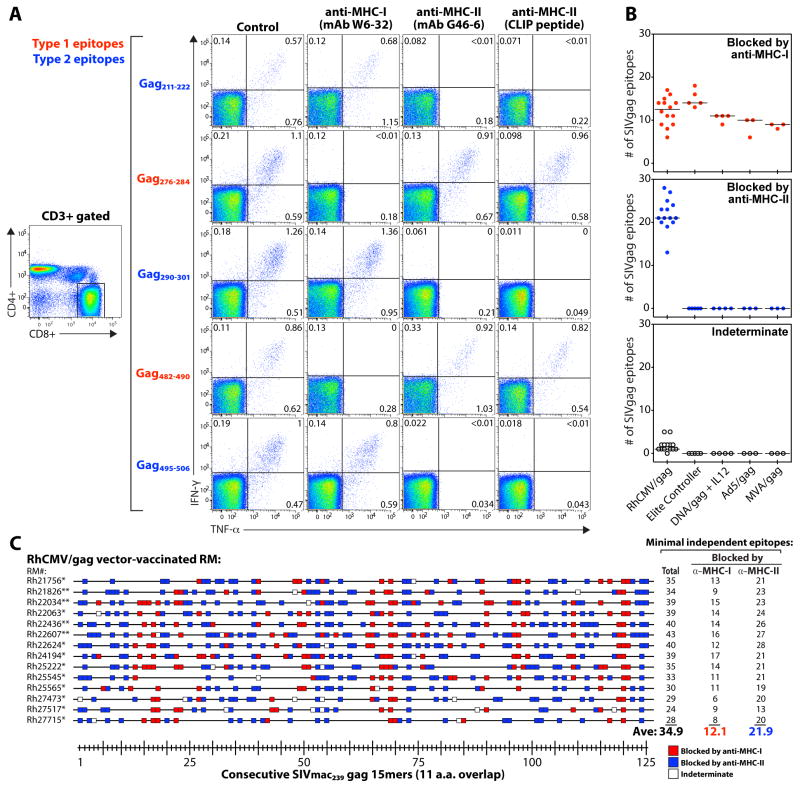
MHC-I-restricted epitopes are typically 8-10 amino acids in length and have position-specific amino acids that engage binding pockets (anchor residues) so as to fit in a “closed end” MHC-I binding groove (13), characteristics consistent with the Type 1 truncation pattern described above. In contrast, the Type 2 truncation pattern is more typical of MHC-II-restricted epitopes [which are typically longer, usually a ≥12mer core, lack specific anchor residues, and are more tolerant of length heterogeneity (14, 15)]. This suggested that the CD8+ T cells recognizing Type 2 SIVgag epitopes in the RhCMV/gag vector-vaccinated RM might be MHC-II-restricted. In this regard, although MHC-II-restricted CD8+ T cell responses are clearly unusual, such responses have been previously reported in both mice (16–21) and humans (22–24), and it has been established that productive TCR signaling does not require specific CD4 or CD8 co-receptor engagement with MHC-II or MHC-I, respectively (25, 26). To investigate the possibility of MHC-II-restricted CD8+ T cell responses in our system, we assessed the ability of “blocking” monoclonal antibodies (mAbs) specific for MHC-I and MHC-II, and the invariant chain-derived, MHC-II-specific binding peptide CLIP (27) to inhibit the Type 1 and Type 2 epitope-specific CD8+ T cell responses in the strain 68-1 RhCMV/gag-vaccinated RM (Fig. 3A; fig. S5). Strikingly, inhibition of the 7 supertope-specific CD8+ T cell responses by these reagents corresponded precisely to the Type 1 vs. Type 2 truncation pattern, with CD8+ T cell recognition of the 4 Type 2 epitopes (Gag41-52, Gag211-222, Gag290-301, Gag495-506) blocked by anti-MHC-II and CLIP, but not anti-MHC-I, and the reverse for CD8+ T cell recognition of the 3 Type 1 epitopes (Gag259-267, Gag276-284, Gag482-490). We then comprehensively characterized the epitope-specific responses mapped in Fig. 2A with respect to MHC-I vs. MHC-II blockade (Fig. 3B,C). All CD8+ T cell responses in the SIV-infected RM and the RM vaccinated with the conventional vaccines were only blocked by anti-MHC-I, whereas in the RhCMV/SIV-vaccinated RM, the CD8+ T cell responses to the majority of the targeted 15mers (63%) were specifically blocked by the MHC-II inhibitors, leaving a minority (35%) blocked only by MHC-I mAbs (with ~2% of responses indeterminate).
Fig. 3.
Most RhCMV vector-elicited CD8+ T cell responses are inhibited by MHC-II blockade. (A) PBMC from a representative strain 68-1 RhCMV/gag-vaccinated RM (of 8 RM similarly analyzed RM, see fig. S5) were stimulated with the designated SIVgag core epitopes (Type 1 vs. Type 2) in the presence of irrelevant isotype control mAbs (IgG1 − clone X40 + IgG2a − clone X39; 10μg each), an anti-MHC-I mAb (W6-32; 10μg), an anti-MHC-II mAb (G46-6; 10μg), or the CLIP peptide (MHC-II-associated invariant chain, amino acids 89-100; 2μg). A typical CD4 vs. CD8 expression profile, showing events gated on CD3+ small lymphocytes, is illustrated on the left, with the CD8 gate used to analyze the CD8+ T cell responses indicated. The IFN-γ vs. TNF-α profiles (right panels) include only events collected through these gates, and thus reflect CD8hi/CD4negative T cells. (B,C) All the SIVgag 15mer peptide responses shown in Fig. 2A were subjected to MHC-I (mAb W6-32) vs. MHC-II (mAb G46-6) blockade and classified as being specifically inhibited (blocked) by anti-MHC-I vs. anti-MHC-II mAbs, or indeterminate. (B) For each RM, the average number of peptide-specific responses in each category are shown, classified by vaccine type. (C) The sensitivity of each SIVgag peptide response in 14 strain 68-1 RhCMV/gag-vaccinated RM [*BAC-derived RhCMV/gag; **non-BAC-derived RhCMV/gag(L)] to blockade by anti-MHC-I (red boxes) vs. anti-MHC-II (blue boxes) mAbs is shown (open boxes indicate indeterminate), with the minimal number of independent epitopes in the MHC-I- and MHC-II-associated categories designated at right (note: the total number of independent epitopes increased in some RM from that listed in Fig. 2A due to increased resolution afforded by the MHC blocking data).
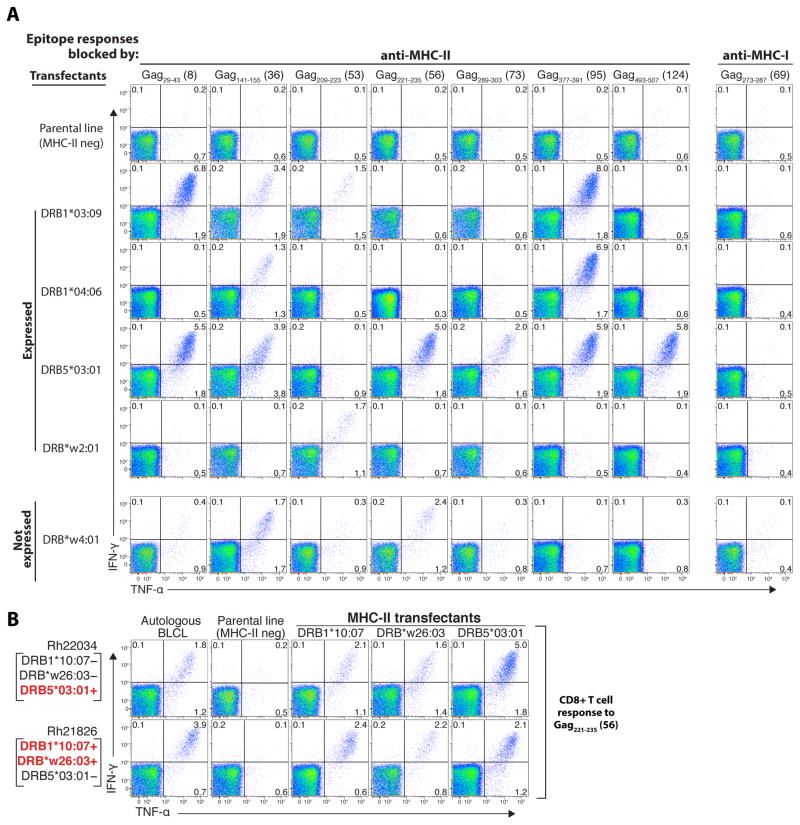
To confirm that the MHC-II-blocked CD8+ T cell responses were MHC-II-restricted – defined as the epitope in question being recognized in the context of an MHC-II surface protein – and to investigate the basis of the promiscuity of these responses across MHC-disparate RM, we constructed cell lines expressing single rhesus MHC-II allomorphs, focusing on MHC-II alleles expressed by 4 RhCMV/gag-vaccinated RM with characterized SIVgag epitope recognition profiles (fig. S6). Flow cytometric ICS assays showed that pulsing of the MHC-II allomorph transfectants, but not the parental MHC-II negative cell line, with individual peptides resulted in robust CD8+ T cell stimulation of only those responses classified as MHC-II-associated by blocking experiments (Fig. 4A; table S3), and these responses could be blocked with anti-MHC-II mAbs and CLIP peptide, but not anti-MHC-I mAbs (fig. S7). Importantly, individual MHC-II allomorphs presented multiple peptides, and individual peptides were frequently presented by multiple MHC-II allomorphs (Fig. 4A; table S3). The ability of individual allomorphs to present multiple gag peptides helps explain the striking breadth of these MHC-II-restricted CD8+ T cell responses. The ability of multiple MHC-II allomorphs to present many of the same individual peptides suggests that the common recognition of these peptides by RhCMV/gag vector-elicited CD8+ T cells across MHC-disparate RM (e.g., their supertope character) is likely due to all RM expressing at least one effective MHC-II allomorph for each response.
Fig. 4.
Multiple MHC-II allomorphs can present type 2 epitopes to RhCMV/gag vector-elicited CD8+ T cells. (A) PBMC from a strain 68-1 RhCMV/gag-vaccinated RM (Rh22034) were incubated with SIVgag peptide-pulsed (and washed) RM3 cells (the MHC-II negative parental cell line) vs. RM3 transfectants expressing single Mamu-DR molecules, and then evaluated for peptide-specific CD8+ T cell recognition using flow cytometric ICS to detect induction of IFN-γ and/or TNF-α production (response frequencies are indicated in each quadrant). The Mamu-DR molecules tested included four that are expressed by Rh22034 (DRB1*0309, DRB1*0406, DRB5*0301, and DRB*w201), and one that is not expressed (DRB*w4:01). The SIVgag 15mer peptides tested corresponded to known MHC-II-blocked CD8+ T cell epitopes (Type 2) in this RM, except for Gag273-287 (15mer #69), which was MHC-I-blocked (Type 1), and therefore used as a negative control. (B) Similar analysis of the presentation of the MHC-II-blocked (Type 2) Gag221-235 (15mer #56) peptide to CD8+ T cells from two strain 68-1 RhCMV/gag vector-vaccinated RM (Rh22034 and Rh21836) by autologous B-lymphoblastoid cells, MHC-II null parental cells and single MHC-II transfectants corresponding to Mamu-DRB alleles that are reciprocally expressed by these 2 RM (expressed alleles denoted in red, non-expressed in black).
As previously reported for MHC-II-restricted CD4+ T cell responses (28), we found that MHC-II-restricted, SIVgag-specific CD8+ T cells elicited by RhCMV/gag vectors can respond to their specific peptide epitope in the context of peptide-binding MHC-II allomorphs that are not expressed by the responding T cell donor (Fig. 4A,B; table S3), indicating that the TCR of these T cells recognize the bound peptide alone or in combination with non-polymorphic structures on the MHC-II molecule. Interestingly, the expression of an MHC-II allomorph capable of binding a given epitope-containing peptide by a given RM did not always result in a response to that peptide [table S3; note, for example, the lack of CD8+ T cell responses to Gag73-87 in Rh22034, to Gag141-155 in Rh21826 and Rh22607, and to Gag377-391 in Rh21826 despite the expression of MHC-II allomorphs that can present these epitopes in other RM]. This suggests that TCR repertoire and/or immunoregulatory mechanisms also play a role in determining the targeting of RhCMV vector-elicited CD8+ T cells, and in particular, whether epitopes are targeted by all vaccinated RM or only a subset.
Phenotype and function of RhCMV/SIV vector-elicited CD8+ T cell responses
The unusual epitope specificity of the SIV-specific CD8+ T cells generated and maintained by RhCMV/SIV vector vaccination raises the question of their functional potential, especially that of the unconventional MHC-II-restricted population that dominates these CD8+ T cell responses. First, in this regard, these supertope-specific CD8+ T cell responses are not an artifact of the high peptide concentrations used in standard ICS assays, as responses to the optimal peptides, both Type 1 and Type 2, can be demonstrated at peptide dilutions of 1:105 and greater (Fig. 5A). Second, both Type 1 and Type 2 supertope-specific CD8+ T cell responses arise immediately after vaccination (Fig. 5B), and are coordinately distributed throughout the body in the pattern previously reported for RhCMV/SIV vector-vaccinated RM (Fig. 5C,D) (10). Third, as previously reported for RhCMV-specific CD8+ T cells and RhCMV/SIV vector-elicited, SIV-specific CD8+ T cells (10, 11), both Type 1 and Type 2 supertope-specific T cells manifest an identical phenotype indicative of dominant effector memory cell differentiation (CCR7−, CD28−) and an identical polyfunctional profile consistent with this effector-memory phenotype: high TNF, IFN-γ, and MIP-1β production, high CD107 externalization (degranulation) and low IL-2 production (Fig. 5E,F). Because effector memory differentiation is thought to be Ag-driven, these data suggest that in vaccinated RM, these SIV-specific CD8+ T cells receive equivalent in vivo exposure to Type 1 and Type 2 epitopes.
Fig. 5.
RhCMV/gag vector-elicited CD8+ T cells show similar function regardless of MHC-I vs. MHC-II restriction. (A) Serial log10 dilutions of 4 core (optimal) SIVgag supertope peptides (2 each MHC-I- and MHC-II-restricted) were used to stimulate PBMC from strain 68-1 RhCMV/gag-vaccinated RM (n = 5) and the response to each peptide dilution was determined by flow cytometric ICS. The frequency of responding CD8+ T cells (TNF-α+ and/or IFN-γ +) at each dilution was normalized to the maximal response at the initial peptide concentration. The figure shows the mean + SEM of the normalized responses for each epitope. (B) Peripheral blood CD8+ T cell responses to total SIVgag 15mer mixes and to 4 core (optimal) SIVgag supertope peptides (2 each MHC-I- and MHC-II-restricted) were quantified by flow cytometric ICS (TNF-α + and/or IFN-γ +) following strain 68-1 RhCMV/gag vaccination (mean + SEM; n = 24) to demonstrate the relative kinetics of induction of the MHC-1 vs. MHC-II-restricted supertope responses. (C,D) CD8+ T cell responses to 2 MHC-I-restricted (C) and 2 MHC-II-restricted (D) core (optimal) SIVgag supertope peptides were quantified by flow cytometric ICS (TNF-α + and/or IFN-γ +) in mononuclear cell preparations from the indicated tissues at necropsy of strain 68-1 RhCMV/gag vector-vaccinated RM (mean + SEM; n = 4). (E) PBMC from strain 68-1 RhCMV/gag-vaccinated RM (n =14) were stimulated with total SIVgag 15mer mixes or the MHC-I- vs. MHC-II-restricted core (optimal) SIVgag supertope peptides shown and the expression of CD28 vs. CCR7 was determined on the responding cells (TNF-α + and/or IFN-γ +) by flow cytometric ICS, allowing delineation of the mean (+ SEM) proportion of the responding cells manifesting the designated central memory (TCM), transitional effector memory (TTrEM) and effector memory (TEM) phenotypes. (F) PBMC from strain 68-1 RhCMV/gag-vaccinated RM (n =14) were stimulated with total SIVgag 15mer mixes or the MHC-I- vs. MHC-II-restricted core (optimal) SIVgag supertope peptides shown and the frequencies of cells within the CD8+ memory compartment producing each cytokine or showing CD107 externalization were determined. The figure shows the mean (+ SEM) of these response frequencies after background subtraction.
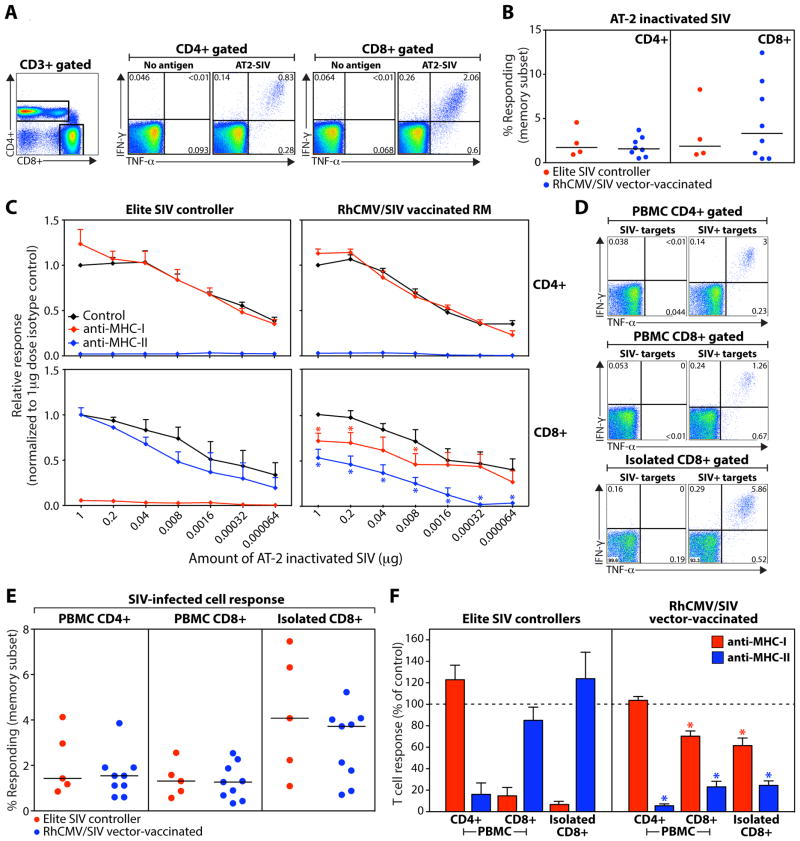
The most important question regarding the function of the RhCMV/SIV vector-elicited CD8+ T cells is the extent to which they can recognize naturally processed SIV Ag, and in particular, SIV-infected cells. To evaluate this question, we compared the ability of RhCMV/SIV vector-elicited, SIV-specific T cells vs. conventional SIV-specific T cells from SIV-infected elite controllers to respond to a) SIV virions chemically inactivated with aldrithiol-2 (AT-2), and b) autologous CD4+ T cells infected with SIVmac239. We then used MHC-I vs. MHC-II blockade to determine the restriction of this recognition. Previous work has established that AT-2-inactivated SIV (AT-2 SIV) is taken up by professional antigen presenting cells (APC) via both MHC-I and MHC-II processing pathways, the former by a fusion-mediated delivery of SIV virion proteins to the cytosol (29). CD4+ and CD8+ T cells from both RhCMV/SIV vector-vaccinated RM and elite controllers responded to AT-2 SIV at concentrations down to <100 picograms p27CA equivalent (Fig. 6A–C). The AT-2 SIV-specific CD4+ and CD8+ T cell responses from the SIV+ elite controllers were, as expected, completely and specifically inhibited with MHC-II and MHC-I blocking reagents, respectively. The AT-2 SIV-specific CD4+ T cell responses from the RhCMV/SIV vector-vaccinated RM were similarly inhibited by MHC-II blockade, but the AT-2 SIV-specific CD8+ T cell responses from these RM were partially inhibited by both the MHC-I and MHC-II blocking reagents, with the MHC-II blocking being most prominent (Fig. 6C; fig. S8). Interestingly, at low AT-2 SIV concentrations (<1.6 ng p27CA equivalent), the MHC-I-restricted component of the RhCMV/SIV vector-elicited CD8+ T cell response (the response present after blocking with anti-MHC-II; blue line in Fig. 6C) disappears entirely, leaving only the MHC-II-restricted component (the response present after blocking with anti-MHC-I; red line in Fig. 6C), indicating that the RhCMV/SIV vector-generated, MHC-II-restricted CD8+ T cells are more sensitive to AT-2 SIV than their MHC-I-restricted counterparts in this assay. This is likely because for AT-2 SIV, the MHC-II antigen presentation pathway processes and presents the MHC-II-restricted epitopes more efficiently than the cytosolic MHC-I pathway and/or cross-presentation can process and present the MHC-I-restricted epitopes.
Fig. 6.
RhCMV vector-elicited CD8+ T cells recognize SIV-infected cells via both MHC-I and MHC-II antigen presentation. (A) Representative flow cytometric profiles of CD4+ and CD8+ T cells in PBMC from an RM vaccinated with strain 68-1 RhCMV/SIV vectors responding to 1 μg (p27CA equivalent) of AT-2 SIV vs. no antigen (quadrant response frequencies indicated). (B) Comparison of AT-2 SIV-specific response frequencies in the CD4+ or CD8+ memory compartment in the blood of SIV+ elite controllers (n = 4) vs. RM vaccinated with strain 68-1 RhCMV/SIV vectors (n = 8). (C) Serial 5-fold dilutions of AT-2 SIV were used to stimulate PBMC from the same strain 68-1 RhCMV/SIV-vaccinated and SIV+ elite controllers shown in panel B, comparing the CD4+ and CD8+ T cell response frequencies in the presence of isotype control mAb vs. MHC-1 blocking mAb W6-32 vs. the MHC-II-blocking CLIP peptide (see Fig. 3). The response frequencies of each subset in each RM were normalized to the unblocked response frequencies at the highest AT-2 SIV dose and the mean + SEM of these normalized response frequencies are shown for each treatment and dose. Asterisks indicate the MHC-I- or MHC-II-blocked CD8+ T cell responses, as fractions of the isotype responses at the same dilution, that are significantly different (p < .05) from 1.0 using a two-tailed Wilcoxon signed rank test (RhCMV/SIV vector group only; see fig. S8). (D) Representative flow cytometric profiles of CD4+ and CD8+ T cells in PBMC and isolated CD8+ T cells from an RM vaccinated with strain 68-1 RhCMV/SIV vectors responding to autologous SIV-infected CD4+ T cells (SIV+ targets) vs. similarly processed and cultured CD4+ T cells that were not SIV-infected (SIV- targets) using an effector to target ratio of 80:1. (E) Comparison of SIV-infected cell-specific response frequencies in the memory subsets of CD4+ and CD8+ T cells in PBMC and isolated CD8+ T cells from SIV+ elite controllers (n = 5) vs. RM vaccinated with strain 68-1 RhCMV/SIV vectors (n = 9). (F) The sensitivity of the SIV+ cell-specific T cell responses shown in panel E to blocking with the MHC-1-blocking mAb W6-32 vs. the MHC-II blocking CLIP peptide is shown. The response frequencies were normalized to the response frequencies in the isotype control and the mean + SEM of these normalized response frequencies are shown for each treatment. Asterisks indicate the normalized MHC-I- or MHC-II-blocked CD8+ T cell responses that are significantly different (p < .05) from 100% using a two-tailed Wilcoxon signed rank test (RhCMV/SIV vector group only; see fig. S9).
We next compared the ability of SIV-specific T cells from RhCMV/SIV vector-vaccinated RM and SIV+ elite controllers to recognize purified, autologous, SIV-infected CD4+ T cells. We evaluated the response of CD4+ and CD8+ T cells in PBMC preparations and the response of isolated CD8+ T cells, the latter to rule out the possibility of indirect presentation of released SIV antigens by professional APC. CD4+ and CD8+ T cells from PBMC and isolated CD8+ T cells from all tested RM specifically recognized purified SIV-infected CD4+ T cells, with the isolated CD8+ T cells showing the most robust response (Fig. 6D,E). In these assays, the CD4+ T cell response to SIV-infected cells was, as expected, completely inhibited by MHC-II blocking reagents in all RM, whereas the CD8+ T cell responses to SIV-infected cells showed the same dichotomy described above for AT-2 SIV responses (Fig. 6F; fig. S9). The recognition of SIV-infected cells by the CD8+ T cells from SIV+ elite controllers was completely MHC-I-dependent, whereas the ability of CD8+ T cells from RhCMV/SIV-vaccinated RM to recognize SIV-infected cells was ~70% blocked by anti-MHC-II reagents and ~30% blocked by anti-MHC-I reagents. This implies that in RhCMV/SIV vector-vaccinated RM, ~70% of the SIV-specific CD8+ T cells capable of recognizing autologous SIV-infected cells are MHC-II-restricted vs. 30% MHC-1-restricted, a 2:1 ratio that closely approximates the ratio of the number of MHC-II- vs. MHC-I-restricted epitopes recognized by CD8+ T cells in RhCMV/gag vector-vaccinated RM (Fig. 3B). Taken together, these data demonstrate that the unconventional epitopes recognized by strain 68-1 RhCMV/SIV vector-elicited CD8+ T cells can be processed and presented by autologous SIV-infected cells, and therefore, that this processing and presentation is independent of CMV gene expression.
CMV genes control targeting of CMV-elicited CD8+ T cell responses
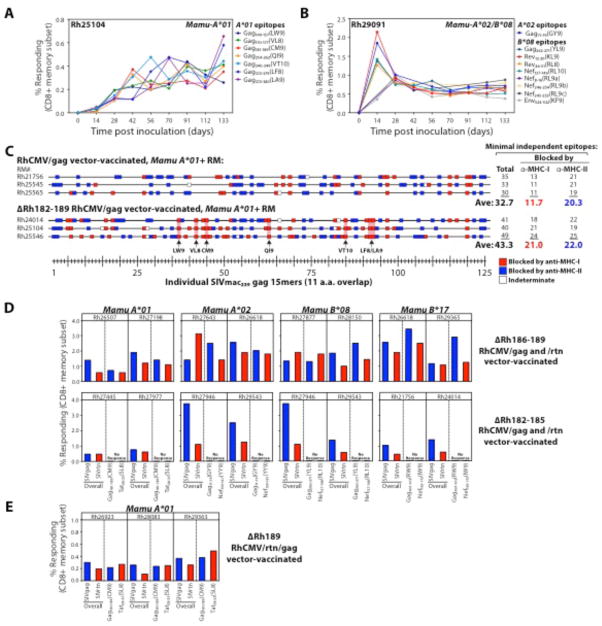
The data described above strongly suggest that CMV has evolved specific mechanisms to modulate CD8+ T cell priming, perhaps to redirect the response away from the internally processed epitopes that make CMV-infected cells vulnerable to CD8+ T cell-mediated cytolysis. Potential candidates for mediating this effect are the Rh182, 184, 185, and 189 genes (the RhCMV orthologues of the US2, 3, 6 and 11 genes in human CMV) that down-regulate MHC-I expression in CMV-infected cells, preventing their recognition by CMV-specific CD8+ T cells (12, 30–32) (fig. S10,11). It is possible that these MHC-I down-regulators might prevent direct priming of CD8+ T cells by CMV-infected cells and thereby modify CD8+ T cell epitope recognition hierarchies. RM fibroblasts infected with RhCMV/SIV vectors with the Rh182-189 region deleted (ΔRh182-189) are readily recognized by CD8+ effector T cells (fig. S11), and due to this susceptibility, these vectors fail to superinfect RhCMV+ RM (12). However, these vectors can persistently infect CMV-naïve RM, and elicit CD8+ T cell responses to SIV inserts in this setting (12). Strikingly, in sharp contrast to the RhCMV/SIV vectors with intact MHC-I down-regulation, ΔRh182-189 RhCMV/SIV vectors elicit CD8+ T cells recognizing all Mamu-A*01-, -A*02-, and B*08-restricted canonical epitopes (Fig. 7A,B). Importantly, the targeting of canonical epitopes by the SIVgag-specific CD8+ T cells elicited by these ΔRh182-189 (strain 68-1-derived) RhCMV vectors was in addition to, not in lieu of, the unconventional MHC-I- and MHC-II-restricted (supertope-directed) CD8+ T cell responses described above (Fig. 7C). It is also noteworthy that in contrast to conventional responses to these canonical epitopes, in which responses to certain epitopes are dominant and others subdominant (33, 34), the canonical epitope-targeted responses elicited by ΔRh182-189 RhCMV/SIV vectors are all-inclusive with each response very similar in magnitude (Fig. 7A,B), indicating that these CMV-based vectors are still fundamentally different in their CD8+ T cell priming characteristics.
Fig. 7.
Rh189 (US11) gene prevents canonical epitope recognition. (A,B) Flow cytometric ICS was used to follow the development of canonical epitope-specific CD8+ T cell responses in the blood of 2 RM, one expressing Mamu-A*01, the other expressing both Mamu-A*02 and -B*08, after vaccination with strain 68-1-derived RhCMV/SIV vectors in which the Rh182-189 (US2-11) genes were deleted [note: these RM were CMV naïve prior to vaccination, as the Rh182-189-deleted vectors cannot superinfect RhCMV+ RM (12)]. (C) CD8+ T cell responses to SIVgag were epitope-mapped and MHC-blocked (as described in Figs. 2 and 3) in 6 Mamu-A*01-expressing (initially RhCMV-naïve) RM, 3 vaccinated with the unmodified strain 68-1 RhCMV/gag vector and 3 vaccinated with the ΔRh182-189 (ΔUS2-11) version of this vector. Responses blocked by anti-MHC-I- vs. anti-MHC-II are indicated by red and blue boxes, respectively, with the 15mers containing canonical Mamu-A*01-restricted epitopes indicated by pink rectangles. (D) The peak acute phase CD8+ T cell responses in blood to whole SIVgag and SIVrtn peptide mixes and canonical SIVgag, SIVnef, and SIVtat epitopes are shown for (initially RhCMV+) RM expressing the designated MHC-I alleles and vaccinated with SIVgag- and SIVrtn-expressing strain 68-1 RhCMV vectors lacking either Rh186-189 (US8-11) or Rh182-186 (US2-6). (E) The peak acute phase CD8+ T cell responses in blood to whole SIVgag and SIVrtn peptide mixes and canonical SIVgag and SIVtat epitopes are shown for 3 (initially RhCMV+) RM expressing Mamu-A*01 and vaccinated with an SIVgag- and SIVrtn-expressing strain 68-1 RhCMV vector lacking only Rh189 (US11).
We next asked whether the appearance of canonical epitope recognition required complete abrogation of MHC-I down-regulation by construction and assessment of sub-region deletant vectors (ΔRh182-185 and ΔRh186-189). These vectors only partially down-regulate MHC-I and partially prevent cytolytic T cell recognition (figs. S10,11), and unlike the ΔRh182-189 vectors, can super-infect CMV+ RM, as evidenced by their ability to elicit SIV-specific T cell responses in this setting (Fig. 7D). If MHC-I down-regulation per se was the mechanism preventing canonical epitope recognition, one might predict that neither of these deletants would elicit such canonical epitope responses, or potentially, such responses would only be seen with the ΔRh182-185 construct (lacking 3 of 4 MHC-I inhibitory genes – the US2, 3 and 6 orthologues), which most closely resembles the full (ΔRh182-189) deletant (fig. S11). However, this was not the case (Fig. 7D) – only the ΔRh185-189 construct (lacking the US11 orthologue) elicited CD8+ T cells against canonical epitopes (and did so for all tested epitopes restricted by 4 different MHC-I alleles), despite the very limited ability of the Rh189 (US11) gene product to down-regulate MHC-1. The specific involvement of the Rh189/US11 gene in preventing canonical epitope recognition by RhCMV vector-elicited CD8+ T cells was confirmed by the development of such responses in RM vaccinated with a RhCMV/SIV vector lacking only the Rh189 gene (ΔRh189; Fig. 7E). Thus, Rh189, the orthologue of human CMV US11, specifically prevents priming of CD8+ T cells to canonical epitopes, almost certainly by a mechanism other than its known ability to target MHC-I molecules for degradation, since Rh189 (US11) alone had only a very modest ability to both down-modulate MHC-I and inhibit T cell recognition of RhCMV infected cells (figs. S10,11).
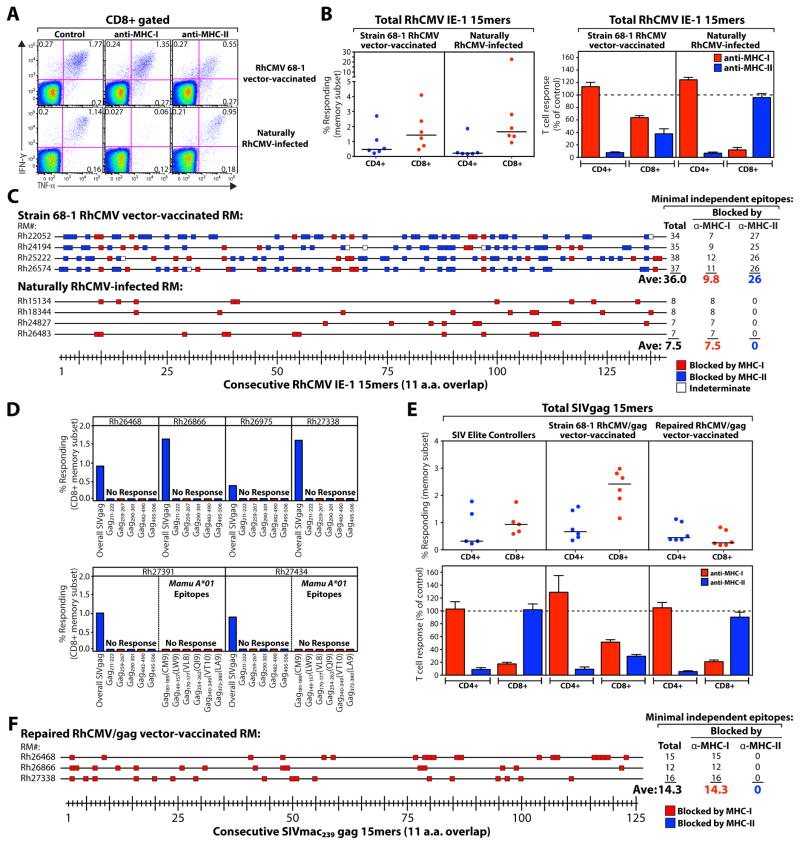
The presence of MHC-I- and MHC-II-restricted, supertope-directed CD8+ T cell responses in RM administered ΔRh182-189 RhCMV vectors indicates that other CMV-encoded mechanisms must be responsible for this unconventional CD8+ T cell targeting. To identify candidate CMV genes associated with, and potentially responsible for, this targeting, we first asked whether CD8+ T cell responses to CMV immediate early (IE) protein target unconventional epitopes (in particular, supertopes restricted by MHC-II), assessing both RM naturally infected with wildtype RhCMV (colony circulating strains) and RM vaccinated with strain 68-1 RhCMV/SIV vectors. Not surprisingly, RM vaccinated with the strain 68-1 RhCMV/SIV vector demonstrated IE-specific CD8+ T cell responses with identical targeting characteristics as the SIVgag-specific CD8+ T cell responses in the same RM: >30 distinct IE epitopes/RM, including a majority of epitope-specific responses that were blocked with anti-MHC-II, and a minority blocked with anti-MHC-I. However, in striking contrast, the IE-specific CD8+ T cell responses in naturally RhCMV-infected RM were much more narrowly targeted (~8 epitopes/RM), and showed no evidence of MHC-II restriction or epitope promiscuity (Fig. 8A–C, fig. S12A), consistent with conventional immunodominance hierarchies. These findings likely account for why unconventionally targeted CMV-specific CD8+ T cell responses have not been reported in naturally exposed CMV+ RM and humans (despite considerable analysis of these responses) and more importantly, implicate genetic differences between the strain 68-1-based RhCMV vectors and wildtype RhCMV in the mechanism(s) responsible for generating the unconventionally targeted CD8+ T cell responses.
Fig. 8.
Unconventional CD8+ T cell targeting is restricted to responses elicited by fibroblast-adapted RhCMV lacking UL128-131 orthologue expression. (A) Representative flow cytometric profiles of CD8+ T cells in PBMC from an unvaccinated, naturally RhCMV-infected RM (colony circulating strain) vs. a strain 68-1 RhCMV/SIV vector-vaccinated RM responding to consecutive 15mer peptides (11 amino acid overlap) comprising the RhCMV IE-1 protein in the presence of isotype control vs. blocking anti-MHC-I vs. blocking anti-MHC-II mAbs. (B) Comparison of the frequency (left panel) and sensitivity to blockade with anti-MHC-I vs. anti-MHC-II mAbs (right panel; mean + SEM) of IE-1-specific CD4+ and CD8+ T cells from naturally RhCMV-infected vs. strain 68-1 RhCMV/SIV vector-vaccinated RM (n = 6 per group; see fig. S12A). (C) CD8+ T cell responses to RhCMV IE-1 in naturally RhCMV-infected and strain 68-1 RhCMV/SIV vector-vaccinated RM (n = 4 each) were epitope-mapped to determine recognition of 137 consecutive 15mer IE-1 peptides and then the MHC association of each response was classified by sensitivity to blockade with anti-MHC-I vs. anti-MHC-II mAbs. (D) The peak, acute phase CD8+ T cell response frequencies in blood to the whole SIVgag 15mer mix, each of the 5 universal RhCMV/gag vector-associated supertopes, and in the 2 Mamu-A*01+ RM (Rh27391 and Rh27434), each of the indicated canonical SIVgag epitopes restricted by this allele, are shown in 6 RM vaccinated with a strain 68-1 RhCMV/gag vector in which expression of RhCMV orthologues of HCMV UL128-131 genes (Rh157.6, 157.4 and 157.5) has been restored. (E) Comparison of the frequency (top panel) and sensitivity to blockade with anti-MHC-I vs. anti-MHC-II mAbs (bottom panel; mean + SEM) of SIVgag-specific CD4+ and CD8+ T cells from SIV+ elite controllers vs. RM vaccinated with the original strain 68-1 RhCMV/SIV vector vs. RM vaccinated with the Rh157.4-.6 (UL128-131)-repaired RhCMV/gag vector (n = 6 per group; see fig. S12B). (F) CD8+ T cell responses to SIVgag in 3 RM vaccinated with the Rh157.4-.6 (UL128-131)-repaired RhCMV/gag vector were epitope-mapped and then the MHC association of each response was classified by sensitivity to blockade with anti-MHC-I vs. anti-MHC-II mAbs (compare to Fig. 3C).
Most of the RhCMV/SIV vectors used in this study were derived from bacterial artificial chromosome (BAC)-cloned RhCMV strain 68-1, which was multiply passaged in fibroblast culture prior to its cloning. Compared to wildtype CMV, BAC-derived RhCMV has deletion or major defects in expression of the Rh13.1, Rh61/Rh60, Rh157.5, Rh157.4 and Rh157.6 genes, corresponding, respectively, to the RL13, UL36, UL128, UL130 and UL131 genes of human CMV (35, 36). To assess the role of these genes in the epitope targeting of CD8+ T cells primed by RhCMV/gag vectors, we inserted SIVgag into the previously constructed recombinant RhCMV68-1.2 in which functional expression of the Rh61/Rh60, Rh157.4, Rh157.5 and Rh157.6 genes were restored (36). Strikingly, when RM were inoculated with this repaired RhCMV/gag vector, the SIVgag-specific CD8+ T cell responses elicited did not include recognition of any of the previously defined MHC-I or MHC-II supertopes, were much more narrowly targeted than the response elicited by the unrepaired 68-1 strain vector, and were entirely MHC-I-associated (Fig. 8D–F; fig. S12B). Of the genes repaired in RhCMV68-1.2, Rh61/Rh60 (UL36) could be ruled out as being responsible for this phenotype since this gene is intact in RhCMV/gagL, a non-BAC derived precursor of recombinant RhCMV (12) that induces T cell responses similar to BAC-derived RhCMV (Fig. 2A). Therefore, we conclude that the RhCMV orthologues of UL128, UL130 and UL131 preclude the priming of supertope-specific and MHC-II-restricted T cells. The proteins expressed by these genes associate with gH/gL to form a pentameric CMV virion surface complex involved in cell tropism [e.g., infection of non-fibroblasts (36)], suggesting a possible role for cell tropism in T cell priming. Notably, the SIVgag-specific response in Mamu-A*01+ RM vaccinated with the repaired RhCMV/SIV vector did not develop responses to canonical Gag epitopes (Fig. 8D, bottom panel), suggesting that US11 still precludes canonical epitope targeting in this setting. Thus, RhCMV encodes 2 independent mechanisms that inhibit the priming of CD8+ T cells targeting distinct epitope types: 1) Rh189 (US11), which inhibits responses to canonical epitopes, and 2) UL128-131 (Rh157.4-.6), which inhibit responses to highly promiscuous, unconventional epitopes, the majority of which are MHC-II-restricted.
DISCUSSION
The CMVs are an extraordinarily successful, ancient family of viruses that have co-evolved with their mammalian hosts. The persistent infections mediated by these viruses have a distinctive biology – a host-parasite relationship in which the virus elicits and maintains high frequency, effector-differentiated T cell responses that stringently control its replication (preventing potentially life-threatening disease), but at the same time establishes immune evasion programs that prevent these responses from either clearing infection or interfering with viral transmission (11, 12, 37–39). Although, theoretically, viral evasion of CD8+ T cell responses might occur at both the afferent and effector phases of the response, the fact that CMV-specific CD8+ T cells comprise 10% or more of the circulating memory compartment in over half of CMV+ individuals (38) would support the conclusion that the strategies used by the CMV to evade these responses focus on protecting infected cells from CD8+ T cell effector function. While it has been proposed that viral-encoded MHC-I down-regulators might prevent direct presentation and thereby inhibit or change the quality of CD8+ T cell priming, in the systems studied to date, the effect of these genes on CD8+ T cell priming has been minor at best (2, 31, 40–43), consistent with the major contribution of these genes being inhibition of CD8+ effector T cell recognition of virally infected cells. Indeed, the process of cross-presentation, the ability of non-infected professional antigen-presenting cells to initiate CD8+ T cell priming, would seem to be the host’s evolutionary counter to viral strategies aimed at inhibiting or substantially modifying CD8+ T cell priming, unless they can act at a distance from infected cells (31). However, the data presented in this report indicate that RhCMV can fundamentally manipulate CD8+ T cell priming with a sophistication and stringency that is both surprising, and to our knowledge, unprecedented for any other viral pathogen. RhCMV does not prevent CD8+ T cell priming overall, but rather, specifically and completely prevents the development of CD8+ T cells capable of recognizing certain types of CMV-encoded epitopes, redirecting the targeting of elicited CD8+ T cell responses to distinct epitope sets. Our results indicate that CD8+ T cell responses to a specific antigen can differ not only by the differentiation state, physiology and functional potential of the responding cells, as previously described (44), but also by differential targeting of non-overlapping epitopes.
Of the 2 components of CMV-mediated manipulation of CD8+ T cell targeting identified – the Rh189 (US11)-associated inhibition of responses to canonical epitopes, and the Rh157.4-.6 (UL128-131)-associated inhibition of diverse MHC-I- and MHC-II-restricted supertope recognition, the former is the easier to conceptualize. Canonical epitopes are pathogen-derived peptides that are efficiently processed and presented by the MHC-I-linked endogenous Ag presentation pathway (given their predominant recognition across vectors that deliver the targeted proteins to the cytosol), and thus are likely to represent the most efficient targets for CD8+ T cell-mediated cytolysis of virally infected cells. Although CMV can actively and efficiently down-regulate MHC-I in infected cells, this activity cannot protect against cytolytic T cell responses recognizing efficiently processed epitopes that are present in the cytoplasm of newly infected cells before these MHC-I down-regulators are expressed, including IE proteins and tegument and other viral proteins brought in with the infecting virion (45, 46). By preventing CD8+ T cell priming to such epitopes, this potential vulnerability is avoided. The ability of Rh189 (US11) to mediate this specific inhibition of canonical epitope priming does not appear to derive from its known ability to target MHC-I molecules for proteolysis in infected cells, as by itself, Rh189 (US11) can exert only modest MHC-I down-regulation, which was insufficient to prevent T cell stimulation. Moreover, this function would not easily explain the complete lack of any canonical epitope priming, which implies that both direct- and cross-presentation of these epitopes is prevented (the latter being independent of MHC-I expression in infected cells). In this regard, it should be noted that although mouse CMV can down-regulate MHC-I, it does not encode a Rh189 (US11) orthologue (47) and mouse CMV has been shown to be capable of eliciting responses to canonical epitopes (albeit by cross-presentation) (40, 42), suggesting that the Rh189 (US11)-mediated mechanism is specific to primate CMVs.
The effect of the UL128-131 orthologues on CD8+ T cell priming by RhCMV is more difficult to conceptualize, because it involves the ability of these genes to suppress priming of an entirely novel set of CD8+ T cell responses characterized by their targeting of diverse, highly promiscuous epitopes, predominantly in the context of MHC-II presentation. The inescapable implication of this observation is that an intrinsic mechanism in RhCMV infection generates these unconventional CD8+ T cell responses, and prompted the evolutionary development of the Rh157.4-.6 (UL128-131)-mediated mechanism to suppress this generation. In both RhCMV and HCMV, these genes (Rh157.4-.6; UL128-131) encode 3 components of an alternative entry receptor for non-fibroblasts (36, 48, 49), but determination of whether the effect of this gene region on CD8+ T cell priming is a consequence of this function, other reported functions of these genes [e.g., chemokine activity by UL128 (50)] or a completely new mechanism will require further analysis, as will understanding of the mechanisms by which RhCMV vectors lacking these genes elicit these unprecedented responses. Almost certainly, these mechanisms will delineate a new pathway of antigen presentation, perhaps one initiated by viral infection of fibroblasts or related stromal cells.
Although the mechanistic origin of the paradigm-breaking CD8+ T cell responses elicited by these strain 68-1 RhCMV vectors remains unclear, their existence indicates that previous understanding of the biology of CD8+ T cell epitope targeting is incomplete. First, there is the issue of the restriction of CD8+ T cell responses by MHC-II, rather than MHC-I, proteins. The specificity, breadth and immediate response kinetics of the MHC-II-restricted CD8+ T cells identified here indicate that thymic selection pathways in primates do not preclude development and peripheral export of naïve CD8+ T cells with the ability to specifically recognize varied, MHC-II-bound peptides. Although this might reflect accidental MHC-II cross-reactivity of TCR selected on MHC-I molecules, as has been suggested for previous observations of CD8+ T cell alloreactivity against MHC-II (17, 22, 23), this cross-reactivity cannot be a rare “happenstance” occurrence, but rather, given the diversity of the MHC-II-restricted, CD8+ T cell responses we observed, must be an intrinsic feature of a degenerate repertoire. In this regard, many of the MHC-II-restricted CD8+ T cell responses studied in detail here, like many CD4+ T cell responses (28), recognize specific peptide epitopes in the context of multiple MHC-II allomorphs, including allomorphs not expressed by the T cells, indicating that allele-specific MHC determinants do not make a major contribution to the recognition specificity of these T cells, and implying an intrinsic element of cross-reactivity in the TCR mediating these responses.
The second paradigm-violating aspect of the strain 68-1 RhCMV/SIV vector-elicited CD8+ T cell responses is the promiscuity of both the MHC-II- and MHC-I-restricted epitope recognition. Although MHC-II-restricted CD4+ T cell responses are intrinsically broader and more promiscuous than conventional MHC-I-restricted CD8+ T responses (14, 15, 51, 52), including descriptions of universal epitopes (53), the number of universal MHC-II-restricted supertopes found in the RhCMV/gag-elicited CD8+ T cell responses would be highly unusual for a CD4+ T cell response, and the identification of multiple universal or nearly universal MHC-I-restricted supertopes is, to our knowledge, unprecedented (54). For the MHC-II-restricted responses, the promiscuity appears to reflect the ability of multiple MHC-II allomorphs to bind and present these supertopes, such that all (or most) RM have at least one MHC-II allomorph that can bind and present the peptide involved in each response. The same mechanism may also apply to the MHC-I-restricted supertopes, although with MHC-I, there is the additional possibility that some or all of these MHC-I supertopes are presented by invariant MHC-I molecules (55). It is clear that the processing and presentation of these epitopes is not restricted to RhCMV-derived antigen, nor is in any way dependent on modification of antigen presentation pathways by the RhCMV vector, as both MHC-I- and MHC-II-restricted RhCMV/SIV vector-elicited T cells were able to specifically recognize naturally processed SIV virions and autologous SIV-infected CD4+ T cells. Given that, as discussed above, the naïve RM CD8+ T cell repertoire must include T cells with unusual specificities described here and that naturally processed SIV antigens lead to presentation of the determinants recognized by these T cells, we must conclude that the failure to identify these CD8+ T cell specificities in the vast majority of CD8+ T cell responses to infectious agents or vaccines reflects peripheral immunoregulatory mechanisms that normally preclude such responses and/or favor conventionally targeted responses, mechanisms that must be bypassed by priming with strain 68-1 RhCMV vectors.
Our findings reveal a heretofore unrecognized flexibility in CD8+ T cell recognition, definitively demonstrate that the established rules of epitope selection and immunodominance are not absolute, and define a completely new kind of CD8+ T cell response, one that has the potential to be more effective than “natural” responses for some pathogens, particularly those, like HIV/SIV, for which natural responses are ineffective. Until efficacy analysis is performed on Rh157.4-.6 (UL128-131)-repaired RhCMV/SIV vectors [with and without Rh189 (US11) deletion], it is unclear whether the distinct targeting of strain 68-1 RhCMV/SIV vector-elicited CD8+ T cell response is specifically responsible for the striking efficacy of these vectors against mucosal challenge with highly pathogenic SIV (10), but these responses almost certainly participate in this protection, as no other SIV-specific CD8+ T cells are available in the >50% of RM that were stringently protected by this strain 68-1 RhCMV/SIV vector-containing vaccine. Moreover, there are good reasons to believe that the unconventional CD8+ T cell responses generated by Rh157.4-.6 (UL128-131)-deficient CMV vectors might have specific advantages for HIV/SIV and perhaps other pathogens. First, there is the enormous breadth of these responses. In the strain 68-1 RhCMV/gag vector-vaccinated RM studied here, the elicited SIVgag-specific CD8+ T cells recognized an average of 22 MHC-II- and 12 MHC-I-restricted, largely non-overlapping core epitopes (comprising 12 and 9 amino acids, respectively). Even allowing for 10% linear overlap, the core epitopes of these SIVgag-specific CD8+ T cell responses would encompass ~335 of the 510 amino acids in the SIVgag sequence, corresponding to 66% coverage of the protein! If recapitulated in an HCMV/HIV vaccine, such breadth would greatly increase the chance that vaccine-elicited CD8+ T cells would robustly recognize the divergent sequences in circulating HIV strains, as well as the likelihood that these responses will recognize vulnerable, conserved epitopes. Second, the protection provided by these unconventional, supertope-targeted CD8+ T cell responses will be independent of specific MHC alleles, and therefore equally applicable to all individuals in all populations, in contrast to vaccines eliciting conventional responses which preferentially protect individuals already predisposed to T cell-mediated viral control by virtue of expression of protective MHC alleles. Third, the fact that the epitope targeting of these unconventional CD8+ T cell responses is entirely distinct from the “natural” CD8+ T cell responses generated by HIV itself has crucial implications for both therapeutic and prophylactic AIDS vaccine development. In the case of a therapeutic vaccine administered to HIV+ individuals on suppressive anti-retroviral therapy (ART), the virus being targeted by the vaccine will in many, if not most, subjects have escaped any effective conventionally targeted CD8+ T cell responses prior to viral suppression (7). A vaccine that simply boosts these natural, conventionally targeted, HIV-specific T cell responses would likely not provide control of such escaped viruses after ART withdrawal, whereas the unconventionally targeted T cells elicited by a CMV/HIV vaccine will not be affected by this prior escape. Given that HIV escape mutations may be transmitted, and therefore can become intrinsic components of circulating virus sequences (56), the same consideration would extend to a prophylactic HIV/AIDS vaccine – the efficacy of the unconventionally targeted CMV/HIV vaccine-elicited CD8+ T cells will not be affected by escape from the conventional targeting of natural CD8+ T cell responses. Finally, the MHC-II restriction of the majority of the unconventional responses elicited by Rh157.4-.6 (UL128-131)-deleted CMV vectors might provide unique advantages for protection against pathogens that, like HIV and Mycobacterium tuberculosis, infect MHC-II+ cells (macrophages, activated T cells, etc.), essentially bringing the robust effector capabilities of the CD8+ T cell lineage (and its freedom from infection by HIV) to bear on MHC-II-expressing target cells (an effector response that would be in addition to that mediated by the CD8+ effector T cells recognizing MHC-I-restricted epitopes).
In conclusion, we have discovered that RhCMV has an intrinsic ability to elicit CD8+ T cell responses to unconventional epitopes, distinct in quality and quantity from all infectious agents studied to date. Moreover, this virus can exert unprecedented control of its own recognition by CD8+ T cells, with wildtype strains using Rh157.4-.6 (UL128-131) expression to divert CD8+ T cell targeting away from these unconventional epitopes, and Rh189 (US11) expression to redirect responses away from the canonical epitopes that likely constitute the most efficient targets for cytolysis. These observations strongly hint that the relationship of this ancient virus and the mammalian cellular immune system is more complicated and intertwined than previously appreciated, and suggest that better understanding of this relationship will offer new insights into fundamental immunologic mechanisms. Moreover, these findings have practical implications for vaccine development, namely the ability to construct different CMV vectors (with or without US11: with or without UL128-131) that specifically elicit CD8+ T cell responses with widely divergent patterns of epitope recognition, including responses that break conventional immunodominance hierarchies. CMV thus constitutes the first programmable viral vaccine platform that allows custom targeting of vaccine-elicited CD8+ T cells to whichever set of epitopes is most appropriate for the pathogen in question.
MATERIALS AND METHODS
Rhesus Macaques
A total of 165 purpose-bred male or female juvenile rhesus macaques (RM) (Macaca mulatta) of Indian genetic background were used in this study, including 110 RM vaccinated with strain 68-1 RhCMV/SIV vectors (wildtype or genetically modified, alone or subsequent to heterologous priming with conventional vaccines or virally suppressed SIV infection), 47 RM with SIV infection alone (SIVmac239 or SIVmac251), and 8 unvaccinated RM that were naturally infected with colony-circulating strains of RhCMV. All RM were used with the approval of the Oregon National Primate Research Center Institutional Animal Care and Use Committee, under the standards of the US National Institutes of Health Guide for the Care and Use of Laboratory Animals. RM used in these experiments were free of cercopithicine herpesvirus 1, D-type simian retrovirus, and simian T-lymphotrophic virus type 1. MHC-1 genotyping for the Mamu-A*01, Mamu-A*02, Mamu-B*08 and Mamu-B*17 alleles was performed by sequence-specific priming PCR, as described (57). Selected RM were DRB-genotyped by deep sequencing. Briefly, amplicons of the Mamu-DRB region were created via amplification of cDNA by PCR with high-fidelity Phusion polymerase (NEBiolabs) and a pair of universal MHC-DRB-specific primers (5′-CGTATCGCCTCCCTCGCGCCATCAG-MID-CTGGTCCTGTCCTGTTCTCC; 5′-CTATGCG CCTTGCCAGCCCGCTCAG-MID-TGGAAGGTCCAGTCTCCATT) using the following thermocycling conditions: 98°C for 3 minutes, (98°C for 5 seconds, 60°C for 10 seconds, 72°C for 20 seconds) for 25 cycles, and 72°C for 5 minutes. The primary cDNA-PCR products were purified using AMpure XP magnetic beads (Beckman Coulter Genomics). Emulsion PCR using a Lib-A kit (Roche/454 Life Sciences), bead purification, and pyrosequencing procedures with the Roche/454 GS Junior instrument were carried out as per the manufacturer’s instructions. Data analysis was performed using a Labkey database in conjunction with Geneious-Pro® bioinformatics software (Biomatters Ltd.) for sequence assembly. Mononuclear cell preparations for immunologic assays were obtained from blood, bone marrow, bronchoalveolar lavage (BAL), lymph nodes, spleen, liver, bone marrow, and intestinal mucosal, as previously described (58, 59). Purified CD8+ T cells (>90% pure) were obtained from PBMC using CD8 microbeads and LS columns (Miltenyi Biotec). Plasma viral loads of SIV+ RM were determined by quantitative real time RT-PCR (60). SIV+ RM were considered SIV controllers if the plasma viral loads were <2.0 x 104 copies/ml, and elite controllers if the plasma viral loads were <3.0 x 103 copies/ml.
RhCMV/SIV Vectors
The construction, characterization and administration of strain 68-1-derived RhCMV/SIV have been previously described in detail (10–12). All recombinant viruses used in this study were derived from strain RhCMV 68-1 BAC except for RhCMV(gagL) which was generated by replacing GFP in RhCMV-EGFP with the SIVgag expression cassette by in vivo recombination in tissue culture (12). Unlike BAC-derived constructs, RhCMVgagL contains an intact ORF Rh61/Rh60 (UL36) as described for RhCMV68-1 (35, 61). Due to tissue culture adaptation, both BAC and non-BAC RhCMV 68-1 constructs contain a deletion of ORF 157.5 and most of ORF Rh157.4 encoding homologs of HCMV UL128 and UL130, respectively (62). In low passage RhCMV, these two ORFs are translated from the same polycistronic mRNA encompassing Rh157.6 (UL131) (36). In RhCMV68-1 this transcript is truncated and the poly-A site is deleted, suggesting that Rh157.6 (UL131) is not expressed even though its coding region is intact. To generate a vector with complete UL128-131 expression, we inserted the SIVgag expression cassette into Rh211 of RhCMV68-1.2, a recombinant virus in which Rh61/Rh60 (UL36), Rh157.4 (UL130) and Rh157.5 (UL128) had been repaired (36). Deletion mutants within the Rh182-189 genomic region, which includes gene products inhibiting MHC-I antigen presentation, are shown in fig. S10. ΔRh182-189 RhCMV/gag has been previously described (12). Similarly, we created ΔRh182-189 RhCMV/rtn and/env by replacing the genomic region encoding Rh182-189 [base pairs (bp) 193,161–199,823, using the BAC genome annotation by (35)] with the EF1α SIVrev/tat/nef or gH SIV/env expression cassettes. The partial deletion mutants ΔRh182-185 RhCMV/gag and/rtn were generated by replacing bp 193,161 and 196,305 with an expression cassette for SIVgag or SIVrev/tat/nef. The partial deletion mutants ΔRh186-189 RhCMV/gag and rtn were constructed by replacing bp 196,593 to 199,823 with SIVgag or SIVrev/tat/nef expression cassette. To generate recombinant RhCMV that only lacks Rh189 (US11) we replaced the Rh189 coding region with that of SIVgag in RhCMVrtn. This vector thus expresses SIVgag under control of the Rh189 promoter and SIVrtn (inserted into Rh211) under control of the EF1α promoter (fig. S10). All of the recombinant viruses were characterized and confirmed by restriction digest and the antigen inserts including their flanking regions were sequence verified. Expression of SIV antigens was verified by immunoblot. Additionally, adjacent gene expression was verified by RT-PCR.
Other Vaccines
The construction, characterization and administration of the Ad5/gag vectors used in this study have been previously described (10). MVA/gag was constructed by insertion of codon-optimized, full-length SIVmac239 gag gene into the MVA shuttle vector, pLW44, under the control of MH5, an early/late vaccinia promoter, to generate the recombinant plasmid, pJV7. Flanking sequences within pLW44 directed insertion of the recombinant construct into the thymidine kinase locus by homologous recombination. Chicken embryonic fibroblast cells were transfected with pJV7 followed by infection with MVA strain 1974 to generate recombinant virus expressing SIVmac239gag (SIVgag expression confirmed by western blot). Recombinant virus was plaque-purified and amplified in large-scale culture. Viral stocks were purified over a 24–40% sucrose gradient followed by pelleting through a 36% sucrose cushion with the pellet then suspended in 1 mM Tris-Cl, pH 9.0. For MVA/gag vaccination, RM were administered 108 plaque-forming units of this vector via intramuscular injection. The DNA/gag + IL-12 vaccines were provided by Inovio Pharmaceuticals. Briefly, codon-optimized, 5′ and 3′ halves of the full-length SIVmac239 gag were cloned into the pVAX backbone (Invitrogen) such that the SIVgag insert expression was controlled by the human CMV (HCMV) promoter/enhancer and the bovine growth hormone polyadenylation signal. The optimized rhesus macaque IL-12 adjuvant was constructed via modification of a previously used unoptimized version of RM IL-12 (63). Modification included codon and RNA optimization of the p35 and p40 insert sequences only, which was carried out by GeneArt (Invitrogen). RM were administered 1 mg of the 2 SIVgag constructs and 0.5mg of IL-12 construct with the DNA being delivered into the quadriceps muscle followed by in vivo electroporation employing the Cellectra constant current device (Inovio Pharmaceuticals, Inc.) as previously described (64).
Antigens and Antigen-Presenting Cells
The synthesis of sequential 15-mer peptides (overlapping by 11 amino acids) comprising the SIVgag, rev, nef, tat, env, pol and RhCMV IE-1 proteins, as well as specific 9–14mer peptides within these proteins, was performed by Intavis AG, based on the SIVmac239 sequence (Genbank Accession #M33262) (65) or the strain 68-1 RhCMV IE-1 sequence (Genbank Accession #AY186194) (61). All peptides are identified by the position of their inclusive amino acids from the n-terminus (e.g., Gagxx-yy). Consecutive 15mers are also designated by their 15mer position starting from the n-terminal 15mer (e.g., Gag1-15 is 15mer #1; Gag4-19 is 15mer #2, etc.). Unless otherwise specified, these peptides were used in T cell assays used at 2μg/ml (whether alone or in pan-protein mixes). Aldrithiol-2 inactivated-SIV (AT-2-SIV; lot P4146, AIDS and Cancer Virus Program, Frederick National Laboratory, Frederick, MD) was produced as previously described (29). Autologous B-lymphoblastoid cell lines (BLCL) were generated by infecting rhesus PBMC with Herpesvirus papio (66). Autologous SIV-infected target cells were produced by spinoculation of activated CD4+ T cells with sucrose-purified SIVmac239, followed by 4 days of culture and then purification with CD4 microbeads and LS columns (Miltenyi Biotec), as previously described (67). Infected cell preparations were >95% CD4+ T cells and >50% SIV-infected following enrichment and were used at an effector:target ratio of 80:1. Construction of single Mamu-DR allomorph transfectants was performed as previously described (68), except that Mamu-DR alleles were inserted into plasmid pCEP4 (Invitrogen) rather than pcDNC3.1. Mamu-DRA*01:05 was paired with DRB1*10:07, DRB1*04:06, DRB1*03:09, DRB5*03:01, DRB*w2:01, and DRB*w26:03, while Mamu-DRA*01:021 was paired with DRB*w4:01. Prior to MHC-II restriction assays, mRNA from these transfectants was extracted using the AllPrep DNA/RNA Mini Kit (Qiagen), amplified by RT-PCR using a universal primer pair (5′-GACACTGATGGTGCTGAGC-3′ and 5′-GCTGCACTGTGAAGCTCTC-3′) spanning the highly polymorphic β1 region of Mamu-DRB, and sequence confirmed. MHC-II transfectants and BLCL were pulsed with the Gag peptide of interest at a final concentration of 5 μg/ml for 90 minutes (37°C) then washed twice with warm PBS and once with warm R10 to remove unbound peptide before being used to stimulate freshly isolated PBMC at an effector:target ratio of 10:1.
T Cell Assays
SIV- and RhCMV-specific CD4+ and CD8+ T cell responses were measured in mononuclear cell preparations from blood and tissues by flow cytometric ICS, as previously described in detail (10–12). Briefly, mononuclear cells or isolated CD8+ T cells were incubated with antigen (peptide, AT-2 SIV, peptide-pulsed BLCL or MHC-II transfectants, or SIV-infected CD4+ T cells) and the co-stimulatory molecules CD28 and CD49d (BD Biosciences) for 1 hr, followed by addition of Brefeldin A (Sigma-Aldrich) for an additional 8 hrs. Co-stimulation without antigen served as a background control. The MHC association (MHC-I vs. MHC-II) of a response was determined by pre-incubating isolated mononuclear cells or antigen-presenting cells for 1hr at room temperature in the presence of 10μg/ml of mAbs against MHC-I (clone–W6-32) vs. MHC-II (HLA-DR; clone–G46-6) or CLIP peptide (MHC-II-associated invariant chain, amino acids 89–100; 2μg/ml) prior to adding peptides or combining effector and target cells and incubating per the standard ICS assay. Stimulated cells were fixed, permeabilized and stained as previously described (10–12), and flow cytometric analysis was performed on an LSR-II instrument (BD Biosciences). Analysis was done using FlowJo software (Tree Star). In all analyses, gating on the light scatter signature of small lymphocytes was followed by progressive gating on the CD3+ population and then the CD4+/CD8− vs. CD4−/CD8+ T cell subsets. Antigen specific response frequencies for CD4+ or CD8+ T cell populations were routinely determined from intracellular expression of CD69 and either or both IFN-γ and TNF-α [in select experiments, responses were also characterized by intracellular CD69 and either IL-2 or MIP-1β production or CD107 externalization (11)]. In other select experiments, Boolean gates of (CD69+/TNF-α+ and/or CD69+/IFN-γ+) were generated and expression of CD28 and CCR7 was determined on the gated (responding) CD8+ T cell population (11). Response frequencies were reported after background subtraction and memory correction, as previously described (11, 58). For epitope deconvolution experiments, stricter response criteria were used to prevent false positives. In these studies, a response to a given 15mer peptide was considered positive if the frequency of events clustered as CD69+, TNF-α+ and IFN-γ+ was > 0.05%, with background <0.01% in at least 2 independent assays. The classification of individual peptide responses as MHC-I- vs. MHC-II-associated was based on >90% inhibition of the response by either MHC-I or MHC-II blockade relative to the isotype control. Responses that did not meet these criteria were considered indeterminate. Minimal independent epitope numbers were estimated from the positive responses identified by testing of consecutive 15mer peptides by the following criteria: single positive peptide = 1 independent epitope; 2 adjacent positive peptides = 1 independent epitope; 3 adjacent positive peptides = 2 independent epitopes; 4 adjacent positive peptides = 2 independent epitopes; and 5 adjacent positive peptides = 3 independent epitopes. These estimations of the minimal number of independent epitopes were initially conducted without the benefit of the MHC association data, but were then revised using the same criteria, applied independently for MHC-I vs. MHC-II blocked responses.
Statistics
For comparisons of independent samples, we applied bivariate Mann-Whitney U tests (69), also known as Wilcoxon Rank Sum tests. For one-sample comparisons to a fixed null-hypothesized value (such as percentages compared to 100%), we applied one-sample Wilcoxon Signed Rank tests (69). All tests were conducted as two-tailed tests with a type-I error rate of 5%. We used the R statistical computing language(70) for all statistical analyses.
Supplementary Material
Fig. S1. RhCMV/gag vector-elicited CD8+ T cells respond to homologous boosting with an increase in peripheral blood response frequencies and induction of proliferation. The CD8+ T cell response to individual SIVgag 15mer peptides was determined in two strain 68-1 RhCMV/gag-vaccinated RM (Rh27473 and Rh27517) using flow cytometric ICS (as described in Fig. 1A). The frequency of the top 10 of these epitope-specific responses in blood and the proliferative status of the responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells) were followed after re-administration of the same RhCMV/gag (308 days post initial vaccination).
Fig. S2. RhCMV/gag vectors do not boost gag-specific CD8+ T cell responses primed with electroporated DNA/gag + IL-12 vaccination. The CD8+ T cell response to individual SIVgag 15mer peptides was determined in a second RM (Rh27520) vaccinated by electroporation of a DNA/gag + IL-12 plasmid using flow cytometric ICS (results from the other RM is shown in Fig. 1E). The frequency of the top 6 epitope-specific responses in blood and the proliferative status of the peptide-responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells) were followed after boosting with strain 68-1 RhCMV/gag (308 days post initial vaccination). This figure also shows induction of 4 SIVgag 15mer-specific CD8+ T cell responses after RhCMV/gag vaccination that were not detectable in the pre-existing SIVgag-specific responses elicited by the DNA/gag + IL-12 vaccine.
Fig. S3. RhCMV/gag vectors do not boost canonical gag-specific CD8+ T cell responses in SIV+ RM on fully suppressive anti-retroviral therapy (ART). Two SIVmac251-infected Mamu-B*08+ RM (Rh25429 and Rh26557; plasma viral loads = 53,000 copies/ml and 6,700 copies/ml, respectively), were treated with combination anti-retroviral therapy consisting of 2 reverse transcriptase inhibitors (20mg/day Tenofovir, 50mg/day Emtricitabine), an integrase inhibitor (2.5 mg/day Dolutegravir) and protease inhibitor (1200 mg/day Darunavir boosted with 200mg/day Ritonavir) at 288 and 260 days post-SIV infection. Plasma viral loads were suppressed below the level of detection (30 copies/ml) within 2 weeks of ART initiation and maintained at undetectable levels for the duration of the experiment. The magnitude of CD8+ T cell responses to the Mamu-B*08-restricted canonical epitopes Nef137-146 (RL10), Nef246-254 (RL9b), Env573-581 (KL9), and Env717-725 (LF9) was determined, as well as the proliferative status of the epitope-responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells), using flow cytometric ICS, and then these parameters were followed after vaccination of these 2 RM with strain 68-1 RhCMV/gag (133 days after the initiation of ART). This figure also shows induction of 4 SIVgag 15mer-specific CD8+ T cell responses after RhCMV/gag vaccination that were not detectable in the pre-existing SIVgag-specific responses elicited by SIV infection.
Fig. S4. Truncation analysis of SIVgag 15mers targeted commonly by RhCMV/gag vector-elicited CD8+ T cells. The core epitopes of selected SIVgag 15mer peptides, designated by amino acid position (and the # of the consecutive 15mer from the amino terminus), targeted by CD8+ T cells from strain 68-1 RhCMV/gag-vaccinated RM were determined by flow cytometric ICS analysis of CD8+ T cell responses to the indicated truncated peptides. The figure shows analysis of 3 different RM per peptide, except for 15mer #s 69 and 73, for which 2 RM each are shown in Fig. 2B. The amino terminal and carboxy terminal truncations showing the highest response above that of the parent 15mer define the core epitopes (yellow). Note the striking similarity in the response pattern to each of these truncated peptides between the 3 RM tested, consistent with the same epitope accounting for the response to the parent 15mer in all animals.
Fig. S5. The recognition of SIVgag supertopes by RhCMV/gag vector-elicited CD8+ T cells can be inhibited with either MHC-I or MHC-II blockade. PBMC from strain 68-1 RhCMV/gag-vaccinated RM (n = 8 for Gag211-222, Gag276-284, Gag290-301, Gag482-490, Gag495-506; n = 5 for Gag41-52, Gag259-267) were stimulated with the designated SIVgag core epitopes (classified as Type 1 vs. Type 2 by the length of the core epitope; see Fig. 2B and fig. S4) in the presence of irrelevant isotype control mAbs (IgG1 – clone X40 + IgG2a – clone X39; 10μg/ml each), an anti-MHC-I mAb (W6-32; 10μg/ml), an anti-MHC-II mAb (G46-6; 10μg/ml), or the CLIP peptide (MHC-II-associated invariant chain, amino acids 89-100; 2μg/ml). The response frequencies were normalized to the response frequencies in the isotype control-treated cultures and the mean + SEM of these normalized response frequencies are shown for each treatment. Note that the responses to the 3 epitopes classified as Type 1 were only blocked with the anti-MHC-I mAb and the 4 epitopes classified as Type 2 were only blocked with the anti-MHC-II mAb and the CLIP peptide.
Fig. S6. Validation of transfected cell lines expressing single Mamu-DR molecules corresponding to MHC-II allomorphs expressed by 4 strain 68-1 RhCMV/gag vector-vaccinated RM. (A) RM #s 21826, 22034, 22436, and 22607 were Mamu-DRB-genotyped by Roche/454 pyrosequencing. Green highlight indicates alleles selected for MHC-II transfectant generation. (B) One Mamu-DRB allele and its paired Mamu-DRA allele were transfected into a parental (MHC-II-negative) cell line (RM3 cells). Cells were stained with a cross-reactive human HLA-DR-PE monoclonal antibody for 15 minutes at room temperature to assess Mamu-DR expression. Cells were washed once with 1X PBS supplemented with 10% fetal bovine serum, fixed with 2% paraformaldehyde, collected on an LSR-II flow cytometer, and analyzed with FlowJo. MHC-II-expressing B-lymphoblastoid cells (BLCL) served as a positive control, while the MHC-II-negative parental cell line (RM3) was used as a negative control. Note that the intensity of Mamu-DR expression on the surface of the single Mamu-DRA/B transfectants was as high or higher than that of the BLCL.
Fig. S7. Inhibition of an MHC-II transfectant-mediated CD8+ T cell response with MHC-II, but not MHC-I, blockade. Autologous B-lymphoblastoid cells (BLCL) and the DRB1*10:07 expressing transfectant were pulsed with the Gag289-303 15mer peptide (15mer #73) in the presence of irrelevant isotype control mAbs (IgG1 – clone X40 + IgG2a – clone X39; 10μg/ml each), an anti-MHC-I mAb (W6/32; 10μg/ml), an anti-MHC-II mAb (G46-6; 10μg/ml), or the CLIP peptide (MHC-II-associated invariant chain, amino acids 89-100; 2μg/ml). Note that these responses are only inhibited with MHC-II blockade.
Fig. S8. Statistical analysis of the MHC association of AT-2 SIV-specific CD8+ T cell responses in SIV+ elite controllers vs. RhCMV/SIV vector-vaccinated RM. The boxplots illustrate the comparison of MHC-I-blocked (and MHC-II-blocked) CD8+ T cell responses between elite SIV controllers and strain 68-1 RhCMV/SIV-vaccinated RM. MHC-I-blocked (and MHC-II-blocked) responses at all AT-2 SIV dilutions, represented as fractions of the isotype response at that dilution, are combined across dilutions. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant.
Fig. S9. Statistical analysis of the MHC association of SIV-infected cell-specific CD8+ T cell responses in SIV+ elite controllers vs. RhCMV/SIV vector-vaccinated RM. The boxplots illustrate the comparison of MHC-I-blocked (and MHC-II-blocked) CD8+ T cell responses between elite SIV controllers and strain 68-1 RhCMV/SIV-vaccinated RM. MHC-I-blocked (and MHC-II-blocked) are represented as fractions of the isotype response. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant.
Fig. S10. Effect of Rh182-189 gene expression on the ability of RhCMV to down-modulate surface expression of MHC-1 in infected fibroblasts. (A) The diagram illustrates the gene modifications made in strain 68-1 RhCMV to determine the role of the Rh182-189 genes (US2-11 orthologues) in the unconventional epitope targeting of SIV-specific CD8+ T cells elicited by RhCMV/SIV vectors. For the vectors lacking the whole Rh182-189 region or the Rh182-185 and Rh186-189 subregions, these regions were replaced with either SIVgag- or SIVrev/tat/nef (rtn)-coding cassettes (using the EF1α promoter) or SIVenv (using the HCMV gH promoter). For the vector lacking Rh189 alone, a strain 68-1 RhCMV/rtn vector (with EF1α–driven RTN coding cassette at a different place in the RhCMV genome) was modified by replacement of the Rh189 coding region with the SIVgag coding region, such that SIVgag expression is driven by the Rh189 promoter (creating a dual gag/rtn-expressing vector). (B) Flow cytometry was used to compare the level of MHC-I expression on rhesus fibroblasts infected with these deletant vectors, compared to the strain 68-1 wildtype vector and mock infection, 48 hours after infection. Infected cells were stained on the cell surface with an Alexafluor647-conjugated, anti-MHC-I mAb (w6/32) and then fixed and permeabilized and stained with an in-house produced RhCMV-specific mAb (biotinylated and then visualized with streptavidin-Cy7). MHC-I levels were analyzed on an LSR-II flow cytometer. Flow cytometric profiles of MHC-I expression on RhCMV antigen-expressing fibroblasts from a representative experiment are shown at left. The bar graph at right shows the average mean fluorescent intensity of MHC-I staining normalized to mock infection (+ SEM) from 4 independent experiments. Note that deletion of the entire Rh182-189 region counters all RhCMV-mediated MHC-I down-regulation, and deletion of the Rh182-185 region has a partial effect. However, deletion of Rh186-189 and of Rh189 alone (both lacking only one known MHC-I down-modulator – Rh189/US11) has little to no effect, with cell surface of MHC-I overlapping that of the wildtype strain 68-1 RhCMV.
Fig. S11. Effect of Rh182-189 gene expression on the ability of RhCMV/gag- or RhCMV/rtn-infected fibroblasts to present canonical epitopes to epitope-specific T cell lines. Rhesus fibroblasts expressing the appropriate MHC-I molecule (Mamu-A*01, Mamu-A*02, or Mamu-B*17) were infected with each of the indicated strain 68-1-derived RhCMV/gag or/rtn vectors (see fig. S10) at a multiplicity of infection of 10. After 24 hours of incubation, the frequency of infected cells in each culture was determined by intracellular staining (using either SIVgag- or RhCMV-specific mAbs), and the fibroblasts were co-cultured with CD8+ T cell lines in an IFN-γ ELISPOT assay at a 1:1 ratio of infected fibroblasts and T cells. The CD8+ T cell lines used were selected for their specificity for the following SIVmac239-derived epitopes: A) Gag181-189 CM9, B) Tat28-35 SL8, C) Nef159-167 YY9, D) Nef221-229 YY9, E) Nef165-173 IW9, or F) Nef159-203 MW9. Untreated fibroblasts and peptide-pulsed fibroblasts were used as the negative and positive controls, respectively.
Fig. S12. Statistical analysis of the MHC association of IE-1- and SIVgag-specific CD8+ T cell responses elicited by RhCMV vectors/viruses with and without Rh157.4-.6 (UL128-131). (A) The boxplots illustrate the comparison of MHC-I- and MHC-II-blocked CD8+ T cell responses to total RhCMV IE-1 15mer peptides between naturally RhCMV-infected RM and RM vaccinated with strain 68-1 RhCMV vectors (see Fig. 8B). (B) The boxplots illustrate the comparison of MHC-I- and MHC-II-blocked CD8+ T cell responses to total SIVgag 15mer peptides between RM vaccinated with the original strain 68-1 RhCMV/gag vector and the Rh157.4-.6 (UL128-131)-repaired RhCMV/gag vector (see Fig. 8E). In both sets of analyses, the MHC-I-blocked (and MHC-II-blocked) are represented as fractions of the isotype response. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant in both A and B.
Table S1. List of canonical epitopes associated with conventional SIVgag-specific CD8+ T cell responses in RM expressing the Mamu-A*01, -A*02, -B*08, and -B*17 MHC-I alleles. The table shows the canonical epitopes restricted by the indicated Mamu alleles analyzed in this study.
Table S2. Comprehensive analysis of CD8+ T cell responses to canonical epitopes in RM vaccinated with RhCMV/SIV vectors and expressing one or more of the Mamu-A*01, A*02, B*08, and B*17 MHC-I alleles. The table shows all of the RhCMV/SIV-vaccinated RM in which CD8+ T cell responses to canonical epitopes restricted by the indicated Mamu alleles were assayed by flow cytometric ICS. No above-background response was identified to any of these canonical epitopes in any “wildtype” strain 68-1 RhCMV/SIV vector-vaccinated RM that expressed the appropriate restricting Mamu allele. BT = Below Threshold
Table S3. Comprehensive analysis of the Mamu DR allomorph specificity of RhCMV/gag-elicited MHC-II-restricted, CD8+ T cell responses in 4 RM. PBMC from the 4 indicated RM (see also fig. S6) were incubated with autologous B-lymphoblastoid cells (BLCL), MHC-II-null RM3, or the indicated single Mamu-DR allomorph transfectants pulsed with the indicated SIVgag peptides and were then analyzed for CD8+ T cell responses by flow cytometric ICS (see Fig. 4). Combinations that resulted in CD8+ T cell responses above background (no peptide) are indicated by + signs (blue boxes); combinations that did not result in CD8+ T cell responses above background are indicated by - signs (open boxes). Mamu-DR alleles that are expressed in each RM are indicated in red; non-expressed alleles are shown in white. Note that CD8+ T cells recognizing a given peptide in the context of an autologous (expressed) Mamu-DR molecule can also respond to that peptide in the context of an allogeneic Mamu-DR molecule, as long as that molecule is also capable of presenting that particular peptide in RM expressing that allele.
Acknowledgments
We are grateful to M. Morrow, A. Khan, K. Broderick, and D. Weiner for providing and administering the electroporated DNA/gag + IL-12 vaccines, G. Spies and B. Moss for the MVA/gag vector, M. Chiuchiolo and C. Parks for the Ad5/gag vector, P. Barry for the original GFP-expressing RhCMV 68-1 virus and the RhCMV 68-1 BAC, and T. Shenk for providing the repaired RhCMV 68-1.2 BAC. We also gratefully acknowledge D. Watkins for MHC-I typing, D. O’Connor and R. Wiseman for MHC-II typing, and R. Geleziunas, J. Hesselgesser and Gilead Science, Inc. for providing anti-retroviral drugs. We thank A. Townsend for help with the graphics. We thank J. Hines, A. Bhusari, R. Derrah, D. Duell, D. Morrow, E. McDonald, C. Kahl, S. Hagen, M. Jarvis, A. Sylwester, and L. Boshears for technical or administrative assistance. The data presented in this paper are tabulated in the main paper and the supplementary materials. This research was supported by the National Institutes of Health (P01 AI094417, RO1 AI060392, RO1 AI059457, P51 OD 011092 and contract #HHSN261200800001E), the Bill and Melinda Gates Foundation (Collaboration for AIDS Vaccine Discovery Program), and the International AIDS Vaccine Initiative (IAVI). LJP, KF, SGH have applied for a patent entitled “CMV Glycoproteins and Recombinant Vectors” (PCT/US2011/029930) and have filed a provisional patent application entitled “Cytomegalovirus Vectors Enabling Control of T Cell Targeting” (application # 61772962). OHSU and Drs. Picker, Hansen, Früh and Nelson have a significant financial interest in TomegaVax, Inc., a company that may have a commercial interest in the results of this research and technology. The potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services or NIH, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES AND NOTES
- 1.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11:823. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 2.Yewdell JW. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity. 2006;25:533. doi: 10.1016/j.immuni.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Irvine K, Bennink J. Factors influencing immunodominance hierarchies in TCD8+-mediated antiviral responses. Expert Rev Clin Immunol. 2006;2:135. doi: 10.1586/1744666X.2.1.135. [DOI] [PubMed] [Google Scholar]
- 4.Assarsson E, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J Immunol. 2007;178:7890. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 5.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picker LJ, Hansen SG, Lifson JD. New paradigms for HIV/AIDS vaccine development. Annu Rev Med. 2012;63:95. doi: 10.1146/annurev-med-042010-085643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol. 2010;10:11. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barouch DH, et al. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J Virol. 2003;77:7367. doi: 10.1128/JVI.77.13.7367-7375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd PA, et al. Vaccine-induced CD8+ T cells control AIDS virus replication. Nature. 2012;491:129. doi: 10.1038/nature11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen SG, et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science. 2010;328:102. doi: 10.1126/science.1185350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rammensee HG, Falk K, Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 14.Southwood S, et al. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363. [PubMed] [Google Scholar]
- 15.Chelvanayagam G. A roadmap for HLA-DR peptide binding specificities. Hum Immunol. 1997;58:61. doi: 10.1016/s0198-8859(97)00185-7. [DOI] [PubMed] [Google Scholar]
- 16.Mizuochi T, Tentori L, Sharrow SO, Kruisbeek AM, Singer A. Differentiation of Ia-reactive CD8+ murine T cells does not require Ia engagement. Implications for the role of CD4 and CD8 accessory molecules in T cell differentiation. J Exp Med. 1988;168:437. doi: 10.1084/jem.168.1.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki H, et al. Origin of a T cell clone with a mismatched combination of MHC restriction and coreceptor expression. J Immunol. 1994;153:4496. [PubMed] [Google Scholar]
- 18.Matechak EO, Killeen N, Hedrick SM, Fowlkes BJ. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 1996;4:337. doi: 10.1016/s1074-7613(00)80247-2. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu T, Takeda S. CD8 T cells from major histocompatibility complex class II-deficient mice respond vigorously to class II molecules in a primary mixed lymphocyte reaction. Eur J Immunol. 1997;27:500. doi: 10.1002/eji.1830270222. [DOI] [PubMed] [Google Scholar]
- 20.Tyznik AJ, Sun JC, Bevan MJ. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II-restricted T cells. J Exp Med. 2004;199:559. doi: 10.1084/jem.20031961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce EL, Shedlock DJ, Shen H. Functional characterization of MHC class II-restricted CD8+CD4− and CD8-CD4− T cell responses to infection in CD4−/− mice. J Immunol. 2004;173:2494. doi: 10.4049/jimmunol.173.4.2494. [DOI] [PubMed] [Google Scholar]
- 22.Heemskerk MH, et al. Dual HLA class I and class II restricted recognition of alloreactive T lymphocytes mediated by a single T cell receptor complex. Proc Natl Acad Sci USA. 2001;98:6806. doi: 10.1073/pnas.111162298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rist M, Smith C, Bell MJ, Burrows SR, Khanna R. Cross-recognition of HLA DR4 alloantigen by virus-specific CD8+ T cells: a new paradigm for self-/nonself-recognition. Blood. 2009;114:2244. doi: 10.1182/blood-2009-05-222596. [DOI] [PubMed] [Google Scholar]
- 24.Hirosawa T, et al. Mismatched human leukocyte antigen class II-restricted CD8(+) cytotoxic T cells may mediate selective graft-versus-leukemia effects following allogeneic hematopoietic cell transplantation. Cancer Sci. 2011;102:1281. doi: 10.1111/j.1349-7006.2011.01949.x. [DOI] [PubMed] [Google Scholar]
- 25.Viola A, et al. Quantitative contribution of CD4 and CD8 to T cell antigen receptor serial triggering. J Exp Med. 1997;186:1775. doi: 10.1084/jem.186.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lustgarten J, Waks T, Eshhar Z. CD4 and CD8 accessory molecules function through interactions with major histocompatibility complex molecules which are not directly associated with the T cell receptor-antigen complex. Eur J Immunol. 1991;21:2507. doi: 10.1002/eji.1830211030. [DOI] [PubMed] [Google Scholar]
- 27.Sette A, Southwood S, Miller J, Appella E. Binding of major histocompatibility complex class II to the invariant chain-derived peptide, CLIP, is regulated by allelic polymorphism in class II. J Exp Med. 1995;181:677. doi: 10.1084/jem.181.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corradin C, Lanzavecchia A. Chemical and functional analysis of MHC class II-restricted T cell epitopes. Int Rev Immunol. 1991;7:139. doi: 10.3109/08830189109061771. [DOI] [PubMed] [Google Scholar]
- 29.Buseyne F, et al. MHC-I-restricted presentation of HIV-1 virion antigens without viral replication. Nat Med. 2001;7:344. doi: 10.1038/85493. [DOI] [PubMed] [Google Scholar]
- 30.Powers C, DeFilippis D, Malouli D, Fruh K. In: Human Cytomegalovirus. Shenk TE, Stinski MF, editors. Springer-Verlag; Berlin Heidelberg: 2008. pp. 333–359. [Google Scholar]
- 31.Yewdell JW, Hill AB. Viral interference with antigen presentation. Nat Immunol. 2002;3:1019. doi: 10.1038/ni1102-1019. [DOI] [PubMed] [Google Scholar]
- 32.Pande NT, Powers C, Ahn K, Fruh K. Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J Virol. 2005;79:5786. doi: 10.1128/JVI.79.9.5786-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen TM, et al. CD8(+) lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J Virol. 2001;75:738. doi: 10.1128/JVI.75.2.738-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loffredo JT, et al. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82:1723. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malouli D, et al. Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J Virol. 2012;86:8959. doi: 10.1128/JVI.01132-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilja AE, Shenk T. Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci USA. 2008;105:19950. doi: 10.1073/pnas.0811063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powers C, DeFilippis V, Malouli D, Fruh K. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol. 2008;325:333. doi: 10.1007/978-3-540-77349-8_19. [DOI] [PubMed] [Google Scholar]
- 38.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Britt WJ. In: Human Cytomegalovirus. Shenk TE, Stinski MF, editors. Springer-Verlag; Berlin Heidelberg: 2008. pp. 333–359. [Google Scholar]
- 40.Busche A, et al. Priming of CD8+ T Cells against Cytomegalovirus Encoded Antigens Is Dominated by Cross Presentation. J Immunol. 2013;190:2767. doi: 10.4049/jimmunol.1200966. [DOI] [PubMed] [Google Scholar]
- 41.Gainey MD, Rivenbark JG, Cho H, Yang L, Yokoyama WM. Viral MHC class I inhibition evades CD8+ T-cell effector responses in vivo but not CD8+ T-cell priming. Proc Natl Acad Sci USA. 2012;109:E3260. doi: 10.1073/pnas.1217111109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munks MW, Pinto AK, Doom CM, Hill AB. Viral interference with antigen presentation does not alter acute or chronic CD8 T cell immunodominance in murine cytomegalovirus infection. J Immunol. 2007;178:7235. doi: 10.4049/jimmunol.178.11.7235. [DOI] [PubMed] [Google Scholar]
- 43.Snyder CM, Allan JE, Bonnett EL, Doom CM, Hill AB. Cross-presentation of a spread-defective MCMV is sufficient to prime the majority of virus-specific CD8+ T cells. PLoS One. 2010;5:e9681. doi: 10.1371/journal.pone.0009681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masopust D, Picker LJ. Hidden memories: frontline memory T cells and early pathogen interception. J Immunol. 2012;188:5811. doi: 10.4049/jimmunol.1102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riddell SR, Rabin M, Geballe AP, Britt WJ, Greenberg PD. Class I MHC-restricted cytotoxic T lymphocyte recognition of cells infected with human cytomegalovirus does not require endogenous viral gene expression. J Immunol. 1991;146:2795. [PubMed] [Google Scholar]
- 46.Manley TJ, et al. Immune evasion proteins of human cytomegalovirus do not prevent a diverse CD8+ cytotoxic T-cell response in natural infection. Blood. 2004;104:1075. doi: 10.1182/blood-2003-06-1937. [DOI] [PubMed] [Google Scholar]
- 47.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 48.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci USA. 2005;102:18153. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79:10330. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao H, Tao R, Zheng Q, Xu J, Shang S. Recombinant HCMV UL128 expression and functional identification of PBMC-attracting activity in vitro. Arch Virol. 2013;158:173. doi: 10.1007/s00705-012-1378-8. [DOI] [PubMed] [Google Scholar]
- 51.Greenbaum J, et al. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rao X, Hoof I, Costa AI, van Baarle D, Kesmir C. HLA class I allele promiscuity revisited. Immunogenetics. 2011;63:691. doi: 10.1007/s00251-011-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander J, et al. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 54.Perez CL, et al. Broadly immunogenic HLA class I supertype-restricted elite CTL epitopes recognized in a diverse population infected with different HIV-1 subtypes. J Immunol. 2008;180:5092. doi: 10.4049/jimmunol.180.7.5092. [DOI] [PubMed] [Google Scholar]
- 55.Hofstetter AR, Sullivan LC, Lukacher AE, Brooks AG. Diverse roles of non-diverse molecules: MHC class Ib molecules in host defense and control of autoimmunity. Curr Opin Immunol. 2011;23:104. doi: 10.1016/j.coi.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crawford H, et al. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J Exp Med. 2009;206:909. doi: 10.1084/jem.20081984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loffredo JT, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pitcher CJ, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 59.Veazey RS, et al. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 60.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 61.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW. Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol. 2003;77:6620. doi: 10.1128/JVI.77.12.6620-6636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oxford KL, et al. Protein coding content of the ULb′ region of wild-type rhesus cytomegalovirus. Virology. 2008;373:181. doi: 10.1016/j.virol.2007.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Egan MA, et al. Priming with plasmid DNAs expressing interleukin-12 and simian immunodeficiency virus gag enhances the immunogenicity and efficacy of an experimental AIDS vaccine based on recombinant vesicular stomatitis virus. AIDS Res Hum Retroviruses. 2005;21:629. doi: 10.1089/aid.2005.21.629. [DOI] [PubMed] [Google Scholar]
- 64.Laddy DJ, et al. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. J Virol. 2009;83:4624. doi: 10.1128/JVI.02335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kestler H, et al. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 66.Voss G, Nick S, Stahl-Hennig C, Ritter K, Hunsmann G. Generation of macaque B lymphoblastoid cell lines with simian Epstein-Barr-like viruses: transformation procedure, characterization of the cell lines and occurrence of simian foamy virus. J Virol Methods. 1992;39:185. doi: 10.1016/0166-0934(92)90137-3. [DOI] [PubMed] [Google Scholar]
- 67.Sacha JB, et al. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol. 2007;178:2746. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Giraldo-Vela JP, et al. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;82:859. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolfe DA, Hollander M. Nonparametric Statistical Methods. Wiley; New York: 1973. [Google Scholar]
- 70.R Development Core Team. R, A Language and Environment for Statistical Computing. 2011 http://www.R-project.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. RhCMV/gag vector-elicited CD8+ T cells respond to homologous boosting with an increase in peripheral blood response frequencies and induction of proliferation. The CD8+ T cell response to individual SIVgag 15mer peptides was determined in two strain 68-1 RhCMV/gag-vaccinated RM (Rh27473 and Rh27517) using flow cytometric ICS (as described in Fig. 1A). The frequency of the top 10 of these epitope-specific responses in blood and the proliferative status of the responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells) were followed after re-administration of the same RhCMV/gag (308 days post initial vaccination).
Fig. S2. RhCMV/gag vectors do not boost gag-specific CD8+ T cell responses primed with electroporated DNA/gag + IL-12 vaccination. The CD8+ T cell response to individual SIVgag 15mer peptides was determined in a second RM (Rh27520) vaccinated by electroporation of a DNA/gag + IL-12 plasmid using flow cytometric ICS (results from the other RM is shown in Fig. 1E). The frequency of the top 6 epitope-specific responses in blood and the proliferative status of the peptide-responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells) were followed after boosting with strain 68-1 RhCMV/gag (308 days post initial vaccination). This figure also shows induction of 4 SIVgag 15mer-specific CD8+ T cell responses after RhCMV/gag vaccination that were not detectable in the pre-existing SIVgag-specific responses elicited by the DNA/gag + IL-12 vaccine.
Fig. S3. RhCMV/gag vectors do not boost canonical gag-specific CD8+ T cell responses in SIV+ RM on fully suppressive anti-retroviral therapy (ART). Two SIVmac251-infected Mamu-B*08+ RM (Rh25429 and Rh26557; plasma viral loads = 53,000 copies/ml and 6,700 copies/ml, respectively), were treated with combination anti-retroviral therapy consisting of 2 reverse transcriptase inhibitors (20mg/day Tenofovir, 50mg/day Emtricitabine), an integrase inhibitor (2.5 mg/day Dolutegravir) and protease inhibitor (1200 mg/day Darunavir boosted with 200mg/day Ritonavir) at 288 and 260 days post-SIV infection. Plasma viral loads were suppressed below the level of detection (30 copies/ml) within 2 weeks of ART initiation and maintained at undetectable levels for the duration of the experiment. The magnitude of CD8+ T cell responses to the Mamu-B*08-restricted canonical epitopes Nef137-146 (RL10), Nef246-254 (RL9b), Env573-581 (KL9), and Env717-725 (LF9) was determined, as well as the proliferative status of the epitope-responding cells (as measured by Ki-67 expression on the TNF-α + and/or IFN-γ + cells), using flow cytometric ICS, and then these parameters were followed after vaccination of these 2 RM with strain 68-1 RhCMV/gag (133 days after the initiation of ART). This figure also shows induction of 4 SIVgag 15mer-specific CD8+ T cell responses after RhCMV/gag vaccination that were not detectable in the pre-existing SIVgag-specific responses elicited by SIV infection.
Fig. S4. Truncation analysis of SIVgag 15mers targeted commonly by RhCMV/gag vector-elicited CD8+ T cells. The core epitopes of selected SIVgag 15mer peptides, designated by amino acid position (and the # of the consecutive 15mer from the amino terminus), targeted by CD8+ T cells from strain 68-1 RhCMV/gag-vaccinated RM were determined by flow cytometric ICS analysis of CD8+ T cell responses to the indicated truncated peptides. The figure shows analysis of 3 different RM per peptide, except for 15mer #s 69 and 73, for which 2 RM each are shown in Fig. 2B. The amino terminal and carboxy terminal truncations showing the highest response above that of the parent 15mer define the core epitopes (yellow). Note the striking similarity in the response pattern to each of these truncated peptides between the 3 RM tested, consistent with the same epitope accounting for the response to the parent 15mer in all animals.
Fig. S5. The recognition of SIVgag supertopes by RhCMV/gag vector-elicited CD8+ T cells can be inhibited with either MHC-I or MHC-II blockade. PBMC from strain 68-1 RhCMV/gag-vaccinated RM (n = 8 for Gag211-222, Gag276-284, Gag290-301, Gag482-490, Gag495-506; n = 5 for Gag41-52, Gag259-267) were stimulated with the designated SIVgag core epitopes (classified as Type 1 vs. Type 2 by the length of the core epitope; see Fig. 2B and fig. S4) in the presence of irrelevant isotype control mAbs (IgG1 – clone X40 + IgG2a – clone X39; 10μg/ml each), an anti-MHC-I mAb (W6-32; 10μg/ml), an anti-MHC-II mAb (G46-6; 10μg/ml), or the CLIP peptide (MHC-II-associated invariant chain, amino acids 89-100; 2μg/ml). The response frequencies were normalized to the response frequencies in the isotype control-treated cultures and the mean + SEM of these normalized response frequencies are shown for each treatment. Note that the responses to the 3 epitopes classified as Type 1 were only blocked with the anti-MHC-I mAb and the 4 epitopes classified as Type 2 were only blocked with the anti-MHC-II mAb and the CLIP peptide.
Fig. S6. Validation of transfected cell lines expressing single Mamu-DR molecules corresponding to MHC-II allomorphs expressed by 4 strain 68-1 RhCMV/gag vector-vaccinated RM. (A) RM #s 21826, 22034, 22436, and 22607 were Mamu-DRB-genotyped by Roche/454 pyrosequencing. Green highlight indicates alleles selected for MHC-II transfectant generation. (B) One Mamu-DRB allele and its paired Mamu-DRA allele were transfected into a parental (MHC-II-negative) cell line (RM3 cells). Cells were stained with a cross-reactive human HLA-DR-PE monoclonal antibody for 15 minutes at room temperature to assess Mamu-DR expression. Cells were washed once with 1X PBS supplemented with 10% fetal bovine serum, fixed with 2% paraformaldehyde, collected on an LSR-II flow cytometer, and analyzed with FlowJo. MHC-II-expressing B-lymphoblastoid cells (BLCL) served as a positive control, while the MHC-II-negative parental cell line (RM3) was used as a negative control. Note that the intensity of Mamu-DR expression on the surface of the single Mamu-DRA/B transfectants was as high or higher than that of the BLCL.
Fig. S7. Inhibition of an MHC-II transfectant-mediated CD8+ T cell response with MHC-II, but not MHC-I, blockade. Autologous B-lymphoblastoid cells (BLCL) and the DRB1*10:07 expressing transfectant were pulsed with the Gag289-303 15mer peptide (15mer #73) in the presence of irrelevant isotype control mAbs (IgG1 – clone X40 + IgG2a – clone X39; 10μg/ml each), an anti-MHC-I mAb (W6/32; 10μg/ml), an anti-MHC-II mAb (G46-6; 10μg/ml), or the CLIP peptide (MHC-II-associated invariant chain, amino acids 89-100; 2μg/ml). Note that these responses are only inhibited with MHC-II blockade.
Fig. S8. Statistical analysis of the MHC association of AT-2 SIV-specific CD8+ T cell responses in SIV+ elite controllers vs. RhCMV/SIV vector-vaccinated RM. The boxplots illustrate the comparison of MHC-I-blocked (and MHC-II-blocked) CD8+ T cell responses between elite SIV controllers and strain 68-1 RhCMV/SIV-vaccinated RM. MHC-I-blocked (and MHC-II-blocked) responses at all AT-2 SIV dilutions, represented as fractions of the isotype response at that dilution, are combined across dilutions. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant.
Fig. S9. Statistical analysis of the MHC association of SIV-infected cell-specific CD8+ T cell responses in SIV+ elite controllers vs. RhCMV/SIV vector-vaccinated RM. The boxplots illustrate the comparison of MHC-I-blocked (and MHC-II-blocked) CD8+ T cell responses between elite SIV controllers and strain 68-1 RhCMV/SIV-vaccinated RM. MHC-I-blocked (and MHC-II-blocked) are represented as fractions of the isotype response. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant.
Fig. S10. Effect of Rh182-189 gene expression on the ability of RhCMV to down-modulate surface expression of MHC-1 in infected fibroblasts. (A) The diagram illustrates the gene modifications made in strain 68-1 RhCMV to determine the role of the Rh182-189 genes (US2-11 orthologues) in the unconventional epitope targeting of SIV-specific CD8+ T cells elicited by RhCMV/SIV vectors. For the vectors lacking the whole Rh182-189 region or the Rh182-185 and Rh186-189 subregions, these regions were replaced with either SIVgag- or SIVrev/tat/nef (rtn)-coding cassettes (using the EF1α promoter) or SIVenv (using the HCMV gH promoter). For the vector lacking Rh189 alone, a strain 68-1 RhCMV/rtn vector (with EF1α–driven RTN coding cassette at a different place in the RhCMV genome) was modified by replacement of the Rh189 coding region with the SIVgag coding region, such that SIVgag expression is driven by the Rh189 promoter (creating a dual gag/rtn-expressing vector). (B) Flow cytometry was used to compare the level of MHC-I expression on rhesus fibroblasts infected with these deletant vectors, compared to the strain 68-1 wildtype vector and mock infection, 48 hours after infection. Infected cells were stained on the cell surface with an Alexafluor647-conjugated, anti-MHC-I mAb (w6/32) and then fixed and permeabilized and stained with an in-house produced RhCMV-specific mAb (biotinylated and then visualized with streptavidin-Cy7). MHC-I levels were analyzed on an LSR-II flow cytometer. Flow cytometric profiles of MHC-I expression on RhCMV antigen-expressing fibroblasts from a representative experiment are shown at left. The bar graph at right shows the average mean fluorescent intensity of MHC-I staining normalized to mock infection (+ SEM) from 4 independent experiments. Note that deletion of the entire Rh182-189 region counters all RhCMV-mediated MHC-I down-regulation, and deletion of the Rh182-185 region has a partial effect. However, deletion of Rh186-189 and of Rh189 alone (both lacking only one known MHC-I down-modulator – Rh189/US11) has little to no effect, with cell surface of MHC-I overlapping that of the wildtype strain 68-1 RhCMV.
Fig. S11. Effect of Rh182-189 gene expression on the ability of RhCMV/gag- or RhCMV/rtn-infected fibroblasts to present canonical epitopes to epitope-specific T cell lines. Rhesus fibroblasts expressing the appropriate MHC-I molecule (Mamu-A*01, Mamu-A*02, or Mamu-B*17) were infected with each of the indicated strain 68-1-derived RhCMV/gag or/rtn vectors (see fig. S10) at a multiplicity of infection of 10. After 24 hours of incubation, the frequency of infected cells in each culture was determined by intracellular staining (using either SIVgag- or RhCMV-specific mAbs), and the fibroblasts were co-cultured with CD8+ T cell lines in an IFN-γ ELISPOT assay at a 1:1 ratio of infected fibroblasts and T cells. The CD8+ T cell lines used were selected for their specificity for the following SIVmac239-derived epitopes: A) Gag181-189 CM9, B) Tat28-35 SL8, C) Nef159-167 YY9, D) Nef221-229 YY9, E) Nef165-173 IW9, or F) Nef159-203 MW9. Untreated fibroblasts and peptide-pulsed fibroblasts were used as the negative and positive controls, respectively.
Fig. S12. Statistical analysis of the MHC association of IE-1- and SIVgag-specific CD8+ T cell responses elicited by RhCMV vectors/viruses with and without Rh157.4-.6 (UL128-131). (A) The boxplots illustrate the comparison of MHC-I- and MHC-II-blocked CD8+ T cell responses to total RhCMV IE-1 15mer peptides between naturally RhCMV-infected RM and RM vaccinated with strain 68-1 RhCMV vectors (see Fig. 8B). (B) The boxplots illustrate the comparison of MHC-I- and MHC-II-blocked CD8+ T cell responses to total SIVgag 15mer peptides between RM vaccinated with the original strain 68-1 RhCMV/gag vector and the Rh157.4-.6 (UL128-131)-repaired RhCMV/gag vector (see Fig. 8E). In both sets of analyses, the MHC-I-blocked (and MHC-II-blocked) are represented as fractions of the isotype response. The depicted p-values, from two-tailed Wilcoxon rank-sum tests, indicate that the differences for both MHC-I-blocked and MHC-II-blocked CD8+ T cell response fractions are statistically significant in both A and B.
Table S1. List of canonical epitopes associated with conventional SIVgag-specific CD8+ T cell responses in RM expressing the Mamu-A*01, -A*02, -B*08, and -B*17 MHC-I alleles. The table shows the canonical epitopes restricted by the indicated Mamu alleles analyzed in this study.
Table S2. Comprehensive analysis of CD8+ T cell responses to canonical epitopes in RM vaccinated with RhCMV/SIV vectors and expressing one or more of the Mamu-A*01, A*02, B*08, and B*17 MHC-I alleles. The table shows all of the RhCMV/SIV-vaccinated RM in which CD8+ T cell responses to canonical epitopes restricted by the indicated Mamu alleles were assayed by flow cytometric ICS. No above-background response was identified to any of these canonical epitopes in any “wildtype” strain 68-1 RhCMV/SIV vector-vaccinated RM that expressed the appropriate restricting Mamu allele. BT = Below Threshold
Table S3. Comprehensive analysis of the Mamu DR allomorph specificity of RhCMV/gag-elicited MHC-II-restricted, CD8+ T cell responses in 4 RM. PBMC from the 4 indicated RM (see also fig. S6) were incubated with autologous B-lymphoblastoid cells (BLCL), MHC-II-null RM3, or the indicated single Mamu-DR allomorph transfectants pulsed with the indicated SIVgag peptides and were then analyzed for CD8+ T cell responses by flow cytometric ICS (see Fig. 4). Combinations that resulted in CD8+ T cell responses above background (no peptide) are indicated by + signs (blue boxes); combinations that did not result in CD8+ T cell responses above background are indicated by - signs (open boxes). Mamu-DR alleles that are expressed in each RM are indicated in red; non-expressed alleles are shown in white. Note that CD8+ T cells recognizing a given peptide in the context of an autologous (expressed) Mamu-DR molecule can also respond to that peptide in the context of an allogeneic Mamu-DR molecule, as long as that molecule is also capable of presenting that particular peptide in RM expressing that allele.