Abstract
Piwi-interacting RNAs (piRNAs) ensure transposable element silencing in Drosophila, thereby preserving genome integrity across generations. Primary piRNAs arise from the processing of long RNA transcripts produced in the germ line by a limited number of telomeric and pericentromeric loci. Primary piRNAs bound to the Argonaute protein Aubergine then drive the production of secondary piRNAs through the “ping-pong” amplification mechanism that involves an interplay with piRNAs bound to the Argonaute protein Argonaute-3. We recently discovered that clusters of P-element-derived transgenes produce piRNAs and mediate silencing of homologous target transgenes in the female germ line. We also demonstrated that some clusters are able to convert other homologous inactive transgene clusters into piRNA-producing loci, which then transmit their acquired silencing capacity over generations. This paramutation phenomenon is mediated by maternal inheritance of piRNAs homologous to the transgenes. Here we further mined our piRNA sequencing data sets generated from various strains carrying transgenes with partial sequence homology at distinct genomic sites. This analysis revealed that same sequences in different genomic contexts generate highly similar profiles of piRNA abundances. The strong tendency of piRNAs for bearing a U at their 5′ end has long been recognized. Our observations support the notion that, in addition, the relative frequencies of Drosophila piRNAs are locally determined by the DNA sequence of piRNA loci.
Keywords: Drosophila melanogaster, argonaute proteins, epigenetics, germline, paramutation, piRNA biogenesis, transposable elements
Repression of Transposable Elements (TEs) in the Drosophila germline by Piwi-interacting RNAs (piRNAs) preserves genome integrity and prevents the transmission to next generations of mutations induced by TE mobilization. Over the past years, major progresses have been made in the understanding of the mechanisms of piRNA biogenesis and activity in flies.1
The Drosophila melanogaster genome carries a limited number of loci (~140) that contain arrays of TE fragments and are most often bi-directionally transcribed in the germ cells to produce both sense and antisense piRNA precursor transcripts. In contrast, a single flamenco TE cluster is uni-directionally transcribed in the ovarian follicle cells surrounding the germ cells, produces antisense piRNA precursors exclusively, and is mainly involved in silencing of a specific class of retrotransposons called errantiviruses. piRNA precursor transcripts in the germ cells reach the nuage, a diffuse structure surrounding the nucleus where a number of components from the piRNA pathway accumulate.2 In the nuage, the cleavage of piRNA precursor transcripts by the nuclease Zucchini and subsequent 3′ shortening give rise to primary 23–28-nt long piRNAs.3,4 Antisense primary piRNAs bound to the PIWI Argonaute Aubergine (Aub) can then enter the so-called “ping-pong” amplification step by pairing with sense piRNA precursors. This results in the slicing of precursors and generates secondary sense piRNAs, which, in turn, associate with the PIWI Argonaute AGO3 and guide the cleavage of more antisense precursors.5,6 Thus, the ping-pong mechanism drives the amplification of a population of antisense piRNAs that overlap by 10 nt with sense piRNAs. Indeed, this 10 nt overlap “signature” reflects the piRNA-guided cleavage of piRNA precursors by PIWI Argonautes, which occurs between nucleotide 10 and 11, relative to the 5′ end of the guide piRNA. As another feature of primary piRNAs is their bias for having a 5′ uridine (1U), which is probably due to the nucleotide preferences of Aubergine and Piwi proteins, the ping-pong mechanism amplifies of population of AGO3-associated secondary piRNAs with a bias for an adenine at position 10 (10A).
Although the ping-pong amplification provides an obvious mechanism of TE transcript degradation, TE silencing by Piwi-bound piRNAs may also operate at transcriptional level (TGS). The Piwi Argonaute protein distributes in both cytoplasm and nucleus.6,7 In ovarian follicle cells, which are devoid of Aub and AGO3, the slicer activity of Piwi protein is not required for TE silencing, whereas a mutation impairing Piwi nuclear localization abolishes TE silencing.8,9 Moreover, in a follicle-derived cell line, Piwi-bound piRNAs target TE sequences dispersed in euchromatin, reduce their Pol II occupancy, and trigger the formation of H3K9me3 repressive marks.10 A recent study suggests that Piwi may similarly mediate TGS in some somatic adult tissues.11
In contrast to the progresses made on the biogenesis and silencing mechanisms by piRNAs, what defines piRNA-producing loci and how the production of piRNAs by these loci is regulated is not well understood. It is likely that the repetitive nature of piRNA loci is an important cis-regulatory feature for the production of primary piRNAs. Besides, the deposition of H3K9me3 mark at TE clusters by dSETDB1 was involved in piRNA production12 and the HP1-family protein Rhino binds to dual strand clusters to promote their transcription and piRNA production.13 However, the mechanisms by which these factors are targeted to piRNA loci are still unknown.
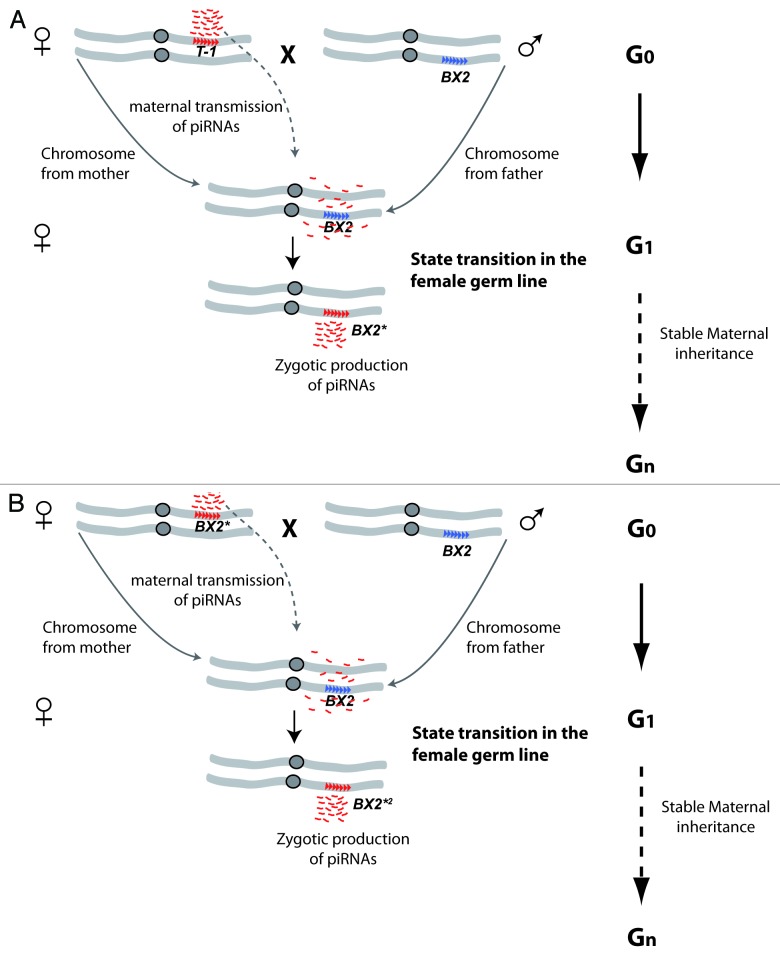
Our recent finding that piRNAs are vectors of a paramutation in Drosophila14 adds another layer of complexity by indicating that maternally deposited piRNAs can trigger trans-generational emergence of some piRNA-producing loci. In previous studies, we had shown that two P{lArB} transgenes inserted in Telomeric Associated Sequences (TAS) and containing the Adh and rosy genes of D. melanogaster and a bacterial lacZ gene, repress germline expression of lacZ reporter transgenes inserted at a distance, through a homology-dependent silencing mechanism called Trans-Silencing Effect (TSE).15,16 These telomeric P{lArB} insertions (hereafter referred to as P-1152) mimic natural P-elements whose insertions in TAS results in the production of P-element-derived piRNAs and establishment of maternally transmitted P-element repression.17- 19 In addition to P-1152, we found that T-1, a repeat cluster of 7 P{lacW} transgenes containing the white and lacZ genes and inserted in the middle of chromosome arm 2R,20 also produces piRNAs and triggers strong TSE.21 In contrast, other P{lacW} clusters inserted at the exact same location, including BX2 that has the same number of P{lacW} repeats as T-1, did not induce detectable TSE. TSE strongly correlates with piRNA production, as small RNA sequencing from P-1152 or T-1 but not from BX2 ovaries revealed numerous transgene-derived piRNAs. Strikingly, when BX2 males were crossed with T-1 females (Fig. 1A), the female progeny containing the BX2 chromosome acquired strong TSE capacity (noted as BX2*). This effect was observed without T-1 chromosome inheritance from the T-1 mother and was then stably inherited over generations. Moreover, when BX2* females were crossed with BX2 males (Fig. 1B), the female progeny containing the “naïve” BX2 chromosome in turn acquired strong TSE capacity (noted as BX2*2). Thus, the BX2 to BX2* transition is a paramutation, previously defined as an epigenetic interaction between two alleles of a locus, through which one allele induces a heritable modification of the other allele without modifying the DNA sequence.22,23 Moreover, the acquired and stable TSE capacities of the BX2* and BX2*2 lines correlated with the production of a high level of BX2-derived piRNAs in ovaries, and were abolished in aubergine but not in Dicer-2 mutants. Altogether, these results imply that piRNAs can play the role of a maternally deposited signal that first triggers and then maintains over generations the production of piRNA from a previously inactive locus (Fig. 1). Interestingly, a recent work suggests that resembling mechanisms may account for the acquisition of I-element repression capacity in Drosophila strains devoid of functional copies of this LINE-like element.24
Figure 1. Paramutation of the BX2 locus involves maternally inherited T-1 piRNAs. (A) Whereas the T-1 transgene cluster produces piRNAs (small red dashes), the BX2 transgene cluster does not; these distinct properties are completely stable over generations. When T-1 females are crossed to BX2 males (G0), the female progeny (G1) that inherited the BX2 chromosome from fathers and T-1-derived piRNAs from the mother (but not the T-1 chromosome) start to zygotically produce high levels of BX2-derived piRNAs. The inactive (blue) to active (red) state transition of the BX2 locus is noted with an asterisk (BX2*). This so-called paramutation can be further maternally inherited in the next generations (Gn). (B) Similarly as in (A), maternal inheritance of BX2*-derived piRNAs triggers the state transition of an inactive BX2 loci in G1, associated to zygotic production of piRNAs. This second-order paramutation is noted as BX2*2 and can be further maternally inherited in the next generations (Gn). The seven repeats of the P{lacW} transgene in the T-1 and BX2 loci are represented by blue or red arrowheads, depending on the states of the loci (active in red, inactive in blue).
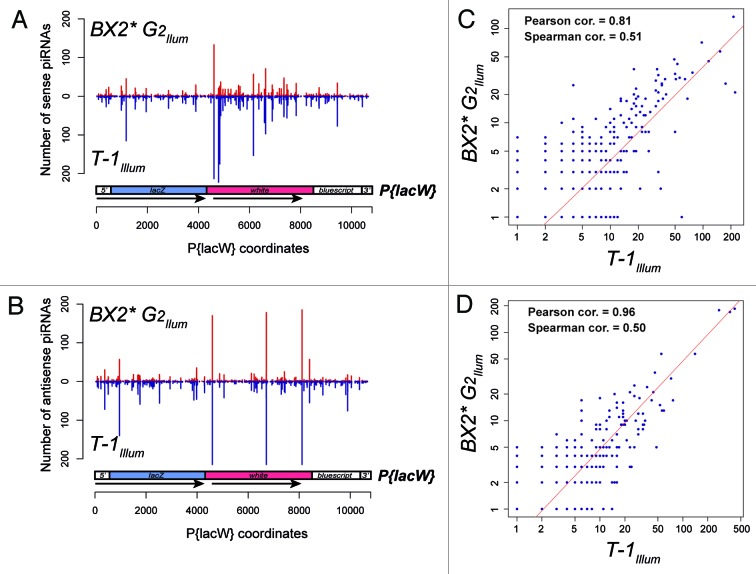
Our small RNA sequencing of small RNA libraries14 prepared using an Illumina set of RNA adaptor (Illum) revealed that piRNA abundance profiles from T-1 and BX2* ovaries after two generations (G2) are quite similar. This similarity is apparent from the observed degree of symmetry when either sense (Fig. 2A) or antisense (Fig. 2B). T-1Illum and BX2*G2Illum piRNA abundances were plotted on the same maps. Accordingly, abundances of sense as well as antisense piRNAs from T-1Illum and BX2*G2Illum showed a strong correlation (Fig. 2C and D). Note that although the Spearman correlation coefficient (based on ranking correlation) is less impressive in these analyses, it is more appropriate and robust than the Pearson correlation coefficient (based on linear regression of the values) when the data do not necessarily come from a bivariate normal distribution, which is likely the case for piRNA abundance variables. Cloning biases impact the small RNA libraries generated,25 thereby altering quantitation and possibly accounting for the strong correlations between the T-1Illum and BX2*G2Illum profiles of piRNA abundances. Indeed, these cloning biases were reflected by the lower correlations between the sense and antisense BX2*G2Illum profiles and the sense and antisense BX2*G2IdT profiles obtained under the same genetic settings but using another IdT set of RNA adapters (Table 1, Pearson cor. 0.38 and 0.26, Spearman cor. 0.28 and 0.27). However, these correlations were still highly significant (P values < 2.2e-16 in both Pearson and Spearman correlation tests). In addition, sense and antisense profiles from BX2*G2IdT and T-1Illum obtained using different set of RNA adapters during library preparations remain also significantly correlated (Table 1, Pearson cor. 0.28 and 0.27, Spearman cor. 0.29 and 0.27, all P values < 2.2e-16). In agreement with a previous report,26 these data suggest that cloning biases in small RNA libraries are not sufficient to explain correlations between profiles of piRNA abundances and that these profiles are in part determined by the DNA sequence of piRNA producing loci. The analysis of small RNA libraries all prepared using the same IdT set of RNA adapters further supports this conclusion, as both sense and antisense piRNA abundance profiles remain strongly correlated in the BX2* line after 42 generations (Table 1, BX2*G2IdT vs BX2*G42IdT) as well as in a BX2*2 line after 36 generations (Table 1, BX2*G2IdT vs BX2*2G36IdT).
Figure 2. The profiles of piRNA abundances in T-1 and BX2* ovaries show strong correlations. The numbers of piRNAs (23–28 nt small RNA reads) matching the sense strand (A) or the antisense strand (B) of P{lacW} in T-1 (blue bars) or BX2* ovaries (red bars) were plotted relatively to the P{lacW} nucleotide coordinates. Number of reads of individual sense (C) or antisense (D) piRNA sequences matching P{lacW} in T-1 (x-axes) or BX2* (y-axes) ovaries were plotted in scatter plots. The Illum index indicates that the small RNA libraries were prepared using the same Illumina set of RNA adapters (see ref. 14). The red line corresponds to the linear regression of the data. The Pearson and Spearman correlation coefficients were computed using the cor.test function in R and the methods “pearson” or “spearman.”
Table 1. Pearson and Spearman correlation coefficients of BX2* and T-1 profiles of piRNA abundances.
| Pearson | s BX2*G2IdT | as BX2*G2IdT | s BX2*G42IdT | as BX2*G42IdT | s BX2*2G36IdT | as BX2*2G36IdT | s BX2*G2Illum | as BX2*G2Illum | s T-1Illum | as T-1Illum |
|---|---|---|---|---|---|---|---|---|---|---|
|
s BX2*G2IdT |
1 |
-0.01 |
0.88 |
-0.01 |
0.91 |
-0.01 |
0.38 |
-0.01 |
0.28 |
-0.01 |
|
as BX2*G2IdT |
|
1 |
-0.01 |
0.89 |
-0.01 |
0.88 |
-0.01 |
0.26 |
0.00 |
0.27 |
|
s BX2*G42IdT |
|
|
1 |
-0.01 |
0.91 |
-0.01 |
0.28 |
-0.01 |
0.22 |
-0.01 |
|
as BX2*G42IdT |
|
|
|
1 |
-0.01 |
0.88 |
-0.01 |
0.17 |
0.00 |
0.18 |
|
s BX2*2G36IdT |
|
|
|
|
1 |
-0.02 |
0.29 |
-0.01 |
0.22 |
-0.01 |
|
as BX2*2G36IdT |
|
|
|
|
|
1 |
-0.01 |
0.20 |
-0.01 |
0.21 |
|
s BX2*G2Illum |
|
|
|
|
|
|
1 |
-0.01 |
0.81 |
-0.01 |
|
as BX2*G2Illum |
|
|
|
|
|
|
|
1 |
0.00 |
0.96 |
|
s T-1Illum |
|
|
|
|
|
|
|
|
1 |
0 |
|
as T-1Illum |
|
|
|
|
|
|
|
|
|
1 |
| |
IdT adaptor set |
Illumina adaptor set |
||||||||
| |
|
|
|
|
|
|
|
|
|
|
|
Spearman |
s BX2*G2IdT |
as BX2*G2IdT |
s BX2*G42IdT |
as BX2*G42IdT |
s BX2*2G36IdT |
as BX2*2G36IdT |
s BX2*G2Illum |
as BX2*G2Illum |
s T-1Illum |
as T-1Illum |
|
s BX2*G2IdT |
1 |
-0.04 |
0.55 |
-0.03 |
0.57 |
-0.03 |
0.28 |
-0.01 |
0.29 |
-0.02 |
|
as BX2*G2IdT |
|
1 |
-0.04 |
0.46 |
-0.05 |
0.50 |
-0.02 |
0.27 |
-0.03 |
0.27 |
|
s BX2*G42IdT |
|
|
1 |
-0.04 |
0.58 |
-0.04 |
0.30 |
-0.01 |
0.31 |
-0.02 |
|
as BX2*G42IdT |
|
|
|
1 |
-0.04 |
0.51 |
-0.02 |
0.27 |
-0.03 |
0.29 |
|
s BX2*2G36IdT |
|
|
|
|
1 |
-0.05 |
0.30 |
-0.01 |
0.30 |
-0.03 |
|
as BX2*2G36IdT |
|
|
|
|
|
1 |
-0.02 |
0.29 |
-0.03 |
0.29 |
|
s BX2*G2Illum |
|
|
|
|
|
|
1 |
-0.02 |
0.51 |
-0.02 |
|
as BX2*G2Illum |
|
|
|
|
|
|
|
1 |
-0.02 |
0.50 |
|
s T-1Illum |
|
|
|
|
|
|
|
|
1 |
0 |
|
as T-1Illum |
|
|
|
|
|
|
|
|
|
1 |
| IdT adaptor set | Illumina adaptor set | |||||||||
Using the indicated sets of RNA adapters (Illum or IdT) to generate small RNA libraries, we had generated 10 sequencing data sets from BX2 and T-1 strains under various genetic settings (see text and ref. 14). Sense (s) and antisense (as) piRNA abundance profiles were handled as vectors of 11,690 variables indexed with the P{lacW} coordinates and taking as values the number of sequenced piRNAs mapped at these coordinates (most 5′ nucleotide location). Pearson (upper table) and Spearman (lower table) correlations between these vectors were then computed in pairwise combinations using the R software. Significant correlations (P value < 2.2e-16) are bolded.
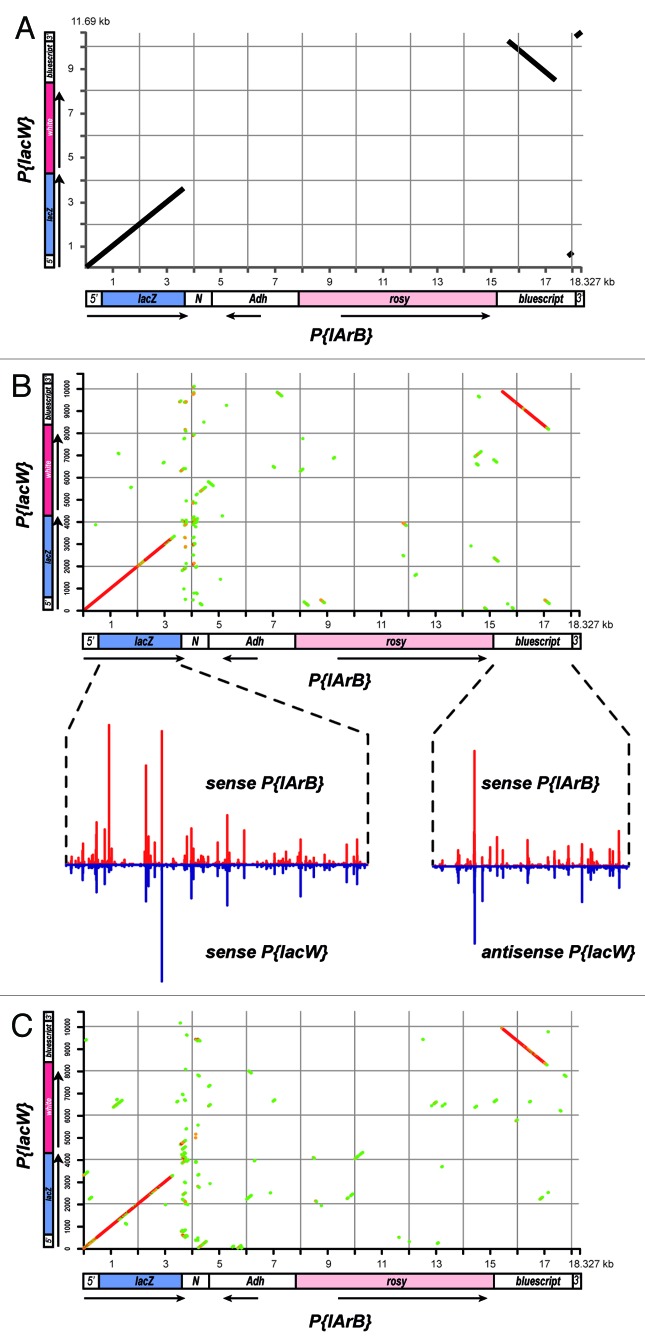
Interestingly, the P{lArB} transgenes of the telomeric P-1152 locus and the P{lacW} transgenes at the BX2 and T-1 loci in the middle of chromosome 2R have some DNA sequences in common, as evidenced by a dot matrix view of a Blast alignment of the two types of transgenes (Fig. 3A). Namely, P{lArB} and P{lacW} share the lacZ gene fused to the 3′ UTR of hsp70 as well as a common pBluescript-derived backbone either in direct or in inverse orientation relatively to lacZ (Fig. 3). In contrast, the Adh coding sequences fused to a 620 bp DNA fragment of unknown origin and the rosy marker gene are specific to P{lArB} whereas the white marker gene is specific to P{lacW}. This situation of two piRNA producing loci located at distinct genomic positions but sharing partial homology allowed us to test whether DNA sequences locally impact the profiles of piRNA abundances. To this aim, we computed the Spearman correlations between all possible 500 nt profile segments generated by the sense strand of P{lArB} in P-1152 ovaries and all possible 500 nt profile segments generated by both sense and antisense strands of P{lacW} in T-1 ovaries (Fig. 3B, two times ~181 millions of correlations were computed). We repeated the same procedure with all possible 500 nt profile segments generated by the antisense strand of P{lArB} (Fig. 3C). As mentioned above, we chose the Spearman correlation because it is more stringent when the data do not necessarily come from a bivariate normal distribution. In addition, the 500 nt size of profile segments was a good compromise between the amount of information contained in segments and the number of pairwise correlations to be computed. If higher than 0.3 (P value < 2.2e-16), the correlation coefficients were then plotted in a dot matrix. As a result, correlation dot matrixes generated with the sense (Fig. 3B) as well as the antisense strand (Fig. 3C) of P{lArB} echoed the dot matrix of Blastn alignment between P{lArB} and P{lacW}. Thus, the piRNA abundance profiles generated by both strands of the lacZ sequences at the P-1152 or T-1 loci strongly correlated with each other. Strikingly, the inverse orientations of the Pbluescript-derived backbones in P{lArB} and P{lacW} were captured in both correlation dot matrixes (Fig. 3B and C), indicating that the profiles of piRNA abundances in the P-1152 and T-1 loci are locally determined by DNA sequences but not by their relative orientation.
Figure 3. Sequences common to P{lacW} and P{lArB} in T-1 and P-1152 loci, respectively, generate highly similar profiles of piRNA abundances. (A) Dot matrix view of a Blastn alignment between P{lArB} and P{lacW}. In these transgenes, the lacZ and pBluescript sequences are 100% identical (black lines) and in the same or inverse orientation relatively to the P-ends (5′ and 3′), respectively. The 620 bp sequence of the region noted N in P{lArB} is unknown. (B) Spearman correlation matrix between profiles of piRNA abundances from the sense strand of P{lArB} and profiles of piRNA abundances from either strands of P{lacW}. All possible correlations between 500 nt sliding windows were computed using an in-house python script (available upon request) and the stats.spearman function from the scipy python module. Spearman correlation coefficients were plotted relatively to the 5′ coordinates of the windows in P{lArB} (x-axis) and P{lacW} (y-axis) only if higher than 0.32: in green if lower than 0.35, in orange when between 0.35 and 0.4, and in red if higher than 0.4. For clarity, piRNA profiles from the indicated strands are shown in insets as in Figure 2 for the lacZ and pBluescript regions with high profile similarities. The antisense pBluescript profile from P{lacW} was reversed before plotting. (C) The Spearman correlation matrix between profiles of piRNA abundances from the antisense strand of P{lArB} and profiles of piRNA abundances from either strands of P{lacW} was computed and displayed as in (B).
The extended analysis of our piRNA sequencing data sets suggests that the relative abundance of piRNAs is locally determined in piRNA producing loci. Further supporting this notion, it was possible to consistently remap transposons and retrotransposons dispersed in the genome by scanning genome-wide piRNA profiles with a sliding window and computing correlations with reference profiles for families of transposons and retrotransposons (data not shown). Several steps in piRNA biogenesis may be sequenced-biased, leading to sequence-dependent profiles of piRNA abundances. Cleavage of long RNA precursors by the Zucchini RNase may involve preferences for local RNA motives and/or local RNA secondary structures. In addition to the established preference of Aubergine and Piwi for piRNAs starting with a 5′U, other sequence motives may be responsible for preferential piRNA loading into these Argonautes. Finally, thermodynamic features and target matches that in turn depend on piRNA sequences may influence the stability of piRNAs and thus their abundances in small RNA libraries. In any case, our data favor a model in which local sequence rather than long-distance chromosomal environment is a primary determinant of the abundance profiles of piRNAs.
Acknowledgements
We thank Armand Taranco for helpful discussions on statistical correlations. This work was supported by fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche to AV and CH, from the Fondation pour la Recherche Médicale to AV, from the Association Nationale de la Rercherche (ANR) to ALB, and by grants from the Foundation ARC pour la Recherche contre le Cancer to SR and from the ANR (project “Nuclear endosiRNAs”) to CA.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25756
References
- 1.Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev. 2013;23:44–52. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Wang J, Xu J, Zhang Z, Koppetsch BS, Schultz N, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151:871–84. doi: 10.1016/j.cell.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishimasu H, Ishizu H, Saito K, Fukuhara S, Kamatani MK, Bonnefond L, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–7. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 4.Ipsaro JJ, Haase AD, Knott SR, Joshua-Tor L, Hannon GJ. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–83. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 6.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 7.Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–14. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 8.Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 9.Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, et al. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci USA. 2011;108:18760–5. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–80. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24:502–16. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, et al. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–9. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–49. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vanssay A, Bougé AL, Boivin A, Hermant C, Teysset L, Delmarre V, et al. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature. 2012;490:112–5. doi: 10.1038/nature11416. [DOI] [PubMed] [Google Scholar]
- 15.Roche SE, Rio DC. Trans-silencing by P elements inserted in subtelomeric heterochromatin involves the Drosophila Polycomb group gene, Enhancer of zeste. Genetics. 1998;149:1839–55. doi: 10.1093/genetics/149.4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josse T, Teysset L, Todeschini AL, Sidor CM, Anxolabéhère D, Ronsseray S. Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation. PLoS Genet. 2007;3:1633–43. doi: 10.1371/journal.pgen.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronsseray S, Lehmann M, Nouaud D, Anxolabéhère D. The regulatory properties of autonomous subtelomeric P elements are sensitive to a Suppressor of variegation in Drosophila melanogaster. Genetics. 1996;143:1663–74. doi: 10.1093/genetics/143.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marin L, Lehmann M, Nouaud D, Izaabel H, Anxolabéhère D, Ronsseray S. P-Element repression in Drosophila melanogaster by a naturally occurring defective telomeric P copy. Genetics. 2000;155:1841–54. doi: 10.1093/genetics/155.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorer DR, Henikoff S. Transgene repeat arrays interact with distant heterochromatin and cause silencing in cis and trans. Genetics. 1997;147:1181–90. doi: 10.1093/genetics/147.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronsseray S, Boivin A, Anxolabéhère D. P-Element repression in Drosophila melanogaster by variegating clusters of P-lacZ-white transgenes. Genetics. 2001;159:1631–42. doi: 10.1093/genetics/159.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–89. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coe EH. A Regular and Continuing Conversion-Type Phenomenon at the B Locus in Maize. Proc Natl Acad Sci USA. 1959;45:828–32. doi: 10.1073/pnas.45.6.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–88. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorefan K, Pais H, Hall AE, Kozomara A, Griffiths-Jones S, Moulton V, et al. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012;3:4. doi: 10.1186/1758-907X-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muerdter F, Olovnikov I, Molaro A, Rozhkov NV, Czech B, Gordon A, et al. Production of artificial piRNAs in flies and mice. RNA. 2012;18:42–52. doi: 10.1261/rna.029769.111. [DOI] [PMC free article] [PubMed] [Google Scholar]