Abstract
The acquisition of antibiotic resistance by human pathogens poses a significant threat to public health. The mechanisms that control the proliferation and expression of antibiotic resistance genes are not yet completely understood. The aminoglycosides are a historically important class of antibiotics that were introduced in the 1940s. Aminoglycoside resistance is conferred most commonly through enzymatic modification of the drug or enzymatic modification of the target rRNA through methylation or through the overexpression of efflux pumps. In our recent paper, we reported that expression of the aminoglycoside resistance genes encoding the aminoglycoside acetyl transferase (AAC) and aminoglycoside adenyl transferase (AAD) enzymes was controlled by an aminoglycoside-sensing riboswitch RNA. This riboswitch is embedded in the leader RNA of the aac/aad genes and is associated with the integron cassette system. The leader RNA can sense and bind specific aminoglycosides such that the binding causes a structural transition in the leader RNA, which leads to the induction of aminoglycoside antibiotic resistance. Specific aminoglycosides induce reporter gene expression mediated by the leader RNA. Aminoglycoside RNA binding was measured directly and, aminoglycoside-induced changes in RNA structure monitored by chemical probing. UV cross-linking and mutational analysis identified potential aminoglycoside binding sites on the RNA.
Keywords: antibiotic resistance, aminoglycoside, riboswitch, antibiotic sensing RNA, induction of antibiotic resistance, integron
Introduction
Riboswitches are non-coding mRNAs that regulate the expression of biosynthetic genes by binding small molecule metabolites or cofactors to control the biosynthetic pathway of the metabolite or cofactor.1-5 They act as biosensors that regulate the concentration of the small molecule in the cell. Networks of riboswitches have an important role in prokaryotic metabolism where they employ diverse mechanisms to interact with transcriptional and/or translational machineries. More than 20 types of riboswitch have been identified that are responsive to a diverse set of chemical classes including purines and their derivatives,3,6 ribonucleotide cofactors,7-11 amino acids,12-14 sugars,15 antibiotics,16 anions,17 and metal ions.18,19
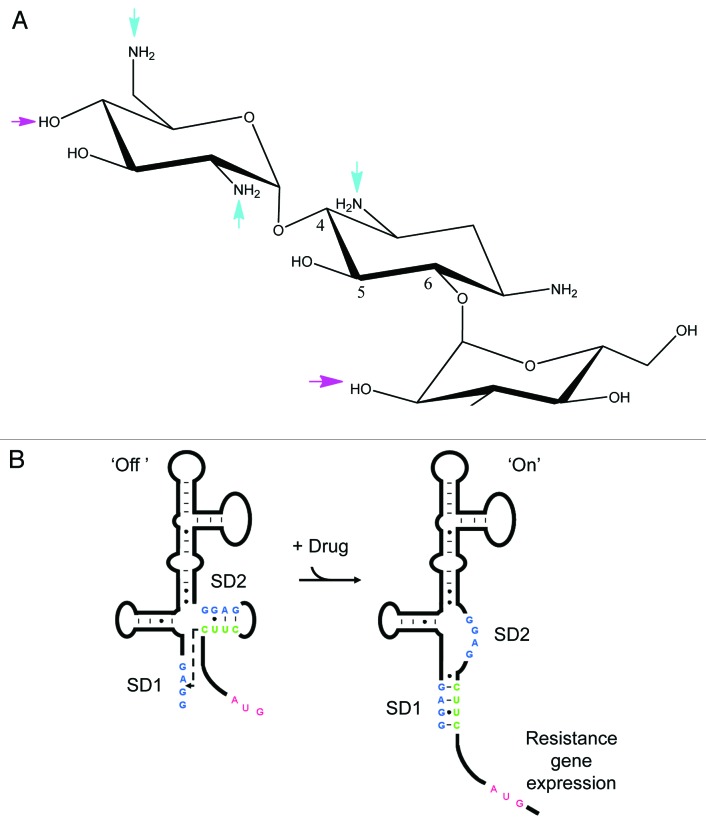
The aminoglycoside antibiotics were originally isolated as natural products and were among the first antibiotics used to treat serious bacterial infections in the clinic (reviewed in refs. 20 and 21). They target the A site in the decoding region of the 30S ribosomal subunit and inhibit translocation.22-24 Aminoglycoside resistance is associated with the mobile elements on plasmids or integrons and appeared soon after their introduction.21,25,26 Resistance to aminoglycosides is mainly conferred by enzymatic modification of the drug, methylation of the target rRNA, or the overexpression of efflux pumps.27 The enzymatic modifications of the aminoglycosides include acetylation, adenylylation, and phosphorylation. Aminoglycoside acetyl transferases (AAC) or aminoglycoside adenyl transferases (AAD) catalyze acetylation or adenylylation of aminoglycoside and confer resistance to the drugs28 (Fig. 1A).
Figure 1. (A) Kanamycin B, the 4, 6 deoxystreptamine aminoglycoside antibiotic that is most susceptible to inactivation by enzymatic modification. The 4, 5, and 6 positions of deoxstreptamine are indicated. Arrows indicate the major sites of inactivation by N-acetylation (cyan) and O-adenylation (magenta). (B) Schematic representation of the proposed model for the induction of aminoglycoside resistance. Aminoglycoside binding to the leader RNA induces a change in the leader RNA structure such that the Anti-SD2 sequence CUUC base-pairs with SD1 consequently unmasking SD2 for ribosomal binding and translation of the resistance gene. SD1 and SD2 are colored blue, the anti-SD2 sequence is green, and the start codon of the resistance gene is red.
We recently found that expression of an aac gene from Pseudomonas fluorescens can be controlled by an aminoglycoside-sensing riboswitch.16 The riboswitch RNA is present in the leader RNAs of acc/aad genes. Our data demonstrate that the leader RNA can specifically bind certain aminoglycosides such that the binding induces a structural change in the RNA which, in turn, leads to the induction of the acc/aad gene expression. We showed the following evidence to support this conclusion:
(1) Specific aminoglycosides induce expression of a reporter gene mediated through the acc/aad leader RNA, in comparison with a control leader RNA. Kanamycin B, sisomycin, and other 4,6 deoxystreptamine aminoglycosides induce expression of the reporter gene. On the other hand, the control molecules; the 4,5 deoxystreptamine derivatives ribostamycin, paromomycin, or neamine failed to induce the reporter gene.
(2) Aminoglycoside binding to the leader RNA can be measured directly by Surface Plasmon Resonance (SPR): We found that the aminoglycosides that induce reporter gene expression in the reporter assays also display the highest affinities for the leader RNA [e.g., kanamycin B and sisomycin (2.78 and 6.8 µM, respectively)]. In contrast, the control antibiotics displayed lower affinities for the leader RNA [e.g., neamine and ribostamycin (47 and 589 µM, respectively)].
(3) Aminoglycoside binding to the leader RNA induces a structural transition in the leader RNA that can be detected by changes in gel electrophoretic mobility and chemical probing. The 4,6 deoxystreptamine aminoglycosides that induce reporter gene expression and bind to the leader RNA with high affinity also cause a conformational change to the RNA upon binding as measured by chemical probing. This correlation suggested that there may be a connection between the induced change in RNA structure and the induction of gene expression. In comparison, the control (4,5 deoxystreptamine derivatives) drugs that fail to induce reporter gene expression also bind to the leader RNA with low affinity and cause no structural change in the RNA.
(4) A specific cross-link can be detected between an inducing aminoglycoside and the leader RNA.
(5) Mutational analysis of the leader RNA confirms the main features of the RNA secondary structure and the importance of the structural elements within it for aminoglycosides binding.
Based on the above data, we proposed a regulatory model (Fig. 1B) to explain how aminoglycoside binding to the RNA can regulate gene expression: In the absence of the antibiotic the ribosome binding site (SD2) of the resistance gene is sequestered by a complementary anti-SD sequence that blocks ribosome binding; antibiotic binding induces a structural transition that unmasks SD2 for ribosome binding leading to translation of the resistance gene (Fig. 1B). There are parallels between this proposed mechanism for the control of translational initiation and that the employed by flavin mononucleotide, SMK S-adenosyl methionine, and the thiamine pyrophosphate-dependent regulatory riboswitches.7,8,29
(6) We further showed by real-time RT PCR that aminoglycoside-leader RNA interactions control gene expression at the level of translation and probably not at the level of transcription.
(7) We then investigated the role of a putative leader peptide embedded within the leader RNA on the induction of the aac/aad gene. In the case of induction of the erythromycin resistance methyltransferase ermC by macrolide antibiotics, the leader peptide encoded by its leader RNA plays an important role in ribosomal stalling.30-32 We made a series of mutations in which the amino acid sequence of the leader peptide was unchanged while the leader RNA sequence was altered and the effects of these mutants were analyzed by reporter assays. We found that, in contrast to ermC, induction of the reporter gene was independent of nascent leader peptide stalling. This suggested that the induction of reporter gene expression that we observed was specific for the sequence of the leader RNA.
(8) We additionally showed that the induction of the reporter genes occurs independently of antibiotic-ribosome interactions i.e., induction of the reporter gene can also take place in the presence of resistant ribosomes.
Analysis of the Conserved Sequence in the aac/aad 5′ Leader RNA
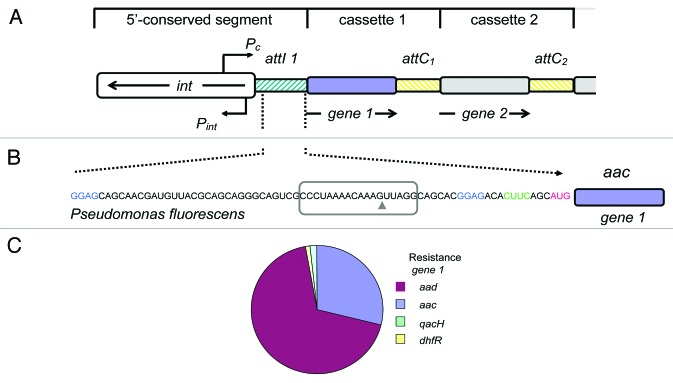
We analyzed the untranslated region mRNAs of 50 aminoglycoside resistance genes, including genes encoding for acetyl transferases, adenyl transferases, phospho transferases, rRNA methyl transferases, and efflux pumps. The leader RNA of two aminoglycoside acetyl transferase and three aminoglycoside adenyl transferase genes were closely related. The 75 nucleotide (nt) leader RNA sequence was later experimentally demonstrated to be the minimal functional unit of an aminoglycoside sensing riboswitch that controls the expression of aac/aad. Part of the DNA sequence corresponding to the 75 nt leader RNA overlaps with the attI1 site of class 1 integrons and includes the 7 bp attI1 insertion site (Fig. 2A and B).33,34 The 75 nt leader RNA is located between the (divergent) integrase and the AAC or AAD resistance proteins of the class 1 integrons of a variety of antibiotic resistant strains. A blast search35 of the 75 nt leader RNA aac/aad sequence shows it to be located in the 5′ conserved segment (Fig. 2A and B). The pie chart (Fig. 2C) shows that the majority of the top 150 annotated resistance genes in the blast search that neighbor the aac/aad sequence encode AAC or AAD resistance proteins. Only two non-aminoglycoside resistance proteins were identified by this search, dihydrofolate reductase (dhfR) and the nonfunctional multidrug exporter (qacH). Integrons were originally discovered through the proliferation of antibiotic resistance36 (reviewed in ref. 26), they have a critical role in plasmid-based capture and expression of antibiotic resistance genes. Antibiotic resistance genes are accumulated by the integron-based site-specific recombination mechanism. The attI1 insertion site is the 5′ site-specific recombination insertion site that is recognized by the integron encoded integrase (int).34,37,38 The integrase gene is expressed from a divergent promoter (Pint) upstream of the attI1 site (Fig. 2).39 The transcription of resistance gene cassettes inserted at the attI1 site is generally driven by a strong upstream promoter (Pc) as the assimilated resistance genes mostly lack their own promoters (Fig. 2).39-41 Thus, the upstream sequences of genes inserted at the attI1 site have an important prerequisite for a putative riboswitch; they are transcribed. In class 1 integrons, the transcript that encompasses the attI1 sequence also includes a conserved putative leader peptide that includes a consensus SD sequence (GGAG) and initiation and stop codons. The stop codon is located in the attI1 site but is created during gene capture and originates from the inserted gene cassette.34,40,42 The downstream sequence (between attI1 and the resistance gene) therefore contains elements of the regulatory sequences of the inserted resistance genes and shows considerable sequence variation.
Figure 2. (A) A schematic of the 5′ conserved segment of class 1 integrons, the inserted gene cassettes (shown here as genes 1 and 2) are transcribed by the strong promoter Pc (derived from refs. 39, 40 and 66). The divergent promoter Pint transcribes the integrase (int) gene. Single gene cassettes are inserted by integrase-mediated recombination between integrase recognition sites within the attI1 (green hatching) and attC elements (yellow hatching), a regulatory RNA is also associated with the attC element.40 Integrase expression is linked to the SOS response that can also be induced by antibiotics. (B) The 75 nucleotide leader aac/aad riboswitch RNA from Pseudomonas fluorescens, the RNA is located between the divergent int and aac genes at the conserved 5′ end of the integron. The leader RNA transcript is produced by the strong promoter Pc. SD1 and SD2 are colored blue, the anti-SD2 sequence green, and the start codon of the resistance gene is red. The equivalent positions of the core attI1 recognition site on the corresponding DNA are indicated by the box, the arrowhead indicates the approximate position of the attC 3′ DNA insertion. (C) Pye chart representing a blast search of the aac/aad riboswitch sequence from Pseudomonas fluorescens. Of the top 150 blast hits 111 were annotated such that the identities of the neighboring genes were known. The majority of neighboring resistance genes encoded aminoglycoside adenyltransferases (aad, 76) and aminoglycoside acetyltransferases (aac, 32). There was also a minor representation of the non-aminoglycoside resistance genes dihydrofolate reductase (dhfR, 1) and a non-functional multidrug exporter (qacH, 2).
Integration and resistance genes captured by integrons are controlled by the expression of the integrase gene. Integrase gene expression has been shown to be controlled by the SOS response, which, in turn, can be induced by sub-inhibitory doses of antibiotics, including aminoglycosides.43,44 We have shown that the leader RNA sequence that includes the attI1 site is preserved at the level of the RNA (and also the DNA) sequence. Because the SOS response is also known to be highly mutagenic,43 the presence of the conserved regulatory RNA sequence would therefore stabilize and protect the attI1 recombination site from accumulating mutations. The association of an antibiotic resistance gene with a site-specific recombination site would also confer a selective advantage to the integron in an antibiotic rich environment. A concurrent inducible riboswitch that responds to a commonly used antibiotic would therefore give an additional selective advantage to the integron cassette.
Specific Aminoglycoside-Induced Reporter Gene Expression Mediated by the aac/aad Leader RNA
We have shown that certain aminoglycosides induce reporter gene expression through interactions with the 75 nt leader RNA of aac/aad by both agar diffusion assays and solution-based assays of β-galactosidase reporter gene expression. The induction of reporter gene expression by aminoglycosides was observed both on agar plates and in solution. Although reproducible, the relative induction measured in solution was relatively low. A number of riboswitches that function at the level of translation display levels of reporter gene expression in this range, including the purine binding riboswitches that bind guanine3 and adenine,45 the riboflavin binding FMN riboswitch,46 and the SMK S-adenosyl methionine binding riboswitch.29,47 Thus, the levels of reporter gene expression measured are close to previously characterized riboswitches, and are consistent with the riboswitch model of translational regulation that we propose7,29,46,47 (Fig. 1B). To further investigate solution-based induction,16 we constructed a new reporter in which the full-length CDS replaced the α-fragment of the β-galactosidase CDS and observed induction of the reporter gene expression to be 2.5- or 3.2-fold by kanamycin B or sisomycin, respectively (Zhang, Chen, and Murchie, unpublished data), confirming our original observations.
Functional Specificity of Aminoglycoside-RNA Interactions
We showed that the 4,6 deoxystreptamine aminoglycosides such as kanamycin B, sisomycin could induce reporter gene expression through the 75 nt leader RNA but the control 4,5 deoxystreptamine derivatives like ribostamycin did not cause induction of reporter gene expression, suggesting that the induction of reporter gene expression through the 75 nt leader RNA requires specific aminoglycosides. To confirm that the induction of the reporter gene by certain aminoglycosides was mediated specifically by the leader RNA, we created an additional reporter in which the leader RNA of aac/aad was replaced by the leader RNA of cat-86.16,48 No induction was observed in agar diffusion assays.16 To further verify this observation, we measured β-galactosidase activity of the cat-86 leader in solution (Fig. 3) in the presence of inducing and control aminoglycosides and no induction was seen. This confirms that the induction of the reporter gene expression by specific aminoglycosides requires the 75 nt leader RNA. The induction of the reporter gene requires both the 75 nt leader RNA and certain aminoglycosides, indicating that interactions between the leader RNA and the aminoglycosides are specific. For inducing aminoglycosides, the correlation between the in vitro binding by SPR and the specificity of the reporter gene induction that we measure in vivo suggests that the drug-RNA interaction is biologically relevant.
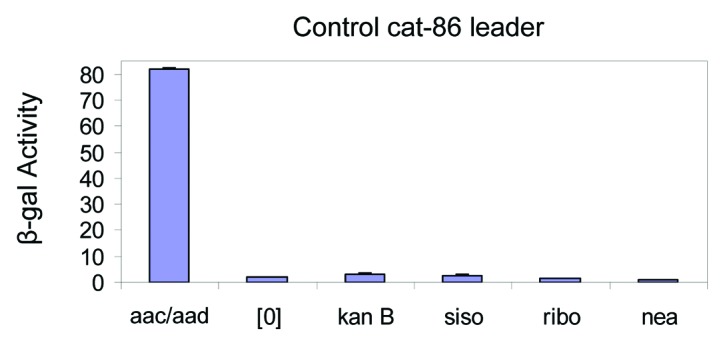
Figure 3. The cat-86 leader RNA does not induce reporter gene expression in response to aminoglycoside antibiotics.16 Miller assay of β-galactosidase activity of the control cat-86 leader RNA in the absence [0] and with the aminoglycoside antibiotics kanamycin B (kanB 10 µM), sisomycin (siso 1.25 µM), ribostamycin (10 µM), and Neamine (50 µM). Typical induction data for aac/aad in the presence of kanamycin B (5 µM) is shown for comparison.
Structured RNAs are comprised of negatively charged pockets to which the cationic amine groups of the aminoglycosides have a propensity to bind,49 additional binding sites have been identified in rRNA.24,50 Binding sites have also been found in the HIV trans-activating-region51 and Rev responsive element52 and in auto catalytic ribozymes.53 However, aminoglycoside binding is not simply associated with a general inhibitory activity.54 For example, in the absence of metal ions, they also promote self-cleavage by the hairpin ribozyme55 in contrast to their inhibitory activity against the hammerhead ribozyme.56 Their potential to shape RNA structures has been used to create aminoglycoside binding RNA aptamers.57,58 An in vitro selected streptomycin binding RNA58 exhibited a novel binding mode59 that led to the prediction that a streptomycin binding aptamer with a regulatory role may be found.60 Subsequently, a combination of in vitro selection combined with genetic screening identified a neomycin responsive synthetic riboswitch that could regulate translational initiation in S. cerevisiae.61,62
The aminoglycosides, like other natural product antibiotics, are secondary metabolites. They are synthesized by multi-step biosynthetic pathways involving multiple gene products.63 Within their natural producer organism they rarely achieve the dosing levels for antibiotic activity. It is therefore perhaps simplistic to regard them solely as inhibitors of bacterial growth, and a role was predicted for them as effector molecules in the regulation of diverse biological activities.64 Novel functionalities are emerging for antibiotics and they can be considered to be modulators as well as inhibitors.21,54,63 For example, sub-inhibitory doses of the aminoglycoside tobramycin have been shown to induce biofilm formation in Pseudomonas aeruginosa and in Escherichia coli.65 In this case, instead of targeting the ribosome, the antibiotic targets a predicted cyclic diguanosine monophosphate (c-di-GMP) phosphodiesterase [the aminoglycoside response regulator gene (arr)] inducing biofilm-specific aminoglycoside resistance. Cyclic diguanosine monophosphate is a second messenger molecule that controls cell surface adhesion.65 Sub-inhibitory concentrations of aminoglycosides (and other antibiotics) are also known to induce the bacterial SOS response through interference with the SOS transcriptional repressor LexA.43,44
The Function of the Putative Leader Peptide on the Induction of Reporter Gene Expression
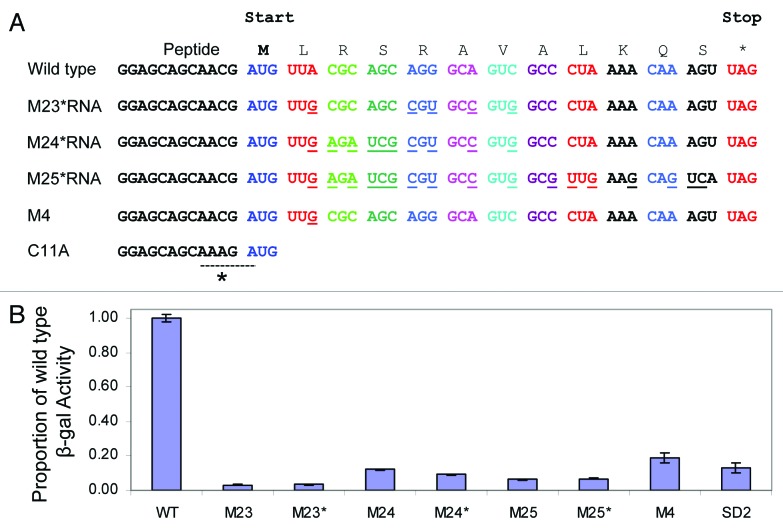
The 75 nt leader RNA encodes a putative short leader peptide and we investigated function for the nascent leader peptide on the induction of the reporter gene expression. We made three mutations (M23–25) in which the amino acid sequence of the leader peptide was retained, but the leader RNA sequence was altered. M23–25 greatly reduced the induction of the reporter gene. Our initial constructs included a point mutation (C11A) outside the peptide coding sequence; this point mutation in the mutants M23–25 also introduces the sequence AAAGA that overlaps the initiation codon of the putative leader peptide (Fig. 4A). The mutation C11A was originally introduced to destabilize a potential competing RNA structure. The sequence AAAGA was proposed to act as a decoy SD sequence (Ronald Breaker personal communication) such that overlap between the proposed decoy SD sequence and the putative leader peptide initiation codon might disrupt expression of the leader peptide. To investigate the possibility that the point mutation C11A was a decoy SD sequence that caused the change in phenotype that we observe for M23–25, we therefore repaired the mutation to the wild-type RNA and made three new mutant constructs M23*‒25*. For comparison, the mutation SD2 (G60C) that inactivates the ribosome binding site of the resistance gene indicates the meaningful lower signal window (Fig. 4B). We found that the mutations M23*‒25* also greatly reduce the induction of the reporter gene and have the same phenotype as M23–25, compared with the mutated SD2, and the phenotype is comparable with the single point mutation M4 (A18) (Fig. 4B) that also retains the leader peptide sequence but does not have the mutation C11A (Fig. 4A). We therefore found no evidence for a decoy SD site. This further confirms our original conclusion on the role of the leader peptide. The induction of the reporter gene by aminoglycosides is dependent on the conserved putative riboswitch RNA sequence and does not depend on leader peptide expression.
Figure 4. (A) The sequence of the wild-type leader peptide (the start and stop codons are indicated). The wild-type leader RNA and the three conservative mutant sequences M23*, M24*, and M25* (mutations underlined) are shown; the mutations retain the wild-type peptide sequence but change the RNA sequence. The mutants M23*, M24*, and M25* correct an additional mutation C11A (marked *) that introduces the sequence AAAGA that was proposed to act as a “decoy” SD sequence (dashed line) as shown. The C11A mutation was originally introduced to destabilize a potential RNA secondary structure in the leader RNA. The point conservative mutation M4 (A18G) that has the wild-type C11 sequence is also shown. (B) Miller assay of β- galactosidase activity of wild-type RNA, the original mutants M23, M24, M25, and M4 and three new mutants M23*, M24*, M25* in the presence of 5 μM KanB. Mutant RNA activity is expressed as a proportion of wild type (WT) RNA activity. The mutation SD2 (G60C) inactivates the ribosome binding site of the reporter gene and indicates the meaningful lower signal window. The error bars correspond to the standard deviation of three independent experiments. This is a confirmation of the original observation and disproves the proposed decoy SD sequence.
While the process of integron insertion has become progressively clarified in terms of the requirements for target sequence recognition and the mechanism of insertion by the integrase protein, new aspects of integron proliferation continue to emerge.66 It is noteworthy that as a component of the class I integrons the aminoglycoside-sensing riboswitch sequence that we have identified is found mostly between the divergent int gene and the acetyl or adenyltransferase genes (Fig. 2 see also Fig. 1 in ref. 66). This preponderance of aminoglycoside genes inserted adjacent to an aminoglycoside-sensing riboregulatory RNA suggest that, while integron integration may occur randomly, at least within class 1 integrons, the association with aminoglycoside resistance confers a selective advantage.66 Cassette insertion at the aatI1 insertion site leads to the incorporation of regulatory features from the inserted gene but also introduces considerable sequence variation in the regulatory regions of the inserted aac/aad genes. Genes inserted at this position are known to be transcribed by a strong promoter.39,41
Although the mechanism of insertion and transcription of integron DNA is becoming better understood, there have been fewer studies of the mechanisms of translation within integron sequences.40,42 Hanau-Bercot et al. studied the expression of the aac(6_)-Ib7 gene cassette from Citrobacter freundii42 and concluded that expression of aac was regulated at the level of translation of the leader peptide, and that the integron transcript functioned to allow initiation of translation at inserted gene cassettes that do not have correct ribosome binding sites42 (reviewed in ref. 40). Although there are some similarities with our own investigations, they are not directly comparable. The inserted Citrobacter freundii sequence lacks the SD2 and anti-SD sequence of Pseudomonas fluorescens; these are important components of our regulatory model (Fig. 1). Perhaps most importantly the experiments in Citrobacter freundii were done in the absence of added aminoglycoside. We also show that the riboswitch RNA has an overriding role in this regulatory region compared with the peptide sequence (Fig. 416). Expression of the resistance gene is delicately balanced; both of the aac/aad reporter constructs display elevated background levels of reporter gene expression in the absence of added aminoglycosides. Our current view is that lower levels of translation through the integron transcript and leader peptide as proposed for Citrobacter freundii42 contribute to the background levels of reporter gene expression that we observe, and that the riboswitch acts as “dimmer switch”67,68 in response to sub-lethal doses of the antibiotic to induce expression of resistance genes. It is intriguing to speculate as to whether the Citrobacter freundii RNA might also act as a riboswitch in response to added aminoglycosides.
Since the 1940s resistance to antibiotics has emerged in response to a highly selective antibiotic-rich environment.21 It has become a major clinical threat and an understanding of the mechanisms of resistance is critical. Induction of the expression levels of the AAC/AAD proteins in response to added aminoglycosides has been a well-known phenomenon for over three decades.28,69,70 Our data go some way toward explaining this long-standing phenomenon.
In summary, we have shown that an aminoglycoside-sensing RNA regulates an aminoglycoside antibiotic resistance gene. Antibiotic-specific riboswitches in the 5′UTRs of resistance genes would constitute a powerful way of controlling resistance gene expression within a bacterial population. We speculate that drug-sensing RNAs may have additional roles in controlling the expression of other drug inactivating enzymes.
Acknowledgments
We thank Ron Breaker for discussion. In a recent review, Julian Davies21 commented that there have been over 200,000 references on the subject of antibiotic resistance; we regret that we could not cite all of the many important publications in this field. This work was supported by National Key 973 grant (2010CB912602). City of Shanghai Key project grant (09DJ1400601). Natural Science Foundation grants 31170749 to AM and 31100543 to WXS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25757
References
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–79. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. From ribosome to riboswitch: control of gene expression in bacteria by RNA structural rearrangements. Crit Rev Biochem Mol Biol. 2006;41:329–38. doi: 10.1080/10409230600914294. [DOI] [PubMed] [Google Scholar]
- 3.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–86. doi: 10.1016/S0092-8674(03)00391-X. [DOI] [PubMed] [Google Scholar]
- 4.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–7. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 7.Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, et al. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–56. doi: 10.1016/S0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 8.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–6. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 9.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043. doi: 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 10.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat Struct Biol. 2003;10:701–7. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 11.McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc Natl Acad Sci USA. 2003;100:3083–8. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–9. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 13.Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA. 2003;100:12057–62. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–97. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–6. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 16.Jia X, Zhang J, Sun W, He W, Jiang H, Chen D, et al. Riboswitch control of aminoglycoside antibiotic resistance. Cell. 2013;152:68–81. doi: 10.1016/j.cell.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–5. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cromie MJ, Shi Y, Latifi T, Groisman EA. An RNA sensor for intracellular Mg(2+) Cell. 2006;125:71–84. doi: 10.1016/j.cell.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 19.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–92. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 20.Armstrong ES, Miller GH. Combating evolution with intelligent design: the neoglycoside ACHN-490. Curr Opin Microbiol. 2010;13:565–73. doi: 10.1016/j.mib.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies J, Davis BD. Misreading of ribonucleic acid code words induced by aminoglycoside antibiotics. The effect of drug concentration. J Biol Chem. 1968;243:3312–6. [PubMed] [Google Scholar]
- 23.Fourmy D, Recht MI, Blanchard SC, Puglisi JD. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–71. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 24.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–8. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 25.Liebert CA, Hall RM, Summers AO. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–22. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–20. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. 2009;78:119–46. doi: 10.1146/annurev.biochem.78.082907.145923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingeot-Leclercq MP, Glupczynski Y, Tulkens PM. Aminoglycosides: activity and resistance. Antimicrob Agents Chemother. 1999;43:727–37. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–33. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 30.Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16:103–32. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- 31.Weisblum B. Inducible resistance to macrolides, lincosamides and streptogramin type B antibiotics: the resistance phenotype, its biological diversity, and structural elements that regulate expression--a review. J Antimicrob Chemother. 1985;16(Suppl A):63–90. doi: 10.1093/jac/16.suppl_A.63. [DOI] [PubMed] [Google Scholar]
- 32.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 33.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. Site-specific insertion of gene cassettes into integrons. Mol Microbiol. 1993;9:41–52. doi: 10.1111/j.1365-2958.1993.tb01667.x. [DOI] [PubMed] [Google Scholar]
- 34.Partridge SR, Recchia GD, Scaramuzzi C, Collis CM, Stokes HW, Hall RM. Definition of the attI1 site of class 1 integrons. Microbiology. 2000;146:2855–64. doi: 10.1099/00221287-146-11-2855. [DOI] [PubMed] [Google Scholar]
- 35.Lopez R, Silventoinen V, Robinson S, Kibria A, Gish W. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003;31:3795–8. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–83. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 37.Bouvier M, Demarre G, Mazel D. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 2005;24:4356–67. doi: 10.1038/sj.emboj.7600898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacDonald D, Demarre G, Bouvier M, Mazel D, Gopaul DN. Structural basis for broad DNA-specificity in integron recombination. Nature. 2006;440:1157–62. doi: 10.1038/nature04643. [DOI] [PubMed] [Google Scholar]
- 39.Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacquier H, Zaoui C, Sanson-le Pors MJ, Mazel D, Berçot B. Translation regulation of integrons gene cassette expression by the attC sites. Mol Microbiol. 2009;72:1475–86. doi: 10.1111/j.1365-2958.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 41.Lévesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 42.Hanau-Berçot B, Podglajen I, Casin I, Collatz E. An intrinsic control element for translational initiation in class 1 integrons. Mol Microbiol. 2002;44:119–30. doi: 10.1046/j.1365-2958.2002.02843.x. [DOI] [PubMed] [Google Scholar]
- 43.Baharoglu Z, Mazel D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother. 2011;55:2438–41. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, et al. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 45.Lemay J-F, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–94. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith AM, Fuchs RT, Grundy FJ, Henkin TM. The SAM-responsive S(MK) box is a reversible riboswitch. Mol Microbiol. 2010;78:1393–402. doi: 10.1111/j.1365-2958.2010.07410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duvall EJ, Williams DM, Mongkolsuk S, Lovett PS. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984;158:784–90. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermann T, Westhof E. Docking of cationic antibiotics to negatively charged pockets in RNA folds. J Med Chem. 1999;42:1250–61. doi: 10.1021/jm981108g. [DOI] [PubMed] [Google Scholar]
- 50.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, et al. Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol. 2007;14:727–32. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 51.Mei HY, Mack DP, Galan AA, Halim NS, Heldsinger A, Loo JA, et al. Discovery of selective, small-molecule inhibitors of RNA complexes--I. The Tat protein/TAR RNA complexes required for HIV-1 transcription. Bioorg Med Chem. 1997;5:1173–84. doi: 10.1016/S0968-0896(97)00064-3. [DOI] [PubMed] [Google Scholar]
- 52.Zapp ML, Stern S, Green MR. Small molecules that selectively block RNA binding of HIV-1 Rev protein inhibit Rev function and viral production. Cell. 1993;74:969–78. doi: 10.1016/0092-8674(93)90720-B. [DOI] [PubMed] [Google Scholar]
- 53.von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–70. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 54.Schroeder R, Waldsich C, Wank H. Modulation of RNA function by aminoglycoside antibiotics. EMBO J. 2000;19:1–9. doi: 10.1093/emboj/19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Earnshaw DJ, Gait MJ. Hairpin ribozyme cleavage catalyzed by aminoglycoside antibiotics and the polyamine spermine in the absence of metal ions. Nucleic Acids Res. 1998;26:5551–61. doi: 10.1093/nar/26.24.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stage TK, Hertel KJ, Uhlenbeck OC. Inhibition of the hammerhead ribozyme by neomycin. RNA. 1995;1:95–101. [PMC free article] [PubMed] [Google Scholar]
- 57.Famulok M, Hüttenhofer A. In vitro selection analysis of neomycin binding RNAs with a mutagenized pool of variants of the 16S rRNA decoding region. Biochemistry. 1996;35:4265–70. doi: 10.1021/bi952479r. [DOI] [PubMed] [Google Scholar]
- 58.Wallace ST, Schroeder R. In vitro selection and characterization of streptomycin-binding RNAs: recognition discrimination between antibiotics. RNA. 1998;4:112–23. [PMC free article] [PubMed] [Google Scholar]
- 59.Tereshko V, Skripkin E, Patel DJ. Encapsulating streptomycin within a small 40-mer RNA. Chem Biol. 2003;10:175–87. doi: 10.1016/S1074-5521(03)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piganeau N, Schroeder R. Aptamer structures: a preview into regulatory pathways? Chem Biol. 2003;10:103–4. doi: 10.1016/S1074-5521(03)00028-0. [DOI] [PubMed] [Google Scholar]
- 61.Weigand JE, Sanchez M, Gunnesch E-B, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weigand JE, Schmidtke SR, Will TJ, Duchardt-Ferner E, Hammann C, Wöhnert J, et al. Mechanistic insights into an engineered riboswitch: a switching element which confers riboswitch activity. Nucleic Acids Res. 2011;39:3363–72. doi: 10.1093/nar/gkq946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sengupta S, Chattopadhyay MK, Grossart H-P. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol. 2013;4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davies J. What are antibiotics? Archaic functions for modern activities. Mol Microbiol. 1990;4:1227–32. doi: 10.1111/j.1365-2958.1990.tb00701.x. [DOI] [PubMed] [Google Scholar]
- 65.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–5. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 66.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baird NJ, Kulshina N, Ferré-D’Amaré AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010;7:328–32. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swiatlo E, Kocka FE. Inducible expression of an aminoglycoside-acetylating enzyme in Providencia stuartii. J Antimicrob Chemother. 1987;19:27–30. doi: 10.1093/jac/19.1.27. [DOI] [PubMed] [Google Scholar]
- 70.Williams JW, Northrop DB. Purification and properties of gentamicin acetyltransferase I. Biochemistry. 1976;15:125–31. doi: 10.1021/bi00646a019. [DOI] [PubMed] [Google Scholar]