Abstract
Genetic variations in the adrenergic receptor (ADR) have been associated with body composition in cross-sectional studies. Recent findings suggest that ADR variants may also modify body composition response to lifestyle. We assessed the role of ADR variants in body composition response to 12 months of resistance training versus control in previously sedentary postmenopausal women. Randomized trial completers were genotyped for A2BGlu9/12 by fragment length analysis, and B2Gln27Glu and B3Trp64Arg by TaqMan (n=148, 54% hormone therapy users). Associations between genotypes and body composition, by dual energy X-ray absorptiometry, were analyzed using univariate models. There was no main effect of individual genes on change in body composition, however, gene × exercise interactions were observed for A2BGlu9/12 and B2Gln27Glu on change in lean soft tissue (LST, p=0.02); exercisers on the A2BGlu9- background gained LST compared to a loss among controls over 12 months (p<0.05), with no significant intervention effect on the A2B Glu9+ background. Similarly, there was a significant LST gain with exercise on the B2Glu27+ background compared to loss among controls and no intervention effect on the B2Glu27- background. A non-significant association between total body fat (TBF) and B3Trp64Arg persisted among sedentary controls only when intervention groups were separated (%TBF gain with B3Arg 64+ carriage, p=0.03); exercisers lost TBF regardless of genotype. In summary, effect modification by lifestyle was demonstrated on ADR A2B, B2, and B3 genetic backgrounds. Individuals with certain ADR genotypes may be more vulnerable to adverse changes in body composition with sedentary behavior, thus these candidate genes warrant further study.
Keywords: body composition, lean soft tissue, resistance training, adrenergic receptor, post-menopausal women, genetics
Introduction
Exercise induced changes in weight and body composition exhibit a wide range of non-random inter-individual variability resulting from both genetic and environmental factors (Bouchard et al. 1993; Bouchard 2008; Bouchard and Agurs-Collins 2008). Allelic variants among members of the adrenergic receptor (ADR) gene family have been studied as biologically plausible genetic modifiers of body composition (e.g., % body fat) with a handful of studies addressing associations between these genes and body composition in the context of physical activity (Garenc et al. 2001; Garenc et al. 2003; Phares et al. 2004; Wolfarth et al. 2005). The alpha- and beta-adrenergic receptors, responsive to catecholamines, adrenaline and noradrenaline, interact on the cell surface of adipocytes to influence fat oxidation through antilipolytic (α-ADR) and lipolytic (β-ADR) pathways (Tarkovacs et al. 1994). Thus, allelic variation in the ADR family may influence the coupling of exercise-induced catecholamines to fat lipolysis and beta oxidation (Hoffstedt et al. 1999; Umekawa et al. 1999; Macho-Azcarate et al. 2002; Macho-Azcarate et al. 2003) along with mediating effects on heart rate, blood pressure and respiration (Wagoner et al. 2000; Kirstein and Insel 2004; McCole et al. 2004; Zhang et al. 2005a; Zhang et al. 2005b). The ADRB2 has also been shown to regulate lipolysis in human skeletal muscle (Hagstrom-Toft et al. 1998). Of particular interest, the ADRB3 is thought to have dual metabolic effects including lipolysis and thermogenesis in white and brown adipose tissue respectively (Krief et al. 1993; Hoffstedt et al. 1999; Collins et al. 2004; Kirstein and Insel 2004). Although brown adipose tissue, rich in ADRB3 and thermogenic uncoupling proteins, was thought to be characteristic of neonatal adipose mass in humans, significant amounts of brown adipocytes have been found to persist in adulthood within typically white adipose depots (Krief et al. 1993), which may be influenced by environment and behavior (Huttunen et al. 1981; Huttunen and Kortelainen 1990), and may have a greater effect on metabolism than previously thought (Collins et al. 2004).
Several polymorphisms have been identified among the ADR gene family members (Kirstein and Insel 2004), with a number of the published studies on body composition and related traits focused on common gene polymorphisms in the ADRB2 and ADRB3 genes that include substitutions at amino acids 16 and 27 in ADRB2 (Arg16→Gly, Gln27→Glu) and amino acid 64 in ADRB3 (Trp64→Arg) (Wolfarth et al. 2005; Rankinen et al. 2006). Early in vitro studies support altered receptor function and differences in ligand responsiveness between the common variant alleles in man (Pietri-Rouxel et al. 1997; Arner and Hoffstedt 1999; Umekawa et al. 1999). Despite early association studies that suggested association of allelic variants with earlier onset of type 2 diabetes, risk of abdominal obesity, insulin resistance, and an increased capacity to gain weight (Kirstein and Insel 2004), the overall significance of these common variants on body composition continues to be debated rcont with the failure to satisfactorily replicate findings across studies and study populations (Rankinen et al. 2006; Brodde 2008).
In the context of response to exercise-based interventions, only a handful of studies have reported on the effects of these variants. The HERITAGE Study found, for example, that carriers of any Glu27 allele in ADRB2 lost less fat, by computed tomography (CT), than the non-carriers following 20 weeks of endurance training (Garenc et al. 2003). In contrast, Phares et al. found that study participants that carried any Glu27 lost more fat, by dual energy X-ray absorptiometry (DXA), with 24-weeks of aerobic training than those who were homozygous for the Gln27 allele (Phares et al. 2004). Phares et al. also found that ADRB3 Arg64 allele carriage enhanced fat loss with training (Phares et al. 2004); similar analysis of ADRB3 within The HERITAGE Study was null (Garenc et al. 2001). The discrepancy between studies is unclear and may be due, in part, to differences in measurement techniques, as well as race and gender stratification conducted in The HERITAGE Study, but not in the study by Phares et al., which was limited by sample size. Nevertheless, these aerobic training trials provide novel data to suggest body composition responsiveness to physical activity based on ADR background, however, the lack of control groups has not allowed for the examination effect modification by lifestyle or exercise behavior.
Unlike the ADRB2 and ADRB3 genes, fewer studies have been conducted on the role of polymorphisms in the α-adrenergic receptors (ADRA) and body composition traits, although there is evidence that the deletion polymorphism in ADRA2B that lacks three glutamic acid residues between amino acid 301-303 (designated Glu9-) demonstrates reduced G protein coupled receptor kinase mediated phosphorylation and diminished agonist promoted desensitization (Small et al. 2001; Small and Liggett 2001; Small et al. 2003). The ADRA2B deletion polymorphism and body composition outcomes are primarily observational, while a few involve diet and/or aerobic activity (Rankinen et al. 2004; Rankinen et al. 2006). In a cross-sectional study, Dionne et al. found no independent effect of ADRA2B on multiple body composition and fitness measures in a wide weight range of healthy Caucasian women (Dionne et al. 2001), which was supported by the lack of association between the variant and change in total body fat or trunk fat with aerobic training in the aforementioned study by Phares et al. (Phares et al. 2004). In contrast, lower basal metabolic rate and weight gain (Heinonen et al. 1999; Sivenius et al. 2001) on the ADRA2BGlu9/9 background has been demonstrated and results from the Finnish Diabetes Prevention Study implied a potential effect modification by lifestyle with differential effects of diet and physical activity on the homozygous wild-type and homozygous variant carriers to decrease risk of type 2 diabetes in the context of weight loss. The infrequent study of this gene related to body composition, although it is a potential antagonist to the frequently studied beta adrenergic receptor variants, coupled with associations between relevant characteristics of obesity and ADRA2B, begs further investigation.
Our previous work on the Bone Estrogen and Strength Training (BEST) Study, which blocked postmenopausal women by hormone therapy (HT) versus no hormone therapy (NHT) and randomized them to exercise (EX) or no exercise (NEX) (Metcalfe et al. 2001; Going et al. 2003; Teixeira et al. 2003; Cussler et al. 2005), demonstrated that body composition (i.e. total body fat, abdominal fat, lean soft tissue), measured by dual energy X-ray absorptiometry, may be significantly altered by resistance training in post-menopausal women (Teixeira et al. 2003) and, although limited, there is evidence suggesting that adrenergic receptor variants may influence this body composition response to resistance training (Yao et al. 2007), as well as aerobic training (Phares et al. 2004). These observations positioned the BEST Study to begin to address recent recommendations by leading scientists recently convened by the National Cancer Institute for the Gene-Nutrition and Gene-Physical Activity Interactions in the Etiology of Obesity Workshop which called for precision measurements of phenotype and candidate gene testing within a well-controlled intervention (Bouchard and Agurs-Collins 2008). Therefore, we genotyped participants in The BEST Study for ADRA2BGlu9/12, ADRB2Gln27Glu, and ADRB3Trp64Arg to explore whether adrenergic receptor candidate gene variants and gene × environment (i.e. exercise) interactions affected individual variability in body composition response to 12 months sedentary, control behavior versus a physically active lifestyle focused on supervised resistance training. We hypothesized that homozygous carriers of the wild-type ADRA2B (Glu12/12), carriers of any ADRB2Glu27, or carriers of any ADRB3Arg64 alleles would demonstrate greater fat loss in response to resistance training.
Methods
Study Design
The BEST study was a block-randomized resistance training trial in sedentary postmenopausal women (n=320), using or not using hormone replacement therapy (HT versus NHT), where participants were randomized to exercise (EX) or no exercise (control—NEX) within each HT group. Participants agreed to maintain HT status, body weight, and diet during the trial (Going et al. 2003; Teixeira et al. 2003). Women who completed 12 months of the BEST Study (n=266) were re-recruited and consented for genetic studies. Differences between completers and drop-outs have been previously described (Going et al. 2003). In brief, dropouts (n=54) were 2 years younger than women who completed the study (53.6±4.5 versus 55.6±4.7 years; p<0.01) by independent t-tests. There were no significant differences in years past menopause, estrogen levels, height, weight, percent fat, lean soft tissue, and total body and regional bone mineral density between drop outs and completers.
A total of 148 (55.6%) returned the consent along with a buccal sample for DNA extraction. Among those who re-consented, 56.8% were from the EX group and 43.2% were from the NEX group. Though the difference between EX and NEX groups was not significant, a greater number of HT users participated in the genetic study, 53% HT use in NEX group and 55% HT use in the EX group. There were no differences in body composition between HT and NHT at baseline reported in the parent study (Teixeira et al. 2003) or in the present analysis. Other baseline differences between HT and NHT in the parent study have been previously reported; the NHT group was significantly older (1.5 yrs) and greater years past menopause (1.6 yrs) at baseline than the HT group in the parent study. The same dynamic was present in the ancillary genetics study at baseline.
In spite of the difference in response based on intervention groups, there were no significant differences between those who re-consented for the ancillary genetic study and those who could not be located or chose not to re-consent for the following at baseline: hormone therapy status, height (m), weight (kg), total body fat (kg and %), abdominal fat (kg), least soft tissue (LST, kg), energy intake (kcal). Age and body mass index (BMI) differences reached statistical significance (main study: 55.5 ± 4.8 yrs, BMI 25.3 ± 3.8 kg/m2; ancillary study: 56.1 ± 4.6 yrs, BMI 24.9 ± 3.8, p<0.05), but the genotyped sub-sample of the parent study was not considered clinically different from those in the main study at baseline due to these small differences. Following the 12 month intervention, there were no differences between those in the genetic study versus those not participating in the genetic study for the following: change in weight, change in BMI, change in total body fat (kg and %), change in abdominal fat, change in LST, and energy intake. Thus, the genetic sub-sample is highly representative of the parent study.
Exercise Program
The 12-month intervention arm engaged in supervised, high intensity resistance training and moderate impact weight-bearing exercise for 75 minutes, 3 days/week. Eight weight training exercises were performed per session, where individuals completed two sets of 6-8 repetitions at 70-80% of the one-repetition maximum loads per exercise. Strength was measured every 6–8 weeks over the 12 months to increase loads and maintain relative intensity. Detailed descriptions of the study design, exclusion criteria, and exercise protocol have been previously published (Metcalfe et al. 2001; Going et al. 2003; Teixeira et al. 2003).
Body Composition Measurements
Standard anthropometric measurements (i.e. height, weight) and body composition, including total and regional body fat and lean soft tissue (dual-energy x-ray absorptiometry, DXA, model DPX-L; Lunar Radiation Corporation, Madison, WI, software version 1.3y Lunar) were obtained at baseline and 12 months. As previously published, all total body scans were completed in medium scan mode to ensure appropriate image resolution. The scanner was calibrated daily against a standard calibration block supplied by the manufacturer and participant positioning for the total body scan was standardized. All body composition measures were performed in duplicate at both time points. The between duplicate CV was <1.8% for lean soft tissue and fat tissue; therefore, means of the duplicates have been used in all statistical analyses (Metcalfe et al. 2001; Going et al. 2003; Teixeira et al. 2003). Since total abdominal fat measures by DXA have been found to be comparable to computed tomography measures of total abdominal fat (Jensen et al. 1995), the total body DXA scans were retrospectively analyzed for abdominal fat by an operator set region of interest extending from the inter-vertebral space between the first and second lumbar vertebrae to the iliac crest.
Genotyping
DNA was obtained from buccal cells collected in mouthwash either during routine lab visits or via a mailed buccal cell collection kit, containing a sealed 44 mL bottle of Scope mouthwash (Proctor & Gamble, Cincinnati, OH), a sealed, sterile collection cup, instructions for collection, and a prepaid return envelope. Participants were asked not to eat or drink for 1 hr prior to sample collection, swish 10 mL mouthwash vigorously for 45 sec, expectorate into the cup, and mail the container in the provided packaging (Garcia-Closas et al. 2001).
DNA extraction was performed by the QIAampDNA Mini Kit (QIAGEN #51104, Valencia, CA) following manufacturer guidelines. DNA quality and quantity were assessed by 558 base pair polymerase chain reaction (PCR), separated on a 2% agarose gel by electrophoresis, and visualized by ethidium bromide staining. DNA quantification was also performed by PicoGreen dsDNA Quantitation Reagent and fluorimetry. The ADRA2BGlu9/12 insertion/deletion variant was genotyped according to previously published methods for PCR and gel electrophoresis based fragment length analysis (Sivenius et al. 2001), using primers designed for the long (112bp) and the short (103bp) variants (forward primer AGGGTGTTTGTGGGGCATCTCC, reverse primer CAAGCTGAGGCCGGAGACACTG, Midland Chemicals, Midland, TX). Four positive (10, 1, 0.1, and 0.01ng DNA) and one negative control were used across all gels. The use of a 10bp ladder clearly discriminated between the 112bp and the 103bp bands. No other fragment lengths were present. Only one failure could not be resolved following repeat genotyping, thus, there is one missing value for the ADRA2B genotype used in these analyses.
Allelic determination for single nucleotide polymorphisms (SNP), ADRB2Gln27Glu and ADRB3Trp64Arg, was performed using primers specific to these variants from Assays-by-Design; current reference SNP accession ID numbers are rs1042714 and rs4994 respectively (Applied Biosystems, Foster City, CA). ADRB2 probes were VIC for Gln and FAM for Glu alleles (forward primer CCTTCTTGCTGGCACCCAAT and reverse primer TGCCCACCACCCACAC). ADRB3 probes were VIC for Trp and FAM for Arg alleles (forward primer GCAACCTGCTGGTCATCGT and reverse primer ACGAACACGTTGGTCATGGT). Reaction components included TaqMan Genotyping Master Mix (12.5 μL), 40x TaqMan ODC SNP Genotyping Assay Mix (probe) (0.63 μL), and genomic DNA (10 ng in 11.87 μL dH2O) to yield 25 μL reactions. Thermal cycler conditions were as follows: an initial step at 95°C for 10 min, followed by 45 cycles of denaturation at 92°C for 15 sec and anneal/extend at 60°C for 1 min. The plate was read and analyzed using an ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA). A human variation panel of DNA (Coriell Institute for Medical Research, Camden, New Jersey) was surveyed to identify the best representatives for discrimination of each allele. These allelic representatives were run in replicates of 8 on each plate as the reference set for the ABI 7700 Sequence Detection System. Replicates of 8 negative controls were also present on each plate. After repeat genotyping, all failures were resolved; thus, there were no missing values for ADRB3 and ADRB2 genotypes used in these analyses.
Statistical Analyses
All statistical analyses were performed using SPSS v14.0 (SPSS Inc., Chicago, IL). All body composition data are expressed as mean ± standard error (SE) and/or p-value. The three possible genotypic frequencies for each gene studied were evaluated by Chi-Squared tests and found to be in Hardy-Weinberg equilibrium.
Baseline participant characteristic differences between intervention groups were evaluated by independent t-tests. Within each intervention group, participant characteristic changes pre- and post-intervention (baseline and 12 months) were evaluated using paired t-tests. Because of small samples size for homozygous minor variant genotypes (Table I), genotypes for each gene were collapsed into carrier versus non carrier groups. ADRA2B genotypes consisted of carriage of any 9 glutamic acids on ADRA2B (Glu9+, i.e. Glu9/12 and Glu 9/9) versus homozygotes for 12 glutamic acids, (i.e. Glu9- or insertion variant Glu12/12). ADRB3 genotypes consisted of any ADRB3Arg64+ (carriage of genotypes Trp/Arg and Arg/Arg) versus ARDB3Arg64- (carriage of Trp/Trp). ADRB2 genotypes consisted of any ADRB2Glu27+ (carriage of genotypes Gln/Glu and Glu/Glu) versus ADRB2Glu27- (carriage of Gln/Gln). Associations between each gene and body composition phenotype (i.e. total body fat, lean soft tissue, etc.) were analyzed separately using univariate models (ANCOVA). Change in the body composition variables of interest served as dependent variables. Models included fixed factors of genotype and exercise which enabled the evaluation these terms individually, as well as their interaction. Covariates included age, hormone replacement therapy status, and baseline body composition values. Estimated marginal means of the gene × exercise interactions, adjusted by the covariates, are presented in figures. Exploratory analyses of variance (ANOVA) were completed within intervention group to determine individual genotype effects. Similarly, to evaluate intervention effects on specific genetic backgrounds, separate exploratory ANOVA models within each genotype were utilized.
Table I.
ADR gene polymorphism frequencies for 1yr completers (N=148).
| ADR gene polymorphisma | Allele Frequency | Genotype Frequencyb | |||
|---|---|---|---|---|---|
| ADRA2B Glu9/Glu12 | Glu12 | Glu9 | Glu12/ Glu12 | Glu12/ Glu9 | Glu9/ Glu9 |
| 0.69 | 0.31 | 0.47 (70) | 0.42 (62) | 0.10 (15) | |
| ADRB3 Trp64Arg | Trp64 | Arg64 | Trp/Trp | Trp/Arg | Arg/Arg |
| 0.93 | 0.07 | 0.85 (126) | 0.15 (22) | 0.00 (0) | |
| ADRB2 Gln27Glu | Gln27 | Glu27 | Gln/Gln | Gln/Glu | Glu/Glu |
| 0.60 | 0.40 | 0.42 (62) | 0.45 (67) | 0.13 (19) | |
One subject could not be genotyped for the ADRA2BGlu9/12 variant.
Values in parentheses are the sample sizes.
Two main effects of interest (gene, exercise) and one interaction (gene × exercise) are presented for each body composition end point. We have set the level of significance at p=0.05, however, for a more cautious interpretation of the results presented herein, the Bonferroni adjustment for multiple comparisons may be applied which would alternatively set the level of significance to p=0.02 for each end point.
Statement of Ethics
We certify that all applicable institutional and governmental regulations concerning the ethical use of human volunteers were followed during this research. The University of Arizona Institutional Review Board approved the study protocol and collection of DNA. All participants included in the analysis provided written informed consent prior to their participation after the nature of the study and procedures had been fully explained to them.
Results
Baseline Characteristics
Descriptive statistics for the genetic sub-sample of the BEST study are presented in Table II. Baseline age, hormone replacement therapy status, weight, and measures of body composition were similar between exercise and control groups in the ancillary genetics study (p>0.4). Among those genotyped, the allelic frequencies were equally distributed across HT versus NHT for ADRB3 and ADRB2 carriers versus non-carriers, but HT users were not equally distributed across ADRA2B carriers versus non-carriers (Chi-Squared Test p=0.03), thus HT status was used as a covariate in all further analyses presented. In agreement with the parent study (Teixeira et al. 2003), there were no differences in body composition between HT and NHT at baseline (data not presented).
Table II.
Baseline subject characteristics and changes post-12 month intervention.
| Characteristics | Total (n=148) |
Control (n=64) |
Exercise (n=84) |
pa |
|---|---|---|---|---|
| Age (years) | 56.1 ± 0.37 | 56.4 ± 0.62 | 55.8 ± 0.46 | 0.398 |
| Hormone Therapy (%) | 54 % | 53% | 55 % | 0.844 |
| Weight (kg) | 66.9 ± 0.96 | 66.0 ± 1.36 | 67.6 ± 1.33 | 0.415 |
| Change | 0.12 ± 0.22 | 0.25 ± 0.37 | 0.02 ± 0.27 | 0.620 |
| Total body fat (%) | 37.7 ± 0.54 | 37.44 ± 0.89 | 37.86 ± 0.68 | 0.707 |
| Change | -0.63 ± 0.21 | 0.09 ± 0.31 | -1.19 ± 0.27b | 0.002 |
| Total body fat (kg) | 25.38±0.68 | 24.95±1.03 | 25.71±0.91 | 0.577 |
| Change | -0.312±0.20 | 0.15±.032 | -0.67±0.26 | 0.046 |
| Abdominal Fat (kg) | 2.73±.010 | 2.72±0.16 | 2.73±0.13 | 0.964 |
| Change | -0.023 | 0.05±0.04 | -0.08±0.04 | 0.021 |
| Lean Soft Tissue (kg) | 38.30±0.38 | 37.95±0.52 | 38.56±0.54 | 0.431 |
| Change | 0.497b | 0.024±0.15 | 0.86±0.11 | 0.001 |
Values are expressed as means ± SE.
Probability for Student’s t test comparing intervention groups.
Indicates significant change within group after intervention p<0.05.
Main Effect of Exercise
In both the parent and ancillary genetic study, the main effect of exercise was significant across body composition measures (p<0.05), with change in percent total body fat and lean soft tissue being strongest, in spite of stable body weight overall. Resistance training significantly reduced total body fat and abdominal fat (p<0.05) and increased lean soft tissue (p<0.05) in both studies; there were no significant differences in body composition results between parent and ancillary studies. Changes in body composition variables for the ancillary genetic study participants are represented in Table II.
ADR Variant Frequencies
Genotyping for the variants of interest, ADRA2BGlu9/12, ADRB3Trp64Arg and ADRB2Gln27Glu demonstrated that allelic variation in our study was within the range of previous studies (Kirstein and Insel 2004; Rankinen et al. 2006). Allelic and genotype frequencies are shown in Table I; 31% carried ADRA2BGlu9+, 7% carried ADRB3Arg64+, and 35% carried ADRB2Glu27+. Although power to evaluate gene × gene interactions is limited, the frequencies of co-carriage may be of interest and are provided as a supplemental table.
Associations between ADR Variants and Body Composition at Baseline
There were no significant associations between body composition and individual genes at baseline. However, non-significant increases in total body fat (% and kg), trunk fat (kg), and abdominal fat (kg) in ADRB2Glu27+ carriers (p<0.1) and a trend toward decreased lean soft tissue on the ADRA2BGlu9- background (p<0.06) were present (data not presented).
Individual ADR Variants as Determinants of Change in Body Composition at 12 months
Table III presents the main effect of exercise, individual gene variant carriage, and the gene × exercise interactions for each of the ADR variants, evaluated by separate univariate models with a single gene of interest and exercise included as fixed factors and covariates of age, HT status, and baseline values for the dependent body composition variable of interest. Univariate models including the other 2 genes as covariates were also run; results from these models were not appreciably different from the single gene models. In addition, although limited by sample size, we evaluated the associations herein by each genotype and found that the overall gene × exercise interaction p-values were unchanged for all body composition outcomes when groups were based on homozygous wild-type, heterozygote, and homozygous variant carriage versus carrier/non-carrier categorization. Results presented in Table III reflect the single gene models for variant carriage versus non-carriage.
Table III.
Probability values for main effect of gene and gene × exercise interactions relative to change in body composition
| %TBF | AbFat (kg) | LST (kg) | ||
|---|---|---|---|---|
|
| ||||
| Model 1 | ||||
| ADRA2B | 0.91 | 0.95 | 0.98 | |
| Exercise | 0.004 | 0.02 | <0.0001 | |
| ADRA2B × exercise | 0.71 | 0.90 | 0.02 | |
|
| ||||
| Adjusted R2 | 3% | 1% | 22% | |
|
| ||||
| Model 2 | ||||
| ADRB3 | 0.13 | 0.52 | 0.19 | |
| Exercise | 0.001 | 0.02 | <0.0001 | |
| ADRB3 × exercise | 0.13 | 0.38 | 0.12 | |
|
| ||||
| Adjusted R2 | 5% | 3% | 21% | |
|
| ||||
| Model 3 | ||||
| ADRB2 | 0.56 | 0.74 | 0.65 | |
| Exercise | 0.002 | 0.01 | <0.0001 | |
| ADRB2 × exercise | 0.47 | 0.17 | 0.02 | |
|
| ||||
| Adjusted R2 | 3% | 3% | 22% | |
Percent total body fat, %TBF; abdominal fat, AbFat; lean soft tissue, LST. Univariate (GLM) model fixed factors were exercise and genotype. Covariates included: age, hormone therapy status, and baseline value of respective body composition outcome.
The effect of exercise was independently significant for each body composition variable across all individual gene models (p≤0.02) (Table III). There was no significant main effect of the individual genes on changes in body composition overall at 12 months. However, ADRA2B × exercise and ADRB2 × exercise were significantly associated with change in lean soft tissue (p=0.02). When the genotypes were split for further exploration, as depicted in Figure 1, we found that only the ADRA2BGlu9- genotype demonstrated a significant difference between exercise and control groups for lean soft tissue (p<0.05). There was no significant effect of the resistance training intervention on lean soft tissue for ADRA2BGlu9+ carriers. In addition, within the exercise group, greater lean soft tissue gain over 12 months occurred among ADRA2BGlu9- carriers (A2BGlu9- : 1.09 ± 0.17kg versus A2BGlu9+: 0.67 ± 0.16kg; p<0.1 within exercisers), although the difference was not significant. Similarly, as shown in Figure 2, the lean soft tissue difference between exercise and control groups was also significant for the ADRB2Glu27+ carriers only. There was no significant effect of the resistance training intervention on lean soft tissue on the ADRB2Glu27- background. Within the sedentary controls, the ADRB2Glu27+ genotype also demonstrated a borderline association with loss of lean soft tissue, compared to the ADRB2Glu27- group (B2Glu27+ : -0.21 ± 0.18 versus B2Glu27-: 0.29 ± 0.20; p<0.1 within controls) which gained a small amount of lean soft tissue over 12 months.
Figure 1. Change in lean soft tissue (kg) by ADRA2BGlu9/12 genotype following 12 months of resistance training versus control, no exercise.
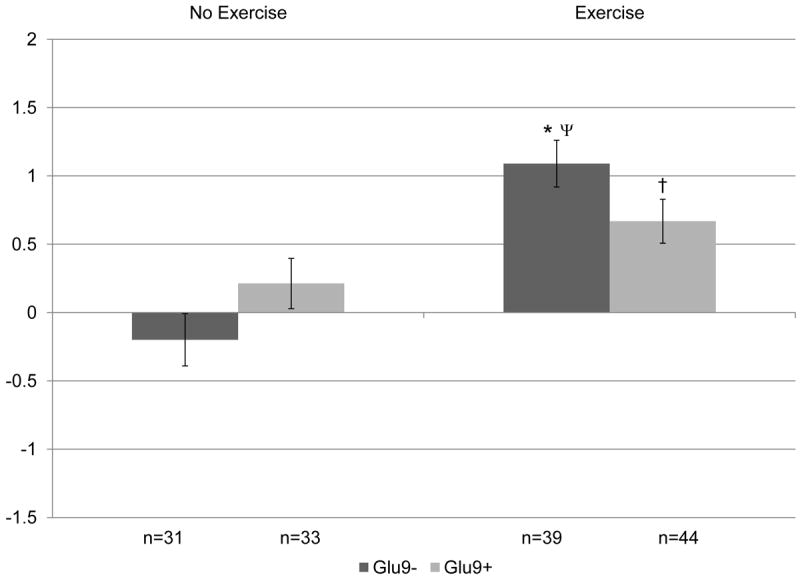
Covariates include: age, hormone therapy status, and baseline lean soft tissue. P=0.02 for overall gene × exercise interaction. When split by genotype in exploratory analyses: *p<0.05 within genotype between no exercise and exercise groups; †p<0.1 within genotype between no exercise and exercise groups. When split by exercise in exploratory analyses: Ψp≤0.1 between genotype difference within intervention group.
Figure 2. Change in lean soft tissue (kg) by ADRB2Gln27Glu genotype following 12 months of resistance training versus control, no exercise.
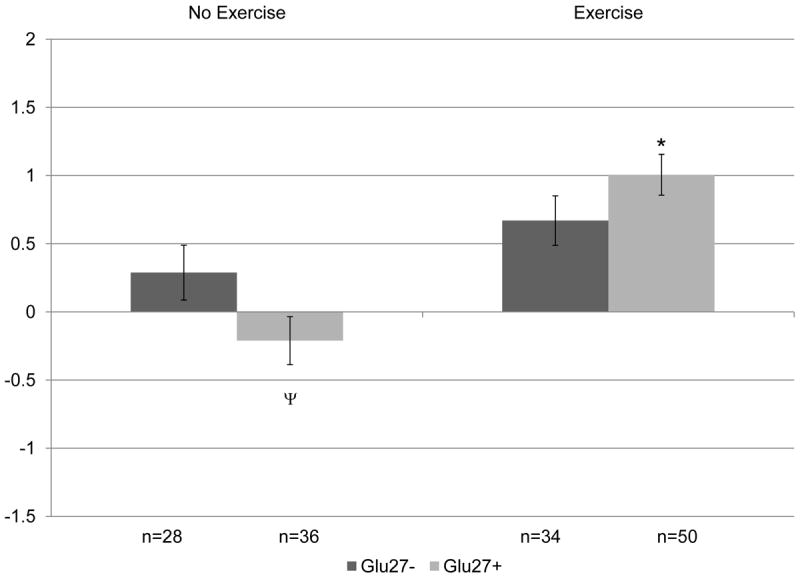
Covariates include: age, hormone therapy status, and baseline lean soft tissue. P=0.02 for overall gene × exercise interaction. When split by genotype in exploratory analyses: *p<0.05 within genotype between no exercise and exercise groups. When split by exercise in exploratory analyses: Ψp≤0.1 between genotype difference within intervention group.
The overall analysis presented in Table III also indicated a potential genetic effect on % total body fat with ADRB3 alone and gene × exercise effect on the same background (p=0.13). When split on intervention group, the effect was limited to sedentary lifestyle; carriers of any ADRB3Arg64+ in the control group gained significantly greater % total body fat over 12 months than the non-carriers (B3Arg64+: 1.62 ± 0.83% versus B3Arg64-: -0.16 ± 0.34%; p= 0.03 within controls), while exercisers lost significant and essentially equivalent % total body fat regardless of the carrier status of the ADRB3Trp64Arg polymorphism (B3Arg64+: -1.18 ± 0.70 versus B3Arg64-: -1.19 ± 0.30; p= 0.94 within exercisers). These % total body fat differences are depicted in Figure 3 by genotype and intervention group. A similar, but not significant, relationship was found between ADRB3Arg64+ and loss of lean soft tissue in the sedentary control group, as well (B3Arg64+: -0.62 ± 0.36 versus B3Arg64-:0.11 ± 0.14; p<0.1 within controls), while exercisers of either genotype gained lean soft tissue (p<0.05 for exercise versus control within each genotype). Lean soft tissue differences between ADRB3 genotype and intervention group are demonstrated in Figure 4.
Figure 3. Change in total body fat (%) by ADRB3Trp64Arg genotype following 12 months of resistance training versus control, no exercise.
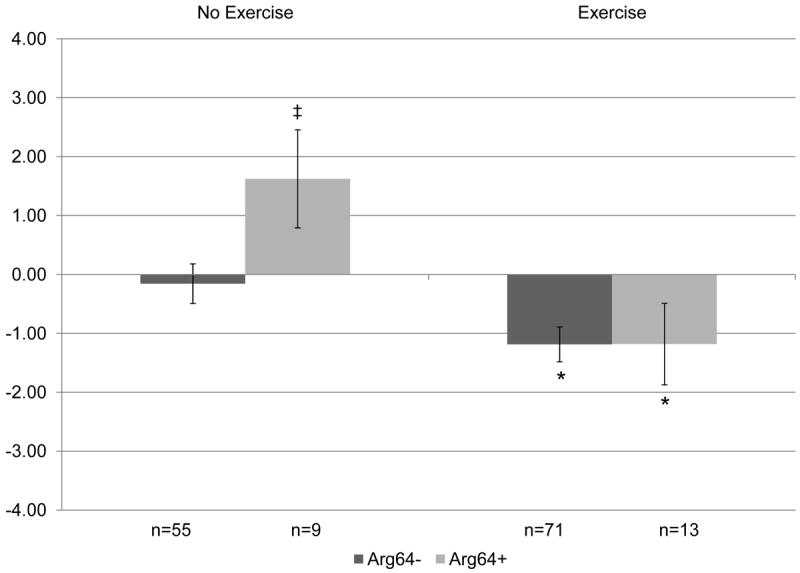
Covariates include: age, hormone therapy status, and baseline total body fat (%). P=0.1 for overall gene × exercise interaction. When split by genotype in exploratory analyses: *p<0.05 within genotype between no exercise and exercise groups. When split by exercise in exploratory analyses: ‡ p<0.05 between genotype difference within intervention group.
Figure 4. Change in lean soft tissue (kg) by ADRB3Trp64Arg genotype following 12 months of resistance training versus control, no exercise.
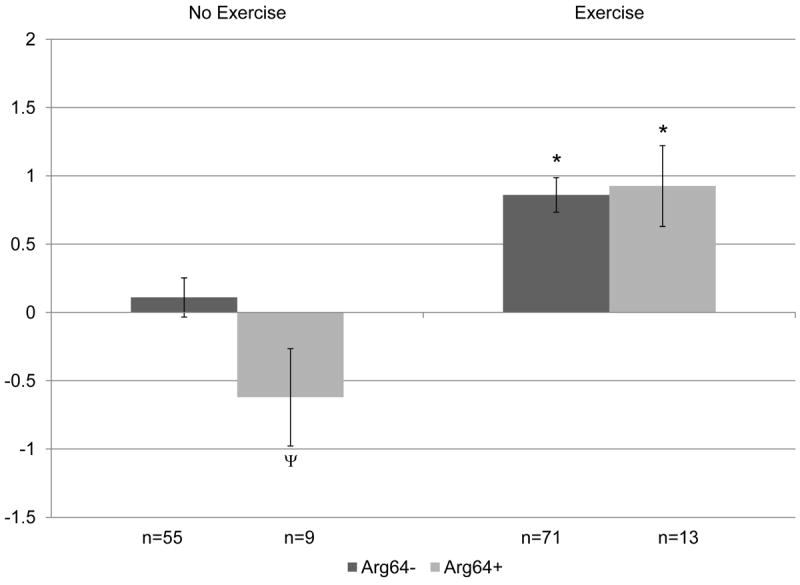
Covariates include: age, hormone therapy status, and baseline lean soft tissue. P=0.1 for overall gene × exercise interaction. When split by genotype in exploratory analyses: *p<0.05 within genotype between no exercise and exercise groups. When split by exercise in exploratory analyses: Ψp≤0.1 between genotype difference within intervention group.
Discussion
Our results demonstrate that a lifestyle change, focused on resistance training exercise, positively influences body composition in postmenopausal women, independent of any strong main effects of the single genetic variants in the studied ADR gene variants. However, the observed gene × exercise interactions in this study with respect to body composition, support a potential modifying effect. When explored, variability in the direction and magnitude of body composition change over 12 months based on genetic background was observed primarily among sedentary controls. We did not observe these same gene effects over the 12 months in those women who were randomized to the exercise intervention.
The impetus to investigate variants in ADR genes came from cross-sectional studies which associated adverse body composition phenotypes with variant carriage of ADRB3Arg64+or ADRB2Glu27+ (Rankinen et al. 2006) and other work associating ADRA2BGlu9/9 genotype with lower basal metabolic rate and weight gain (Heinonen et al. 1999; Sivenius et al. 2001). These results are in contrast with a more recent study of enhanced fat loss with physical activity on these same beta adrenergic backgrounds and no association with the alpha adrenergic variant alone (Phares et al. 2004). The opposing effects of the same genotypes demonstrated by cross-sectional studies versus a lifestyle intervention alluded to gene × environment interactions.
In cross-sectional analyses from our baseline data, none of the individual ADR variants studied were associated with obesogenic phenotypes (data not shown). However, higher total body fat (% and kg), trunk fat (kg) and abdominal fat(kg) in ADRB2Glu27+ carriers (p<0.10), although non-significant, was consistent with the literature (Rankinen et al. 2006), and a trend towards lower lean soft tissue on the ADRA2BGlu9- background, was present (p<0.06); these associations did not persist with the intervention. In addition, while no baseline relationship was demonstrated for the ADRB3 variant, continued sedentary behavior (control) was associated with significant increases in percent total body fat over the 12 months in ADRB3Arg64+ carriers. Polymorphisms in ADRA2B, ADRB3, and ADRB2 all emerged as potential modifiers of lean soft tissue when we controlled for exposure to exercise behavior. Prior cross-sectional studies of these ADR variants, including our baseline evaluation, have not captured gene × activity interactions, which we believe importantly influence body composition change over time, such that the modest gene effects are lost.
The hypothesis that genetic influences on body mass or composition may be modifiable and dependent on environment and behavior is becoming more widely accepted (Booth et al. 2002; Bouchard 2008). Twin studies have demonstrated that genetic influences on BMI and waist circumference, a measure of central adiposity, are attenuated by environmental exposures such as physical activity (McCaffery et al. 2009; Mustelin et al. 2009). However, the lack of adequate evaluation of environmental exposures to physical activity persist in the literature, including investigations of the ADR gene family, and may be partially responsible for inconsistent results relating individual ADR genes to obesity and body composition. For example, the Arg64 polymorphism in ADRB3 as the ‘low activity variant’ has been extensively studied as a candidate risk allele for obesity and lifetime weight gain (Arner 1995; Arner and Hoffstedt 1999; Kirstein and Insel 2004). Yet a meta-analysis suggests that if there is a role for the Arg64 variant in obesity and risk of weight gain, it is modest (Allison et al. 1998), which may be due, in part, to sedentary and physically active individuals combined regressing towards the mean.
Supportive of our results, a Spanish study found that carriage of the ADRB3Arg64+ variant was a risk factor for obesity in sedentary individuals, but not in the active individuals when leisure time physical activity was assessed (Marti et al. 2002). Similarly, Meirhaeghe found a significant association between obesity and the ADRB2Gln27Glu variant among the sedentary (Meirhaeghe et al. 1999). The 24 week clinical trial of aerobic training by Phares et al also support a role for an interaction between physical activity and the ADRB3Trp64Arg and ADRB2Gln27Glu variants (Phares et al. 2004). In contrast, the large HERITAGE Family Study with a similar study design did not find an association between ADRB3Trp64Arg or ADRB2Gln27Glu carriage and change in body composition with endurance training in women (Garenc et al. 2001; Garenc et al. 2003). However, in Caucasian women, an Arg to Gly at amino acid 16 in ADRB2, which tends to co-localize with the ADRB2Gln27Glu (Brodde 2008), was associated with change in adiposity with training (Garenc et al. 2003).
The control group in our study allowed for comparisons between active and non-active individuals, which was not possible in the two prior aerobic training trials, and which may partially account for differences between our study and the HERITAGE Family Study, in addition to technological differences in the evaluation of body composition change (DXA versus CT). We also evaluated training that was primarily resistance based, which may interact differently with the ADR gene variants than aerobic training to influence body composition change. Population characteristics, especially racial/ethnic diversity, may also influence reported outcomes. Our population was limited to postmenopausal women who were primarily Caucasian, while both aerobic training trials included male and female participants (Garenc et al. 2001; Garenc et al. 2003; Phares et al. 2004), with significant diversity represented in the HERITAGE Family Study (Garenc et al. 2001; Garenc et al. 2003).
Studies of other obesity-related outcomes also support a role for gene × environment interactions and point toward improvements in study design to further elucidate these interactions, i.e. more specific evaluation of overall and regional body composition, careful quantification of exposure to lifestyle or environmental variation, randomized controlled exposures to environmental stimulus or lifestyle change, and the potential for these and other genes to act epistatically on the highly redundant and regulated physiological mechanisms related to energy balance (Lenard and Berthoud 2008). For example, the only other resistance training investigation of similar ADR variants focused on CT derived intramuscular leg fat (IMF) and the effect of ADRA2B and ADRB2 gene and gene × gene interaction. Yao et al found that midthigh IMF responded more favorably to exercise on the ADRB2Glu27+ background (p=0.01), on the ADRA2BGlu9+ (p=0.04) background, and on the ADRA2BGlu9+ × ADRB2Glu27+ background (p=0.02) compared to non-carriers of both variants, with the combination explaining the greatest variance in change in IMF (7.4%). These results lend additional support for the role of these specific genes and their combinations as important determinants of individual response to strength training exercise (Yao et al. 2007).
Although we did not measure IMF, univariate models assessing the gene × exercise interactions seen for ADRA2B and ADRB2 for change in lean soft tissue accounted for 22% of the variance. Lean soft tissue measurements or associations with adrenergic genetic variants are rarely reported in the literature. In addition, common individual gene variants studied as candidates or identified in genome wide association studies typically account for <10% of the variance in obesity phenotypes (Frazer et al. 2009). Based on the increased oxidative capacity within individuals with increased lean soft tissue (McArdle et al. 2007), it is possible that some of the gene effects attributed to genetic influences at the adipocyte may be indirect effects of increased energy requirements at the muscular level due to genetic influences on muscle tissue or physiological processes that enhance muscle tissue performance. We must also consider the role of genotype on behaviors that may affect body composition, such as exercise compliance due to genetic differences in exercise tolerance or food choice in response to exercise.
The ancillary nature of the study, the relatively homogenous population structure (postmenopausal, primarily non-Hispanic White women), and our limited sample were limitations of this study. Additionally, although body composition outcomes at baseline and post-intervention were not different between hormone therapy users and non-users, we statistically controlled for hormone therapy status in all analyses due to the potential for hormone therapy to interact with body composition response to exercise and genotype. We were unable to specifically evaluate the contribution of hormone therapy in this study because it was not randomly assigned and we were underpowered to evaluate the 3-way interaction of gene × exercise × hormone therapy. Future efforts to improve measurement of environmental exposures or behavior changes with larger sample sizes are needed to confirm the observed gene × environment interactions seen here and to evaluate 3-way interactions, to include hormone therapy as a potential modulator of effects demonstrated in this analysis among postmenopausal women.
Additionally, co-carriage of the candidate genes may have influenced our results, however, this trial was not sufficiently powered to investigate gene × gene and gene × gene × environment interactions which are of great interest; nor were we powered to evaluate individual genotypes (homozygous wild-type versus heterozygotes versus homozygous variants). Thus, due to concern over false discovery and lack of a replication set we have focused on individual gene and gene × environment interactions. However, exploratory analyses of our data suggest an overall association between the ADRA2B × ADRB3 × exercise and change in % total body fat (p=0.03) and abdominal fat (p=0.05), with significant intervention effects on the ADRA2BGlu9- × ADRB3Arg64+ background, but not other genetic backgrounds. In addition, within the non-exercising, control group, the ADRA2BGlu9- × ADRB3Arg64+ genetic background gained significantly more % total body fat relative to gene combination ADRA2BGlu9- × ADRB3Arg64- over the 12 month intervention. The suggestion of a more plastic body composition phenotype on this genetic background is supported by results from Phares et al. who reported greater total and trunk fat loss in this background (ADRA2BGlu9- on the ADRB3Arg64+) with aerobic exercise training in men and women over 24 weeks (Phares et al. 2004). Although our exploratory analyses were limited by sample size, the shared direction of effect with the aerobic trial increases the significance of our study and supports the need to further evaluate these genes and their interactions with exercise as predictors of tissue specific response in larger sample sizes enriched for rare alleles.
In summary, the main effect of exercise on body composition (i.e., fat loss and lean soft tissue gain) was significant in both the overall BEST trial (Teixeira et al. 2003) and this ancillary study independent of ADR genotypes. Largely consistent with previous studies, we found that genetic influences on change in body composition at 12 months in individuals who were sedentary localized to the ADR gene loci. Given our limited sample size, the main effect of exercise may have masked any underlying, modest acting, effect on body composition by ADR gene variants with the exercise intervention. Our results highlight the strong positive modulating effect of exercise on body composition across all genotypes, the vulnerability to adverse body composition changes with sedentary behavior among postmenopausal women, and suggest that these changes with inactivity may be more profound in certain genetic backgrounds. Thus, exercise may play a key protective role against adverse gene effects on obesity. Additional studies are needed to confirm our findings.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health Grant AR39559 and the 2005 Gatorade Sports Science Institute Student Grant Award. Scope mouthwash was generously donated by Proctor and Gamble. We thank Betsy Wertheim and Elizabeth Jacobs for their critical review of the manuscript and participants and staff of the Bone Estrogen and Strength Training Study. We also thank Dr. Michael Hogan and staff for the use of their equipment and genotyping expertise. The supporters of this research will not benefit from the results of the present study.
References
- Allison DB, Heo M, Faith MS, Pietrobelli A. Meta-analysis of the association of the Trp64Arg polymorphism in the beta3 adrenergic receptor with body mass index. Int J Obes Relat Metab Disord. 1998;22(6):559–66. doi: 10.1038/sj.ijo.0800625. [DOI] [PubMed] [Google Scholar]
- Arner P. The beta 3-adrenergic receptor--a cause and cure of obesity? N Engl J Med. 1995;333(6):382–3. doi: 10.1056/NEJM199508103330612. [DOI] [PubMed] [Google Scholar]
- Arner P, Hoffstedt J. Adrenoceptor genes in human obesity. J Intern Med. 1999;245(6):667–72. doi: 10.1046/j.1365-2796.1999.00495.x. [DOI] [PubMed] [Google Scholar]
- Booth FW, Chakravarthy MV, Spangenburg EE. Exercise and gene expression: physiological regulation of the human genome through physical activity. J Physiol. 2002;543(Pt 2):399–411. doi: 10.1113/jphysiol.2002.019265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C. Gene-environment interactions in the etiology of obesity: defining the fundamentals. Obesity (Silver Spring) 2008;16(Suppl 3):S5–S10. doi: 10.1038/oby.2008.528. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Agurs-Collins T. Studying gene-behavior interactions: summary of recommendations. Obesity (Silver Spring) 2008;16(Suppl 3):S95–6. doi: 10.1038/oby.2008.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Despres JP, Mauriege P. Genetic and nongenetic determinants of regional fat distribution. Endocr Rev. 1993;14(1):72–93. doi: 10.1210/edrv-14-1-72. [DOI] [PubMed] [Google Scholar]
- Brodde OE. Beta-1 and beta-2 adrenoceptor polymorphisms: functional importance, impact on cardiovascular diseases and drug responses. Pharmacol Ther. 2008;117(1):1–29. doi: 10.1016/j.pharmthera.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Collins S, Cao W, Robidoux J. Learning new tricks from old dogs: beta-adrenergic receptors teach new lessons on firing up adipose tissue metabolism. Mol Endocrinol. 2004;18(9):2123–31. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- Cussler EC, Going SB, Houtkooper LB, Stanford VA, Blew RM, Flint-Wagner HG, Metcalfe LL, Choi JE, Lohman TG. Exercise frequency and calcium intake predict 4-year bone changes in postmenopausal women. Osteoporos Int. 2005;16(12):2129–41. doi: 10.1007/s00198-005-2014-1. [DOI] [PubMed] [Google Scholar]
- Dionne IJ, Turner AN, Tchernof A, Pollin TI, Avrithi D, Gray D, Shuldiner AR, Poehlman ET. Identification of an interactive effect of beta3- and alpha2b-adrenoceptor gene polymorphisms on fat mass in Caucasian women. Diabetes. 2001;50(1):91–5. doi: 10.2337/diabetes.50.1.91. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10(4):241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, Bender PK, Beck JC, Le Marchand L, Lum A, Alavanja M, Hayes RB, Rutter J, Buetow K, Brinton LA, Rothman N. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10(6):687–96. [PubMed] [Google Scholar]
- Garenc C, Perusse L, Chagnon YC, Rankinen T, Gagnon J, Borecki IB, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. Effects of beta2-adrenergic receptor gene variants on adiposity: the HERITAGE Family Study. Obes Res. 2003;11(5):612–8. doi: 10.1038/oby.2003.88. [DOI] [PubMed] [Google Scholar]
- Garenc C, Perusse L, Rankinen T, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. The Trp64Arg polymorphism of the beta3-adrenergic receptor gene is not associated with training-induced changes in body composition: The HERITAGE Family Study. Obes Res. 2001;9(6):337–41. doi: 10.1038/oby.2001.43. [DOI] [PubMed] [Google Scholar]
- Going S, Lohman T, Houtkooper L, Metcalfe L, Flint-Wagner H, Blew R, Stanford V, Cussler E, Martin J, Teixeira P, Harris M, Milliken L, Figueroa-Galvez A, Weber J. Effects of exercise on bone mineral density in calcium-replete postmenopausal women with and without hormone replacement therapy. Osteoporos Int. 2003;14(8):637–43. doi: 10.1007/s00198-003-1436-x. [DOI] [PubMed] [Google Scholar]
- Hagstrom-Toft E, Enoksson S, Moberg E, Bolinder J, Arner P. beta-Adrenergic regulation of lipolysis and blood flow in human skeletal muscle in vivo. Am J Physiol. 1998;275(6 Pt 1):E909–16. doi: 10.1152/ajpendo.1998.275.6.E909. [DOI] [PubMed] [Google Scholar]
- Heinonen P, Koulu M, Pesonen U, Karvonen MK, Rissanen A, Laakso M, Valve R, Uusitupa M, Scheinin M. Identification of a three-amino acid deletion in the alpha2B-adrenergic receptor that is associated with reduced basal metabolic rate in obese subjects. J Clin Endocrinol Metab. 1999;84(7):2429–33. doi: 10.1210/jcem.84.7.5818. [DOI] [PubMed] [Google Scholar]
- Hoffstedt J, Poirier O, Thorne A, Lonnqvist F, Herrmann SM, Cambien F, Arner P. Polymorphism of the human beta3-adrenoceptor gene forms a well-conserved haplotype that is associated with moderate obesity and altered receptor function. Diabetes. 1999;48(1):203–5. doi: 10.2337/diabetes.48.1.203. [DOI] [PubMed] [Google Scholar]
- Huttunen P, Hirvonen J, Kinnula V. The occurrence of brown adipose tissue in outdoor workers. Eur J Appl Physiol Occup Physiol. 1981;46(4):339–45. doi: 10.1007/BF00422121. [DOI] [PubMed] [Google Scholar]
- Huttunen P, Kortelainen ML. Long-term alcohol consumption and brown adipose tissue in man. Eur J Appl Physiol Occup Physiol. 1990;60(6):418–24. doi: 10.1007/BF00705030. [DOI] [PubMed] [Google Scholar]
- Jensen MD, Kanaley JA, Reed JE, Sheedy PF. Measurement of abdominal and visceral fat with computed tomography and dual-energy x-ray absorptiometry. Am J Clin Nutr. 1995;61(2):274–8. doi: 10.1093/ajcn/61.2.274. [DOI] [PubMed] [Google Scholar]
- Kirstein SL, Insel PA. Autonomic nervous system pharmacogenomics: a progress report. Pharmacol Rev. 2004;56(1):31–52. doi: 10.1124/pr.56.1.2. [DOI] [PubMed] [Google Scholar]
- Krief S, Lonnqvist F, Raimbault S, Baude B, Van Spronsen A, Arner P, Strosberg AD, Ricquier D, Emorine LJ. Tissue distribution of beta 3-adrenergic receptor mRNA in man. J Clin Invest. 1993;91(1):344–9. doi: 10.1172/JCI116191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenard NR, Berthoud HR. Central and peripheral regulation of food intake and physical activity: pathways and genes. Obesity (Silver Spring) 2008;16(Suppl 3):S11–22. doi: 10.1038/oby.2008.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho-Azcarate T, Marti A, Calabuig J, Martinez JA. Basal fat oxidation and after a peak oxygen consumption test in obese women with a beta2 adrenoceptor gene polymorphism. J Nutr Biochem. 2003;14(5):275–9. doi: 10.1016/s0955-2863(03)00035-4. [DOI] [PubMed] [Google Scholar]
- Macho-Azcarate T, Marti A, Gonzalez A, Martinez JA, Ibanez J. Gln27Glu polymorphism in the beta2 adrenergic receptor gene and lipid metabolism during exercise in obese women. Int J Obes Relat Metab Disord. 2002;26(11):1434–41. doi: 10.1038/sj.ijo.0802129. [DOI] [PubMed] [Google Scholar]
- Marti A, Corbalan MS, Martinez-Gonzalez MA, Martinez JA. TRP64ARG polymorphism of the beta 3-adrenergic receptor gene and obesity risk: effect modification by a sedentary lifestyle. Diabetes Obes Metab. 2002;4(6):428–30. doi: 10.1046/j.1463-1326.2002.00227.x. [DOI] [PubMed] [Google Scholar]
- McArdle WD, Katch FI, Katch VL. Human energy expenditure during rest and physical activity. In: Keifer R, Kerrins R, editors. Exercise physiology: energy, nutrition, and human performance. Baltimore (MD): Lippincott, Williams, and Wilkins; 2007. pp. 195–208. [Google Scholar]
- McCaffery JM, Papandonatos GD, Bond DS, Lyons MJ, Wing RR. Gene × environment interaction of vigorous exercise and body mass index among male Vietnam-era twins. Am J Clin Nutr. 2009;89(4):1011–8. doi: 10.3945/ajcn.2008.27170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCole SD, Shuldiner AR, Brown MD, Moore GE, Ferrell RE, Wilund KR, Huberty A, Douglass LW, Hagberg JM. Beta2- and beta3-adrenergic receptor polymorphisms and exercise hemodynamics in postmenopausal women. J Appl Physiol. 2004;96(2):526–30. doi: 10.1152/japplphysiol.00498.2003. [DOI] [PubMed] [Google Scholar]
- Meirhaeghe A, Helbecque N, Cottel D, Amouyel P. Beta2-adrenoceptor gene polymorphism, body weight, and physical activity. Lancet. 1999;353(9156):896. doi: 10.1016/S0140-6736(99)00251-2. [DOI] [PubMed] [Google Scholar]
- Metcalfe L, Lohman T, Going S, Houtkooper L, Ferreira D, Flint-Wagner H, Guido T, Martin J, Wright J, Cussler E. Postmenopausal women and exercise for the prevention of osteoporosis: The Bone, Estrogen, and Strength Training (BEST) study. ACSM’s Health and Fitness Journal. 2001;5(3):6–14. [Google Scholar]
- Mustelin L, Silventoinen K, Pietilainen K, Rissanen A, Kaprio J. Physical activity reduces the influence of genetic effects on BMI and waist circumference: a study in young adult twins. Int J Obes (Lond) 2009;33(1):29–36. doi: 10.1038/ijo.2008.258. [DOI] [PubMed] [Google Scholar]
- Phares DA, Halverstadt AA, Shuldiner AR, Ferrell RE, Douglass LW, Ryan AS, Goldberg AP, Hagberg JM. Association between body fat response to exercise training and multilocus ADR genotypes. Obes Res. 2004;12(5):807–15. doi: 10.1038/oby.2004.97. [DOI] [PubMed] [Google Scholar]
- Pietri-Rouxel F, St John Manning B, Gros J, Strosberg AD. The biochemical effect of the naturally occurring Trp64-->Arg mutation on human beta3-adrenoceptor activity. Eur J Biochem. 1997;247(3):1174–9. doi: 10.1111/j.1432-1033.1997.01174.x. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Perusse L, Rauramaa R, Rivera MA, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2003 update. Med Sci Sports Exerc. 2004;36(9):1451–69. doi: 10.1249/01.mss.0000139902.42385.5f. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14(4):529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Sivenius K, Lindi V, Niskanen L, Laakso M, Uusitupa M. Effect of a three-amino acid deletion in the alpha2B-adrenergic receptor gene on long-term body weight change in Finnish non-diabetic and type 2 diabetic subjects. Int J Obes Relat Metab Disord. 2001;25(11):1609–14. doi: 10.1038/sj.ijo.0801798. [DOI] [PubMed] [Google Scholar]
- Small KM, Brown KM, Forbes SL, Liggett SB. Polymorphic deletion of three intracellular acidic residues of the alpha 2B-adrenergic receptor decreases G protein-coupled receptor kinase-mediated phosphorylation and desensitization. J Biol Chem. 2001;276(7):4917–22. doi: 10.1074/jbc.M008118200. [DOI] [PubMed] [Google Scholar]
- Small KM, Liggett SB. Identification and functional characterization of alpha(2)-adrenoceptor polymorphisms. Trends Pharmacol Sci. 2001;22(9):471–7. doi: 10.1016/s0165-6147(00)01758-2. [DOI] [PubMed] [Google Scholar]
- Small KM, McGraw DW, Liggett SB. Pharmacology and physiology of human adrenergic receptor polymorphisms. Annu Rev Pharmacol Toxicol. 2003;43:381–411. doi: 10.1146/annurev.pharmtox.43.100901.135823. [DOI] [PubMed] [Google Scholar]
- Tarkovacs G, Blandizzi C, Vizi ES. Functional evidence that alpha 2A-adrenoceptors are responsible for antilipolysis in human abdominal fat cells. Naunyn Schmiedebergs Arch Pharmacol. 1994;349(1):34–41. doi: 10.1007/BF00178203. [DOI] [PubMed] [Google Scholar]
- Teixeira PJ, Going SB, Houtkooper LB, Metcalfe LL, Blew RM, Flint-Wagner HG, Cussler EC, Sardinha LB, Lohman TG. Resistance training in postmenopausal women with and without hormone therapy. Med Sci Sports Exerc. 2003;35(4):555–62. doi: 10.1249/01.MSS.0000058437.17262.11. [DOI] [PubMed] [Google Scholar]
- Umekawa T, Yoshida T, Sakane N, Kogure A, Kondo M, Honjyo H. Trp64Arg mutation of beta3-adrenoceptor gene deteriorates lipolysis induced by beta3-adrenoceptor agonist in human omental adipocytes. Diabetes. 1999;48(1):117–20. doi: 10.2337/diabetes.48.1.117. [DOI] [PubMed] [Google Scholar]
- Wagoner LE, Craft LL, Singh B, Suresh DP, Zengel PW, McGuire N, Abraham WT, Chenier TC, Dorn GW, 2nd, Liggett SB. Polymorphisms of the beta(2)-adrenergic receptor determine exercise capacity in patients with heart failure. Circ Res. 2000;86(8):834–40. doi: 10.1161/01.res.86.8.834. [DOI] [PubMed] [Google Scholar]
- Wolfarth B, Bray MS, Hagberg JM, Perusse L, Rauramaa R, Rivera MA, Roth SM, Rankinen T, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2004 update. Med Sci Sports Exerc. 2005;37(6):881–903. [PubMed] [Google Scholar]
- Yao L, Delmonico MJ, Roth SM, Hand BD, Johns J, Conway J, Douglass L, Hurley BF. Adrenergic receptor genotype influence on midthigh intermuscular fat response to strength training in middle-aged and older adults. J Gerontol A Biol Sci Med Sci. 2007;62(6):658–63. doi: 10.1093/gerona/62.6.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li X, Huang J, Li Y, Thijs L, Wang Z, Lu X, Cao K, Xie S, Staessen JA, Wang JG. Cardiovascular and metabolic phenotypes in relation to the ADRA2B insertion/deletion polymorphism in a Chinese population. J Hypertens. 2005a;23(12):2201–7. doi: 10.1097/01.hjh.0000189869.48290.91. [DOI] [PubMed] [Google Scholar]
- Zhang HF, Li XL, Xie SF, Zhu J, Wang ZZ, Liang LR, Cao KJ, De W, Yuan L, Huang J. ADRA2B gene insertion/deletion polymorphism and artery compliance. Chin Med J (Engl) 2005b;118(21):1797–802. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


