Abstract
Background
Accurate assessment of CYP2D6 phenotypes from genotype is inadequate in patients taking CYP2D6 substrate together with CYP2D6 inhibitors. We propose a novel CYP2D6 scoring systems that incorporates the impact of concomitant medications with the genotype in calculating the CYP2D6 activity score.
Method
Training (n=159) and validation (n=81) data sets were obtained from a prospective cohort tamoxifen pharmacogenetics registry. Two inhibitor factors were defined: one genotype-independent and one genotype-based. Three CYP2D6 gene score systems, and their combination with the inhibitor factors, were compared. These three scores were based on Zineh, Zanger, and Gaedigk's approaches. Endoxifen/NDM-Tam plasma ratio was used as the phenotype.
Results
The overall performance of the three gene score systems without consideration of CYP2D6 inhibiting medications in predicting CYP2D6 phenotype was poor both in the training (R2=0.24, 0.22 and 0.18) and the validation set, (R2=0.30, 0.24 and 0.15). Once the CYP2D6 genotype-independent inhibitor factor was integrated into the score calculation, the R2 values in the training and validation data sets are nearly twice as high as the genotype only scoring model: (0.44, 0.43, 0.38) and (0.53, 0.50, 0.41) respectively.
Conclusion
The integration of the inhibitory effect of concomitant medications with the CYP2D6 genotype into the composite CYP2D6 Activity Score doubled our ability to predict the CYP2D6 phenotype. However, endoxifen phenotypes still varied substantially, even with incorporation of CYD2D6 genotype and inhibiting factors, suggesting that other, yet unidentified, factors must be involved in tamoxifen activation.
Keywords: activity score, CYP2D6, inhibition, tamoxifen
Introduction
Cytochrome P450 2D6 (CYP2D6) is involved in the metabolism of approximately 20% of all drugs in clinical use, including β-blockers, antiarrhytmics, antidepressants and antipsychotics (1;2). It is also responsible for the metabolic activation of tamoxifen and opioids like codeine, hydrocodone and oxycodone (3;4).
The metabolic activity of CYP2D6 varies up to 1000-fold among individuals and ethnic population (5-7). The genetic contribution to this variability in CYP2D6 activity has been well documented (8). CYP2D6 is a highly polymorphic gene with more than 70 allelic variants identified (http://www.cypalleles.ki.se/), which encode for proteins with full (CYP2D6*1, *2, *33, *35), reduced (CYP2D6 *9, *10, *17, *29, *37, *41, *45, *46) or no enzymatic activity (CYP2D6*3-*8, *11-*16, *18-*21, *38, *40, *42, *44) (9). Multiple copies of CYP2D6 alleles (i.e., *1, *2, *35) have been reported in subjects with the ultra-rapid metabolism (UM) phenotype (10;11). This large inter-individual variability in CYP2D6 capacity has important clinical consequences (1;4). The poor metabolism (PM) status has repeatedly been associated with an increased risk of adverse effects in patients taking drugs whose metabolism is largely dependent on CYP2D6 (12-17). PMs also show decreased efficacy when treated with drugs that require metabolic activation by CYP2D6 (16;18). On the other hand, UMs have been reported to have a poor therapeutic response to certain CYP2D6 substrates (13;19-21) or increased opiates toxicity including fatalities (22-26). Furthermore, the cost of treating patients with extremes in CYP2D6 activity (PMs and UMs) has been estimated to be $4,000 to $6,000 per year greater than the cost of treating patients in the extensive metabolism (EM) and intermediate metabolism (IM) groups (27).
In addition, CYP2D6 activity can be affected by non-genetic factors. For example, many commonly prescribed medications are known to inhibit the catalytic activity of CYP2D6 (28). Pregnancy and disease conditions also affect CYP2D6 activity. As with CYP2D6 genetic variations, drug interactions may have important clinical consequences as shown recently for tamoxifen by our group (18;29;30).
A growing body of evidence is now available showing that inter-subject variability of CYP2D6 caused by genetic and non-genetic factors is indeed a determinant of response to some of the drugs primarily metabolized by CYP2D6. Despite this, it is still difficult to integrate such information to the drug development process and to guide decisions in clinical practice. It follows that precise knowledge of the CYP2D6 metabolic status might aid to design appropriate trials or identify a subgroup of patients that are at high risk for failure of therapy or adverse effects during initiation of therapy with CYP2D6 substrates. Traditionally, different groups of phenotypes have been used to describe range of CYP2D6 activity (e.g. PMs, EMs) and these phenotypes could be measured through administration of a probe drug or predicted from genotypes. However, given the complexity of the CYP2D6 gene structure, large numbers of unique CYP2D6 diplotypes with uncertain phenotype outcome have been identified (30-32). Accurate assessment of CYP2D6 phenotypes from genotype is inadequate in patients taking a CYP2D6 substrate together with inhibitors of the enzyme. Because of these limitations, researchers have now recognized the need to improve traditional ways of assessing CYP2D6 metabolic status. Several such approaches have been proposed recently, among them the genotype-phenotype relationship described by Zanger et al (8) the CYP2D6 activity score system proposed by Zineh et al (33) and Gaedigk et al (34). There are distinct advantages of using an activity score for grouping CYP2D6 genotypes. In the research arena, it permits combinations of many different diplotypes into a few activity scores, therefore decreasing the number of groups for comparison in genotype-phenotype association studies. In the clinical practice, clinical laboratories can easily calculate a CYP2D6 activity score, thus simplifying the interpretation of the genotype results. However, these approaches have not incorporated the impact of concomitant medications in calculating the activity score. The impact of the inhibition on CYP2D6 activity varies with the CYP2D6 genotype and the potency of the CYP2D6 inhibitor (weak, moderate and strong inhibition) (29;30).
Our prior work (29;30;35) has demonstrated that the conversion of tamoxifen to its active metabolite, endoxifen, is dependent on CYP2D6 activity. The purpose of this study is to utilize the rich genotype, concomitant medication and phenotype data from a prospective tamoxifen pharmacogenetics cohort study to: 1) develop a CYP2D6 scoring system that accounts for CYP2D6 genotype and CYP2D6-mediated drug interactions; 2) compare the performance of our scoring method with other previously published methods.
Methods
Data on medication history, genotype for 29 CYP2D6 alleles and plasma concentrations of tamoxifen and its metabolites were obtained from 159 women who participated in our tamoxifen pharmacogenetics trial published elsewhere (30). This sample was used as the training data set for this work. A second group of 81 patients who were enrolled at a later stage in the same study was used as a validation dataset. In the referred study on tamoxifen pharmacogenetics the endoxifen/N-desmethyltamoxifen (endoxifen/NDM) plasma ratio reflected better the CYP2D6 activity than the endoxifen plasma concentration alone. In the current study, we utilized the endoxifen/NDM plasma ratio as an index measure of CYP2D6 metabolic capacity.
CYP2D6 Gene Score
Based on the available phenotypic data, CYP2D6 alleles were assigned a value that reflects the expected activity of the CYP2D6 enzyme for which they code. In table 1, fully functional CYP2D6 alleles (e.g. *1, *2, *33, *35) were assigned a score of 1. CYP2D6 alleles associated with reduced enzyme activity (e.g. *9,*10, *17, *29, *37, *41, *45, *46) were scored as 0.5. The null CYP2D6 alleles (e.g.*3-*8, *11-*16, *18-*20, *38, *40, *42, *44, *56) and their duplications received a value of 0. Duplications of fully active alleles (e.g. *1×N, *2×N, *35×N) were assigned a value +1, rendering a score of 2 for each of these duplicated variants. Duplicated reduced activity alleles (e.g. *41×N and *45×N) were assigned a value of +0.5, yielding a score of 1. This proposed gene score is similar to that described by Gaedigk's et al (34).
Table 1.
CYP2D6 star allele score definition.
| CYP2D6 Alleles | Function | System 1 (Zineh et al.) | System 2 (Zanger et al.) | Proposed Gene Score System |
|---|---|---|---|---|
| *3-*8, *11-*16, *18-*20, *36, *38, *40, *42, *44, *56, *4×N | PM | 0 | 0 | 0 |
| *10, *17, *37 | IM | 0.5 | 1 | 0.5 |
| *9, *29, *41, *45, *46 | IM | 0.75 | 1 | 0.5 |
| *1, *2, *33, *35 | EM | 1 | 2 | 1 |
| *41×N, *45×N | UM | 1.5 | 3 | 1 |
| *1×N, *2×N, *35×N | UM | 2 | 3 | 2 |
Note: proposed gene score system is similar to Gaedigk's system (34). The addition is assigning *41×N as 1.
The system 1 (Table 1) is defined by Zineh's notions (33): PM = 0, IM (*10, *17, *37) = 0.5, IM (a*9, *29, *41, *45, *46) = 0.75, EM = 1, UM (*41×N, *45×N) = 1.5, and UM (*1×N, *2×N, *35×N) = 2. System 2 (Table 1) is defined by Zanger's approach (8): PM = 0, IM = 1, EM=2, and UM = 3. The score of a bi-allelic CYP2D6 genotype is defined as:
| [1] |
CYP2D6 Inhibitor Factor
Once we calculated the expected activity of the CYP2D6 enzyme according to the genotype, we incorporated the possible effects of known CYP2D6 inhibitors on the performance of the CYP2D6 protein. CYP2D6 inhibitors were classified as either weak or strong in accordance to the FDA definition (36). We used two different methods to calculate the magnitude of the reduction of CYP2D6 activity caused by the concomitant use of a CYP2D6 inhibitor. First, we considered the inhibitory effect to be independent of the genotype, now represented by the CYP2D6 Gene Score. We assumed that weak CYP2D6 inhibitors would decrease by half the CYP2D6 metabolic capacity in all individuals, and strong CYP2D6 inhibitors would completely abolish the CYP2D6 activity, resulting in phenocopy (converting all individuals into poor metabolizers). Therefore, weak and strong CYP2D6 inhibitors were assigned an inhibitor factor of 0.5 and 0, respectively, with no consideration of the CYP2D6 Gene Score (Table 2, no CYP2D6 genotype input). In the second method, we took into account that individuals with high CYP2D6 enzyme capacity experience a larger inhibitory effect than individuals with decreased CYP2D6 enzyme activity when exposed to CYP2D6 inhibitors. Following this premise, we modified the inhibitor factor according to the individual CYP2D6 Gene Scores, assigning low inhibitor factor values to individuals with high CYP2D6 Gene Scores and vice-versa.
Table 2.
CYP2D6 inhibitor factor definition
| CYP2D6 Inhibitor Factor Calculation Method | CYP2D6 Inhibitor Factor | |
|---|---|---|
|
| ||
| No CYP2D6 genotype input | No CYP2D6 inhibitor | 1 |
| CYP2D6 Weak inhibitor* | 0.5 | |
| CYP2D6 Strong inhibitor** | 0 | |
|
| ||
| With CYP2D6 genotype input | No CYP2D6 inhibitor | 1 |
| Weak CYP2D6 inhibitor* | ||
| • CYP2D6 Gene score ≥1.75 | 0.5 | |
| • CYP2D6 Gene score 0.5-1.5 | 0.75 | |
| • CYP2D6 Gene score 0 | 1 | |
| Strong CYP2D6 inhibitor** | ||
| • CYP2D6 Gene score >2 | 0.25 | |
| • CYP2D6 Gene score 0.5-2 | 0 | |
| • CYP2D6 Gene score 0 | 1 | |
Note:
strong inhibitors include paroxetine, amiodarone, and metoclopromide.
Weak inhibitors include sertraline, venlafaxine, citalopram, fluoxetine, buproprion, razadone, and celecoxib.
For example, participants taking a CYP2D6 weak inhibitor received a CYP2D6 inhibitor factor of 0.5 if they had a CYP2D6 Gene Score of 2 and of 0.75 if their CYP2D6 Gene Score was 1. We again took into account that most individuals taking strong CYP2D6 inhibitors are converted into the poor metabolizer status, therefore receiving a CYP2D6 inhibitor factor of 0. The exceptions to this rule were participants with CYP2D6 Gene Scores >2 (i.e. ultra-rapid metabolizers) who tend to maintain some residual CYP2D6 activity despite the exposure to strong CYP2D6 inhibitors (30;37). These individuals were assigned a CYP2D6 inhibitor factor of 0.25 (Table 2, with CYP2D6 genotype input).
CYP2D6 Activity Score
The CYP2D6 Activity Score is expressed as the product of multiplying the CYP2D6 Gene Score by the Inhibitor factor, which is defined in equation [2]:
| [2] |
where the gene score is defined in Table 1, and inhibition factor is defined in Table 2. The CYP2D6 Activity Score represents an estimate of the actual performance of the CYP2D6 enzyme in vivo as determined by the CYP2D6 genotype and the effect of concomitant use of CYP2D6 inhibitors.
Statistical Analysis
Descriptive analysis of demographic variables is performed and presented as sample mean, standard deviation, median, range, or in categories. The predictive performance of various CYP2D6 scores on the log(endoxifen/NDM) are assessed with R2, and their significances are evaluated with p-values through linear regressions. All of the analyses are conducted in SAS 9.1 (SAS Institute Inc., Cary, NC).
Results
Demographics
The training and validation data sets were composed of 159 and 81 participants, respectively. Demographic characteristics such as age, body mass index, frequency of CYP2D6 genotypes and ethnicity in both data sets are presented in Table 3. Both training and validation data sets have similar age distributions, with mean ages between 53 and 54 years, so are their BMIs: 28.5 kg/m2 in the training set and 27.59 kg/m2 in the validation set. In both groups, the majority of patients were white: 90.6% and 96.3% respectively. Most importantly, both data sets have balanced PM/PM and EM/EM frequencies: 4.4% vs 2.5% in PM/PM, and 40.3% vs 44.5% in EM/EM. Therefore, demographic characterstics among the training and validation data sets were similar.
Table 3.
Demographics of participants in a training data set, a validation data set and both sets combined
| Data Sets | |||
|---|---|---|---|
|
| |||
| Training n=159 |
Validation n=81 |
Combined n=240 |
|
| Age (years) | |||
| • Mean | 53.23 | 53.40 | 53.32 |
| • Median (range) | 54 (56) | 55 (52) | 54 (58) |
|
| |||
| Mean Body Mass Index, | 28.53 | 27.59 | 28.17 |
| kg/m2 (range) | 39.34 | 24.36 | 40.09 |
|
| |||
| Ethnicity, n (%) | |||
| • White | 146 (91.83) | 78 (96.30) | 224 (93.33) |
| • Black | 9 (5.66) | 2 (2.47) | 11 (4.58) |
| • Hispanic | 1 (0.63) | 0 (0.00) | 1 (0.42) |
| • Unknown | 3 (1.89) | 1 (2.47) | 4 (1.67) |
|
| |||
| CYP2D6 Genotype, n (%) | |||
| • PM/PM | 7 (4.40) | 2 (2.47) | 9 (3.75) |
| • EM/EM | 64 (40.25) | 36 (44.45) | 100 (41.67) |
CYP2D6 Gene Score as a predictor of CYP2D6 phenotype
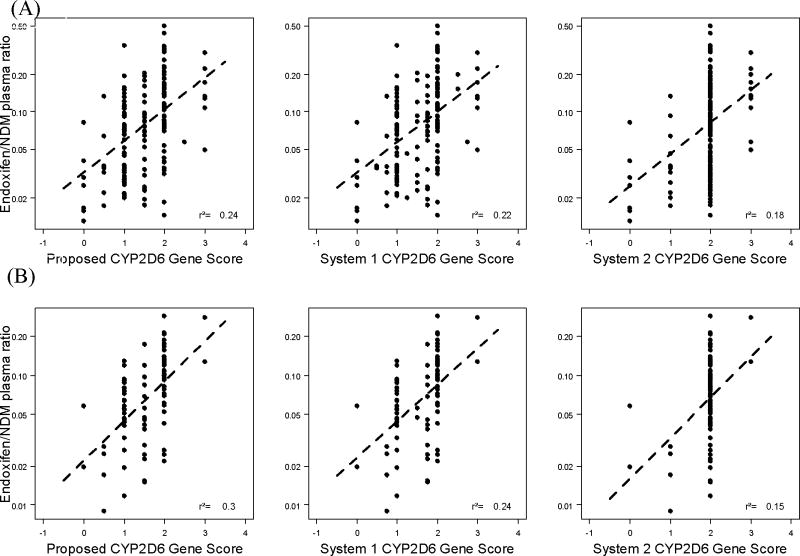
All three scoring systems, i.e. our proposed CYP2D6 Gene Score and the Systems 1 and 2 Gene Scores, showed a weak correlation with the endoxifen/NDM plasma ratio in the training data set (R2=0.24, 0.22 and 0.18, respectively; p < 0.00001), though our proposed score system is slightly better than Systems 1 and 2 Gene Scores. In the validation set, our proposed CYP2D6 Gene Score consistently showed the strongest association with the CYP2D6 index measure (R2=0.30 vs. 0.24 and 0.15, respectively; p <0.00001). In the validation set, the magnitude of the correlation remained low when CYP2D6 Gene Score alone is used (Figure 1).
Figure 1.
Gene score alone A=training, B=validation
CYP2D6 Activity Score as a predictor of CYP2D6 phenotype
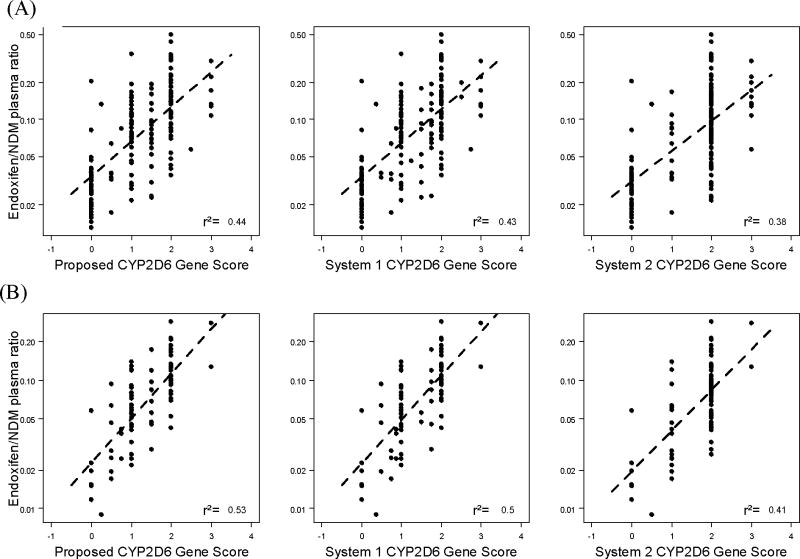
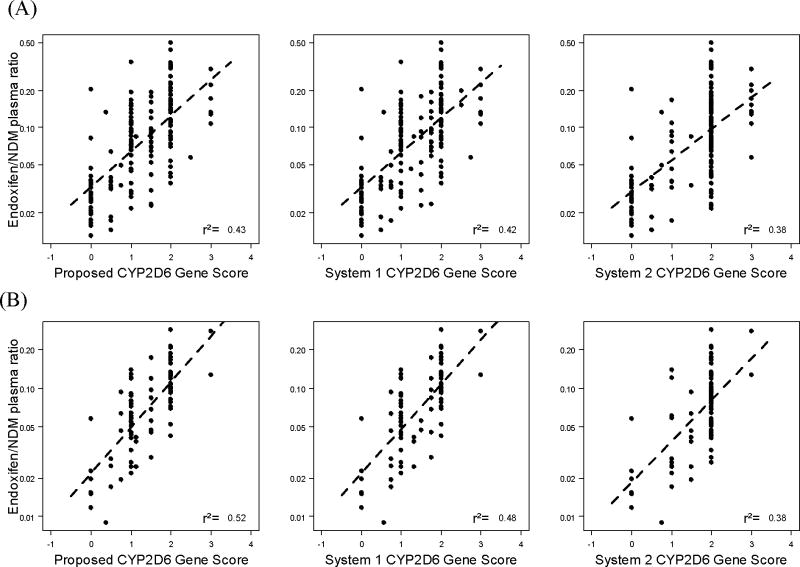
We evaluated the ability of our proposed CYP2D6 Activity Score to estimate the in vivo CYP2D6 activity as measured by the endoxifen/NDM plasma ratio. It is important to note that, as a result of using two different methods to calculate the CYP2D6 inhibitor factor (with and without CYP2D6 genotype input), there are two groups of CYP2D6 Activity Scores for all three scoring systems. In figure 2 we present the correlation of endoxifen/NDM plasma ratio with the three CYP2D6 Activity Score systems when the CYP2D6 Inhibitor Factor is calculated without considering the CYP2D6 genotype. As expected, when information on the concomitant use of CYP2D6 inhibitors is incorporated in the calculation of the CYP2D6 Activity Score, the association of all three scores (proposed system, system 1, and system 2) with the endoxifen/NDM plasma ratio almost doubled in the training data set (R2=0.44, 0.43 and 0.38 vs. 0.24, 0.22 and 0.18, respectively). In the validation data set, a similar pattern was observed (R2=0.53, 0.50 and 0.41 vs. 0.30, 0.24 and 0.15). When the CYP2D6 genotype is considered in the determination of the CYP2D6 Inhibitor Factor, the performance of the CYP2D6 Activity Score did not improve compared to the genotype independent Inhibition Factor (Figure 3), showing R2 values almost identical to those depicted in figure 2 for the Proposed System, System 1 and System 2 in the training (R2=0.43, 0.42 and 0.38, respectively) and the validation (R2=0.52, 0.48 and 0.38, respectively) data sets. Table 4 shows a summary of the performance of the Proposed CYP2D6 Gene and Activity Scores compared to Systems 1 and 2 in the training and validation data sets and in both sets combined.
Figure 2.
Activity score no gen input
Figure 3.
Activity score gen input
Table 4.
Correlation of the proposed CYP2D6 Gene and Activity Scores with endoxifen/NDM plasma ratio compared to System 1 (31) and System 2 (24) in a derivation data set, a validation data set and both sets combined. Results are presented in R2 values.
| Data set | |||
|---|---|---|---|
|
| |||
| Derivation n=159 |
Validation n=81 |
Combined n=240 |
|
|
| |||
|
CYP2D6 Gene Scores Alone (no input from CYP2D6 Inhibitor Factor) | |||
|
| |||
| Proposed CYP2D6 Gene Score System | 0.24 | 0.30 | 0.25 |
| System 1 | 0.22 | 0.24 | 0.22 |
| System 2 | 0.18 | 0.15 | 0.16 |
|
| |||
|
CYP2D6 Activity Scores (input from CYP2D6 Inhibitor Factor not genotype-derived) | |||
|
| |||
| Proposed CYP2D6 Gene Score System | 0.44 | 0.53 | 0.45 |
|
| |||
| System 1 | 0.43 | 0.50 | 0.43 |
|
| |||
| System 2 | 0.38 | 0.41 | 0.36 |
|
| |||
|
CYP2D6 Activity Scores (input from genotype-derived CYP2D6 Inhibitor Factor) | |||
|
| |||
| Proposed CYP2D6 Gene Score System | 0.43 | 0.52 | 0.44 |
|
| |||
| System 1 | 0.42 | 0.48 | 0.42 |
|
| |||
| System 2 | 0.38 | 0.38 | 0.34 |
Note: all the p-values are less than 0.00001 for these correlations.
Discussion
CYP2D6 genetic variation and the effect of CYP2D6 inhibitors on the pharmacokinetics of drugs that utilize this metabolic route have been intensively studied. However, some characteristics of the CYP2D6 genetic polymorphism and the CYP2D6 gene-environment interactions have become important limitations to the clinical use of CYP2D6 genetic information. The increasing number of CYP2D6 alleles described and their many allelic combinations make it difficult for most clinicians to interpret the CYP2D6 genotype results. In addition, with the probable exception of the CYP2D6 poor metabolizers, the functional consequence of the CYP2D6 genetic variation is modulated by other factors, such as physiological states like pregnancy, some herbal products and concomitant use of certain drugs. Therefore, in integrating CYP2D6 genetic information into the decision-making process in clinical practice, the CYP2D6 genotype should be interpreted in the clinical context of the individual patient, taking into account all possible determinants of CYP2D6 metabolic activity. Although some of these determinants are unknown or poorly understood, the CYP2D6 genotype and the concomitant use of CYP2D6 inhibitors are well documented and are readily available for patients and their doctors. Physicians often identify the importance of CYP2D6 genetic variation and drug interactions when prescribing a CYP2D6 substrate. However, there is no current instrument to quantify this interaction. Some progress has been made in grouping CYP2D6 genotypes by expected phenotypic expression (8;30;33;34), but this research has not considered concomitant use of medications.
In the present study, we analyzed for the first time the utility of a CYP2D6 scoring system that incorporates CYP2D6 genetic variation and CYP2D6 mediated drug-drug interactions. In our previous studies, we have shown that tamoxifen is sequentially metabolized by the cytochrome P450 system to endoxifen. Specifically, we have shown that endoxifen is formed from N-desmethyltamoxifen, the major circulating metabolite of tamoxifen, by CYP2D6. Therefore, the ratio of endoxifen to N-desmethyltamoxifen was used as a marker of CYP2D6 activity. We have developed a CYP2D6 Gene Score based on the anticipated effect of genetic variations on the CYP2D6 enzymatic capability and evaluated its correlation with the endoxifen/NDM plasma ratio as an index measure of CYP2D6 activity. As expected, the CYP2D6 Gene Score alone was poorly associated with the CYP2D6 phenotype. That was also the case for the other CYP2D6 scoring approaches (Systems 1 and 2) that utilized genetic information alone. However, when the expected CYP2D6 inhibitory effect was integrated with the CYP2D6 genotype into the composite CYP2D6 Activity Score, our ability to estimate the CYP2D6 phenotype doubled. Interestingly, a similar increase in the correlation of the score with the CYP2D6 index measure was seen with the inclusion of the CYP2D6 Inhibitor Factor into the genotype-based scores developed by other authors. These results highlight the heterogeneity of the CYP2D6 phenotype and the importance of considering other factors in addition to the genotype in the estimation of the CYP2D6 performance in vivo. In the process of integrating the CYP2D6 inhibitors information into the CYP2D6 Activity Score, we hypothesized that taking into account that the CYP2D6 genotype influences the capacity of CYP2D6 inhibitors to decrease the metabolic activity of the CYP2D6 enzyme would allow a more accurate estimation of this activity. However, the CYP2D6 genotype input did not seem to alter the ability of the CYP2D6 Activity Score to predict the CYP2D6 phenotype. A possible explanation is that the great impact of the addition of the CYP2D6 Inhibitor Factor to the Gene Score dilutes the small effect that the genotype input has on the calculation of the CYP2D6 Inhibitor Factor. Therefore, the definition of the CYP2D6 Inhibitor Factor without considering the CYP2D6 genotype appears to be a simple and reliable method. Right now, the selection of inhibition factor values is subjective. A better strategy could be a pharmacokinetics drug interaction model based prediction, which integrates genetics and inhibition mechanisms in score construction. In this paper, we group weak inhibitor and moderate inhibition together, because of their subtle difference. The definition of weak or moderate inhibitor sometimes depends on the substrates from various published in-vitro/in-vivo studies. It needs further exploration.
Our proposed CYP2D6 Activity Score is an improved tool to estimate the CYP2D6 performance in vivo. More importantly, our findings have been replicated in a second sample of individuals. The R2 values in the validation data set were comparable to the training data set when testing the CYP2D6 Gene Score alone and both CYP2D6 Activity Scores. In the validation sample the correlation of the endoxifen/NDM plasma ratio with the CYP2D6 Activity Score was also twice as high as with the CYP2D6 Gene Score alone across all three scoring systems tested. This consistency in the results in two data sets reflects the strength of the correlation between the CYP2D6 Activity Score and the CYP2D6 phenotype. However, other factors that influence the CYP2D6 activity are not being considered making the correlation still imperfect. For example, it has been shown that CYP2D6 genotypes have a different phenotypic expression in Caucasians as compared to African Americans (34). An ethnicity factor could not be tested in our samples due to lack of ethnic diversity in our study population. In the future, ethnicity, pregnancy, herbal products and other variables known to influence CYP2D6 activity could be included in the calculation of the CYP2D6 Activity Score. Another limitation, which applies to any scoring system using CYP2D6 genetic information, is the fact that the functional consequence of many of the described CYP2D6 alleles is unknown, hence the uncertainty in the accurate estimation of the CYP2D6 phenotype from genotype. Despite these limitations, the CYP2D6 Activity Score presented in this work shows an important correlation with the CYP2D6 phenotype and represent a significant improvement over the use of CYP2D6 genotype alone.
Acknowledgments
This study was supported in part by a Pharmacogenetics Research Network grant, 2U-01 GM61373 (DAF, DFH, VS), K24RR020815 (DAF), GM74217 (LL), a clinical pharmacology training grant 5T32-GM-08425 (DAF) from the National Institute of General Medical Sciences, Bethesda, MD, Grant M01-RR000042 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Damon Runyon-Lilly Clinical Investigator award CI-3 from the Damon Runyon Cancer Research Foundation (VS), Fashion Footwear Foundation/QVC Presents Shoes on Sale (DFH)
References
- 1.Ingelman-Sundberg M, Johansson I, Persson I, Oscarson M, Hu Y, Bertilsson L, et al. Genetic polymorphism of cytochrome P450. Functional consequences and possible relationship to disease and alcohol toxicity. EXS. 1994;71:197–207. doi: 10.1007/978-3-0348-7330-7_20. [DOI] [PubMed] [Google Scholar]
- 2.Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- 4.Tyndale RF, Droll KP, Sellers EM. Genetically deficient CYP2D6 metabolism provides protection against oral opiate dependence. Pharmacogenetics. 1997;7:375–9. doi: 10.1097/00008571-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Dahl ML, Johansson I, Palmertz MP, Ingelman-Sundberg M, Sjoqvist F. Analysis of the CYP2D6 gene in relation to debrisoquin and desipramine hydroxylation in a Swedish population. Clin Pharmacol Ther. 1992;51:12–7. doi: 10.1038/clpt.1992.2. [DOI] [PubMed] [Google Scholar]
- 6.Hou ZY, Pickle LW, Meyer PS, Woosley RL. Salivary analysis for determination of dextromethorphan metabolic phenotype. Clin Pharmacol Ther. 1991;49:410–9. doi: 10.1038/clpt.1991.48. [DOI] [PubMed] [Google Scholar]
- 7.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–95. [PMC free article] [PubMed] [Google Scholar]
- 8.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- 9.Ingelman-Sundberg M, Daly AK, Nebert DW. Human Cytochrome P450 (CYP) Allele Nomenclature Committee. Web site. Available from: www.imm.ki.se/CYPalleles.
- 10.Dahl ML, Johansson I, Bertilsson L, Ingelman-Sundberg M, Sjoqvist F. Ultrarapid hydroxylation of debrisoquine in a Swedish population. Analysis of the molecular genetic basis. J Pharmacol Exp Ther. 1995;274:516–20. [PubMed] [Google Scholar]
- 11.Lundqvist E, Johansson I, Ingelman-Sundberg M. Genetic mechanisms for duplication and multiduplication of the human CYP2D6 gene and methods for detection of duplicated CYP2D6 genes. Gene. 1999;226:327–38. doi: 10.1016/s0378-1119(98)00567-8. [DOI] [PubMed] [Google Scholar]
- 12.Michelson D, Read HA, Ruff DD, Witcher J, Zhang S, McCracken J. CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J Am Acad Child Adolesc Psychiatry. 2007;46:242–51. doi: 10.1097/01.chi.0000246056.83791.b6. [DOI] [PubMed] [Google Scholar]
- 13.Rau T, Wohlleben G, Wuttke H, Thuerauf N, Lunkenheimer J, Lanczik M, et al. CYP2D6 genotype: impact on adverse effects and nonresponse during treatment with antidepressants-a pilot study. Clin Pharmacol Ther. 2004;75:386–93. doi: 10.1016/j.clpt.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 14.Schillevoort I, de BA, van der WJ, Steijns LS, Roos RA, Jansen PA, et al. Antipsychotic-induced extrapyramidal syndromes and cytochrome P450 2D6 genotype: a case-control study. Pharmacogenetics. 2002;12:235–40. doi: 10.1097/00008571-200204000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Shams ME, Arneth B, Hiemke C, Dragicevic A, Muller MJ, Kaiser R, et al. CYP2D6 polymorphism and clinical effect of the antidepressant venlafaxine. J Clin Pharm Ther. 2006;31:493–502. doi: 10.1111/j.1365-2710.2006.00763.x. [DOI] [PubMed] [Google Scholar]
- 16.Somogyi AA, Barratt DT, Coller JK. Pharmacogenetics of opioids. Clin Pharmacol Ther. 2007;81:429–44. doi: 10.1038/sj.clpt.6100095. [DOI] [PubMed] [Google Scholar]
- 17.Wuttke H, Rau T, Heide R, Bergmann K, Bohm M, Weil J, et al. Increased frequency of cytochrome P450 2D6 poor metabolizers among patients with metoprolol-associated adverse effects. Clin Pharmacol Ther. 2002;72:429–37. doi: 10.1067/mcp.2002.127111. [DOI] [PubMed] [Google Scholar]
- 18.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–21. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 19.Kawanishi C, Furuno T, Kishida I, Matsumura T, Kosaka K. A patient with treatment-resistant schizophrenia and cytochrome P4502D6 gene duplication. Clin Genet. 2002;61:152–4. doi: 10.1034/j.1399-0004.2002.610211.x. [DOI] [PubMed] [Google Scholar]
- 20.Kawanishi C, Lundgren S, Agren H, Bertilsson L. Increased incidence of CYP2D6 gene duplication in patients with persistent mood disorders: ultrarapid metabolism of antidepressants as a cause of nonresponse. A pilot study. Eur J Clin Pharmacol. 2004;59:803–7. doi: 10.1007/s00228-003-0701-4. [DOI] [PubMed] [Google Scholar]
- 21.Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9:442–73. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 22.Dalen P, Frengell C, Dahl ML, Sjoqvist F. Quick onset of severe abdominal pain after codeine in an ultrarapid metabolizer of debrisoquine. Ther Drug Monit. 1997;19:543–4. doi: 10.1097/00007691-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 23.de Leon J, Dinsmore L, Wedlund P. Adverse drug reactions to oxycodone and hydrocodone in CYP2D6 ultrarapid metabolizers. J Clin Psychopharmacol. 2003;23:420–1. doi: 10.1097/01.jcp.0000085421.74359.60. [DOI] [PubMed] [Google Scholar]
- 24.Gasche Y, Daali Y, Fathi M, Chiappe A, Cottini S, Dayer P, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med. 2004;351:2827–31. doi: 10.1056/NEJMoa041888. [DOI] [PubMed] [Google Scholar]
- 25.Kirchheiner J, Schmidt H, Tzvetkov M, Keulen JT, Lotsch J, Roots I, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2007;7:257–65. doi: 10.1038/sj.tpj.6500406. [DOI] [PubMed] [Google Scholar]
- 26.Koren G, Cairns J, Chitayat D, Gaedigk A, Leeder SJ. Pharmacogenetics of morphine poisoning in a breastfed neonate of a codeine-prescribed mother. Lancet. 2006;368:704. doi: 10.1016/S0140-6736(06)69255-6. [DOI] [PubMed] [Google Scholar]
- 27.Chou WH, Yan FX, de LJ, Barnhill J, Rogers T, Cronin M, et al. Extension of a pilot study: impact from the cytochrome P450 2D6 polymorphism on outcome and costs associated with severe mental illness. J Clin Psychopharmacol. 2000;20:246–51. doi: 10.1097/00004714-200004000-00019. [DOI] [PubMed] [Google Scholar]
- 28.Flockhart DA. Cytochrome P450 Drug Interactions. Available at http://medicine.iupui.edu/flockhart/
- 29.Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 30.Borges S, Desta Z, Li L, Skaar TC, Ward BA, Nguyen A, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Cai WM, Nikoloff DM, Pan RM, de LJ, Fanti P, Fairchild M, et al. CYP2D6 genetic variation in healthy adults and psychiatric African-American subjects: implications for clinical practice and genetic testing. Pharmacogenomics J. 2006;6:343–50. doi: 10.1038/sj.tpj.6500378. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Pan RM, Porter TD, Jensen NS, Silber P, Russo G, et al. New cytochrome P450 2D6*56 allele identified by genotype/phenotype analysis of cryopreserved human hepatocytes. Drug Metab Dispos. 2006;34:1411–6. doi: 10.1124/dmd.106.009548. [DOI] [PubMed] [Google Scholar]
- 33.Zineh I, Beitelshees AL, Gaedigk A, Walker JR, Pauly DF, Eberst K, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther. 2004;76:536–44. doi: 10.1016/j.clpt.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–42. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- 35.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95:1758–64. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 36.Food and Drug Administration. Guidance for Industry Drug Interaction Studies - Study Design, Data Analysis, and Implications for Dosing and Labeling (DRAFT GUIDANCE) 2006. September, 2006. Available from http://www.fda.gov/cder/guidance/6695dft.pdf.
- 37.Laine K, Tybring G, Hartter S, Andersson K, Svensson JO, Widen J, et al. Inhibition of cytochrome P4502D6 activity with paroxetine normalizes the ultrarapid metabolizer phenotype as measured by nortriptyline pharmacokinetics and the debrisoquin test. Clin Pharmacol Ther. 2001;70:327–35. [PubMed] [Google Scholar]