Abstract
The exon junction complex (EJC) participates in the regulation of many post-transcriptional steps of gene expression. EJCs are deposited on messenger RNAs (mRNAs) during splicing and their core consists of eIF4A3, MLN51, Y14, and MAGOH. Here, we show that two genes encoding MAGOH paralogs (referred to as MAGOH and MAGOHB) are expressed in mammals. In macrophages, the expression of MAGOHB, but not MAGOH mRNA, increases rapidly after LPS stimulation. Both MAGOH proteins interact with other EJC components, incorporate into mRNA-bound EJCs, and activate nonsense-mediated decay. Furthermore, the simultaneous depletion of MAGOH and MAGOHB, but not individual depletions, impair nonsense-mediated decay in human cells. Hence, our results establish that the core composition of mammalian EJCs is more complex than previously recognized.
Keywords: EJC, mago nashi homolog, MAGOHB, NMD, gene duplication
Introduction
The process of gene expression in eukaryotic cells is organized as a series of events, which are all performed by large multi-protein machineries.1 After the generation of pre-mRNAs in the nucleus, the transcripts are subjected to processing and export into the cytoplasm, where they undergo translation, quality control, and finally decay. All these processes are linked with each other via interacting molecular machines and are subject to regulation.2
A multi-protein complex, termed the exon junction complex (EJC), is deposited on mRNAs during splicing by the spliceosomal protein CWC22.3-5 The binding of the EJC to mRNA occurs in a sequence-independent manner at its canonical binding site 20–24 nts upstream of exon-exon boundaries, but also at alternative, non-canonical sites within exons.6,7 Components of the EJC (eIF4A3, MLN51, Y14, and MAGOH) are involved in splicing, mRNA export, nonsense-mediated mRNA decay (NMD), and translation initiation.8-13 The two proteins Y14 (RBM8A) and MAGOH (mago-nashi homolog) form a stable heterodimer that binds tightly to eIF4A3 (DDX48) and, thereby, stabilize the binding of eIF4A3 on the mRNA.14-16
Recent studies in mouse have demonstrated that a mutation in the EJC component MAGOH causes microcephaly and reduction of brain size, because MAGOH regulates the levels of microcephaly associated protein Lis1 during neurogenesis.17 Reduced expression levels of Y14, the heterodimerization partner of MAGOH, cause thrombocytopenia with absent radii (TAR) in humans.18 Interestingly, some TAR cases are associated with intellectual disability possibly caused by cerebral dysgenesis.19 Aberrations of the chromosomal region containing Y14 are also frequently associated with intellectual disability and brain malformations.19 Hence, data from mice and humans indicate that physiological expression levels of MAGOH and Y14 might be required for normal brain development.
The function of MAGOH as a component of the EJC is well understood; however, the existence of a duplicated MAGOH gene in mammals is not widely known. Here, we present a comparative characterization of both MAGOH genes, referred to as MAGOH and MAGOHB in humans. Both MAGOH and MAGOHB are expressed in various tissues from adult mice and in human cell lines. While the expression of MAGOHB mRNA in macrophages is strongly increased after LPS stimulation, the levels of MAGOH mRNA remain unchanged. Cells individually depleted of either MAGOH or MAGOHB remained fully competent to execute nonsense-mediated mRNA decay (NMD), whereas the simultaneous depletion of both MAGOH proteins drastically diminished NMD. Hence, two genes (four alleles) coding for MAGOH are present in mammalian cells and contribute to the expression and function of MAGOH proteins.
Results and Discussion
MAGOHB, a duplicated MAGOH gene is evolutionarily conserved
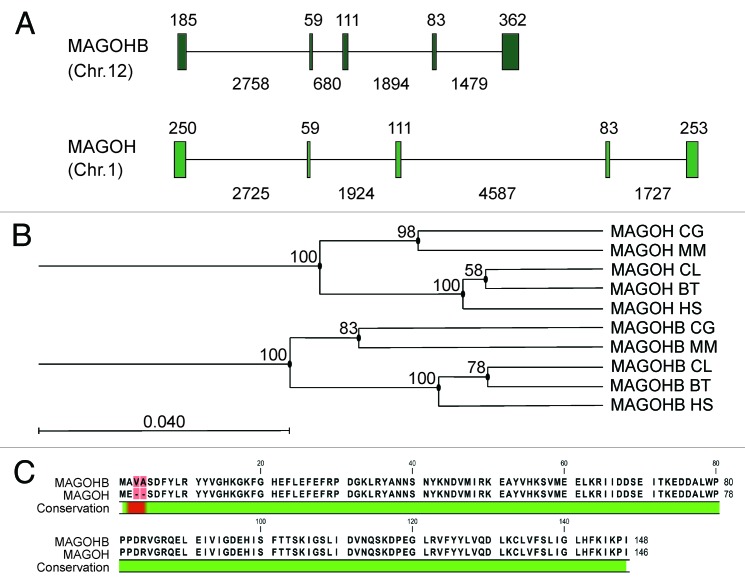
We previously aimed to functionally characterize MAGOH in human cell culture. Notably, we were unsuccessful to deplete MAGOH protein levels by transfecting siRNAs specific for MAGOH (data not shown, see also Fig. 4B, lane 1–6). Following this observation, we noticed that a second MAGOH gene is present in the human genome (NCBI: NM_018048; UniProt: Q96A72), referred to as “MAGOHB” (HUGO nomenclature database; http://www.genenames.org/), as well as two MAGOH pseudogenes (referred to as MAGOH2 and MAGOH3P; HUGO nomenclature database). The two MAGOH genes are located on different chromosomes in humans (MAGOH: Chr. 1; MAGOHB: Chr. 12), both comprise five exons with a conserved length and structure (Fig. 1A). The coding sequences (CDs) of both MAGOH genes share 87% identity at the RNA- and DNA-level. The coding sequence of MAGOHB is two codons longer (148 amino acids) than the MAGOH coding sequence (146 amino acids) and the encoded proteins are identical except for amino acid residues one through four of MAGOHB and residues one and two of MAGOH (Fig. 1C).
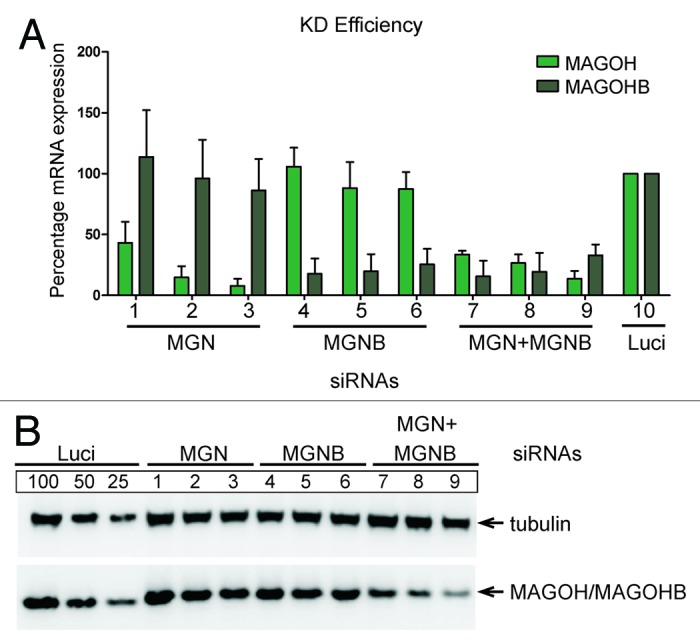
Figure 4. Specific knockdown of MAGOHB and MAGOH. (A) HeLa cells were transfected with siRNAs targeting MAGOH (siRNAs 1, 2, and 3), MAGOHB (siRNAs 4, 5, and 6) or both MAGOH and MAGOHB (siRNA combinations 7, 8, and 9, see Materials and Methods). Expression levels of MAGOH or MAGOHB mRNAs were determined by quantitative RT-PCR and normalized to TATA binding protein (TBP). Reduction of mRNA levels of MAGOH and MAGOHB compared with Luc-siRNA transfected cells (100%) is depicted; data represent the mean percentage (± standard deviation) from three independent experimets. (B) Western blot of HeLa cells that were transfected with the indicated siRNA or siRNA combinations. The knockdown efficiency was assessed with MAGOH-specific antibodies.
Figure 1. Alignment and conservation of the two MAGOH genes/proteins. (A) Human MAGOHB and MAGOH gene structures, exons, and introns are shown as boxes and lines, respectively. The sizes of exons and introns in base pairs (bp) are depicted above and below the diagram, respectively. (B) Maximum likelihood phylogeny estimated with the PhyML algorithm using the ORF sequences of MAGOH and MAGOHB from Homo sapiens (HS), Mus musculus (MM), Bos taurus (BT), Canis lupus (CL), and Cricetulus griseus (CG). The scale bar represents the measure of phylogenetic distance and corresponds to four base substitutions per 100 positions. Numbers at nodes represent bootstrap values. See also Figure S1C. Alignment of the two human MAGOH proteins.
We aimed to investigate the phylogenetic significance of our finding and searched for duplicated MAGOH genes in other species. Strikingly, we found two MAGOH genes in all mammalian genomes that were analyzed. In contrast, there is no evidence for a second MAGOH gene in birds, reptiles, amphibian, or fish. Hence, the existence of duplicated MAGOH genes is restricted to the mammalian clade. We collected the reference sequences of both MAGOH and MAGOHB genes from five different species and generated multiple sequence alignments (CLC Main Workbench, CLC bio). The alignments showed that the coding sequences of the MAGOH and MAGOHB genes in Homo sapiens (HS), Mus musculus (MM), Bos taurus (BT), Canis lupus (CL), and Cricetulus griseus (CG) are well conserved (Fig. S1). Interestingly, we did not find a single amino acid alteration in these mammalian MAGOH and MAGOHB genes, despite a remarkable variability in the codons used by different species. Hence, mutations in the coding region of MAGOH and MAGOHB that have occurred during evolution did not alter the encoded proteins. Further analysis of the MAGOH genes of the five selected species using “estimated maximum likelihood phylogenies” (PHYML,20 CLC Main Workbench) showed that MAGOH and MAGOHB genes cluster within a MAGOH- and a MAGOHB-specific group (Fig. 1B). This suggests that MAGOH genes and MAGOHB genes represent orthologs that putatively evolved from a common ancestor after a gene duplication event.
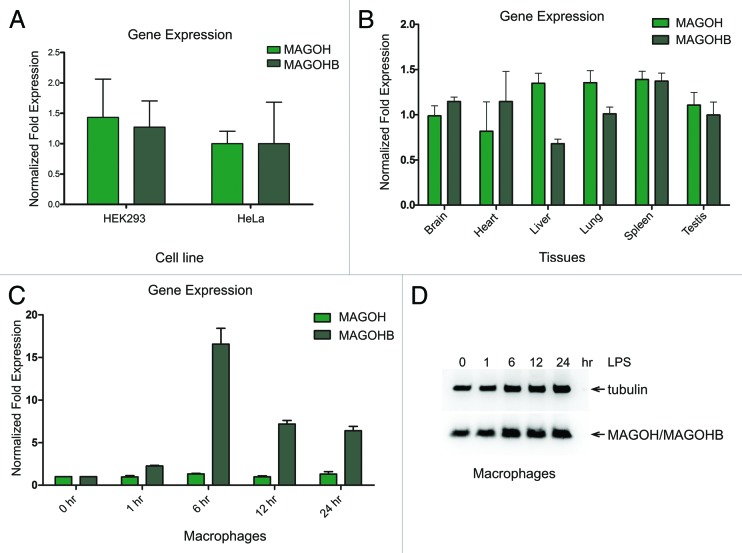
The high degree of sequence conservation of the coding regions of both MAGOH genes indicates that strong evolutionary pressure preserved the primary protein sequence. This suggests that both MAGOH and MAGOHB genes are expressed and both proteins are required for normal cellular function. Therefore, we next investigated if both MAGOH and MAGOHB transcripts are expressed in cultured human cells. To this end, we performed quantitative real-time PCR (qRT-PCR) on cDNA obtained from HEK293 and HeLa cell lines. Noteworthy, MAGOH and MAGOHB mRNAs were detectable in both cell lines (Fig. 2A). Variable mRNA levels of MAGOH and MAGOHB in different samples led us to hypothesize that the expression of two MAGOH genes could be regulated in a cell type-specific manner. We isolated RNA from various tissues of wild-type C57BL/six mice (aged 8 wk) and determined the expression levels of MAGOH and MAGOHB by qRT-PCR. Interestingly, the expression levels of MAGOH were significantly higher (2.0-fold, P < 0.01) than MAGOHB in liver of all three mice tested (Fig. S2), average expression levels of three mice is shown in Figure 2B. Both MAGOH and MAGOHB were equally expressed in the brain, heart, lung, spleen, and testis (Fig. 2B). To gain additional insights into a potential differential expression regulation of the two MAGOH genes, we followed the expression levels of MAGOH and MAGOHB mRNAs in mouse macrophages after lipopolysaccharide (LPS) stimulation. Macrophages have been shown previously to rapidly up- and downregulate a subset of mRNAs in response to LPS stimulation and, therefore, represent a well-established model system to study rapid changes of gene expression.21 We observed a significant increase (2.3-fold, P < 0.001) of MAGOHB mRNA levels already 1 h after LPS stimulation (Fig. 2C), whereas MAGOH mRNA levels were not significantly altered at any time point (Fig. 2C). The mRNA expression of MAGOHB peaked after 6 h (12.5-fold, P < 0.001) and declined at later time points (Fig. 2C), suggesting that the upregulation of MAGOHB expression occurs only for a short period of time. The robust increase in MAGOHB mRNA expression did not lead to a corresponding increase of total MAGOH protein levels, which remained largely unchanged after LPS treatment (Fig. 2D). Taken together, these data show that the MAGOH and MAGOHB genes are evolutionary well conserved among different species and are ubiquitously expressed. Furthermore, the expression of MAGOH and MAGOHB in LPS-stimulated macrophages indicates that the two genes are differentially regulated under physiological conditions at the transcriptional level.
Figure 2. Expression of MAGOH and MAGOHB transcripts. (A) Quantitative RT-PCR analysis of endogenous MAGOHB and MAGOH transcripts in HEK293 and HeLa cells. The expression levels were normalized to TATA binding protein (TBP). (B) Quantitative RT-PCR analysis of the MAGOHB and MAGOH transcripts levels in various tissues of three adult mice (data of individual mice are shown in Fig. S2). (C) Quantitative RT-PCR analysis of the MAGOHB and MAGOH transcripts levels in murine derived macrophages that were stimulated with LPS (10 ng/mL) for 0, 1, 6, 12, and 24 h. Data presented in (A–C) represent the mean value (± standard deviation) obtained from three independent experiments. (D) Western blot of macrophages that were stimulated with LPS (10 ng/mL) for 0, 1, 6, 12, and 24 h. MAGOH protein levels were assessed with MAGOH-specific antibodies.
MAGOHB forms a heterodimer with Y14 and is a part of the EJC
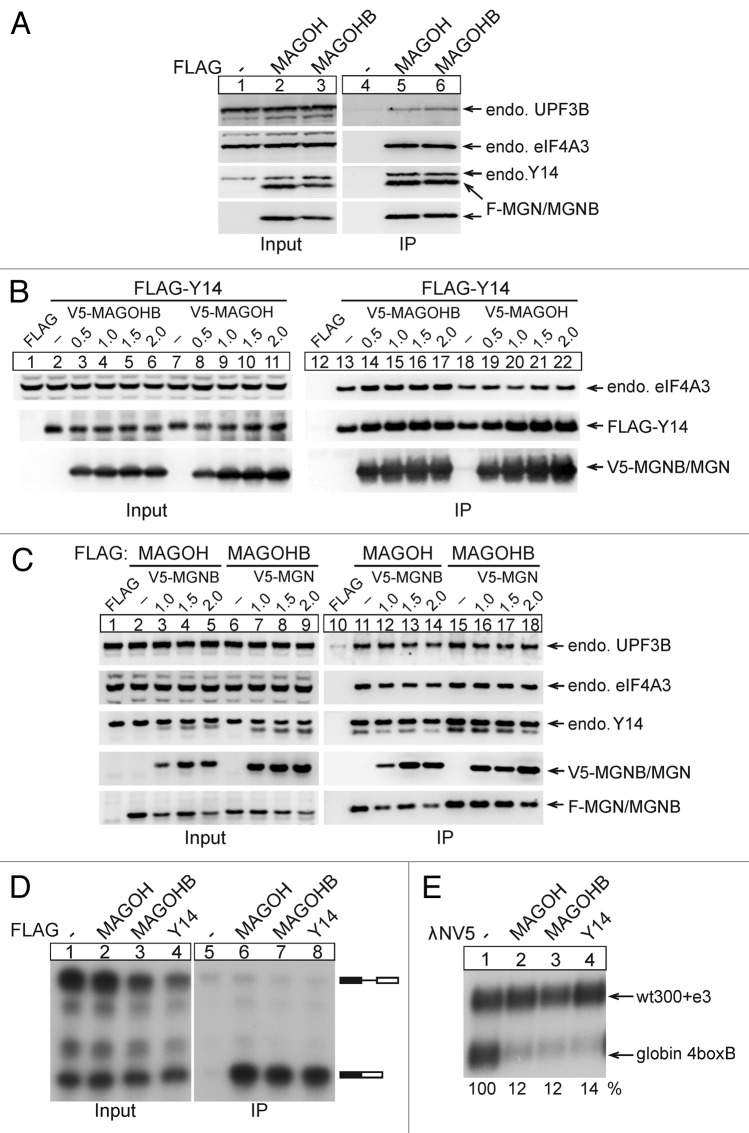
It is well known that MAGOH forms a heterodimer with Y14, which represents a core component of the EJC. Because of the almost identical amino acid sequence of MAGOHB and MAGOH we reasoned that both proteins have identical binding partners, i.e., Y14 and eIF4A3, and are both efficiently incorporated into EJCs. To test this notion, we transfected HeLa cells with FLAG-MAGOHB and performed FLAG immunoprecipitations. FLAG-MAGOHB co-precipitated the EJC core component eIF4A3 and the EJC-interacting protein UPF3B with a similar efficiency as FLAG-MAGOH (Fig. 3A, compare lanes 5 and 6), providing evidence that MAGOHB is incorporated into the EJC in living cells. Unfused FLAG served as a negative control (Fig. 3A, lane 4). Additionally, both FLAG-MAGOH and FLAG-MAGOHB co-precipitated Y14 (Fig. 3A, lanes 5 and 6) and, therefore, are able to form a heterodimer with Y14.
Figure 3. MAGOHB and MAGOH interact with the same proteins and elicit NMD. (A) FLAG-MAGOH and FLAG-MAGOHB were expressed in HeLa cells. Complexes were immunoprecipitated using FLAG-agarose beads. Western blotting with specified antibodies was used to visualize co-precipitated endogenous proteins. (B) Equal amounts of FLAG-Y14 were transfected together with increasing amounts of V5-MAGOHB or V5-MAGOH. Immunoprecipitations were performed using FLAG beads and co-precipitated components of the EJC were detected by immunoblotting. (C) Equal amounts of FLAG-MAGOH and FLAG-MAGOHB were transfected together with increasing amounts of V5-MAGOHB and V5-MAGOH respectively. Immunoprecipitations were performed using FLAG beads and immunoblotting was performed to visualize co-precipitated proteins. (D) A 32P bodylabeled splicing substrate (MINX RNA) was incubated in HeLa nuclear extracts and HEK293 whole cell extracts expressing FLAG-MAGOHB, FLAG-MAGOH, or FLAG-Y14. FLAG-tagged proteins were immunoprecipitated and co-precipitated RNA was separated by denaturing PAGE. (E) Northern blot of HeLa cells co-transfected with the β-globin 4boxB reporter plasmid, a control and the indicated λNV5-fusion proteins. Percentages represent the mean calculated from three independent experiments.
To rule out the possibility that either MAGOH or MAGOHB are more efficiently incorporated into EJCs, we transfected HeLa cells with increasing amounts of V5-tagged MAGOH (Fig. 3B, lanes 8–11 and 19–22) or MAGOHB (Fig. 3B, lanes 3–6 and 14–17) together with FLAG-tagged Y14. Protein complexes were immunoprecipitated via FLAG and co-precipitated V5-MAGOH/V5-MAGOHB and endogenous eIF4A3 detected by immunoblotting. Both MAGOH and MAGOHB co-precipitated efficiently with Y14. Notably, the levels of co-precipitated endogenous eIF4A3 did not change upon MAGOH or MAGOHB overexpression, indicating that either of the MAGOH proteins can be incorporated into the EJC with similar efficiency.
Because both MAGOH proteins interact with a very similar set of proteins, we reasoned that MAGOH and MAGOHB may be competing with each other for cellular interaction partners. To test this possibility, equal amounts of FLAG-tagged MAGOH (Fig. 3C, lanes 2–5 and 11–14) or MAGOHB (Fig. 3C, lanes 6–9 and 15–18) were transfected together with increasing amounts of V5-tagged MAGOHB (Fig. 3C, lanes 3–5 and 12–14) or V5-tagged MAGOH (Fig. 3C, lanes 7–9 and 16–18). FLAG-immunoprecipitated complexes were analyzed by immunoblotting. Notably, the expression of FLAG-MAGOH decreased with increasing amounts of transfected V5-MAGOHB. A similar inverse correlation was observed for FLAG-MAGOHB expression levels and co-transfected V5-MAGOH. The decreasing amounts of FLAG-MAGOH and FLAG-MAGOHB also resulted in a reduced co-precipitation of UPF3B, eIF4A3, and Y14. Nonetheless, this slight effect likely represents an overload of the cellular expression machinery rather than a specific competition between the two MAGOH proteins.
Previously, we have utilized the technique of in vitro splicing reactions combined with immunoprecipitation, to analyze the splicing-dependent deposition of EJCs on a spliced RNA substrate.22 We used a similar approach to determine if MAGOHB is incorporated into EJCs assembled by in vitro splicing. To this end, the MINX pre-mRNA was incubated in HeLa nuclear extracts supplemented with splicing-competent whole cell extract from HEK293 cells expressing different FLAG-tagged proteins (Fig. 3D) under splicing conditions. The FLAG-tagged proteins (MAGOH, MAGOHB, and Y14) were immunoprecipitated and their associated mRNAs visualized by denaturing PAGE. FLAG-MAGOHB, like FLAG-MAGOH and FLAG-Y14, co-immunoprecipitated fully spliced mRNAs (Fig. 3D, compare lane 6, 7 and 8), illustrating that both MAGOH and MAGOHB are efficiently incorporated into EJCs. In summary, our data indicate that both MAGOHB and MAGOH interact with a comparable set of EJC proteins and efficiently incorporate into EJCs assembled during splicing.
MAGOHB elicits nonsense-mediated mRNA degradation
One of the biological functions of the EJC is the detection of PTC-containing transcripts and their subsequent decay by NMD. We performed tethering experiments and used a plasmid expressing β-globin with 4boxB sites in its 3′ UTR as a NMD reporter.23 MAGOH and MAGOHB were expressed as λNV5 fusion proteins to enable their tethering to the reporter mRNAs. As suggested by the almost identical amino acid sequence of MAGOHB and MAGOH and their ability to incorporate into EJCs, tethering of λNV5-MAGOH or λNV5-MAGOHB strongly reduced the abundance of the reporter mRNA (Fig. 3E, compare lane 2 and 3) as compared with the unfused λNV5 (negative control, Fig. 3E, lane 1). This indicates that both MAGOH proteins operate in the NMD pathway and recruit NMD-activating factors to the 3′UTR of a tethering reporter mRNA with similar efficiency.
Effect of MAGOH depletion on cellular NMD substrates
To evaluate if the two MAGOH genes have redundant functions, e.g., during the recognition of NMD targets, we designed a knockdown strategy to deplete cells individually of MAGOH and MAGOHB gene products. Because of their almost identical size and amino acid sequence, the levels of MAGOH and MAGOHB protein cannot be assessed individually, which is reflected by the single band that is detected on MAGOH-specific immunoblots (Fig. 4B, panel MAGOH/MAGOHB). Hence, we refer to the protein band detected by MAGOH-specific antibodies as MAGOH/MAGOHB. For the depletion of MAGOH and MAGOHB, three different siRNAs [referred to as 1–3 (MAGOH) and 4–6 (MAGOHB)] were used, respectively (Fig. 4A, lanes 1–3 and 4–6). Individual transfections with siRNAs 1–3 or 4–6 to deplete MAGOH or MAGOHB gene products, respectively, only partially reduced the levels of total MAGOH/MAGOHB protein (Fig. 4B, compare lane 1–6 with luciferase). However, when we targeted both MAGOH and MAGOHB mRNAs by a combination of different MAGOH and MAGOHB siRNAs, we were able to deplete the majority of the total MAGOH/MAGOHB proteins (Fig. 4B, lane 7, 8, and 9 in comparison with lane 1–6). This establishes that the expression of both MAGOH genes can be reduced with specific siRNAs. However, the efficient depletion of MAGOH/MAGOHB proteins from cells requires the combined knockdown of both genes.
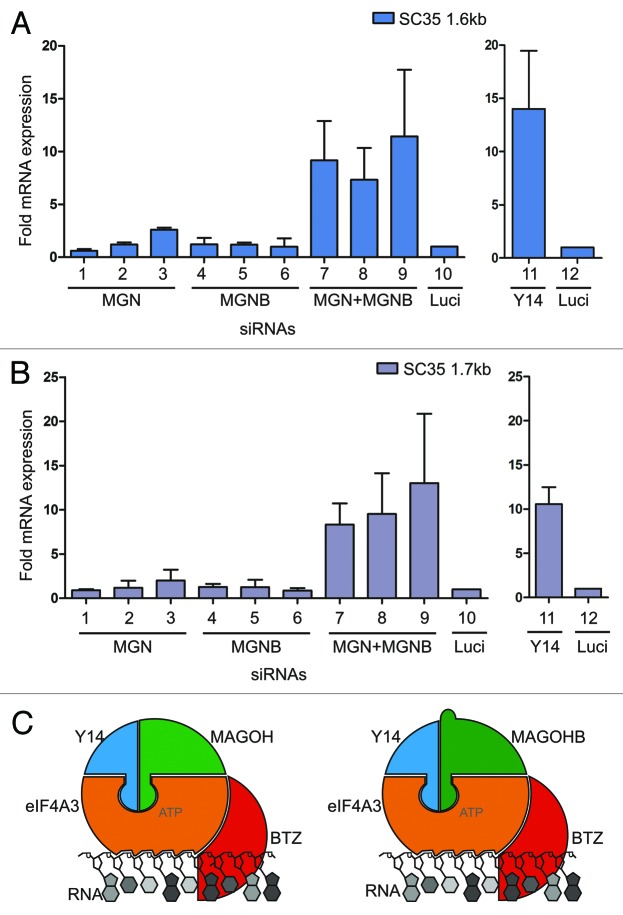
Furthermore, the effects of MAGOH and MAGOHB depletion on the well-established cellular NMD target SC35 (1.6 kb and 1.7 kb) were assessed.24,25 We found that NMD-sensitive transcripts of SC35 (1.6 kb and 1.7 kb) were still efficiently degraded after the depletion of either MAGOH or MAGOHB (Fig. 5A and B, lane 1–3 and 4–6). In contrast, the simultaneous depletion of both MAGOH and MAGOHB proteins caused a drastic increase in SC35 (1.6 kb and 1.7 kb) transcript levels (Fig. 5A and B, lane 7–9), indicating that NMD is strongly impaired in these cells. Depletion of Y14 also upregulated the expression levels of cellular NMD substrates to a similar extent as the combined depletion of MAGOH and MAGOHB and was used as a positive control (Fig. 5A and B, lane 11).
Figure 5. MAGOH and MAGOHB collectively support NMD. (A and B) The effect of single or combined knockdowns of MAGOH and MAGOHB on NMD was determined. The levels of the known cellular NMD-substrates SC35 1.6 kb (A) SC35 1.7 kb (B) were measured by quantitative RT-PCR and normalized to TBP. Fold upmodulation compared with Luc-siRNA transfected cells is depicted. Values for Y14-depleted cells are shown as separate panel. (C) Both MAGOH and MAGOHB are incorporated into the EJC core. Hence, two different types of core EJCs (MAGOH-containing and MAGOHB-containing) are formed in mammalian cells.
Taken together, our results demonstrate that for the efficient and functional depletion of MAGOH, both MAGOH proteins have to be simultaneously depleted by RNAi.
Conclusions
In the current study, we have characterized a second MAGOH gene in human cells, referred to as MAGOHB. In 2009, it was reported that MAGOHB is the only gene whose expression changed after the histone 3 lysine 4 methyltransferase (MLL2) knockout.26 In that study, MAGOHB mRNA levels were reduced in concordance with decreased MLL2 protein levels, whereas MAGOH mRNA levels were unchanged. The existence of a second MAGOH gene in mouse has also been reported by Silver et al.,17 but the functional importance of the two genes has not been addressed until now. Here, we present a detailed analysis of MAGOHB in human cell culture and provide evidence for the existence of MAGOHB in mammals. We detected constitutive expression of both MAGOH and MAGOHB mRNAs in all mouse tissues and cell lines tested. Nonetheless, expression levels of both genes were variable in different tissues and MAGOHB mRNA levels were upregulated in macrophages after LPS stimulation. Therefore, the existence of specific cell types that express only one of the two genes is conceivable. Since a previous report on MAGOH demonstrated its role in brain development,17 cells in the central nervous system (CNS) might display a specific requirement for MAGOH or MAGOHB during development. It will be very interesting to identify such cells in the future, to explore why the loss of a single allele of MAGOH has such a dramatic effect on brain development and size.
In our current study, we report the inclusion of MAGOHB in the EJC (Fig. 3A and D) and its functional involvement in the NMD pathway (Fig. 3E). Furthermore, we address the functional consequences of the depletion of both MAGOH proteins on natural cellular NMD substrates i.e., SC35 1.6 kb and SC35 1.7 kb. Additionally, we confirm that targeting both MAGOH genes by siRNAs is required for the complete knockdown of MAGOH proteins17 and analyze the functional effects of the combined knockdown. This will be important for the design and interpretation of siRNA experiments, as the MAGOHB gene also contributes to the expression of functional MAGOH proteins. Since the amino acid sequence of MAGOH and MAGOHB are very similar, we could not differentiate between the individual contributions of either of the two MAGOH/MAGOHB genes to the overall MAGOH/MAGOHB protein levels. Indeed, MAGOH and MAGOHB expression levels can only be estimated using qPCR quantification of mRNA levels. It is surprising that the mammalian genome contains four alleles encoding two almost identical proteins with highly redundant functions. Nonetheless, it is possible that the two proteins act in different cellular pathways or during different times of development. We currently favor the hypothesis that MAGOH functions as a housekeeping gene, whereas MAGOHB is regulated and will be used at specific time points, under certain cellular conditions, or to compensate for reduced levels of MAGOH. Our finding that the expression of MAGOHB is inducible in mouse macrophages corroborates this hypothesis. Nonetheless, the molecular mechanism for the differential regulation of both MAGOH genes remains elusive. Further work, such as the generation of MAGOHB-knockout mice, will be needed to understand the physiological function associated with either of the MAGOH genes.
Materials and Methods
Plasmids, cell culture, and siRNA transfections
HeLa cells were cultured and transfected as described previously.23 For tethering assays, β-globin 4boxB, the transfection control (wt300+e3), GFP expression vector, and expression plasmids pCI-λNV5, pCI-λNV5-MAGOH, and pCI-λNV5-Y14 were described previously.23 The MAGOHB cDNA was obtained by reverse transcription (RT)-PCR using total HeLa cell RNA and inserted into pCI-λNV5. FLAG- and V5-expression plasmids for immunoprecipitation and MINX for in vitro splicing experiments, were described.22,23 For knockdown assays, 1 × 105 HeLa cells per well were seeded in 6-well plates 24 h before transfection with siRNAs. SiRNAs were transiently transfected using lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer’s recommendations using 1.5 μl of siRNAs (20 μM stock) and 5 μl RNAiMAX reagent. Seventy-two hours post-transfection cells were harvested for analysis of RNA and protein expression. Three siRNAs (IDT) were utilized to deplete MAGOH: [target sequences (1) AGATTAAACC AATCTAGACT GAATA; (2) CGGGAAGTTA AGATATGCCA A; (3) AAGCGTGATG GAGGAACTGA A] and to deplete MAGOHB: [target sequences (4) GCACAAGAGT GTAATGGAAG AACTG; (5) CGGACGGAAA GCTTAGATAT GCCAA; (6) CAGGCTGTTT GTATATTTAA T]. In addition, combinations of siRNAs to deplete both MAGOH as well as MAGOHB: [(7) siRNA1+siRNA4; (8) siRNA1+siRNA5; and (9) siRNA2+siRNA6] were utilized. The control siRNA (Luc: AACGUACGCG GAAUACUUCG ATT) was described previously.27 Cellular NMD substrates SC35 of 1.6 kb and 1.7 kb were described previously.23-25
Isolation of bone marrow derived macrophages (BMDMs)
Macrophages were derived from murine bone marrow as described in reference 28. BMDMs were differentiated in L929 medium and plated in L929 deficient medium on day seven. 2.5 × 105 cells per well were seeded in 12-well plates in triplicates. Macrophages were stimulated the next day with 10 ng/ml LPS (TLR grade, ENZO Life Science). All samples were stimulated at the same time and harvested after different time points.
RNA extraction
Total RNA was extracted with TRI reagent and analyzed by northern blotting as described before.23
Quantitative real-time PCR
Quantitative RT-PCR measuring SYBR Green incorporation was used to quantify mRNA expression levels of MAGOH and MAGOHB. The expression levels of MAGOH were normalized to a housekeeping gene, i.e., TATA binding protein (TBP). The primers used for qPCR were as following: m-MAGOH fwd (ACAAAGGCAA GTTCGGTCAT), m-MAGOH rev (GGCCACAGAG CATCATCTTC), m-MAGOHB fwd (CAAGTTTGGG CACGAGTTTT), m-MAGOHB rev (AGAGCGTCGT CCTCTTTTGT), m-TBP fwd (CCCCACAACT CTTCCATTCT), m-TBP rev (GCCAGGAGTG ATAGGGGTCA T), h-MAGOH fwd (TTCAGGCTCG GTTGTCTTTT), h-MAGOH rev (CAGTTCCTCC ATCACGCTTT), h-MAGOHB fwd (GCGATTTCTA CCTGCGCTAC), h-MAGOHB rev (GAGGCCACAA AGCATCATCT), h-TBP fwd (TGCACAGGAG CCAAGAGTGA A), h-TBP rev (CACATCACAG CTCCCCACCA), SC35 1.6 kb fwd (CGGTGTCCTC TTAAGAAAAT GATGTA), SC35 1.7 kb fwd (GGCGTGTATT GGAGCAGATG TA), SC35 1.6 kb and 1.7 kb rev (CTGCTACACA ACTGCGCCTT TT).
Immunoblot and immunoprecipitation
For immunoblot analysis, equal amounts (20 μg) of total extracts were subjected to SDS-PAGE. Complexes of FLAG-tagged proteins were immunoprecipitated from RNaseA-treated (20 μg/ml) lysates.23
In vitro transcription, in vitro splicing, and RNP immunoprecipitation
Preparation of capped MINX transcripts, in vitro splicing followed by RNP immunoprecipitation were performed as described previously.22
Antibodies
The antibodies against FLAG (F7425 and F1804), Y14 (HPA018403), and Tubulin (T6074) were from Sigma, the MAGOH antibody (sc-271365) was purchased from Santa Cruz and the V5 antibody (18870) from QED. Polyclonal eIF4A3 antibody was raised in rabbits and affinity-purified by Genscript with a KLH-conjugated peptide (ATSGSARKRLLKEED29). UPF3B antiserum was raised in rabbits by Eurogentech against a C-terminal fragment of UPF3B (300–483) and affinity purified.
Supplementary Material
Acknowledgments
We thank Manolis Pasparakis for wild-type C57BL/6 mice, the Leptin and Uhlirova labs for sharing equipment. We are grateful to Eva Tsaousidou for dissecting out mouse brain, Simona Ciriello and Volker Boehm for critical reading of the manuscript, and members of the Gehring lab for useful discussions. This research was funded by grants from the Fritz Thyssen Stiftung and the Deutsche Forschungsgemeinschaft (GE2012/2-1) to NHG.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/rnabiology/article/23855/
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25827
References
- 1.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Alexandrov A, Colognori D, Shu MD, Steitz JA. Human spliceosomal protein CWC22 plays a role in coupling splicing to exon junction complex deposition and nonsense-mediated decay. Proc Natl Acad Sci USA. 2012;109:21313–8. doi: 10.1073/pnas.1219725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, et al. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat Struct Mol Biol. 2012;19:983–90. doi: 10.1038/nsmb.2380. [DOI] [PubMed] [Google Scholar]
- 5.Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH. CWC22 connects pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2012;2:454–61. doi: 10.1016/j.celrep.2012.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, et al. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–64. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saulière J, Murigneux V, Wang Z, Marquenet E, Barbosa I, Le Tonquèze O, et al. CLIP-seq of eIF4AIII reveals transcriptome-wide mapping of the human exon junction complex. Nat Struct Mol Biol. 2012;19:1124–31. doi: 10.1038/nsmb.2420. [DOI] [PubMed] [Google Scholar]
- 8.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–97. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–9. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 10.Ma XM, Yoon SO, Richardson CJ, Jülich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–13. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 11.Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–50. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrançois M, Chagnon P, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–62. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Chazal PE, Daguenet E, Wendling C, Ulryck N, Tomasetto C, Sargueil B, et al. EJC core component MLN51 interacts with eIF3 and activates translation. Proc Natl Acad Sci USA. 2013;110:5903–8. doi: 10.1073/pnas.1218732110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–9. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 15.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–25. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–72. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 17.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–8. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albers CA, Paul DS, Schulze H, Freson K, Stephens JC, Smethurst PA, et al. Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat Genet 2012; 44:435-9, S1-2. [DOI] [PMC free article] [PubMed]
- 19.Rosenfeld JA, Traylor RN, Schaefer GB, McPherson EW, Ballif BC, Klopocki E, et al. 1q21.1 Study Group Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur J Hum Genet. 2012;20:754–61. doi: 10.1038/ejhg.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki T, Hashimoto S, Toyoda N, Nagai S, Yamazaki N, Dong HY, et al. Comprehensive gene expression profile of LPS-stimulated human monocytes by SAGE. Blood. 2000;96:2584–91. [PubMed] [Google Scholar]
- 22.Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res. 2005;33:5965–77. doi: 10.1093/nar/gki905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–96. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glaser S, Lubitz S, Loveland KL, Ohbo K, Robb L, Schwenk F, et al. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2:5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–49. doi: 10.1016/S1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 28.Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc 2008; 2008:pdb prot5080. [DOI] [PubMed]
- 29.Li Q, Imataka H, Morino S, Rogers GW, Jr., Richter-Cook NJ, Merrick WC, et al. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–46. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.