Abstract
The human cytomegalovirus (HCMV) genome was sequenced 20 years ago. However, like other complex viruses, our understanding of its protein coding potential is far from complete. Here we use ribosome profiling and transcript analysis to experimentally define the HCMV translation products and follow their temporal expression. We identified hundreds of previously unidentified open reading frames and confirmed a fraction by mass spectrometry. We found that regulated use of alternative transcript start sites plays a broad role in enabling tight temporal control of HCMV protein expression and allowing multiple distinct polypeptides to be generated from a single genomic locus. Our results reveal an unanticipated complexity to the HCMV coding capacity and illustrate the role of regulated changes in transcript start sites in generating this complexity.
The herpesvirus, human cytomegalovirus (HCMV) infects the majority of humanity, leading to severe disease in newborns and immunocompromised adults (1). The HCMV genome is ~240kb with estimates of between 165-252 open reading frames (ORFs)(2, 3). These annotations likely do not capture the complexity of the HCMV proteome (4) as HCMV has a complex transcriptome (5, 6) and genomic regions studied in detail reveal noncanonical translational events including regulatory (7) and overlapping ORFs (8-11). Defining the full set of translation products- both stable and unstable, the latter with potential regulatory/antigenic function (12)-is critical for understanding HCMV.
To identify the range of HCMV translated ORFs and monitor their temporal expression, we infected human foreskin fibroblasts (HFF) with the clinical HCMV strain Merlin and harvested cells at 5, 24 and 72 hours post infection (hpi) using four approaches to generate libraries of ribosome-protected mRNA fragments (Fig.1a and table S1). The first two measured the overall in vivo distribution of ribosomes on a given message; infected cells were either pre-treated with the translation elongation inhibitor cycloheximide or, to exclude drug artifacts, lysed without drug pre-treatment (no-drug). Additionally, cells were pre-treated with harringtonine or lactimidomycin (LTM) two drugs with distinct mechanisms, which lead to strong accumulation of ribosomes at translation initiation sites and depletion of ribosomes over the body of the message (Fig.1a and (13-15)). A modified RNA-Seq protocol allowed quantification of RNA levels as well as identification of 5′ transcript ends by generating a strong overrepresentation of fragments that start at the 5′ end of messages (Fig.S1 and (16)).
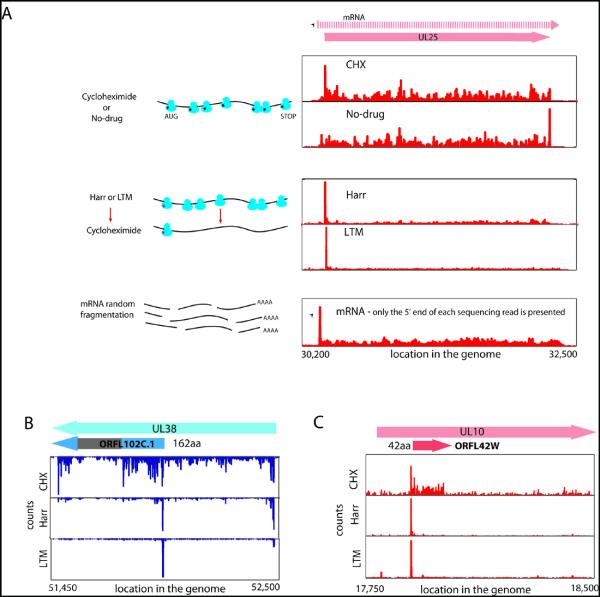
Fig. 1.
Ribosome profiling of HCMV infected cells.
(A) Ribosome occupancies following various treatments (illustrated on the left); cycloheximide (CHX), No-drug, harringtonine (Harr) and LTM together with mRNA profiles of the UL25 gene at 72 hpi. An arrow marks the mRNA start.
(B,C) Ribosome occupancy profiles for UL38 (B) and UL10(C) genes that contain internal initiations. The grey area symbolizes a low complexity region.
The ability of these approaches to provide a comprehensive view of gene organization is illustrated for the UL25 ORF: A single transcript start site is found upstream of the ORF (Fig.1a, mRNA panel). Harringtonine and LTM mark a single translation initiation site at the first AUG downstream of the transcript start (Fig.1a, Harr and LTM panels). Ribosome density accumulates over the ORF body ending at the first in-frame stop codon (Fig.1a, CHX and no-drug panels). In the no-drug sample, excess ribosome density accumulates at the stop codon (Fig.1a, no-drug panel and (14)).
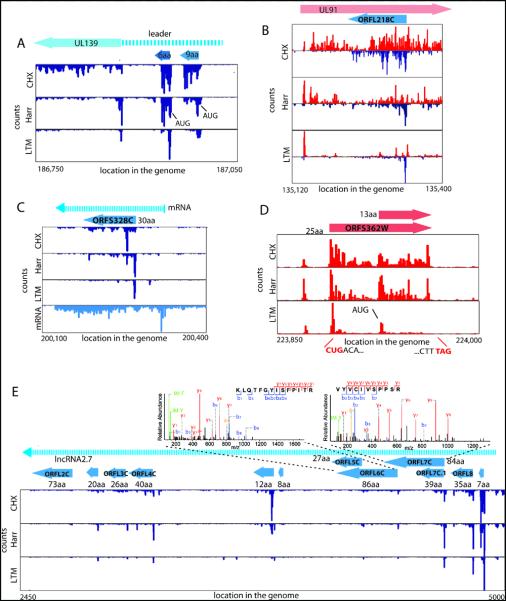
Examination of the full range of HCMV translation products, as reflected by the ribosome footprints, revealed many putative novel ORFs: internal ORFs lying within existing ORFs either in-frame, resulting in N-terminally truncated translation products (Fig.1b), or out of frame, resulting in entirely novel polypeptides (Fig.1c); short uORFs lying upstream of canonical ORFs (Fig.2a); ORFs within transcripts anti-sense to canonical ORFs (Fig.2b); novel short ORFs encoded by distinct transcripts (Fig.2c). For all of these categories, we also observed ORFs starting at near-cognate codons (i.e., codons differing from AUG by one nucleotide), especially CUG (Fig.2d).
Fig. 2.
Many ribosome footprints do not correspond to previously annotated ORFs.
(A) Ribosome occupancy profiles for the leader region of UL139 gene.
(B) Ribosome occupancy profiles of plus and minus strands (red and blue respectively) for the UL91 gene.
(C) mRNA and ribosome occupancy profiles for a novel short ORF.
(D) Ribosome occupancies around a short ORF that initiates at a CUG codon.
(E) Ribosome occupancy profiles for RNA β2.7. The upper panels show the annotated MS/MS spectra of two unique peptides originating from ORFL6C and ORFL7C.
HCMV expresses several long RNAs lacking canonical ORFs, including β2.7, an abundant RNA, which inhibits apoptosis(17). In agreement with β2.7’s observed polysome association (18) multiple short ORFs are translated from this RNA (Fig.2e and Fig.S2) and the corresponding proteins for two of these ORFs were detected by high-resolution mass spectrometry (Fig. 2e). Although the translation efficiency of these ORFs is low, four of them are highly conserved across HCMV strains (Table S2). We found three similar polycistronic coding RNAs (including RNA1.2 and RNA4.9) and two short proteins encoded by these RNAs were confirmed by mass spectrometry (Fig.S3).
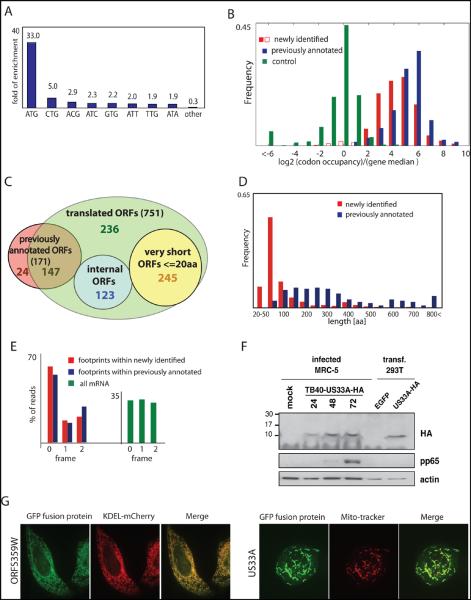
To define systematically the HCMV translated ORFs using the ribosome profiling data, we first annotated HCMV splice junctions identifying 88 splice sites (Table S3). We then exploited the harringtonine-induced accumulation of ribosomes at translation start sites to identify ORFs using a support vector machine (SVM)-based machine learning strategy(14, 19). We observed a strong enrichment for AUG (33-fold) and near cognate codons in the translation initiation sites identified by this analysis (Fig.3a). Visual inspection of the ribosome profiling data confirmed the SVM-identified ORFs and suggested an additional 53 putative ORFs (Table S4). The large majority (86%) of the SVM identified ORFs, and all of the manually identified ones, where identified by SVM analysis of an independent biological replicate (Table S5 and Fig.S4). The observed initiation sites where not caused by harringtonine as LTM treatment also induced ribosome accumulation at the vast majority (>98%) of these positions (Fig.3b).
Fig. 3.
Annotating the HCMV translated ORFs.
(A) Fold enrichment of AUG and near-cognate codons at predicted sites of translation initiation compared to their genomic distribution.
(B) The ribosome footprints occupancy after LTM treatment at each start codon (relative to the median density across the gene) is depicted for the previously annotated ORFs (blue) and newly identified ORFs (red and empty red for ORFs that were removed). The occupancy at a codon five positions downstream of the start codon is depicted as a control (green).
(C) Venn diagram summarizing the HCMV translated ORFs. Note 53 ORFs were initially identified by manual inspection (see text).
(D) The lengths distribution of newly identified ORFs (red) and previously annotated ORFs (blue).
(E) Position of 30-nt ribosome footprints relative to the reading frame in the newly identified ORFs (red) and previously annotated ORFs (blue).
(F) MRC-5 cells were mock-treated or infected with TB40-US33A-HA and protein lysates were analyzed by western blotting with indicated antibodies.
(G) HeLa cells were transfected with GFP fusion proteins together with an ER marker (KDEL-mCherry) or stained with MitoTracker Red and imaged by confocal microscopy.
In total we identified 751 translated ORFs that were supported by both the LTM and harringtonine data (Table S5, Table S6 and file S1). The footprint density measurements for these ORFs were reproducible between biological replicates (Fig.S5 and S6). 147 of these ORFs were previously suggested to be coding (Fig.3c). We did not find strong evidence of translation for 24 previously annotated ORFs (Table S7) although these proteins may well be expressed under different conditions.
Many newly identified ORFs are very short (245 ORFs≤20 codons, Fig.3c) and are found upstream of longer ORFs. We also identified 239 novel short ORFs (21-80 codons, Fig.3d). Lastly, we identified 120 novel ORFs that are longer than 80 amino acids. These are primarily ORFs that contain splice junctions or alternative 5′ ends of previous annotations.
Several lines of evidence support the validity of the ORFs we identified. First, as seen for the previously annotated ORFs, newly identified ORFs showed a significant (P<10−70, Ks test) excess of ribosome footprints at the predicted stop codon similar to what is seen for the previously annotated ORFs (Fig.1a and Fig.S7). Because our ORF predictions were based on translation initiation sites found in the harringtonine and LTM samples, the observation that these accurately predicted downstream stop codons in an untreated sample provides independent support for our approach. Second, ribosome-protected footprints displayed a 3-nt periodicity that was in phase with the predicted start site both globally (Fig.3e) and in specific ORFs that contain internal out of frame ORFs (Fig.S8). Third, brief inhibition of translation initiation using an eIF4A inhibitor Pateamine A(20) led to depletion of ribosome density from the body of the large majority of the predicted ORFs (Fig.S9) indicating that the ribosomes were engaged in active elongation. The newly identified ORFs also exhibited a distribution of expression levels similar to that of previously annotated canonical ORFs (Fig.S10). Finally many of the newly identified ORFs are conserved in other HCMV strains (Table S2).
High-resolution tandem mass spectrometric measurements on virally infected cells using stringent criteria and manual validation (files S2, S3 and (16, 21)) unambiguously detected 53 novel proteins out of the 96 genomic loci that are not overlapping with annotated ORFs and contain at least one unique novel protein that is longer than 55aa (Table S8). For classes of new ORFs that were difficult to monitor by mass spectrometry (i.e., truncated forms of longer proteins or short proteins), we used a tagging approach. For two N-terminally truncated proteins (derived from UL16 and UL38), we confirmed the appearance of alternative shorter transcripts and detected the expected full length and truncated tagged protein products (Fig.S11). The truncated protein derived from UL16 was also observed in the context of the native virus (Fig.S12) and we confirmed a splice variant of UL138 using an antibody (Fig.S12). For five short ORFs (including two initiated at near cognate start sites), we fused the ORFs in frame to a GFP coding region in their otherwise native transcript context. We identified protein products of the expected sizes and confirmed that we correctly identified the translation start sites (Fig.S13). We also showed that one of these short proteins (US33A-57aa), which was not identified by mass spectrometry but was recently predicted to be coding by transcript analysis (6), is expressed in the context of the native virus (Fig. 3f and Fig.S12). Additionally, we focused on the very short, near cognate driven uORFs that lie directly upstream of UL119 and US9, whose inclusion changes during infection as a result of changes in the 5′ end of the transcripts. We found that these uORFs modulated the translation efficiency of a downstream reporter gene (Fig.S14).
Finally, we examined the subcellular localization for 18 newly identified ORFs (11 of which were detected by mass-spectrometry, table S9) using transient expression of GFP-tagged proteins. We detected 15 proteins, ten of which showed specific subcellular localization patterns: six in mitochondria, three in the ER and one in the nucleus (Fig.3g and Fig.S15). Immunoprecipation and mass spectrometry experiments on two of these GFP-tagged proteins; ORF359W (ER localized) and US33A (mitochondrially localized) identified a few specific interacting proteins. Western blot analysis confirmed the interactions with TAP1 (ORF359W) and the mitochondrial inner membrane transport TIM machinery (US33A) (Fig.S16).
HCMV genes are expressed in a temporally regulated cascade. Our data provides an opportunity to monitor viral protein translation throughout infection. Notably, most of the viral genes, including newly identified ORFs, showed tight temporal regulation of protein synthesis levels; 82% of ORFs varied by at least 5-fold. Hierarchical clustering of viral coding regions by their footprints densities during infection (a measure of the relative translation rates) revealed several distinct temporal expression patterns (Fig.S17).
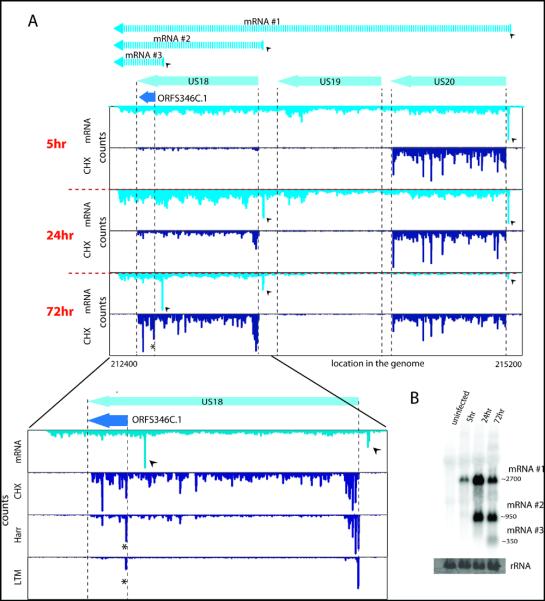
As was seen previously for a limited numbers of genomic loci (8-11, 22), examination of viral transcripts during infection revealed a pervasive use of alternative 5′ ends that is critical to the tight temporal regulation of viral genes expression and production of alternate protein products during infection. For example at the US18-US20 locus, 5 hpi, there is one main transcript starting just upstream of US20 enabling US20 translation. At 24 hpi, a shorter version of the transcript is detected starting immediately upstream of US18 enabling its translation. A third novel transcript isoform starting within the US18 coding sequence, emerges at 72 hpi, resulting in translation of a truncated version of US18 (ORFS346C.1) at this time point (Fig.4a and b). Another example is detailed in Fig. S18 and we identified reproducible temporal regulation of 5′ ends in 61 viral loci (encompassing ~350 ORFs) (Fig.S19, Fig.S20 and table S10) six of which we confirmed by Northern blot analysis (Fig. 4b, Fig.S11 and Fig.S21). Thus our studies reveal a pervasive mode of viral gene regulation in which dynamic changes in 5′ ends of transcripts control protein expression from overlapping coding regions. Just as alternative splicing (a process in which a single gene codes for multiple proteins) expands protein diversity, alternative transcript start sites may provide a broadly used mechanism for generating complex proteomes.
Fig. 4.
A major source of ORFs diversity during infection originates from alternative transcripts starts.
(A) The mRNA and ribosome occupancy profiles around US18-US20 loci at different infection times (marked on the left). Small arrows denote the different mRNA starts and the corresponding mRNAs are illustrated (upper part). The lower panel shows an expanded view of the US18 locus at 72 hpi and includes the harringtonine and LTM profiles (the internal initiation is marked with a star).
(B) Total RNA extracted at different time points during infection was subjected to Northern blotting for ORFS346C.1
The genomic era began with the sequencing of the bacterial DNA virus, phi X, in 1977 (23) and the mammalian DNA virus, Simian virus 40 (24), the following year. Since then, extraordinary advances in sequencing technology have enabled the determination of a vast array of viral genomes. Deciphering their protein coding potential, however, remains challenging. Here we present an experimentally-based analysis of translation of a complex DNA virus, HCMV, using both next generation sequencing and high-resolution proteomics. It is possible that many of the short ORFs we have identified are rapidly degraded and do not act as functional polypeptides. Nonetheless, these could still have regulatory function or be an important part of the immunological repertoire of the virus as MHC class I bound peptides are generated at higher efficiency from rapidly degraded polypeptides(25). Our work yields a framework for studying HCMV by establishing the viral proteome and its temporal regulation, providing a context for mutational studies and revealing the full range of HCMV functional and antigenic potential.
Supplementary Material
Acknowledgments
We thank O. Mandelboim, D. Wolf, M. Trilling, A. Lauring, S. Karniely and Weissman lab members for critical reading of the manuscript; C. Chu for assistance with sequencing; Pelletier, J. for providing Pateamine A; N.S-G is supported by human frontier science program postdoctoral fellowship. This work was supported by HHMI (JSW) and the Max-Planck Society (MM).
References and Notes
- 1.Mocarski ES, Shenk T, Pass RF. Fields BN, Knipe DM, Howley PM, editors. Fields Virology. 2007.
- 2.Davison AJ, et al. J Gen Virol. 2003;84:17. doi: 10.1099/vir.0.18606-0. [DOI] [PubMed] [Google Scholar]
- 3.Murphy E, Rigoutsos I, Shibuya T, Shenk TE. Proc Natl Acad Sci USA. 2003;100:13585. doi: 10.1073/pnas.1735466100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy E, Shenk T. Curr Top Microbiol Immunol. 2008;325:1. doi: 10.1007/978-3-540-77349-8_1. [DOI] [PubMed] [Google Scholar]
- 5.Zhang GJ, et al. Journal of Virology. 2007;81:11267. doi: 10.1128/JVI.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gatherer D, et al. Proc Natl Acad Sci USA. 2011;108:19755. [Google Scholar]
- 7.Cao J, Geballe AP. Mol Cell Biol. 1996;16:7109. doi: 10.1128/mcb.16.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stamminger T, et al. Journal of Virology. 2002;76:4836. doi: 10.1128/JVI.76.10.4836-4847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biegalke BJ, Lester E, Branda A, Rana R. J Virol. 2004;78:9579. doi: 10.1128/JVI.78.17.9579-9583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian Z, Xuan B, Hong TT, Yu D. J Virol. 2008;82:3452. doi: 10.1128/JVI.01964-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grainger L, et al. J Virol. 2010;84:9472. doi: 10.1128/JVI.00855-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Starck SR, et al. Science. 2012;336:1719. doi: 10.1126/science.1220270. [DOI] [PubMed] [Google Scholar]
- 13.Robert F, et al. PLoS One. 2009;4:e5428. doi: 10.1371/journal.pone.0005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT, Lareau LF, Weissman JS. Cell. 2011;147:789. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S, et al. Proc Natl Acad Sci USA. 2012.
- 16. Materials and methods are available as supporting material.
- 17.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Science. 2007;316:1345. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 18.Lord PC, Rothschild CB, DeRose RT, Kilpatrick BA. J Gen Virol. 1989;70:2383. doi: 10.1099/0022-1317-70-9-2383. [DOI] [PubMed] [Google Scholar]
- 19.Joachim T. Making large scale SVM learning practical. Advances in Kernel Methods - Support Vector Learning. 1998.
- 20.Bordeleau ME, et al. Chem Biol. 2006;13:1287. doi: 10.1016/j.chembiol.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Cox J, et al. J Proteome Res. 2011;10:1794. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 22.Leach FS, Mocarski ES. J Virol. 1989;63:1783. doi: 10.1128/jvi.63.4.1783-1791.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci USA. 1977;74:5463. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiers W, et al. Nature. 1978;273:113. [Google Scholar]
- 25.Yewdell JW. Curr Opin Immunol. 2007;19:79. doi: 10.1016/j.coi.2006.11.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.