Abstract
RNA, at the forefront of biochemical research due to its central role in biology, is recognized by proteins through various mechanisms. Analysis of the RNA-protein interface provides insight into the recognition determinants and function. As such, there is a demand for developing new methods to characterize RNA-protein interactions. Saturation transfer difference (STD) NMR can identify binding ligands for proteins in a rather short period of time, with data acquisitions of just a few hours. Two RNA-protein systems involved in RNA modification were studied using STD NMR. The N6-threonylcarbamoyltransferase, YrdC, with nucleoside-specific recognition, was shown to bind the anticodon stem-loop of tRNALysUUU. The points of contact on the RNA were assigned and a binding interface was identified. STD NMR was also applied to the interaction of the archaeal ribosomal protein, L7Ae, with the box C/D K-turn RNA. The distinctiveness of the two RNA-protein interfaces was evident. Both RNAs exhibited strong STD signals indicative of direct contact with the respective protein, but reflected the nature of recognition. Characterization of nucleic acid recognition determinants traditionally involves cost and time prohibitive methods. This approach offers significant insight into interaction interfaces fairly rapidly, and complements existing structural methods.
Keywords: RNA modification, nuclear magnetic resonance, YrdC, L7Ae, modification enzymes, lysyl-tRNA, snoRNA
Introduction
RNA is responsible for performing numerous essential and diverse functions within living organisms. As more RNA molecules are discovered to have vital roles in cellular functions, there is an increasing need to characterize their interactions with proteins. An understanding of the RNA-protein interface provides insight into the recognition determinants and the mechanism of interaction. Nuclear magnetic resonance (NMR) spectroscopy, with its minimal environmental effect on molecules, provides advantages over other biochemical techniques that can disrupt non-covalent binding and is particularly useful when observing transient or weak interactions. NMR spectroscopy also has the ability to observe the interactions at an atomic level; therefore, providing information on specific contacts at the interface of an interaction. RNA-protein interactions are detectable by several NMR-based techniques, such as cross-saturation1,2 and chemical shift perturbation experiments.3 Unfortunately, these only provide data on the protein’s amino acids at the interface with no information about the RNA. RNA chemical shift changes can also be observed during protein titration with nuclear Overhauser effect spectroscopy (NOESY), total correlation spectroscopy (TOCSY), or heteronuclear single quantum coherence (HSQC) experiments. However, these methods have accompanying drawbacks, including long acquisition times, molecular weight limitations, and 15N- or 13C-labeling requirements.
Saturation transfer difference (STD) NMR has proven to be a very powerful method in elucidating receptor-ligand interactions. To date, STD NMR has been most extensively used to study protein interactions with small molecules and carbohydrates; however, this technique can be applied in many areas for detecting intermolecular interactions. It was first developed to screen small molecule libraries for binding activity to proteins.4 Since then, it has been utilized in a variety of systems. For instance, it has been used to identify drug-like compounds that bind RNA receptors,5-7 to investigate interactions between dispersant molecules and molecular nanoparticles,8 and to detect the binding of proteins with 6- and 8-nucleotide DNA sequences.9
Several advantages of STD NMR address the shortcomings of other NMR methods by affording low receptor concentration, short acquisition time, no labeling requirements, and applicability to large proteins.4,10 It also identifies the contact area of the ligand directly from the NMR spectra. A major advantage of STD NMR over traditional binding detection techniques is that the proton(s) of the ligand with the strongest interaction with the receptor will display the most intense NMR signal, thus providing the ability to map the ligand’s binding epitope.10 This method is especially advantageous when protein expression is low or isotope labeling is unavailable, because it requires small amounts of protein. As such, STD NMR complements existing structural, molecular, and biochemical techniques by providing data that may otherwise require significant amounts of time and material.
In the STD experiment, the protein molecule is selectively saturated with a radiofrequency pulse applied to a small frequency window in which no ligand resonances reside. This saturation propagates across the protein to the RNA ligand that is bound. The bound RNA is saturated via intermolecular 1H-1H cross-relaxation at the interface. As would be expected, the persistence of the saturation is determined by the dissociation rate, koff, and is a function of the RNA-protein dissociation constant, Kd. The amount of protein is small; thus, each protein molecule is involved in multiple binding events over the course of an experiment. Over time the population of ligands affected is large, resulting in reduced ligand intensity. When the saturation is applied outside of the frequency region for both ligand and receptor, the resonance intensities are left intact. The difference spectrum results in nonzero intensities, ISTD = Isat,off - Isat,on, representative of RNA directly interacting with the protein and will be the only visible peaks in the difference spectrum.4,10,11
Here, we investigate two distinct RNA-protein interactions that lead to post-transcriptional RNA modifications. Escherichia coli YrdC is one enzyme required for the biosynthesis of a ubiquitous tRNA (tRNA) modification.12 YrdC is an ATPase that binds l-threonine and tRNAs that respond to codons beginning with adenosine.13 It has recently been shown to produce the activated L-threonylcarbamate intermediate for modification of adenonsine to N6-threonylcarbamoyladenosine, (t6A).14 This modification is found 3′-adjacent to the anticodon in many tRNA species,15 and is critical for accurate decoding and maintenance of the translational reading frame.16 Methanocaldococcus jannaschii L7Ae is a ribosomal protein of the large subunit and is among a family of proteins that recognize the RNA kink-turn motif (K-turn).17 K-turns are found in the box C/D and H/ACA small nucleolar RNAs (snoRNAs), which direct 2’-O-methylation and pseudouridylation, respectively, of ribosomal RNAs (rRNAs) in archaea and eukaryotes.18 These two proteins with similar functions recognize RNA with distinct strategies that seemingly require unique research approaches for us to understand the RNA-protein interaction. To detail this distinction, we have applied an adaption of saturation transfer STD NMR to both modification enzymes.
Results and Discussion
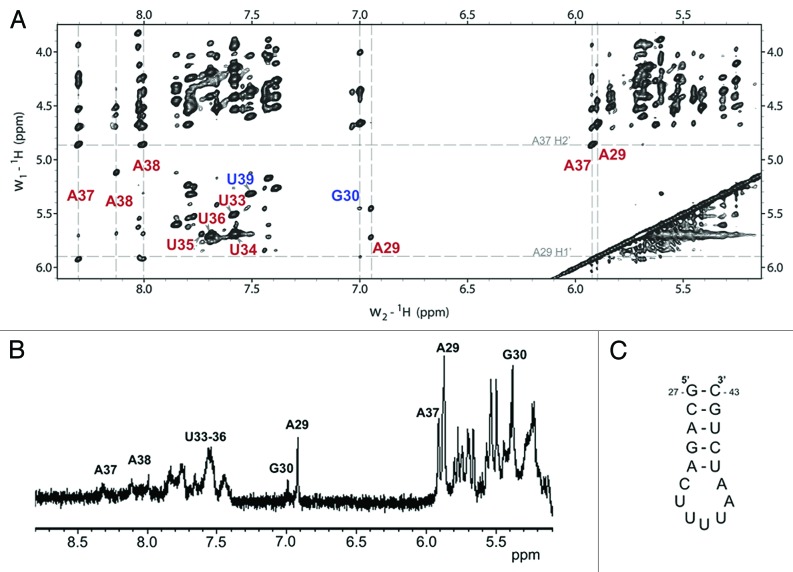
YrdC recognizes the heptadecamer anticodon stem and loop of tRNALysUUU (ASLLysUUU) with a Kd of 270 nM.13 It preferentially binds the unmodified ASLLysUUU in contrast to the modified counterpart and RNAs without the target sequence. This indicates that the recognition elements are very specific and the enzyme is able to discern its substrate tRNA from all other tRNA species. We have shown that this RNA-protein interaction is detectable by STD NMR.13 In order to probe this interaction further, chemical shifts were identified using 2D 1H-1H NOESY spectra collected for complete proton assignment of ASLLysUUU (Fig. 1A). With this data, the signals present in the STD spectrum were identified by their chemical shifts (Fig. 1B). These signals represent the protons that are in direct contact with YrdC; therefore, the nucleosides at the interface of the interaction were determined. We observe strong signals from A29H2, A29H1’, G30H1’, and A37H1’, as well as weak signals from A37H8, G30H8, A38H2, and A38H8. There are additional signals between 7.4 and 7.8 ppm that could not be unambiguously determined in the 1D spectrum due to the high density of resonances in this region. All of the H6 resonances from the loop uridines are present in this region, which would suggest YrdC is binding at least one, but the spectral resolution does not afford the ability to specifically identify each STD intensity.
Figure 1. NMR spectra of 300 μM ASLLysUUU with 3 μM YrdC. (A) 1H-1H NOESY spectrum demonstrating the unambiguous identification of nucleoside resonances with the signals observed in the STD spectrum labeled. (B) STD spectrum with several of the strongest signals assigned are indicative of the YrdC binding region. (C) The secondary structure of ASLLysUUU.
It appears that YrdC binds ASLLysUUU at four nucleotides—A29, G30, A37, A38—in the stem and loop, which, due to the helical turn, are found on one side of the molecule, and potentially U33-U36 (Fig. 2). The distance between A29H2 to A37H1’ is 19.8 Å in the NMR structure of unmodified ASLLys characterized by Durant and Davis.19 According to the crystal structure, E. coli YrdC contains a large concave, positively charged pocket of ~20 Å,20 which could reasonably accommodate this portion of the ASL. Also, YrdC has been shown to bind double-stranded RNA20 and is able to discern t6A-modified from unmodified RNA.13,21 This predicted orientation, with A37 in the binding pocket, would enable YrdC to perform such a function.
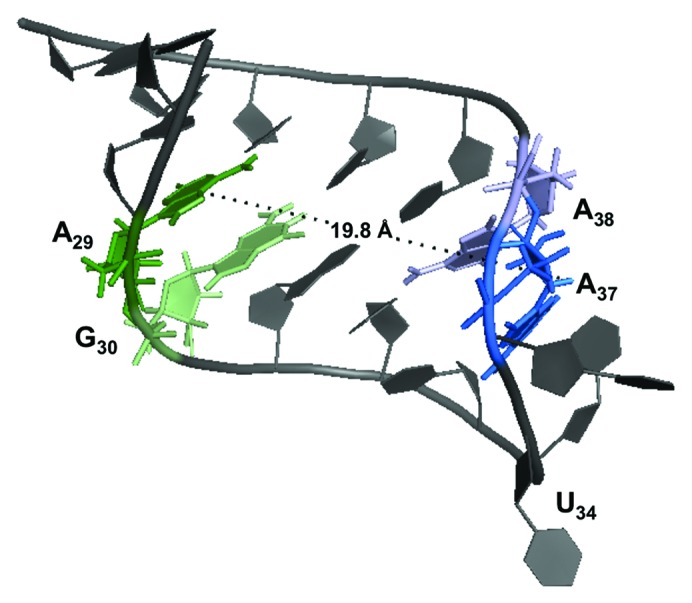
Figure 2. Three-dimensional structure of unmodified ASLLysUUU.19 The nucleotides with STD signals are heighted: A27 (green), G30 (light green), A37 (blue), and A38 (light blue). The distance from A29 to A37 was calculated to be 19.8 Å and is shown (PDB: 1BZ2).
The sequence U36A37A38 is common to all tRNA species with t6A or its derivatives,15 and at least two of these nucleosides appear to be recognized by YrdC. In addition to the peaks we can identify, there are surely more that are within the binding pocket that are not as easily identified in the 1D 1H spectrum. However, this result provides significant, readily achieved insight into the RNA binding interface and depicts the applicability of STD NMR for identifying specific nucleosides recognized by a protein.
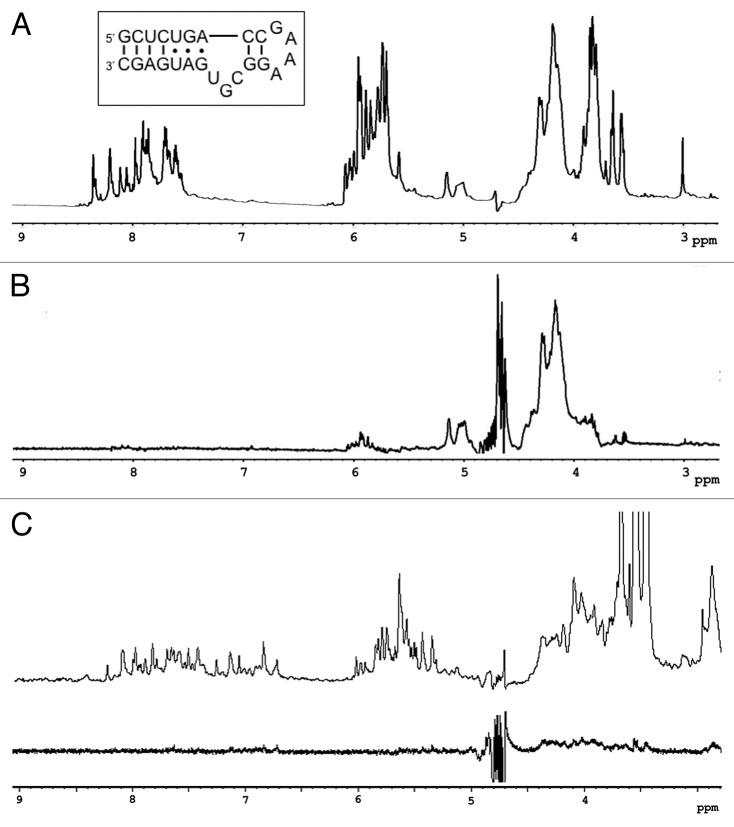
L7Ae is one of the components of the archaeal box C/D small ribonucleoprotein particles (snoRNPs) responsible for rRNA methylation.22 The M. jannaschii and Archaeoglobus fulgidus L7Ae proteins have been shown to bind the structurally unique 25-nt K-turn box C/D snoRNA.22-24 In the STD spectrum, the protein appears to be directly interacting with the backbone ribose (Fig. 3A and B). This is demonstrated by the largest signal intensity between 4 and 4.5 ppm. Since L7Ae specifically recognizes K-turn motifs in RNA sequences but not specific nucleosides,17 it is plausible that specific contacts would be made through bonds with the backbone. A high-resolution co-crystal structure of a homolog, A. fulgidus L7Ae, and the 25-nt box C/D RNA depicts the protein in close proximity to the RNA backbone with only two contacts for bulge nucleobases. The remainder of the nucleosides base-paired and concealed within the helical structure.24 In the co-crystal of M. jannaschii L7Ae with a different K-turn sequence, box H/ACA RNA, the protein engages the RNA in about 10 hydrogen bonds with backbone atoms and direct contacts with a uracil and three guanines.25 The base-specific contacts seen in the crystal structures are not represented in the STD data even though transfer should be detected if the protons are within 3 Å of the protein. This may be due to the dynamics of the molecules that are not observable in static crystal structures, but are reflected in NMR methods. Since the STD signals for ligand protons in contact with the protein are restricted by koff, some contacts may not be visible due to the dynamics of the interaction. However, this may better reflect the natural state of the interface. As a control, the same experiment was performed with an 11-nt double-strand RNA duplex that does not form a K-turn. The RNA did not have any signals in the STD spectrum indicating that it was not bound by M. jannaschii L7Ae (Fig. 3C).
Figure 3. STD spectra of M. jannaschii L7Ae and RNA. (A) 1D 1H reference spectrum and secondary structure (inset) of the 25-nt box C/D RNA and (B) STD spectrum depicting specific binding of RNA ribose protons (3.5–4.5, 4.9–5.2, and 5.7–6.1 ppm). No strong signal intensities are observed > 6.1 ppm. (C) STD NMR spectra of 200 μM 11-nt dsRNA and 2 μM L7Ae performed as a negative control. 1H 1D reference spectrum (top) and STD (bottom). No significant recovery over background was observed.
RNA modification enzymes are an example of a class of proteins that recognize substrates through various mechanisms. Both of the proteins studied here participate the in the recognition of RNA for post-transcriptional nucleoside modification; however, the RNA-protein interaction interfaces greatly differ. The ASLLysUUU-YrdC interaction is sequence-specific, in which the protein recognizes a signature sequence specific only to the target RNAs. By STD NMR the base-specific contacts were observable indicating that the protein distinguishes its target by recognizing certain nucleotides. Conversely, the L7Ae interaction with box C/D RNA is not specific to a sequence, but to a structural motif: the K-turn.22 These differences are reflected in the STD NMR data presented here.
In conclusion, we have demonstrated the utility of STD NMR experiments for investigating RNA-protein interactions. This method provides an additional technique to complement the existing library of methods for studying RNA-protein interactions. It is unique in its ability to produce quantitative data on the binding interface of the ligand in just a few hours. Further analysis with 1D or 2D experiments modified to have an STD component, such as TOCSY or NOESY,4 can also be useful for more complex systems. Even without additional NMR experiments or high-resolution structures, STD NMR offers significant insight into the interface and dynamics of RNA-protein interactions, providing a tool for identifying binding determinants.
Materials and Methods
Sample preparation
RNA samples were chemically synthesized by Dharmacon (Thermo Fisher) and were deprotected per manufacturer’s instructions and dialyzed against the appropriate buffer using a 3500 MWCO membrane (Pierce, Thermo Fisher). ASLLysUUU and recombinant E. coli YrdC were prepared as previously described,13 in 99% D2O.
M. jannaschii L7Ae was a kind gift from Dr E.S. Maxwell (North Carolina State University). The 25-nt box C/D RNA was dialyzed against 25 mM potassium phosphate, pH 7.2, 100 mM NaCl, and lyophilized to dryness then resuspended in 99% D2O to a final concentration of 300 μM. The 11-nt complementary dsRNA (equimolar 5′-GACGUGCGAA G-3′ and 5′-CUUCGCACGU C-3′) was prepared similarly.
NMR experiments
Data were acquired on a Bruker Avance II 700 MHz spectrometer at UAlbany equipped with an ultra-sensitive triple resonance cryoprobe capable of applying pulsed field gradients along the z-axis. NOESY spectra were recorded at 25 °C with a mixing time of 400 ms. Data was processed using NMRPipe.26 Spectra were displayed and analyzed using SPARKY.27
The STD pulse was applied using WATERGATE suppression as previously described,10 with optimization of the saturation time to 2 sec and the on-resonance irradiation to selectively saturate the protein. Experiments were performed at 30 °C on 25-nt box C/D RNA or 11-nt dsRNA before and after the addition L7Ae. A total of 1,028 scans with 16 dummy scans were collected for each.
Acknowledgments
The authors would like to thank Drs E.S Maxwell, K.T. Gagnon, and S. Biswas for providing L7Ae and RNA samples, and the Agris group for their support. This work was supported by grants from the National Science Foundation (MCB1101859) and the National Institutes of Health (5-R01GM23037-25) to PFA.
Glossary
Abbreviations:
- NMR
nuclear magnetic resonance
- STD
saturation transfer difference
- ASL
anticodon stem-loop
- NOESY
nuclear Overhauser effect spectroscopy
- sRNP
small ribonucleoprotein particle
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25948
References
- 1.Lane AN, Kelly G, Ramos A, Frenkiel TA. Determining binding sites in protein-nucleic acid complexes by cross-saturation. J Biomol NMR. 2001;21:127–39. doi: 10.1023/A:1012486527215. [DOI] [PubMed] [Google Scholar]
- 2.Ramos A, Kelly G, Hollingworth D, Pastore A, Frenkiel TA. Mapping the interfaces of protein-nucleic acid complexes using cross-saturation. J Am Chem Soc. 2000;122:11311–4. doi: 10.1021/ja002233w. [DOI] [Google Scholar]
- 3.Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FHT. Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2011;58:1–61. doi: 10.1016/j.pnmrs.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Mayer M, Meyer B. Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew Chem Int Ed. 1999;38:1784–8. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1784::AID-ANIE1784>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 5.Mayer M, James TJ. Detecting Ligand Binding to a Small RNA Target via Saturation Transfer Difference NMR Experiments in D2O and H2O. J Am Chem Soc. 2003;124:13376–7. doi: 10.1021/ja027526z. [DOI] [PubMed] [Google Scholar]
- 6.Mayer M, James TL. NMR-based characterization of phenothiazines as a RNA binding scaffold. J Am Chem Soc. 2004;126:4453–60. doi: 10.1021/ja0398870. [DOI] [PubMed] [Google Scholar]
- 7.Mayer M, Lang PT, Gerber S, Madrid PB, Pinto IG, Guy RK, et al. Synthesis and testing of a focused phenothiazine library for binding to HIV-1 TAR RNA. Chem Biol. 2006;13:993–1000. doi: 10.1016/j.chembiol.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Szczygiel A, Timmermans L, Fritzinger B, Martins JC. Widening the view on dispersant-pigment interactions in colloidal dispersions with saturation transfer difference NMR spectroscopy. J Am Chem Soc. 2009;131:17756–8. doi: 10.1021/ja905637y. [DOI] [PubMed] [Google Scholar]
- 9.Xia Y, Zhu Q, Jun KY, Wang J, Gao X. Clean STD-NMR spectrum for improved detection of ligand-protein interactions at low concentration of protein. Magn Reson Chem. 2010;48:918–24. doi: 10.1002/mrc.2687. [DOI] [PubMed] [Google Scholar]
- 10.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc. 2001;123:6108–17. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 11.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed Engl. 2003;42:864–90. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 12.Deutsch C, El Yacoubi B, de Crécy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem. 2012;287:13666–73. doi: 10.1074/jbc.M112.344028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris KA, Jones V, Bilbille Y, Swairjo MA, Agris PF. YrdC exhibits properties expected of a subunit for a tRNA threonylcarbamoyl transferase. RNA. 2011;17:1678–87. doi: 10.1261/rna.2592411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauhon CT. Mechanism of N6-threonylcarbamoyladenonsine (t(6)A) biosynthesis: isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry. 2012;51:8950–63. doi: 10.1021/bi301233d. [DOI] [PubMed] [Google Scholar]
- 15.Jühling F, Mörl M, Hartmann RK, Sprinzl M, Stadler PF, Pütz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, et al. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39:13390–5. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- 17.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henras AK, Dez C, Henry Y. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr Opin Struct Biol. 2004;14:335–43. doi: 10.1016/j.sbi.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Durant PC, Davis DR. Stabilization of the anticodon stem-loop of tRNALys,3 by an A+-C base-pair and by pseudouridine. J Mol Biol. 1999;285:115–31. doi: 10.1006/jmbi.1998.2297. [DOI] [PubMed] [Google Scholar]
- 20.Teplova M, Tereshko V, Sanishvili R, Joachimiak A, Bushueva T, Anderson WF, et al. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci. 2000;9:2557–66. doi: 10.1110/ps.9.12.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, et al. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn JF, Tran EJ, Maxwell ES. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002;30:931–41. doi: 10.1093/nar/30.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner B, Melcher SE, Wilson TJ, Norman DG, Lilley DMJ. Induced fit of RNA on binding the L7Ae protein to the kink-turn motif. RNA. 2005;11:1192–200. doi: 10.1261/rna.2680605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004;12:807–18. doi: 10.1016/j.str.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 25.Hamma T, Ferré-D’Amaré AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem. 2010;285:805–9. doi: 10.1074/jbc.R109.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Goddard TD, Kneller DG. SPARKY 3. University of California, San Francisco, 2008. [Google Scholar]