Abstract
Many, perhaps most, proteins, are capable of forming self-propagating, β-sheet (amyloid) aggregates. Amyloid-like aggregates are found in a wide range of diseases and underlie prion-based inheritance. Despite intense interest in amyloids, structural details have only recently begun to be revealed as advances in biophysical approaches, such as hydrogen-deuterium exchange, X-ray crystallography, solid-state nuclear magnetic resonance (SSNMR), and cryoelectron microscopy (cryoEM), have enabled high-resolution insights into their molecular organization. Initial studies found that despite the highly divergent primary structure of different amyloid-forming proteins, amyloids from different sources share many structural similarities. With higher-resolution information, however, it has become clear that, on the molecular level, amyloids comprise a wide diversity of structures. Particularly surprising has been the finding that identical polypeptides can fold into multiple, distinct amyloid conformations and that this structural diversity can lead to distinct heritable prion states or strains.
Keywords: cryoEM, hydrogen-deuterium exchange, prion strains, solid-state NMR
1. INTRODUCTION
As early as the 1700s, observations of “lardaceous” or “waxy” substances in the spleen, liver, and kidneys of deceased patients began to arise (1). These deposits were named amyloid after being incorrectly identified as starchy material. We now know that amyloids are actually self-propagating β-sheet-rich protein aggregates and that many (and perhaps most) proteins are able to form structurally similar amyloid-like aggregates (2). Interest in understanding the structural basis of amyloid formation has been strongly motivated by their association with a number of diseases, such as Alzheimer’s (AD), Parkinson’s, and prion diseases. More recently, it has become clear that there is a role of amyloid formation in normal (nondisease) biology. Here the self-propagating, stable nature of amyloid provides a mechanism for controlling diverse phenomena ranging from epigenetic traits in fungi, to pigment deposition and hormone release in humans (3–5).
Structural studies on amyloid started in the 1930s with a fortuitous observation by Astbury & Dickinson (6). When the structure of stretched unfolded protein aggregates was investigated by X-ray diffraction, a common pattern was observed with two major equatorial bands at around 4.7 and 10 Å. This was interpreted to be the result of backbone and side chain spacing in the aggregates, respectively, essentially describing elongated peptide chains running parallel to each other along the axis of the stretched aggregate. However, for poached egg-white films, the backbone 4.7-Å reflection was now on the meridian, whereas the 10-Å side chain reflection remained equatorial. This became known as the “cross-β” pattern and soon came to represent a diagnostic hallmark of amyloid structure, interpreted as short peptide chains that were parallel to each other but aligned perpendicular to the direction of the stretched aggregate.
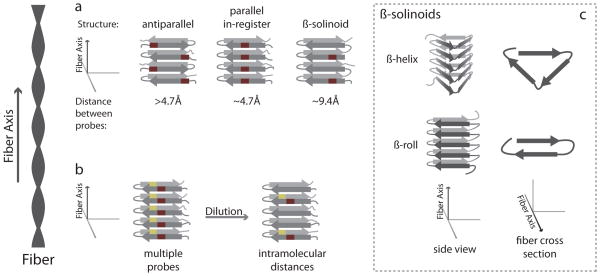
Since the initial observation of the ubiquitous cross-β diffraction pattern in amyloid structure, many more structural traits have been characterized as components of the common amyloid fold. Amyloids are typically long, unbranched fibrous structures formed through the templated homotypic polymerization of hundreds to thousands of monomeric peptides. The fibers are typically 5–15 nm in width and several micrometers in length, and these fibers have similar tinctorial properties, binding the dye congo red and displaying fluorescence bifringence when bound to the dye thioflavin T. Amyloid fibers are also often composed of multiple thin fibers, or protofilaments, that are entwined around each other, giving a twisted ultrastructural appearance with regular periodicity. The protofiliment’s structure involves multiple β-sheets that run parallel to the fiber axis, with individual β-strands perpendicular to the fiber axis. These sheet structures may be arranged in an antiparallel or parallel fashion, although the parallel orientation is most common (Figure 1a)
Figure 1.
Classification of different amyloid folds and techniques to distinguish between them. (a) Amyloid β-sheet structures have largely fallen into three broad categories, all of which can be distinguished from each other by the introduction of a single probe (red rectangle) in identical positions for every monomer and by measuring the distance between probes. In one antiparallel arrangement (left), the probe alternates positions in the fiber structure, resulting in a distance between probes greater than 4.7 Å. In the parallel in-register sheet, the probes are directly adjacent to each other, with a probe distance of ~4.7 Å. In a β-solenoid in which a single monomer forms two layers of the fiber structure, the distance between probes is ~9.4 Å. (b) In systems in which two probes have been introduced (yellow and red rectangles), intramolecular distances can be distinguished from intermolecular distances by diluting labeled protein with unlabeled protein. Dilution increases intermolecular distances while preserving intramolecular distances (right). (c) Types of β-solenoids are depicted schematically. A β-solenoid is a structure in which a monomer loops around in a coil-like manner and forms multiple layers of the fiber structure. Two types of β-solenoid include the β-helix (top), where three β-sheets form a triangular interface, and the β-roll, where two sheets form an interface akin to the β-sandwich.
The recent advances in our understanding of the diversity of amyloid structures have been made possible by the development of complementary structural techniques. Owing to the technique dependency of this field, the section two of this review provides an overview of the common structural techniques used to study amyloid structure. The precise details of how monomers, β-strands, and β-sheets coalesce into fibers, however, can differ greatly between amyloid systems. Thus, the third section provides a more detailed perspective of some of the best-characterized amyloid structures. Finally, as our understanding of amyloid structure has come into focus, it has become more and more evident that amyloid structures can in fact differ within a single amyloid system. This has been particularly significant in the prion field, where the existence of different strains of prion disease has proven to be a formidable hurdle in the acceptance of a causative “protein-only” infectious agent. Structural diversity has provided a plausible and convincing explanation for this phenomenon and is discussed in depth in section four.
2. TECHNIQUES TO STUDY AMYLOID STRUCTURE
2.1. Introduction
Amyloids are typically large (megadalton) elongated structures with varying lengths and often varying ultrastructural appearances. These properties make solution nuclear magnetic resonance (NMR) and X-ray crystallography difficult. In this section, we describe some of the most commonly used techniques employed in amyloid structure characterization, with an emphasis on the higher-resolution methodologies, focusing on the specific contribution of each technique as well as its limitations.
2.2. Fiber Diffraction
Fiber X-ray diffraction was one of the first structural techniques that provided a substantial clue to the overall fold of amyloid fibers (7). In this method, fibrous samples are bombarded with X-ray radiation, and diffraction patterns result from interference patterns from any regularly spaced structural features present in the fibers. As alluded to above, this technique established the cross-β diffraction pattern interpreted as β-sheets parallel to the fiber axis, with β-strands perpendicular (Figure 1a) (6, 7). It has also aided in testing the validity of particular model structures. Here, a theoretical X-ray fiber diffraction pattern is calculated on the basis of a particular model and then is compared to an actual diffraction pattern (8, 9). Despite limitations that make de novo structure determination difficult, fiber X-ray diffraction has played a crucial role in demonstrating the common cross-β nature of the amyloid fold (10) and distinguishing between different structural models.
2.3. Electron and Atomic Force Microscopy
Electron microscopy (EM) and atomic force microscopy (AFM) are the two most widely used microscopy techniques employed in the study of amyloids. Both EM and AFM provide a nanometer-resolution perspective of the ultrastructural characteristics of amyloids. This includes amyloid fiber length and width, morphology such as curvature and persistence length, surface characteristics such as periodic twists, and higher-order assembly. EM and AFM helped establish many of the common ultrastructural characteristics of amyloid fibers, such as the long, relatively straight and unbranched nature of the fibers; the typical fiber width of 5–15 nm; the periodic twist often observed; and the conclusion that many amyloid fibers are made up of the bundling of thinner protofibrils (11–13). To better understand the higher-order assembly of amyloid fibers, scanning transmission electron microscopy (STEM) and tilted-beam transmission electron microscopy, both forms of EM, have provided data that describe the amount of mass-per-unit length (MPL) of amyloids (14, 15). These data are particularly valuable in determining how many monomers make up a single layer of the fiber structure. AFM has also been used to determine the relative fiber rigidity by monitoring its propensity to bend (16). EM and AFM both have the advantage of being single-fiber approaches, which are useful in visualizing structural heterogeneity within a single-fiber preparation.
2.4. Fourier Transform Infrared Spectroscopy and Circular Dichroism
Fourier transform infrared (FTIR) spectroscopy and circular dichroism (CD) are absorptive spectroscopic techniques that measure nonsymmetrical or chiral molecular systems in bulk, FTIR spectroscopy measuring molecular bond vibrational frequencies and CD the differential absorption of left versus right circular polarized light. Both of these properties are highly sensitive to secondary structure, and therefore, deconvoluted FTIR (17) and CD (18) spectra can provide accurate estimations of the contribution of β-sheets, α-helices, and loops to the overall structure. FTIR spectroscopy and CD have thus provided evidence for the transition to a largely β-sheet structure and have been useful in establishing that there are differences in β-sheet content in different fiber preparations of the same protein (19–22).
2.5. Mutagenesis and Introduction of Conjugated Probes
Introduction of single mutations within a sequence opens up a new level of resolution for studying amyloid structure. By scanning a sequence with individual point mutations, one can distinguish regions of the sequence that are important for amyloid structure from those that are not. As amyloids consist mostly of β-sheet structure, the β-sheet breaker, proline, is a natural choice of amino acid. Many such studies have utilized proline scanning to successfully identify regions of structure, and data from these approaches have been highly complementary to other techniques, discussed below, that map out the locations of β-sheets (23–28).
An alternative approach involving mutagenesis introduces a single cysteine residue to provide a unique, chemically active site to couple to a specific structural probe. This cysteine is often labeled with a paramagnetic spin label for use in electron paramagnetic resonance (EPR) (29–36) or a fluorescence probe, such as pyrene, for fluorescence spectroscopy (37, 38). The labeled proteins can then be assembled into the amyloid form, and the probe can report on the structural environment at that precise residue. When performed with an array of singlely labeled proteins, one can get structural information across the entire peptide. With this approach, two major types of structural information can be obtained. First, EPR or fluorescence spectra can indicate the presence or absence of structure at that residue. Second, both EPR and pyrene labeling can be used to measure intra- and intermolecular distances between probes.
Mutagenesis and conjugated probes have two significant drawbacks. First, resolution is limited by the number of residues examined. Thus, to get extensive coverage of a peptide, numerous individual mutations must be made, and data are acquired one by one. More fundamentally, introducing a mutation and a conjugated probe runs a substantial risk of altering or perturbing the very same structure that is trying to be elucidated.
2.6. Hydrogen-Deuterium Exchange
Hydrogen-deuterium (H/D) exchange is based on the principle that a protein possesses many hydrogens that freely exchange with solvent (usually H2O) hydrogens. These exchangeable hydrogens include the amide NH hydrogen that is present in the peptide backbone at every amino acid excluding proline. These backbone amide hydrogens readily exchange with the solvent (39), but this exchange is strongly inhibited when an amide hydrogen is engaged in a hydrogen bond, such as that seen in β-sheets or α-helices. This feature makes H/D exchange particularly well suited for defining regions of a polypeptide that are part of the β-sheet amyloid structure.
One can monitor H/D exchange by placing a protein or amyloid of interest into a D2O-based buffer and following the exchange of protons with deuterons. The rate of exchange is typically monitored through mass spectrometry (HXMS), taking advantage of the fact that deuterons are heavier than protons, or through NMR (HXNMR), taking advantage of the fact that, unlike protons, deuterons do not give an NMR signal. Although HXMS is often simpler and faster, its resolution is limited by the ability to digest a protein into small peptides. In contrast HXNMR potentially gives residue-specific information for each peak assigned in the spectrum. Amyloids, however, are far too large (megadaltons) to be visible by conventional NMR (<70 kDa is optimal). To circumvent this problem, Goto and coworkers developed a novel solvent dissolution strategy using largely anhydrous dimethyl sulfoxide, which prevents further exchange while dissolving the fibers into more NMR-amenable monomers (40, 41)
With spectrum assignments, HXNMR can provide residue-level resolution of the presence of secondary structure in wild-type (WT) and unperturbed protein. It has also illuminated the extreme stability of the amyloid fold (25, 26, 40, 42). This technique has been successful in a number of systems, such as the yeast prion proteins Het-s and Sup35, Aβ, β2-microglobulin, and SH3 domain fibers (25, 26, 40, 42, 43). A significant limitation of HXNMR is the requirement to obtain an assigned and well-dispersed heteronuclear single quantum coherence spectrum, which is often challenging because of the low-sequence complexity (e.g., presence of Asn/Gln-rich repeats) of many amyloid-forming proteins. A more fundamental limitation, however, is the lack of distance restraints as HXNMR only reports on the presence or absence of structure. Thus, HXNMR alone typically cannot provide atomic-resolution molecular models of amyloid structure.
2.7. Cryoelectron Microscopy
Like H/D exchange, cryoEM is a technique that positions itself as a capable alternative to conventional X-ray crystallography and solution NMR in amyloid structure determination. Unlike X-ray crystallography, proteins of interest need not be in a crystalline form for cryoEM; rather, structural data can be acquired on single particles. Furthermore, amyloid preparations do not need to be labeled with stable isotopes, as is the case with NMR, nor do they even need to be highly pure preparations. This is particularly advantageous owing to the inherent heterogeneous nature of the amyloid conformation. Technological and data processing advancements have continuously pushed the resolution of cryoEM making it possible to get high-resolution structural models of amyloid fibers by taking advantage of their helical symmetry through helical reconstruction (44–46).
CryoEM structures of various amyloid conformations have provided a number of different models with common characteristics. They all display a helical twist and have electron densities that are consistent with an extensive β-sheet structure, where individual β-strands run perpendicular to the fiber axis (47–52). In some cases, existing models of the β-sheet structure, determined through other methods, can fit within the cryoEM electron densities, providing consistency across different methodologies (51). Although the structures thus far have provided detailed images, the resolution is close but not quite at the point where the polypeptide can be unambiguously positioned within the electron density. Futhermore, the low contrast of cryoEM images can limit structure elucidation.
2.8. Solid-State Nuclear Magnetic Resonance
Although advances in instrumentation and pulse sequences, such as TROSY, have increased the size limit of what proteins can and cannot be studied by solution NMR (53), amyloid fibers typically still far exceed this threshold. There has been limited success in using solution NMR on amyloids as the only visible peaks in the spectrum correspond to only the most highly mobile, and thus unstructured, regions of the protein (26, 54). Solid-state NMR (SSNMR) provides a viable solution for the study of amyloids as it is tailored for the analysis of noncrystalline solid or solid-like materials, including amyloid fibers. SSNMR studies have resulted in a wealth of new structural data through the use of magic angle spinning, where the SSNMR sample is rotated at high frequency at a particular angle, averaging out chemical shift anisotropy and dipole-dipole couplings, resulting in an increase in sensitivity and resolution (55). This enables a range of high-resolution analyses, including approaches that report on the polypeptide mobility and interresidue distances. Of particular value are approaches that monitor dipolar couplings as couplings between two nuclei are dependant on the distance that separates them. These dipolar couplings are typically averaged out with magic angle spinning; however, they can be recoupled using specific pulse sequences. The ability to recouple these dipoles is dependent on the dipole coupling strength and thus can be used to measure the distance between nuclei within 6 Å (56). This technique has successfully distinguished antiparallel from “in-register” parallel β-sheet structures because distances between identical nuclei in these two models differ substantially (Figure 1a) (57–70). Dilution experiments can also help distinguish between intra- and intermolecular distances, providing quaternary structure information (Figure 1b).
Even in the absence of complete structural analysis, SSNMR can provide key information on the secondary structure of specific residues in an amyloid as carbon nuclei, particularly the Cα, Cβ, and CO, have characteristic chemical shifts based upon the secondary structure and torsion angles in which a particular nucleus is engaged (55). Thus, chemical shifts at each residue of the peptide can map out the location of β-strands and unstructured or loop regions. The information gleaned from this technique is thus similar to that acquired from HXNMR, and as such, the two techniques have been successful at confirming each other’s results (25). Additional recent techniques similarly identify the presence or absence of structure using pulse sequences that selectively eliminate mobile or immobile residues (62, 63)
Because SSNMR is an NMR technique, like HXNMR, it requires the protein of interest to be labeled with an appropriate NMR-visible nucleus, typically 15N or 13C. This can be both a blessing and a challenge. As many amyloids are made up of short peptides, the individual peptides can be synthesized chemically with 15N or 13C residues inserted into specific parts of the sequence, allowing the uncluttered study of only those labeled nuclei. This has been most helpful in SSNMR experiments that measure very specific qualities, such as the distances between two nuclei of interest. Consequently, however, this requires some prior knowledge about where to place labels that will yield positive and productive information. Other amyloids are composed of larger proteins that cannot be chemically synthesized, and the entire protein needs to be labeled, or at least one amino acid type (25, 57, 60–62, 64, 71). The large increase in the number of labels can result in multiple peaks that cannot be resolved nor assigned to particular residues, making these uniformly labeled experiments sometimes less helpful, although there are examples where uniformly labeled samples have not been a problem (25, 57). A possible solution to this difficulty has emerged from chemical ligation, where a chemically synthesized peptide is ligated to a biosynthetically produced fragment of any size, allowing specific incorporation of single or multiple labels into the synthesized peptide (72). Although laborious, protein ligation allows detailed structural SSNMR analyses of longer proteins.
2.9. X-Ray Crystallography
Although the large and heterogeneous nature of amyloid fibers would seem to be incompatible with X-ray crystallography, Eisenberg’s group (73) has enjoyed some remarkable success with this technique. Eisenberg and colleagues have been able to acquire adequate diffraction data from microcrystals of short peptides (6–7 residues) that formed amyloid-like structures (73). The resulting structures bore strong resemblance to many existing models of amyloid fibers, suggesting they may in fact represent the structure of the amyloid fold. Since this initial study, multiple amyloidogenic peptides have been successfully crystallized and their structures solved (74–76).
X-ray crystallography of these short peptides has provided the most detailed and high-resolution view of amyloid fiber structure. A major limitation of this technique is similar to conventional X-ray crystallography in that not all peptides form crystals. More significantly, however, is that current structures are only of 4–7 residue peptides that are far smaller than their parental full-length proteins. Thus, the wealth of information needs to be validated by other methods that can analyze these sequences in the context of the full-length protein.
3. AMYLOID STRUCTURES
3.1. Introduction
Using these structural approaches in concert, a highly detailed perspective of several amyloid structures has begun to emerge. Although these structures share common core characteristics as described in Section 2, there are significant differences in the details of each fold. In Section 3, we introduce different classes of amyloid structure. We then describe three representative structures that are some of the best-characterized amyloid systems to date. The first is the fungal prion [Het-s] from Podospora anserine, next the AD–associated aggregate composed of the peptide Aβ, and finally the X-ray crystallography structure of short amyloidogenic peptides from a variety of amyloid systems. These structures represent our most detailed view of the spectrum of amyloid folds and provide a template that other amyloid structures are likely to resemble.
3.2. Elements of the Amyloid Fold
The primary structural characteristic of amyloid fibers is a predominantly β-sheet structure. These β-sheets, however, can come in two major forms: parallel and antiparallel (Figure 1a). The parallel β-sheet has another important designation, which is based on whether it is in-register or out of register. In-register refers to the instance where, within a β-sheet, each β-strand is composed of identical residues on different molecules that are aligned in register with respect to each other such that each residue is adjacent to the identical residue on each neighboring molecule (Figure 1a). The majority of amyloids studied to date appear to have an in-register parallel β-sheet structure; however, there are some that are parallel but pseudo-in-register (25, 57), and others that are antiparallel (70, 77, 78).
β-sheets can coalesce into a tertiary fold in a number of ways that are found in amyloid structures. The first is termed a β-sandwich, where two β-sheets come face-to-face often separated by a loop hinge region. If this pattern is continued, it can form a superpleated β-sheet structure, where several β-sheets form stacks aligned in the same direction. An alternative cross β-sheet structure found in amyloid is the “β-solinoid,” where the same polypeptide chain forms a single 4.7-Å layer, or rung, of the fiber and then loops back around and forms a second 4.7-Å layer on top of the first. Because a single polypeptide makes up two or more rungs of the structure, it cannot be in register as the neighboring β-strands come from a different part of the same or neighboring protein. However, they can be pseudo in register if the neighboring β-strands share high sequence similarity. The β-solinoid can come in two major flavors (Figure 1c). The first is the classic β-helix, where three β-strands often form a triangular layer and loop back to form an additional layer. The other is the β-roll, where a single layer is composed of two β-strands and resembles the β-sandwich but loops back to form a second identical layer. The first described structure, the [Het-s] prion, represents an example of a β-solinoid structure, whereas Aβ can be thought of as a type of β-sandwich.
3.3. Het-s Structure
[Het-s] is a prion found in the filamentous fungus Podospora anserina, participating in a functional role in heterokaryon incompatibility (79). The genes responsible for this heterokaryon incompatibility are referred to as het genes, and in Podospora, come in two forms, Het-s and Het-S. Only the 289 residue protein Het-s can exist in both a nonprion ([Het-s*]), and prion ([Het-s]) state, and only the prion-positive [Het-s] state confers the heterokaryon incompatibility function (79). [Het-s] is a true prion with a gain-of-function physiologically productive role (80). Despite the fact that this prion is not toxic and even “native” to the host organism, its structure displays all the hallmarks of its toxic and nonnative amyloid cousins.
Initial structural studies on Het-s fibers began with the demonstration that recombinant Het-s proteins would spontaneously form amyloid-like fibers in vitro 81) and that these fibers were indeed the physiologically relevant prion form because their introduction into [Het-s*] cells caused conversion to the [Het-s] state (80). FTIR spectroscopy and CD showed that Het-s fibers were enriched in β-sheet content (81), and protease treatment of the fibers produced a small protease-resistant ~7-kDa fragment that mapped to a C-terminal domain between residues 218–289 (82). Subsequent experiments showed that the N-terminal region of Het-s had a globular fold, and the 218–289 domain was natively unstructured in the nonfibrillar form and was necessary for the prion activity, implicating this region as a putative prion aggregation domain.
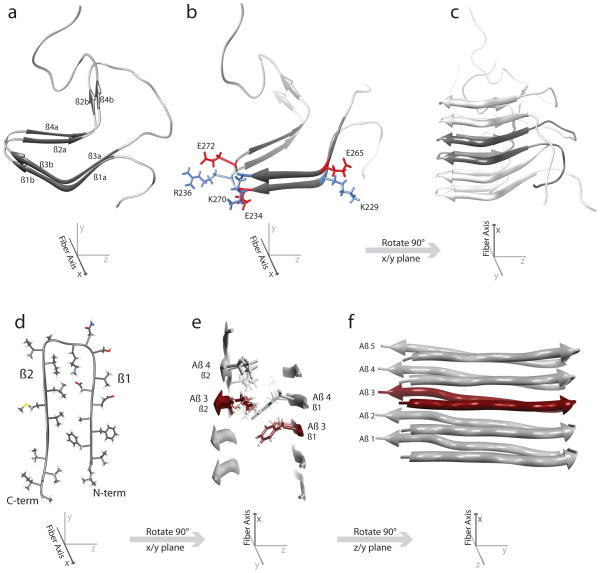
Further work concentrated on the Het-s 218–289 domain, showing through HXMS that this domain became substantially protected, and thus structured, in the fiber form (83, 84). Although the resolution of the HXMS was high, precise mapping of secondary structure did not come until a study that employed HXNMR (25). This study identified four β-strands within the 218–289 domain: residues 226–234 (β1), 237–245 (β2), 262–270 (β3), and 273–282 (β4). SSNMR on these Het-s fibers produced beautiful spectra with well-dispersed peaks; most of these peaks could be unambiguously assigned to specific nuclei on Het-s 218–289. On the basis of chemical shifts, the SSNMR largely confirmed the existence and location of the four β-strands identified by HXNMR, which were again confirmed through proline mutagenesis. With the identification of the four β-strands along with the observed sequence similarity between the strands β1/β3 and β2/β4, a model was postulated that positioned the four strands in a β-roll structure, where a β1-loop-β2 motif formed a single layer and a β3-loop-β4 motif formed a second layer directly on top and in-line with the first layer. This structure predicted a MPL value of one Het-s unit per 0.94 nm, which was precisely what was observed in a later STEM study (85). Though a putative model for Het-s fibers had been made, it was formulated without the insight of any distance restraints. A follow-up SSNMR study provided these needed measurements, resulting in significant, but not radical, modifications to the β-roll model (57). This study used a combination of labeling techniques and dilution experiments to establish 134 total distance restraints, 23 hydrogen bond restraints, and 74 dihedral angle restraints. With these data and previous observations in hand, an atomic model was formulated (Figure 2c). This model revealed a similar overall structure to the original model; however, each previously proposed β-strand actually represented two β-strands (now called a and b). Six of these strands contributed to a β-solenoid structure (β1a,b,β2a and β3a,b, β4a) and two lay outside the solenoid (β2b and β4b) (Figure 2a). The inside of the solenoid formed a dry, hydrophobic core, with most charged and polar side chains pointing outward. There were also several salt bridges (Figure 2b), both intra- and intermolecular, and two asparagine “ladders,” where vertically stacked asparagine side chains form a ladder of hydrogen bonds, all providing substantial stability to the structure. The salt bridges, in particular, were suggested through molecular dynamics simulations to be dynamic and provide considerable thermostability to the structure (86).
Figure 2.
(a–c) Model structure of the Het-s prion fiber [Protein Data Bank (PDB): 2RNM]. (a) A single Het-s monomer is shown in the fiber conformation down the fiber axis. Here, the eight β-strands and the β-helix structure are clearly seen. β1a/β 3a, β1b/β3b, and β2a/β4a for the three β-sheets, respectively, with β2b/β4b lying outside the helix structure. (b) Similar view of the Het-s fiber structure with specific acidic side chains (red) and basic side chains (blue) shown to illustrate the salt bridges that form in the fiber structure. (c) The full fiber structure from a view perpendicular to the fiber axis. A single monomer is highlighted in dark gray. (d–f) Model structure of Aβ1-42 (PDB: 2BEG). (d) A single Aβ1–42 monomer in the fiber conformation is shown down the fiber axis. The N-terminal β-strand and the C-terminal β-strand are labeled β1 and β2, respectively. (e) A view of Aβ1-42 fiber structure from the side, perpendicular to the fiber axis. A single Aβ1-42 monomer is shown in red, with the neighboring monomer in gray. This illustrates how the side chains from one unit in this structure (Aβ 3, strand β2) form an interface with the neighboring molecule (Aβ 4, strand β1). (f) Full structure of Aβ1-42 viewed perpendicular to the fiber axis. A single monomer is displayed in red, with the others in gray.
3.4. Aβ Structure
Two types of aggregates are typically found in the brains of AD patients. Intracellular neurofibrillary tangles are aggregates composed of the hyperphosphorylated protein tau, whereas extracellular plaques are made up of aggregates of the Aβ peptide (87, 88). Aβ is a 39–42-residue peptide that is derived from the membrane protein amyloid-β precursor protein following proteolytic cleavage by β- and γ-secretases (89). Mutations in and around the Aβ sequence have been found to be associated with early-onset AD, as are mutations that result in an elevated extracellular Aβ concentration, although Aβ production seems to be a normal occurrence in healthy individuals (90). Of the multiple forms, Aβ1–42 (where subscripts denote the residues the peptide spans) has been found to be the most toxic and aggregation prone (91, 92). Like other amyloids, Aβ aggregates through a templated polymerization model both in vitro and in vivo. Despite an incomplete understanding of the role of the amyloid form of Aβ in AD, the structures of these aggregates are nonetheless of great interest and have motivated the use of the full arsenal of structural techniques employed in the study of amyloid structure.
Even before the protein responsible for the extracellular amyloid plaques in AD patients was identified, X-ray fiber diffraction had revealed the common cross-β diffraction pattern from extracellular plaques isolated from AD patients (93). Once the peptides from these inclusion bodies were identified, aggregates formed in vitro from these sequences were also found to produce the cross-β diffraction pattern (94, 95). Structural studies continued on the most abundant variants, Aβ1–39, Aβ1–40, and the more aggregation-prone Aβ1–42, which differed simply by the addition of amino acids on the C terminus (87, 91, 92, 96–98). FTIR spectroscopy and CD showed that these truncated Aβ peptide aggregates were composed of primarily β-sheet structure, with FTIR spectroscopy suggesting that these β-sheets are in an antiparallel arrangement (70).
SSNMR has played a critical role in the high-resolution structural determination of Aβ fibers. Initial studies on the Aβ34–42 peptide amyloid suggested the peptide was in an extended β-sheet conformation consisting of a single straight strand per molecule, positioned in an antiparallel arrangement similar to that suggested by FTIR spectroscopy (70). An alternative study, however, found Aβ10–35 had very short distances between single 13C nuclei, an observation only possible if the peptides were in an in-register parallel sheet arrangement (67). Similar studies were performed on other truncations, with Aβ16–22, Aβ11–25, and Aβ14–23 all displaying an antiparallel β-sheet structure (77, 99, 100). When SSNMR was finally performed on full-length Aβ1–40 and Aβ1–42 fibers, these structures were found to be in an in-register parallel β-sheet pattern (67, 101). What could account for these discrepancies? A follow-up study observed that the C terminus of the longer peptides (10–35, 1–40, and 1–42) is hydrophobic and amphiphilic and that the juxtaposition of this region with a nonhydrophobic N terminus, as would occur in an antiparallel β-sheet, would be disfavored (99). When the authors then acylated Aβ16–22 to make it more amphiphilic, they found that this peptide then adopted an in-register parallel β-sheet structure. Thus, the antiparallel β-sheet may have been an artifact resulting from the truncation of hydrophobic and amphiphilic regions on Aβ.
Although full-length Aβ peptides seemed to adopt an in-register parallel β-sheet structure, the location, organization, and packing of this structure was not known. HXMS showed that approximately 50% of the peptide quickly exchanged with the solvent, providing a rough estimate of the structure content (102). Analysis of SSNMR spectra revealed that the N-terminal residues had broader line widths, suggestive of disordered or nonexistent structure in this region (103). HXMS, HXNMR, proline-scanning mutagenesis, and EPR spectroscopy similarly all pointed toward the disordered N terminus and two, or three, C-terminal β-strands (27, 35, 104, 105). Further refinement of the location of these strands, however, was still needed.
Numerous SSNMR and HXNMR studies on full-length Aβ peptides provided the resolution and refinement necessary to provide much of the missing details (42, 69, 106–108). Chemical shift values of the labeled residues with predicted torsion angles identified two regions that corresponded to β-strands, 12–24 and 30–40, with a loop region in between (69). Meanwhile, HXNMR studies on Aβ1–42 provided slightly shifted regions of β-strands, spanning roughly from 18–26 and 31–42 (42). Both of these structures contained two β-strand segments separated by a loop region. The width of Aβ fibers as determined by EM, however, is far too narrow to allow both β-strand segments to extend along the same line; thus, the β-strands must fold back on each other.
Determining how these β-strands folded back was achieved through two independent methods. First, SSNMR on Aβ1–40 was used to determine torsion angles at the predicted loop regions (69). Inter- and intramolecular distance restraints were also made, including the identification of a salt bridge between D23 and K28 (109). This positions the peptide in a β-turn-β structure with the polypeptide chain forming a 180° loop back on itself. Also in this model, odd residues in β-strand 1 (β1) form an intramolecular interface with the even residues in β-strand 2 (β2). An alternative approach introduced the point mutations K28D, D23K, F19G, and G38F into the Aβ1–42 sequence (42). When assayed for fiber-forming ability, none of the mutants formed fibers with identical appearance to WT fibers. It was found, however, that the double mutants K28D/D23K and F19G/G38F, and the quadruple mutant F19A/A21F/V36G/G38F, all restored the ability to form WT-like fibers. This suggested β-strand/β-strand contacts between K28/D23, F19/G38, and A21/V36. When these mutants were tested for their ability to coaggregate with WT Aβ, however, none did. Only when pairs of single-point mutants were mixed in equal ratios with WT (e.g., K28D, D23K, WT) was coaggregation observed. These data suggested a β-sandwich similar to that observed by SSNMR with the odd residues in β1 forming an interface with the even residues on β2, but also indicated domain swapping between peptides, where the β2 on one peptide interfaces with β1 on the next peptide (Figure 2d–f). This is in contrast to the SSNMR structure that advocated a β-sandwich formed by a single peptide lying in the same plane as one layer of the fiber structure (109). Regardless, for both Aβ1–40 and Aβ1–42, the basic fold of an Aβ monomer appears to be a β-strand followed by a hairpin loop and then another β-strand that folds back on itself.
How each of these individual Aβ units come together to form the full amyloid fiber would still require further refinement. As described above, the fiber is arranged in an in-register β-sheet structure. Thus, each Aβ monomer stacks on top of each other exactly in line. MPL data suggested two or three Aβ units per 4.7-Å layer, requiring some sort of horizontal packing of Aβ monomers (15, 67, 110). SSNMR data again provided distance restraints, resulting in a model where the odd residue sides of β2 strands from two Aβ monomers form an intermolecular interface, lying in the same plane (109). Determining the higher-order assembly of an Aβ monomer into the fiber structure has also benefited greatly from cryoEM. A number of studies have produced multiple structures that were constructed from images of a homogeneous population of fibers and even from single fibers (49–51, 111). Though the resolution of these studies is not yet high enough to unambiguously fit the Aβ peptide into the model densities, they do provide enough information to suggest how each Aβ monomer might interface with one other. Some resulting models of Aβ1–40 closely resemble those suggested by the above SSNMR studies (49). Another cryoEM model of Aβ1–42, however, had yet another alternative arrangement of Aβ monomers, suggesting pairs of β-sheets separated by a significant distance (51). This structure differs significantly from other models in that the distance between the pairs of sheets results in a hollow core, which is something predicted and seen in other amyloids, and observed for Aβ in the form of an “electron-luscent” core (47, 112–114). The proposed models of a β-sandwich formed by a single Aβ monomer does, however, fit into the densities of this model.
Through the use of the full spectrum of techniques available for exploring amyloid structure, our understanding of Aβ fiber structure has become one of the most comprehensive of all amyloids. Nonetheless, there remain important issues to resolve. The precise locations of β-strands and loops, although in general agreement, are perhaps not fully settled. Furthermore, how these β-strands and loops position themselves internally and in conjunction with other Aβ units also has not been unequivocally established. Despite these open questions, the current view of Aβ structure, in-register parallel β-strands following a serpentine pattern, embodies many aspects of a canonical structure that many other amyloids are likely to resemble.
3.5. Peptide Structures by X-Ray Crystallography
As briefly described above, X-ray crystallography has experienced limited success in the study of amyloid structure involving long polypeptides. In contrast, crystallographic studies of amyloid-forming peptides have provided a wealth of structural information and insight (73–76). X-ray crystallography on amyloids began in earnest with an observation that six- and seven-residue peptides from the yeast prion protein Sup35 spontaneously formed microcrystals in solution (115). These crystals had some properties of amyloid fibers, such as the cross-β diffraction pattern, and it was known that the peptide on its own could polymerize off of full-length Sup35 fibers. These crystals, however, were far too small for data acquisition on conventional X-ray sources and required a microfocus beam line for data acquisition.
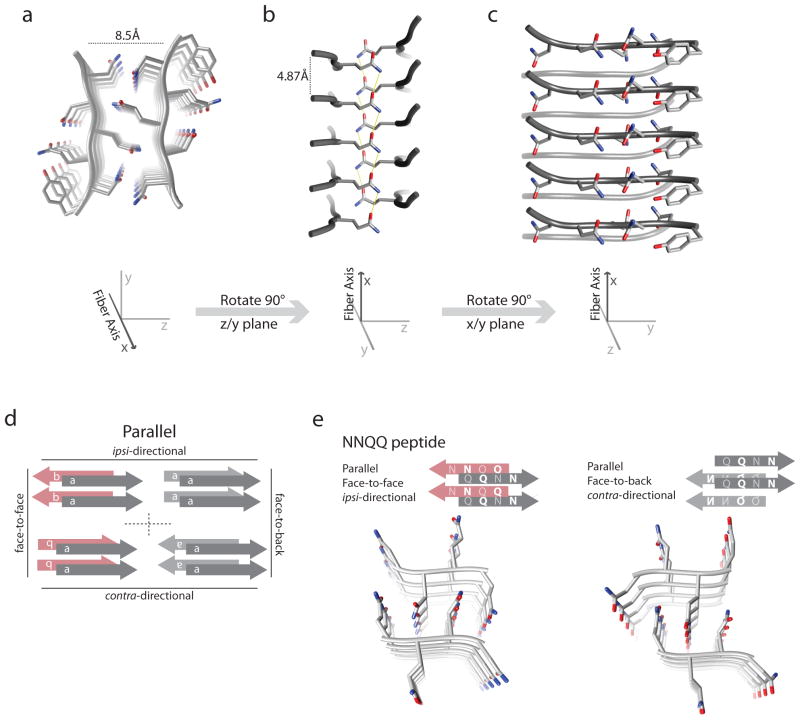
The resulting atomic structures of these two short peptides confirmed a number of observations and predictions observed in other amyloid systems (Figure 3) (73). The main unit of the structure was two β-sheets that ran along the axis of the fiber, separated by a distance of 8.5 Å (Figure 3a). Individual β-strands within each sheet were aligned perpendicular to the fiber axis and spaced 4.87 Å apart, precisely what the cross-β diffraction pattern suggested (Figure 3b). Similar to the SSNMR and HXNMR models for Aβ, the two sheets came together along the long edge of the sheet; however, the two sheets were independent of each other rather than being hinged by loop regions as seen in Aβ.
Figure 3.
(a–c) X-ray structure of the peptide GNNQQNY. (a) A view of the peptide structure down the fiber axis. Visible is the interdigitation of side chains from opposing sheets forming a dry interface devoid of water. (b) The peptide structure viewed from the side, perpendicular to the fiber axis. Visible is the interdigitation of one set of side chains between side chains of opposed peptides above and below. Hydrogen bonds (yellow) that form between asparagine side chains are shown, forming the asparagine ladder structure. (c) Peptide structure viewed face on, perpendicular to the fiber axis. (d–e) Different orientations of the peptide structures. (d) A schematic of some of the possible peptide structures. Peptide structures can differ in three main criteria: parallel versus antiparallel β-sheets, sheets that are face-to-face versus face-to-back, and sheets that are ipsidirectional versus contradirectional. Only parallel arrangements are displayed, where the individual β-strands within a sheet are all oriented in the same direction. Face-to-face versus face-to-back depends on the face of the sheets that forms the interface between the sheets. If this interface is formed by identical faces, it is classified as face-to-face, whereas if the interface is formed by opposite faces, it is classified as face-to-back. The different faces in the schematic are represented by a letter and color (face 1 is labeled a and gray, and face 2 is b and red). Ipsi- versus contradirectional is distinguished by the direction of a particular reference hydrogen bond within a sheet. If the direction of this hydrogen bond is the same for both opposing sheets, it is classified as ipsidirectional, whereas if the hydrogen bond is in opposing directions, it is classified as contradirectional. The direction of each sheet is illustrated by the orientation of the letter of each strand (correct side up versus upside down). (e) The NNQQ peptide in two different conformations, with the corresponding schematic to illustrate the orientations. On the left is NNQQ in a parallel, face-to-face ipsidirectional orientation. On the right is NNQQ peptide in a parallel, face-to-back contradirectional orientation.
Beyond these observations, a number of characteristics not before seen were discovered in the peptide structure. First, the interface between the two β-sheets was very tight, completely excluding any water molecules and was thus referred to as a dry interface. Second, the β-strands in one sheet were slightly offset from the opposing β-sheet such that the side chains from one sheet nestled in between opposing side chains from the opposite β-sheet (Figure 3b). This side chain packing pattern was largely devoid of any hydrogen bonds, relying more on van der Waals interactions with the exception of stacks of Asn and Gln where hydrogen bonds were formed to equivalent side chains above and below on neighboring peptides (Figure 3b). The tight interface in which these side chains carefully interdigitated with each other was termed a steric zipper and has similarities to a polar zipper, previously postulated by Perutz (116).
X-ray crystallography structures of various amyloidogenic peptides have clearly contributed a level of resolution that no other technique has been able to achieve. This advancement in resolution provides new insights into possible fiber structure, including the dry interface and steric zipper. What remains to be seen is if the use of this technique can be extended to peptides longer than seven residues. Most amyloidogenic proteins are in excess of 40 residues. How well the short peptide structures translate into the context of the full-length protein remains to be seen.
4. STRUCTURAL HETEROGENEITY
4.1. Introduction
One of the earliest lessons learned from the structural characterization of amyloids is that there exists a great diversity in the conformations the aggregates can adopt (12). This was particularly crucial in the mammalian prion field, where the existence of different prion disease strains provided a formidable challenge to the protein-only prion hypothesis (117, 118). How could a protein-only infectious agent cause different disease phenotypes in genetically identical animals? Within the context of the protein-only hypothesis, the only explanation for this strain phenomenon was that there existed multiple infectious conformations of the prion protein, each corresponding to a specific disease manifestation (119, 120). As radical as this hypothesis seemed at the time, there is now compelling evidence that this is in fact the case (32, 121–123). The yeast prion system [PSI+], which results from self-propagating aggregates of Sup35, has been particularly valuable for studying the connection between amyloid conformation and heritable differences in prion strain phenotypes (26, 32, 124). For [PSI+], it has been possible to create distinct prion conformations of Sup35 in vitro and to show that infection with these different conformations leads to different prion strains. These studies demonstrated that strains are “enciphered” in self-propagating differences in the conformation of the infectious amyloid of Sup35. Additionally, this ability to create distinct infectious conformations has made it possible to study the structural differences underlying the different strain conformations and reveal the mechanism by which structural and physical properties of the different aggregates relate to their corresponding in vivo phenotypes (26, 124).
Conformational diversity is not limited to prion aggregates and has appeared as recurring theme in a number of amyloid systems. In fact, in the first characterization of amyloid structure, ultrastructural differences between different fibers of a single preparation were observed (12). Despite the difficulty of studying these nonprion amyloids, the gamut of possible conformations these amyloidogenic peptides can form is now coming into focus. As we begin to characterize this spectrum of structures, it will be interesting to see, as in the prion amyloids, what physiological consequences structural diversity imparts in various disease contexts. The next sections detail a subset of some of the better-characterized examples of structural heterogeneity in mammalian prions, yeast prions, and nonprion amyloids.
4.2. Conformational Diversity in Mammalian Prions
Conformational heterogeneity has perhaps been most significant in the field of prions. A prion, as described by Stanley Prusiner in 1982, is “a small proteinaceous infectious particle which is resistant to inactivation by most procedures that modify nucleic acids” (125). Initially allowing the presence of a small amount of nucleic acid, the prion hypothesis evolved to more narrowly describe the infectious agent as composed solely of protein (the prion protein, Prp) and as completely devoid of nucleic acid (126). As a genome-less pathogen was a lengthy divergence from known infectious agents at the time, such as bacteria and viruses, the prion protein-only hypothesis experienced a tumultuous journey to widespread acceptance.
Perhaps one of the more challenging arguments against the protein-only hypothesis was the existence of strains in prion diseases. Strains were first observed as different isolates of the infectious agent that gave rise to different disease phenotypes in the form of incubation times and histopathology when introduced into genetically identical animals (117). Different disease strains are easily explained by differences in the infectious agent’s genome. However, in the absence of any nucleic acid genome, as was proposed to be the case with prions, how could different strains possibly arise? This was a large part of an argument against the protein-only hypothesis when in 1987 Bruce & Dickinson wrote regarding the scrapie prion in sheep, “The considerable strain diversity in scrapie, together with the evidence for mutational change presented here and elsewhere, offer compelling arguments that scrapie has its own independently replicating genome” (118, p. 88). Experiments with two different strains of prion-based transmissible mink encephalopathy, however, offered a possible explanation for this phenomenon. Bessen & Marsh (119, 120) found that the infectious agent that caused the drowsy and hyper strains in transmissible mink encephalopathy had different sensitivities to proteases despite being made up of the same prion protein. This suggested that prion strains were the result of conformational heterogeneity; a model that bypassed the need for a nucleic acid genome.
Despite this study and increasing evidence from others, there remained resistance to not only the protein-only prion hypothesis, but also to the possibility that prion strains result from distinct infectious conformations of the prion protein. How could a protein fold into multiple stable conformations? How is the information encoding these distinct conformations accurately propagated to new hosts? The answers to these questions and consensus on the protein-only hypothesis could not be satisfied without a better understanding of the structures of these infectious particles, as pointed out by Robertson et al. when they wrote that “Final judgment about the nature of the scrapie agent will not be possible until the structures of the prion protein and any associated nucleic acid can be related to scrapie strain specificity” (127, p. 727). Thus, structural insights into prion particles were not only a matter of curiosity; rather, they held the possible solution to the prion strain enigma as well as bore some of the burden of proof for the protein-only hypothesis.
Since the protein-only hypothesis was first introduced in 1982, much progress has been made in the structural characterization of these proteinaceous infectious particles. As described above, prions adopt elongated, fibrous structures that result from the templated polymerization of the prion protein. The ultrastructural characteristics and templating mechanism of prion aggregation closely mirror that of other amyloids and as such have followed a similar trajectory of structure elucidation (128). Although the first indications that prion strains relied on an aggregate conformation were found over 15 years ago, high-resolution insight into different conformations of prion aggregates and a detailed molecular understanding of the consequences of these structural deviations on prion strain phenotypes, for mammalian prions, remain a critical goal in the field.
One of the primary difficulties with studying mammalian prion strain structures is isolating appropriate particles that represent the true infectious material. A given prion strain aggregate may not represent a single infectious conformation; rather, it may be composed of a spectrum of structures, not all of which may act as a true templating and toxic aggregate. Despite the difficulty in producing biochemical amounts of structurally homoegenous physiologically accurate infectious mammalian prions (129), remarkable progress has been made into providing a structural link between prion conformation and disease strains. An early step toward this goal was demonstrated using human prions when two different prion strains were successfully propagated in genetically identical mice, mirroring the initial results seen earlier by Bessen & Marsh (123). More recent studies have utilized “synthetic prions” wherein the prion particles originate from an in vitro source (130) and a method called protein misfolding cyclic amplification, which uses infectious material to convert noninfecious brain homogenates (131, 132). These infectious preparations have proven to be competent for structural studies and have provided strong evidence for the conformation/disease-strain relationship (122, 130, 133–135). As these new amplification techniques will enable more comprehensive structural analyses, it will be exciting to see what conformational diversity lies in mammalian prions.
4.3. Conformational Diversity in Yeast Prions
Although prions were first hypothesized as the infectious agent for a class of diseases known collectively as transmissible spongiform enchphalopathies, they were later proposed to be the underlying causative agent for a number of epigenetic traits in fungi (136, 137). The yeast prion systems have provided a powerful platform for studying the conformational basis of prion strain variants (138–141). The yeast [PSI+] prion state can be monitored through a convenient color-based phenotypic readout wherein [psi−] (nonprion) cells adopt a red color phenotype, and [PSI+] (prion) cells have a white color phenotype (142). [PSI+] strains are manifested in the form of different shades of color, ranging from dark pink to white, and these color phenotypes remain stable upon multiple passages (138). Precise control over which prion strain is adopted was demonstrated for two strains named Sc4 (white) and Sc37 (pink) (32, 121), where polymerization at 4°C and 37°C resulted in aggregates that conferred the Sc4 and Sc37 [PSI+] strains, respectively, when introduced into [psi−] yeast (32). Owing to the loss-of-Sup35p-function nature of the [PSI+] prion, the actual differences in color phenotypes could be explained by differing residual quantities of soluble, and therefore functional, Sup35p in the [PSI+] backgrounds. What could influence the levels of unaggregated protein in these cells? Subsequent biophysical studies began to shed light on this as it was demonstrated that these two fiber preparations differed in their physical characteristics, such as in their ability to be broken and fiber polymerization rate. In the case of Sc4, it is the propensity of the prion to be broken, thereby generating new seeds, which is the key determinant of how strong an impact it will have on a cell. Thus, less-stable prions paradoxically lead to more intense phenotypes (124). Although these observations provided convincing evidence that prion strains were caused by different aggregate conformations as well as by a possible molecular mechanism of phenotype manifestation, they did not provide direct insight into the actual structural differences of these aggregates.
Cysteine mutants with coupled EPR or fluorescent probes as well as FTIR spectroscopy provided early glimpses of the conformational differences between Sc4 and Sc37, albeit at a relatively low resolution (32, 37, 143). FTIR spectroscopy suggested less structure in the Sc4 conformation (143), whereas EPR probe mobility and coupled fluorescent probes at single cysteine residues similarly suggested that Sc37 had a more ridged and extended structure compared to Sc4 (32, 37, 143). Another study that used cysteine-coupled fluorescent probes found that the Sc4 conformation had a smaller core structure and melted at a lower concentration of guanidine than the Sc37 conformation (37). Identification of the “head” and “tail” regions also revealed potentially different tail-tail contacts between these two conformations.
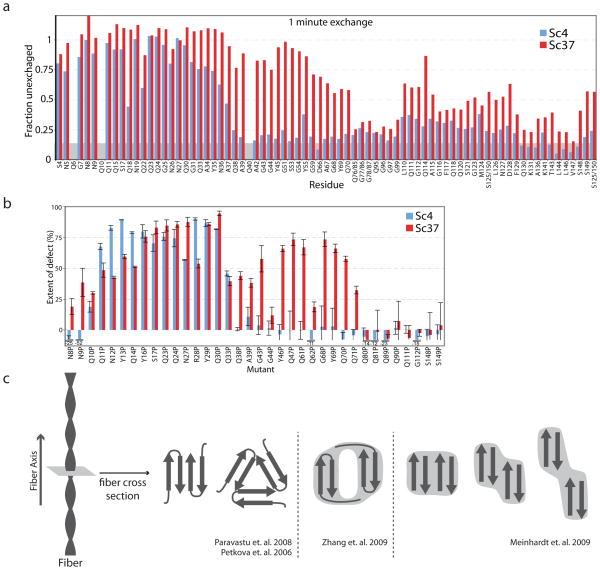
A more detailed picture emerged with a study that utilized HXNMR to probe the Sc4 and Sc37 structures (26). This study found that Sc4 exhibited an extremely stable structure from residues 8–37, whereas Sc37 was structured from residues 5–70 (Figure 4a). This represents an almost doubling of structure in the Sc37 conformation, with evidence that the region of overlapping structure (residues 8–37) represented distinct conformations. Thus, Sc4 and Sc37 most likely form two different folds on the monomeric level, rather than having an identical “amyloid core” with Sc37 displaying an expansion of structure beyond that core. Subsequent mutagenesis analysis from this study, and another work, corroborated this overlapping amyloid core and strain-specific expansion of structure (Figure 4b) (23, 26).
Figure 4.
(a–b) Demonstration of the conformational differences between the yeast prion, strong and weak, derived from the fibers Sc4 and Sc37, respectively. (a) HXNMR data after 1 min of exchange for SupNM residues 4–150 in the Sc4 (blue) and Sc37 (red) conformations. (b) Mutation analysis of Sc4 and Sc37 conformations. Indicated mutants on the horizontal axis were made, and their ability to form the Sc4 (blue) and Sc37 (red) conformations evaluated. The larger the defect in forming these conformations, the higher the value on the y-axis. (c) Conformational diversity of Aβ fibers. Shown are the cross-sectional views of several models of Aβ fiber structures based on SSNMR (left: 108, 109) and cryoEM (middle: 51, right: 49) Light gray areas approximate the electron density observed in the cryoEM structures.
A different and complementary perspective on strain differences has come from the Eisenberg group’s (74, 75) X-ray structural analysis of a number of peptide microcrystals derived from the Sup35 sequence and other amyloids similar to the previously described peptide structure. Sawaya and coworkers (74) successfully solved the structures of peptides with sequences derived from a variety of amyloid sequences including Tau, Aβ, insulin, and amylin. The structure of all these peptides shared a number global similarities to the original Sup35 peptide structure, such as the cross-β sheet structures, the dry interface between sheets, and the steric zippers. They differed, however, through three main criteria that highlighted the possible permutations of amyloid packing structure (Figure 3d). The first was the orientation of individual β-strands within a β-sheet, aligned antiparallel or parallel. The second criterion was in which faces of the sheets face each other. The last criterion was the directionality of the β-sheets, or how the two β-sheets are oriented with respect to each other, defined as ipsidirectional (both sheets are up or down) or contradirectional (one sheet is up while the other is down) (Figure 3d).
The combinations of these characteristics thus allows eight possible orientations of the peptide within the fiber structure. For example, the original Sup35 peptide structure would be classified as parallel, face-to-face, and ipsidirectional. With the addition of all the new peptide structures, five of these eight classifications have now been observed. Significantly, one peptide from Sup35, NNQQ, was found to be able to adopt both a face-to-face and face-to-back orientation (Figure 3e). This may be exactly what is occurring with the Sc4 and Sc37 structures. Residues 40–70 represent a region that is structured in the Sc37 but not the Sc4 conformation, and residues 1–40, although structured in both conformations, have their β-sheets packed in two unique arrangements. The different orientations of β-sheets seen in the peptide structures could thus be representative of these unique conformations between residues 1–40 in Sup35.
With the above advances in our structural understanding, we can now start to piece together the sequences of characteristics that produce the prion strain phenomenon and begin to answer Robertson’s challenge. The disparity in the length of the amyloid fold between the Sc4 and Sc37 conformations provides a structural rationale for the differences in the physical characteristics of the respective fibers (26, 124). Sc37 fibers are less fragile than Sc4 fibers owing to the increased number of residues that participate in the fiber fold. Thus, the smaller amyloid fold of Sc4 results in weaker, more breakable fibers, resulting in many small fibers that are more effective at recruiting and sequestering natively folded protein, which results in a minimal population of soluble and function Sup35p, causing the white color phenotype. The larger amyloid fold of Sc37 results in stronger fibers, which more effectively resist shearing, resulting in fewer fibers to help recruit and sequester natively folded protein; this allows more soluble and functional Sup35p to persist, causing the pink color phenotype. There is also evidence of a strong correlation between aggregate stability and in vivo phenotype in the mammalian system (122). Here, it was demonstrated that the incubation time of the disease correlated with the stability of the aggregate, where the more easily the aggregate melted, the shorter the disease incubation time. Perhaps now, over 20 years later, endeavors into the study of prion structures have finally allowed us to demonstrate how the “structures of the prion protein…can be related to scrapie strain specificity,” satisfying a lingering challenge to the protein-only prion hypothesis.
4.4. Conformational Diversity in Nonprion Amyloids
Conformational heterogeneity has also been of great interest in nonprion amyloids and has enjoyed an even lengthier history. Starting from the initial qualitative observations by EM of variations in amyloid protofibril composition from the same preparation, much progress has been made in identifying these structural differences at high resolution. The usual suspects of SSNMR, HXNMR, and CryoEM have contributed greatly to identifying structural diversity, from differences in quaternary and tertiary packing of Aβ fibers to identifying single residues that participate in a different fold in Aβ, α-synuclein, and β2-microglobulin fibers. These studies have demonstrated that conformational diversity is not a phenomenon confined to the field of prion strains; rather, it is a common feature of almost all amyloids, and may have significant physiological consequences.
4.4.1. β2-Microglobulin
Although the initial observations of structural diversity in amyloids were on tissue-derived material, subsequent higher-resolution studies have concentrated on synthetic systems in which amyloids are made in vitro from peptides or recombinant protein. Like prions, multiple conformations of these amyloids formed either spontaneously or specifically under particular polymerization conditions. β2-Microglobulin is one such example, where multiple types of morphologically different fibers can form under conditions of varying pH and salt (144–146). AFM and EM on synthetic β2-microglobulin fibers have been able to initially qualitatively distinguish a number of morphologically different structures on the basis of fiber length, curvature, height, height modulation, and periodicity (144, 145, 147). H/D exchange on two types of fibers, called mature and immature aggregates, revealed that they indeed represent distinct folds, with the ridged mature fiber structure occupying almost 80% of the residues in the protein, whereas the immature fibers consisted of only 30% (148). This disparity in structure content is consistent with the ridged and flexible morphologies of the respective fibers and the reduced structural stability of immature fibers as revealed by guanidine hydrochloride unfolding. This large conformational variance with predicted structural consequences served as a precedence for the similar results later observed in the [PSI+] prion.
4.4.2. α-Synuclein
α-Synuclein constitutes a major component of intracellular deposits of lipids and proteins called Lewy bodies found in neurons in Parkinson’s disease patients and has a strong genetic correlation to disease. α-Synuclein is also a natively unfolded protein that can form aggregates that display all the hallmarks of amyloids. Synthetic fibers of α-synuclein aggregates have allowed the observation of conformational heterogeneity in this system as well. Unlike β2-microglobulin, however, conditions necessary to form homogeneous conformations in α-synuclein have not yet been worked out, and as such, one of two types of aggregates will form randomly under identical conditions. These types of fibers were studied by SSNMR, revealing that they indeed were structurally distinct based upon chemical shift differences for individual residues (71). However, when secondary structure predictions were performed on the basis of chemical shift values, very similar regions of β-sheets and loops were identified. Furthermore, the most significant chemical shift differences were observed in loop regions, suggesting that the two forms of α-synuclein fibers have similar overall structure but may differ more in loop conformation and packing of β-strands. Unlike Sup35 and β2-microglobulin, the formation of structural heterogeneity in α-synuclein seems much more stochastic and the resulting conformational differences much more subtle.
4.4.3. Aβ
Of all the nonprion amyloids, conformational diversity has been arguably best understood, and most widely studied, in Aβ. In this largely synthetic system, multiple conformations of Aβ fibers have been similarly formed by differing polymerization conditions. One such example is Aβ11–25 fibers that form an antiparallel structure (149). When polymerization was conducted at pH 7.4, SSNMR determined that the β-strand register was 17+k ⇔ 20−k, where k represents integer values, and the two numbers are the residue numbers of adjacent positions on neighboring β-strands. When fibers were formed at pH 2.4, however, the register changed to 17+k ⇔ 22−k. These data provided a clear example of conformational heterogeneity that arises from secondary structure packing rather than from quaternary structure differences.
Work on the full-length Aβ1–40 peptide revealed two more Aβ forms that clearly represented qualitatively different structures. These fibers, called “quiescent” and “agitated,” were formed under still and stirring conditions, respectively (108, 110). The quiescent fibers had a “twisted pair” appearance by EM wherein multiple protofilaments seemed to twist around each other with a specific periodicity. The agitated fibers, conversely, took on a “striated ribbon” morphology, where protofilaments appeared to associate laterally, forming ribbon-like structures. Subsequent SSNMR studies with STEM MPL data on both of these fiber conformations resulted in two distinct potential three-dimensional models. In these models, agitated fibers displayed 2 Aβ1–40 units per β-strand width, which with SSNMR data suggested twofold symmetry, where the previously described β-turn-β motif of a single Aβ1–40 sat next to and in the same plane as second β-turn-β monomer (Figure 4c) (69, 109). The quiescent fibers, alternatively, had three Aβ1–40 units per β-strand width with threefold symmetry (108). Each Aβ monomer in this structure also displayed a similar β-turn-β motif but had three such monomers on a single plane come together, forming a triangular cross-sectional area (Figure 4c). The actual conformations of the β-sheet regions on the residue level did not differ greatly between agitated and quiescent fibers; rather, the primary structural differences laid in the overall twofold versus threefold symmetry as well as the corresponding differences in quaternary contacts between each monomer.
Conservation of the basic β-turn-β motif while modulating the packing of these individual units is a theme also observed by others. In two cryoEM studies, a spectrum of Aβ fiber conformations was observed, varying in increasing fiber widths and crossover lengths, defined as the distance between the thin modulations in the fiber appearance (49, 51). When 12 such conformations were reconstructed and their densities determined, many of them were consistent with two monomers of the familiar β-turn-β motif of Aβ1–40 fitting together to form each 4.7-Å layer of the fiber structure. The models differed in how the two β-turn-β Aβ1–40 monomers came together, varying from the long edges of each monomer being perfectly aligned, forming a rectangular cross section, to the monomers being parallel but offset by almost one entire length of the monomer, forming an elongated “S” cross section (Figure 4c). The cryoEM studies, as well as those of quiescent and agitated fibers, show the diversity of structures that can be achieved by varying the quaternary structural arrangements of each monomeric unit and show how each unique structure can present a distinct surface to the environment. Intriguingly, the initial study on quiescent and agitated fibers suggested not only divergent structures, but also unequal levels of toxicity, with quiescent fibers being more toxic than agitated fibers (110).
Further structural heterogeneity was also achieved not through differing polymerization conditions, rather, through the use of slightly different, but physiologically relevant, alternative peptides. For example, one study investigated the conformational differences between synthetic Aβ1–40 fibers and Aβ1–42 fibers, both of which are present in AD patients (106). Despite only a two–amino acid addition, the authors observed an extension of protection for the N-terminal region stretching out the first β-strand. A second example of an alternative peptide is Iowa mutant Aβ, which harbors a D23N mutation. A study that investigated this peptide found that it yielded a mixed population of two different synthetic fibers as observed by EM (78). When SSNMR was performed on these fiber preparations, investigators found that one fiber form was consistent with the typical in-register parallel β-sheet structure observed for WT fibers, and the other appeared to be in an antiparallel β-sheet structure. This was significant because it represents an entirely different fold for the Aβ peptide and is the first observation of an Aβ peptide over 15 residues long in an antiparallel orientation. The link of this particular peptide to a familial form of AD potentially provides another interesting avenue in the effort to understand physiological consequences of structural diversity.
5. The Next Steps
From the inceptive qualitative observations of structural diversity in amyloid structure, the molecular details of the spectrum of structures a single polypeptide chain can aggregate into and the consequences of this ability are now beginning to be revealed. This connection between structure and function has been most readily and compellingly established in the yeast prion systems, where the causative agent is unequivocally known, can be precisely made in vitro, and has clear and specific phenotypic readouts for each form. The mammalian prions and amyloidoses are inherently more difficult to study. Yet even with these technical challenges, remarkable progress is being made.
SUMMARY POINTS.
Amyloids share a common architecture forming long, unbranched fibers, which are rich in β-sheets, wherein each β-strand lies perpendicular to the fiber axis.
Each peptide in the fiber structure often, but not always, resides in an in-register, parallel fashion, allowing a templating mechanism for fiber polymerization.
High-resolution techniques have enabled a detailed structural view of the Aβ and Het-s amyloids.
We are beginning to obtain molecular understandings of the structural diversity in prion and nonprion amyloids, where conformation differences can exist as distinct folds on the monomer level, rather than subtle differences in peptide packing.
A full understanding of the physiological consequences of conformational diversity has become clear in yeast prions, wherein more compact amyloid folds result in physically weaker fibers, which are able to fragment more easily in vivo, resulting in a “stronger” amyloid phenotype.
FUTURE ISSUES.
What other peptide arrangements might we see in other amyloid systems?
Are the wet and dry interfaces seen in the X-ray structures of short peptides present and representative of the structures of amyloids composed of native and full-length peptides?
What are the structures of amyloids from physiologically relevant (in vivo or tissue) sources, and how do they differ from the structures of synthetic or in vitro–made amyloids?
Do conformational differences in mammalian prions resemble differences seen in the yeast systems?
What are the exact physiological consequences of conformational diversity in mammalian and nonprion amyloid systems?
Acknowledgments
B.H. Toyama is now at the Molecular and Cell Biology Laboratory, at the Salk Institute for Biological Studies, in La Jolla, California.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Brandon H. Toyama, Email: btoyama@salk.edu.
Jonathan S. Weissman, Email: weissman@cmp.ucsf.edu.
LITERATURE CITED
- 1.Kyle RA. Amyloidosis: a convoluted story. Br J Haematol. 2001;114:529–38. doi: 10.1046/j.1365-2141.2001.02999.x. [DOI] [PubMed] [Google Scholar]
- 2.Dobson CM. The structural basis of protein folding and its links with human disease. Philos Trans R Soc Lond B. 2001;356:133–45. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–32. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–41. doi: 10.1146/annurev.micro.56.013002.100603. [DOI] [PubMed] [Google Scholar]
- 6.Astbury WT, Dickinson S. The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem J. 1935;29:2351–60.1. doi: 10.1042/bj0292351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eanes ED, Glenner GG. X-ray diffraction studies on amyloid filaments. J Histochem Cytochem. 1968;16:673–77. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 8.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc Natl Acad Sci USA. 2005;102:315–20. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wille H, Bian W, McDonald M, Kendall A, Colby DW, et al. Natural and synthetic prion structure from X-ray fiber diffraction. Proc Natl Acad Sci USA. 2009;106:16990–95. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CC. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273:729–39. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 11.Cohen AS, Calkins E. Electron microscopic observations on a fibrous component in amyloid of diverse origins. Nature. 1959;183:1202–3. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 12.Shirahama T, Cohen AS. Structure of amyloid fibrils after negative staining and high-resolution electron microscopy. Nature. 1965;206:737–38. doi: 10.1038/206737a0. [DOI] [PubMed] [Google Scholar]
- 13.Boere H, Ruinen L, Scholten JH. Electron microscopic studies on the fibrillar component of human splenic amyloid. J Lab Clin Med. 1965;66:943–51. [PubMed] [Google Scholar]
- 14.Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci USA. 2009;106:14339–44. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsbury CS, Wirtz S, Muller SA, Sunderji S, Wicki P, et al. Studies on the in vitro assembly of a beta 1–40: implications for the search for a beta fibril formation inhibitors. J Struct Biol. 2000;130:217–31. doi: 10.1006/jsbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 16.Knowles TP, Smith JF, Craig A, Dobson CM, Welland ME. Spatial persistence of angular correlations in amyloid fibrils. Phys Rev Lett. 2006;96:238301. doi: 10.1103/PhysRevLett.96.238301. [DOI] [PubMed] [Google Scholar]
- 17.Berthomieu C, Hienerwadel R. Fourier transform infrared (FTIR) spectroscopy. Photosynth Res. 2009;101:157–70. doi: 10.1007/s11120-009-9439-x. [DOI] [PubMed] [Google Scholar]
- 18.Ranjbar B, Gill P. Circular dichroism techniques: biomolecular and nanostructural analyses--a review. Chem Biol Drug Des. 2009;74:101–20. doi: 10.1111/j.1747-0285.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 19.Gasset M, Baldwin MA, Fletterick RJ, Prusiner SB. Perturbation of the secondary structure of the scrapie prion protein under conditions that alter infectivity. Proc Natl Acad Sci USA. 1993;90:1–5. doi: 10.1073/pnas.90.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Termine JD, Eanes ED, Ein D, Glenner GG. Infrared spectroscopy of human amyloid fibrils and immunoglobulin proteins. Biopolymers. 1972;11:1103–13. doi: 10.1002/bip.1972.360110512. [DOI] [PubMed] [Google Scholar]
- 21.Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, Caughey WS. Secondary structure analysis of the scrapie-associated protein PrP 27–30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–80. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 22.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci USA. 1993;90:10962–66. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang HY, Lin JY, Lee HC, Wang HL, King CY. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci USA. 2008;105:13345–50. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba T, Hagihara Y, Higurashi T, Hasegawa K, Naiki H, Goto Y. Amyloid fibril formation in the context of full-length protein: effects of proline mutations on the amyloid fibril formation of beta2-microglobulin. J Biol Chem. 2003;278:47016–24. doi: 10.1074/jbc.M304473200. [DOI] [PubMed] [Google Scholar]
- 25.Ritter C, Maddelein ML, Siemer AB, Luhrs T, Ernst M, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–48. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–37. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 27.Williams AD, Portelius E, Kheterpal I, Guo JT, Cook KD, et al. Mapping Abeta amyloid fibril secondary structure using scanning proline mutagenesis. J Mol Biol. 2004;335:833–42. doi: 10.1016/j.jmb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Wood SJ, Wetzel R, Martin JD, Hurle MR. Prolines and amyloidogenicity in fragments of the Alzheimer’s peptide beta/A4. Biochemistry. 1995;34:724–30. doi: 10.1021/bi00003a003. [DOI] [PubMed] [Google Scholar]
- 29.Cobb NJ, Sönnichsen FD, McHaourab H, Surewicz WK. Molecular architecture of human prion protein amyloid: a parallel, in-register beta-structure. Proc Natl Acad Sci USA. 2007;104:18946–51. doi: 10.1073/pnas.0706522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Der-Sarkissian A, Jao CC, Chen J, Langen R. Structural organization of alpha-synuclein fibrils studied by site-directed spin labeling. J Biol Chem. 2003;278:37530–35. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 31.Margittai M, Langen R. Side chain–dependent stacking modulates tau filament structure. J Biol Chem. 2006;281:37820–27. doi: 10.1074/jbc.M605336200. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–28. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 33.Chen M, Margittai M, Chen J, Langen R. Investigation of alpha-synuclein fibril structure by site-directed spin labeling. J Biol Chem. 2007;282:24970–79. doi: 10.1074/jbc.M700368200. [DOI] [PubMed] [Google Scholar]
- 34.Jayasinghe SA, Langen R. Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J Biol Chem. 2004;279:48420–25. doi: 10.1074/jbc.M406853200. [DOI] [PubMed] [Google Scholar]
- 35.Torok M, Milton S, Kayed R, Wu P, McIntire T, et al. Structural and dynamic features of Alzheimer’s Abeta peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277:40810–15. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 36.Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO. Arrangement of subunits and ordering of beta-strands in an amyloid sheet. Nat Struct Biol. 2002;9:734–39. doi: 10.1038/nsb838. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–72. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohhashi Y, Ito K, Toyama BH, Weissman JS, Tanaka M. Differences in prion strain conformations result from non-native interactions in a nucleus. Nat Chem Biol. 2010;6:225–30. doi: 10.1038/nchembio.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoshino M, Katou H, Hagihara Y, Hasegawa K, Naiki H, Goto Y. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat Struct Biol. 2002;9:332–36. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 41.Hirota-Nakaoka N, Hasegawa K, Naiki H, Goto Y. Dissolution of beta2-microglobulin amyloid fibrils by dimethylsulfoxide. J Biochem. 2003;134:159–64. doi: 10.1093/jb/mvg124. [DOI] [PubMed] [Google Scholar]
- 42.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, et al. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–47. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carulla N, Caddy GL, Hall DR, Zurdo J, Gairi M, et al. Molecular recycling within amyloid fibrils. Nature. 2005;436:554–58. doi: 10.1038/nature03986. [DOI] [PubMed] [Google Scholar]
- 44.Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–55. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- 45.Toyoshima C, Unwin N. Ion channel of acetylcholine receptor reconstructed from images of postsynaptic membranes. Nature. 1988;336:247–50. doi: 10.1038/336247a0. [DOI] [PubMed] [Google Scholar]
- 46.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–50. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 47.Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–21. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jimenez JL, Nettleton EJ, Bouchard M, Robinson CV, Dobson CM, Saibil HR. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Abeta(1–40) fibril polymorphism implies diverse interaction patterns in amyloid fibrils. J Mol Biol. 2009;386:869–77. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sachse C, Fandrich M, Grigorieff N. Paired beta-sheet structure of an Abeta(1–40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci USA. 2008;105:7462–66. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang R, Hu X, Khant H, Ludtke SJ, Chiu W, et al. Interprotofilament interactions between Alzheimer’s Abeta1–42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci USA. 2009;106:4653–58. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmidt M, Sachse C, Richter W, Xu C, Fandrich M, Grigorieff N. Comparison of Alzheimer Abeta(1–40) and Abeta(1–42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci USA. 2009;106:19813–18. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900 K GroEL GroES complex. Nature. 2002;418:207–11. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 54.Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, et al. Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J Am Chem Soc. 2006;128:13224–28. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 55.Heise H. Solid-state NMR spectroscopy of amyloid proteins. ChemBioChem. 2008;9:179–89. doi: 10.1002/cbic.200700630. [DOI] [PubMed] [Google Scholar]
- 56.Griffin RG. Dipolar recoupling in MAS spectra of biological solids. Nat Struct Biol. 1998;5(Suppl):508–12. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 57.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–26. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 58.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry. 2007;46:13505–22. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaroniec CP, MacPhee CE, Bajaj VS, McMahon MT, Dobson CM, Griffin RG. High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc Natl Acad Sci USA. 2004;101:711–16. doi: 10.1073/pnas.0304849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci USA. 2008;105:2403–8. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shewmaker F, McGlinchey RP, Thurber KR, McPhie P, Dyda F, et al. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–76. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan JC, Oyler NA, Yau WM, Tycko R. Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–80. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baxa U, Wickner RB, Steven AC, Anderson DE, Marekov LN, et al. Characterization of beta-sheet structure in Ure2p1–89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–62. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 64.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–59. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lim KH, Nguyen TN, Damo SM, Mazur T, Ball HL, et al. Solid-state NMR structural studies of the fibril form of a mutant mouse prion peptide PrP89–143(P101L) Solid State Nucl Magn Reson. 2006;29:183–90. doi: 10.1016/j.ssnmr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 66.Benzinger TL, Gregory DM, Burkoth TS, Miller-Auer H, Lynn DG, et al. Propagating structure of Alzheimer’s beta-amyloid(10–35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–12. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer’s beta-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry. 2002;41:15436–50. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 68.Iwata K, Fujiwara T, Matsuki Y, Akutsu H, Takahashi S, et al. 3D structure of amyloid protofilaments of beta2-microglobulin fragment probed by solid-state NMR. Proc Natl Acad Sci USA. 2006;103:18119–24. doi: 10.1073/pnas.0607180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, et al. A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA. 2002;99:16742–47. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lansbury PT, Jr, Costa PR, Griffiths JM, Simon EJ, Auger M, et al. Structural model for the beta-amyloid fibril based on interstrand alignment of an antiparallel-sheet comprising a C-terminal peptide. Nat Struct Biol. 1995;2:990–98. doi: 10.1038/nsb1195-990. [DOI] [PubMed] [Google Scholar]
- 71.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA. 2005;102:15871–76. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–89. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 73.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, et al. Structure of the cross-beta spine of amyloid-like fibrils. Nature. 2005;435:773–78. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447:453–57. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 75.Wiltzius JJ, Landau M, Nelson R, Sawaya MR, Apostol MI, et al. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol. 2009;16:973–78. doi: 10.1038/nsmb.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wiltzius JJ, Sievers SA, Sawaya MR, Cascio D, Popov D, et al. Atomic structure of the cross-beta spine of islet amyloid polypeptide (amylin) Protein Sci. 2008;17:1467–74. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Balbach JJ, Ishii Y, Antzutkin ON, Leapman RD, Rizzo NW, et al. Amyloid fibril formation by Abeta16–22, a seven-residue fragment of the Alzheimer’s beta-amyloid peptide, and structural characterization by solid state NMR. Biochemistry. 2000;39:13748–59. doi: 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- 78.Tycko R, Sciarretta KL, Orgel JP, Meredith SC. Evidence for novel beta-sheet structures in Iowa mutant beta-amyloid fibrils. Biochemistry. 2009;48:6072–84. doi: 10.1021/bi9002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saupe SJ. A short history of small s: a prion of the fungus Podospora anserina. Prion. 2007;1:110–15. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dos Reis S, Coulary-Salin B, Forge V, Lascu I, Begueret J, Saupe SJ. The HET-s prion protein of the filamentous fungus Podospora anserina aggregates in vitro into amyloid-like fibrils. J Biol Chem. 2002;277:5703–6. doi: 10.1074/jbc.M110183200. [DOI] [PubMed] [Google Scholar]
- 82.Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–81. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nazabal A, Dos Reis S, Bonneu M, Saupe SJ, Schmitter JM. Conformational transition occurring upon amyloid aggregation of the HET-s prion protein of Podospora anserina analyzed by hydrogen/deuterium exchange and mass spectrometry. Biochemistry. 2003;42:8852–61. doi: 10.1021/bi0344275. [DOI] [PubMed] [Google Scholar]
- 84.Nazabal A, Maddelein ML, Bonneu M, Saupe SJ, Schmitter JM. Probing the structure of the infectious amyloid form of the prion-forming domain of HET-s using high resolution hydrogen/deuterium exchange monitored by mass spectrometry. J Biol Chem. 2005;280:13220–28. doi: 10.1074/jbc.M413185200. [DOI] [PubMed] [Google Scholar]
- 85.Sen A, Baxa U, Simon MN, Wall JS, Sabate R, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–50. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 86.Lange A, Gattin Z, Van Melckebeke H, Wasmer C, Soragni A, et al. A combined solid-state NMR and MD characterization of the stability and dynamics of the HET-s(218–289) prion in its amyloid conformation. ChemBioChem. 2009;10:1657–65. doi: 10.1002/cbic.200900019. [DOI] [PubMed] [Google Scholar]
- 87.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–90. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 88.Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc Natl Acad Sci USA. 1986;83:4044–48. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, et al. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–36. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 90.Selkoe DJ. Physiological production of the beta-amyloid protein and the mechanism of Alzheimer’s disease. Trends Neurosci. 1993;16:403–9. doi: 10.1016/0166-2236(93)90008-a. [DOI] [PubMed] [Google Scholar]
- 91.Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, et al. Assembly and aggregation properties of synthetic Alzheimer’s A4/beta amyloid peptide analogs. J Biol Chem. 1992;267:546–54. [PubMed] [Google Scholar]
- 92.Prelli F, Castano EM, van Duinen SG, Bots GT, Luyendijk W, Frangione B. Different processing of Alzheimer’s beta-protein precursor in the vessel wall of patients with hereditary cerebral hemorrhage with amyloidosis-Dutch type. Biochem Biophys Res Commun. 1988;151:1150–55. doi: 10.1016/s0006-291x(88)80486-8. [DOI] [PubMed] [Google Scholar]
- 93.Kirschner DA, Abraham C, Selkoe DJ. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-beta conformation. Proc Natl Acad Sci USA. 1986;83:503–7. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Inouye H, Fraser PE, Kirschner DA. Structure of beta-crystallite assemblies formed by Alzheimer beta-amyloid protein analogues: analysis by X-ray diffraction. Biophys J. 1993;64:502–19. doi: 10.1016/S0006-3495(93)81393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kirschner DA, Inouye H, Duffy LK, Sinclair A, Lind M, Selkoe DJ. Synthetic peptide homologous to beta protein from Alzheimer disease forms amyloid-like fibrils in vitro. Proc Natl Acad Sci USA. 1987;84:6953–57. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–49. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prelli F, Castano E, Glenner GG, Frangione B. Differences between vascular and plaque core amyloid in Alzheimer’s disease. J Neurochem. 1988;51:648–51. doi: 10.1111/j.1471-4159.1988.tb01087.x. [DOI] [PubMed] [Google Scholar]
- 98.Joachim CL, Duffy LK, Morris JH, Selkoe DJ. Protein chemical and immunocytochemical studies of meningovascular beta-amyloid protein in Alzheimer’s disease and normal aging. Brain Res. 1988;474:100–11. doi: 10.1016/0006-8993(88)90673-7. [DOI] [PubMed] [Google Scholar]
- 99.Gordon DJ, Balbach JJ, Tycko R, Meredith SC. Increasing the amphiphilicity of an amyloidogenic peptide changes the beta-sheet structure in the fibrils from antiparallel to parallel. Biophys J. 2004;86:428–34. doi: 10.1016/S0006-3495(04)74119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bu Z, Shi Y, Callaway DJ, Tycko R. Molecular alignment within beta-sheets in Abeta(14–23) fibrils: solid-state NMR experiments and theoretical predictions. Biophys J. 2007;92:594–602. doi: 10.1529/biophysj.106.091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Antzutkin ON, Balbach JJ, Leapman RD, Rizzo NW, Reed J, Tycko R. Multiple quantum solid-state NMR indicates a parallel, not antiparallel, organization of beta-sheets in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci USA. 2000;97:13045–50. doi: 10.1073/pnas.230315097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kheterpal I, Zhou S, Cook KD, Wetzel R. Abeta amyloid fibrils possess a core structure highly resistant to hydrogen exchange. Proc Natl Acad Sci USA. 2000;97:13597–601. doi: 10.1073/pnas.250288897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Balbach JJ, Petkova AT, Oyler NA, Antzutkin ON, Gordon DJ, et al. Supramolecular structure in full-length Alzheimer’s beta-amyloid fibrils: evidence for a parallel beta-sheet organization from solid-state nuclear magnetic resonance. Biophys J. 2002;83:1205–16. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang SS, Tobler SA, Good TA, Fernandez EJ. Hydrogen exchange-mass spectrometry analysis of beta-amyloid peptide structure. Biochemistry. 2003;42:9507–14. doi: 10.1021/bi0342766. [DOI] [PubMed] [Google Scholar]
- 105.Whittemore NA, Mishra R, Kheterpal I, Williams AD, Wetzel R, Serpersu EH. Hydrogen-deuterium (H/D) exchange mapping of Abeta 1–40 amyloid fibril secondary structure using nuclear magnetic resonance spectroscopy. Biochemistry. 2005;44:4434–41. doi: 10.1021/bi048292u. [DOI] [PubMed] [Google Scholar]
- 106.Olofsson A, Lindhagen-Persson M, Sauer-Eriksson AE, Ohman A. Amide solvent protection analysis demonstrates that amyloid-beta(1–40) and amyloid-beta(1–42) form different fibrillar structures under identical conditions. Biochem J. 2007;404:63–70. doi: 10.1042/BJ20061561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Olofsson A, Sauer-Eriksson AE, Ohman A. The solvent protection of Alzheimer amyloid-beta-(1–42) fibrils as determined by solution NMR spectroscopy. J Biol Chem. 2006;281:477–83. doi: 10.1074/jbc.M508962200. [DOI] [PubMed] [Google Scholar]
- 108.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer’s beta-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–54. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s beta-amyloid fibrils. Science. 2005;307:262–65. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 111.Sachse C, Xu C, Wieligmann K, Diekmann S, Grigorieff N, Fändrich M. Quaternary structure of a mature amyloid fibril from Alzheimer’s Abeta(1–40) peptide. J Mol Biol. 2006;362:347–54. doi: 10.1016/j.jmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 112.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci USA. 2002;99:5591–95. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Serpell LC, Smith JM. Direct visualisation of the beta-sheet structure of synthetic Alzheimer’s amyloid. J Mol Biol. 2000;299:225–31. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 114.Serpell LC, Sunde M, Fraser PE, Luther PK, Morris EP, et al. Examination of the structure of the transthyretin amyloid fibril by image reconstruction from electron micrographs. J Mol Biol. 1995;254:113–18. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 115.Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc Natl Acad Sci USA. 2001;98:2375–80. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Perutz M. Polar zippers: their role in human disease. Protein Sci. 1994;3:1629–37. doi: 10.1002/pro.5560031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dickinson AG, Meikle VM. A comparison of some biological characteristics of the mouse-passaged scrapie agents, 22A and ME7. Genet Res. 1969;13:213–25. doi: 10.1017/s0016672300002895. [DOI] [PubMed] [Google Scholar]
- 118.Bruce ME, Dickinson AG. Biological evidence that scrapie agent has an independent genome. J Gen Virol. 1987;68(Part 1):79–89. doi: 10.1099/0022-1317-68-1-79. [DOI] [PubMed] [Google Scholar]
- 119.Bessen RA, Marsh RF. Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J Gen Virol. 1992;73(Part 2):329–34. doi: 10.1099/0022-1317-73-2-329. [DOI] [PubMed] [Google Scholar]
- 120.Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol. 1994;68:7859–68. doi: 10.1128/jvi.68.12.7859-7868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–23. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 122.Legname G, Nguyen HO, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci USA. 2006;103:19105–10. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–82. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 124.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–89. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 125.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–44. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 126.Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 127.Robertson HD, Branch AD, Dahlberg JE. Focusing on the nature of the scrapie agent. Cell. 1985;40:725–27. doi: 10.1016/0092-8674(85)90328-9. [DOI] [PubMed] [Google Scholar]
- 128.Maji SK, Wang L, Greenwald J, Riek R. Structure-activity relationship of amyloid fibrils. FEBS Lett. 2009;583:2610–17. doi: 10.1016/j.febslet.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 129.May BC, Govaerts C, Prusiner SB, Cohen FE. Prions: so many fibers, so little infectivity. Trends Biochem Sci. 2004;29:162–65. doi: 10.1016/j.tibs.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 130.Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, et al. Synthetic mammalian prions. Science. 2004;305:673–76. doi: 10.1126/science.1100195. [DOI] [PubMed] [Google Scholar]
- 131.Castilla J, Saá P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 132.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–13. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 133.Colby DW, Giles K, Legname G, Wille H, Baskakov IV, et al. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci USA. 2009;106:20417–22. doi: 10.1073/pnas.0910350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Legname G, Nguyen HO, Baskakov IV, Cohen FE, Dearmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci USA. 2005;102:2168–73. doi: 10.1073/pnas.0409079102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Smirnovas V, Kim JI, Lu X, Atarashi R, Caughey B, Surewicz WK. Distinct structures of scrapie prion protein (PrPSc)-seeded versus spontaneous recombinant prion protein fibrils revealed by H/D exchange. J Biol Chem. 2009;284:24233–41. doi: 10.1074/jbc.M109.036558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–69. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 137.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–18. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–86. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci USA. 2002;99(Suppl. 4):16392–99. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. J Biol Chem. 2010;285:20748–55. doi: 10.1074/jbc.M110.115303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–46. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–56. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 143.Tanaka M, Chien P, Yonekura K, Weissman JS. Mechanism of cross-species prion transmission: an infectious conformation compatible with two highly divergent yeast prion proteins. Cell. 2005;121:49–62. doi: 10.1016/j.cell.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 144.McParland VJ, Kad NM, Kalverda AP, Brown A, Kirwin-Jones P, et al. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–46. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 145.Kad NM, Thomson NH, Smith DP, Smith DA, Radford SE. Beta2-microglobulin and its deamidated variant, N17D form amyloid fibrils with a range of morphologies in vitro. J Mol Biol. 2001;313:559–71. doi: 10.1006/jmbi.2001.5071. [DOI] [PubMed] [Google Scholar]
- 146.Hiramatsu H, Lu M, Matsuo K, Gekko K, Goto Y, Kitagawa T. Differences in the molecular structure of beta2-microglobulin between two morphologically different amyloid fibrils. Biochemistry. 2010;49:742–51. doi: 10.1021/bi901536j. [DOI] [PubMed] [Google Scholar]
- 147.Kad NM, Myers SL, Smith DP, Smith DA, Radford SE, Thomson NH. Hierarchical assembly of beta2-microglobulin amyloid in vitro revealed by atomic force microscopy. J Mol Biol. 2003;330:785–97. doi: 10.1016/s0022-2836(03)00583-7. [DOI] [PubMed] [Google Scholar]
- 148.Yamaguchi K, Katou H, Hoshino M, Hasegawa K, Naiki H, Goto Y. Core and heterogeneity of beta2-microglobulin amyloid fibrils as revealed by H/D exchange. J Mol Biol. 2004;338:559–71. doi: 10.1016/j.jmb.2004.02.067. [DOI] [PubMed] [Google Scholar]
- 149.Petkova AT, Buntkowsky G, Dyda F, Leapman RD, Yau WM, Tycko R. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335:247–60. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]