Abstract
Cancer cell metabolism differs from normal cells, yet the regulatory mechanisms responsible for these differences are incompletely understood, particularly in response to acute changes in the tumor microenvironment. HuR, an RNA-binding protein, acts under acute stress to regulate core signaling pathways in cancer through post-transcriptional regulation of mRNA targets. We demonstrate that HuR regulates the metabolic phenotype in pancreatic cancer cells and is critical for survival under acute glucose deprivation. Using three pancreatic cancer cell line models, HuR-proficient cells demonstrated superior survival under glucose deprivation when compared with isogenic cells with siRNA-silencing of HuR expression (HuR-deficient cells). We found that HuR-proficient cells utilized less glucose, but produced greater lactate, as compared with HuR-deficient cells. Acute glucose deprivation was found to act as a potent stimulus for HuR translocation from the nucleus to the cytoplasm, where HuR stabilizes its mRNA targets. We performed a gene expression array on ribonucleoprotein-immunoprecipitated mRNAs bound to HuR and identified 11 novel HuR target transcripts that encode enzymes central to glucose metabolism. Three (GPI, PRPS2 and IDH1) were selected for validation studies, and confirmed as bona fide HuR targets. These findings establish HuR as a critical regulator of pancreatic cancer cell metabolism and survival under acute glucose deprivation. Further explorations into HuR’s role in cancer cell metabolism should uncover novel therapeutic targets that are critical for cancer cell survival in a metabolically compromised tumor microenvironment.
Keywords: HuR, post-transcriptional gene regulation, cancer metabolism, pancreatic cancer
Introduction
Pancreatic cancer remains one of the most lethal cancer types and is particularly resistant to conventional chemotherapeutic agents.1 Mathematical modeling of cancer biology suggests that harsh conditions in the tumor microenvironment impose profound selection pressures on cancer cells, leading to the development of aggressive and invasive features.2 These selection forces may be particularly important for pancreatic cancer tumorigenesis, since as much as 90% of the tumor volume is composed of stroma.3 This biologic feature adversely affects the delivery of oxygen and nutrients to tumor cells,4-6 and has been correlated with poor survival in pancreatic7 and other cancers.8,9 It is not surprising, therefore, that pancreatic cancers are particularly tolerant to nutrient deprivation in vitro, compared with other aggressive gastrointestinal malignancies.10
The “Warburg effect” posits that cancer cells exhibit a preference for glycolytic metabolism, even in the presence of oxygen.11 Recent studies reveal that additional biosynthetic pathways are upregulated in cancer cells.12-15 While metabolomic and other modern molecular assays provide new insights into the metabolic phenotype of cancer cells, the regulatory mechanisms that govern metabolic transformation in cancer cells remain poorly understood. A prevailing theory suggests that dysregulation (primarily through somatic mutations) of oncogenes and tumor suppressor genes is the principle mechanism for metabolic reprogramming,11,16 and has been validated in a variety of tumor systems with multiple cancer genes (e.g., KRAS, PI3K, PTEN, p53).13,16,17 However, the theory fails to adequately address how cancer cells acutely adapt to metabolic stress. Somatic mutations are random events and are selected for over time; they do not occur on demand by cancer cells in response to acute metabolic stress. Moreover, genetic changes are irreversible.18 A more flexible regulatory strategy, such as post-transcriptional regulation of cancer genes, is better suited to navigate the unpredictable, dynamic, and heterogeneous tumor microenvironment.
RNA-binding proteins (RBPs) regulate biologic processes through post-transcriptional control of target mRNAs. These proteins regulate mRNA stability and protein translation. Through these mechanisms, RBPs can efficiently and reversibly alter the proteome of a cell. RBPs typically have thousands of mRNA targets, which encode proteins with critical biologic functions.19 HuR (ELAVL1 or embryonic lethal abnormal vision-like protein 1) was one of the first RBPs studied in cancer cells, and remains one of the best characterized to date.20-22 HuR binds to mRNAs primarily at U- or AU-rich sequences, which are typically, but not exclusively, located in the untranslated regions of target transcripts.23 In response to cellular stressors (e.g., UV light, chemotherapeutics or hypoxia24,25), cytoplasmic levels of HuR increase and post-transcriptional regulatory functions of HuR are exerted on its bound mRNA cargo. The importance of HuR biology in tumorigenesis is supported by numerous clinicopathologic studies across diverse tumor types, which consistently reveal a correlation between cytoplasmic HuR expression (i.e., “activated” HuR) and tumor aggressiveness.26
Targets of RBPs often cluster into discrete functional groups (called “RNA regulons”), which enable a single regulatory molecule (such as HuR) to coordinate an entire signaling pathway through post-transcriptional regulation of multiple pathway components.19 For instance, yeast studies demonstrate that multiple mRNAs in a common metabolic pathway (e.g., glycolysis) have similar decay rates, presumably due to a shared post-transcriptional regulator.27 Cancer cells exploit this regulatory feature by hijacking HuR to promote numerous pro-survival pathways such as angiogenesis (targets include VEGF), anti-apoptosis (BCL2, MCL1, DR5), cell cycle progression (cyclin A, B1, cyclin E1) and drug resistance (DR5).28,29 Based on this previous body of work, we hypothesized that acute metabolic stress, such as glucose deprivation (as a model of the compromised tumor microenvironment), activates a pro-survival metabolic program in pancreatic cancer cells mediated by HuR.
Results
HuR expression affects pancreatic cancer cell survival under glucose deprivation
Experimental cell lines (control, HuR silenced with siRNA oligos and HuR overexpressed by plasmid transfection) are designated throughout the manuscript by the parental cell line along with the transfected plasmid, as detailed in Table S1 (e.g., HuR silenced MiaPaCa2 cells are designated MiaPaCa2.siHuR and controls are MiaPaCa2.siCTRL).
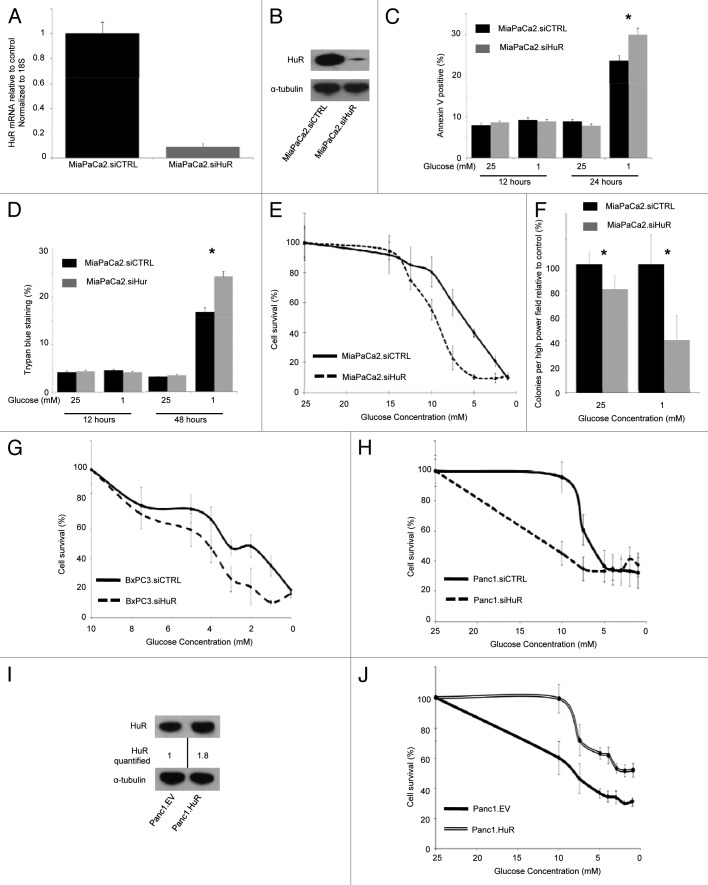
The effect of HuR on apoptosis and cell viability in normal (25 mM glucose) and glucose deprived (1 mM glucose) media was assessed in HuR proficient (MiaPaCa2.siCTRL) and deficient (MiaPaCa2.siHuR) cells. Representative quantitative PCR (qPCR) and immunoblot (Fig. 1A and B) experiments after HuR silencing demonstrated greater than 90% reduction in mRNA and 70% reduction in protein expression, respectively. Apoptosis was assessed by Annexin V staining and no differences were observed between MiaPaCa2.siCTRL and MiaPaCa2.siHuR cells at 12 h (Fig. 1C). However, cells incubated with 1 mM glucose for 24 h showed an increase in apoptotic signaling, with the greatest amount of apoptosis occurring in the MiaPaCa2.siHuR cells. These findings suggest that HuR depletion sensitizes pancreatic cancer cells to glucose deprivation. A similar pattern was observed in an analysis of cell death by Trypan blue staining. Death was comparable between MiaPaCa2.siCTRL and MiaPaCa2.siHuR cells after just 12 h of glucose deprivation, but increased in both groups of cells by 48 h in 1 mM glucose. As with apoptosis, the proportion of affected cells was greatest for MiaPaCa2.siHuR cells (Fig. 1D).
Figure 1. HuR protects pancreatic cancer cells against glucose-deprivation. In bar graphs: siCTRL, black bars; siHuR, gray bars (*p < 0.05). In survival curves (unless indicated): siCTRL are solid lines; siHuR are dashed lines. (A) Representative qPCR 24 h after transfection in MiaPaCa2 cells. (B) Representative immunoblot 48 h after transfection in MiaPaCa2 cells. (C) Annexin V staining in MiaPaCa2 cells. (D) Trypan blue staining in MiaPaCa2 cells. (E) PicoGreen assays at 7 d in MiaPaCa2 cells. (F) Three-week soft-agar colony formation assays in MiaPaCa2 cells. PicoGreen assays at seven days in (G) BXPC3 and (G) PANC1 cells. (I) Representative immunoblot 72 h after transfection in PANC1 cells. (J) PicoGreen assay at seven days in PANC1.HuR (double line-top) vs. PANC1.EV (single line-bottom).
The effect of HuR silencing in the setting of glucose deprivation for longer time intervals was assessed using a PicoGreen assay at 7 d. MiaPaCa2.siCTRL cells had improved survival when the starting glucose concentration was 12.5 mM or less, as compared with MiaPaCa2.siHuR cells (Fig. 1E). In these experiments, glucose concentrations in the media typically decreased by 20–50% per day. The glucose concentration IC50 equivalents were 6 mM (MiaPaCa2.siCTRL) and 10 mM (MiaPaCa2.siHuR), respectively (Fig. 1E).
To measure in vitro tumorigencity, we performed an anchorage independent growth assay over a 3 wk time period in 25 mM glucose and under glucose deprivation (1 mM). MiaPaCa2.siCTRL cells exhibited increased colony growth (both size and number of colonies) as compared with MiaPaCa2.siHuR cells (Fig. 1F) in both glucose conditions. However, the greatest difference occurred between cells incubated with 1 mM starting glucose. Long-term cell survival assays (over 7 d with PicoGreen) were performed for two other pancreatic cancer cell lines, BxPC3 (Fig. 1G) and PANC1 cells (Fig. 1H). As with MiaPaCa2 cells, BxPC3.siCTRL and PANC1.siCTRL cells both had superior survival, as compared with BxPC3.siHuR and PANC1.siHuR cells under glucose deprivation.
Complementary to HuR silencing studies, the impact of HuR overexpression on cell survival under glucose deprivation was tested, and PANC1.HuR cells had superior survival to PANC1.EV cells (Fig. 1I). A similar survival advantage was not observed with MiaPaCa2.HuR cells, as compared with MiaPaCa2.EV (data not shown). Taken together, these data provide convincing evidence, for the first time, that HuR provides a survival advantage to pancreatic cancer cells in the setting of acute glucose deprivation.
HuR translocates from the nucleus to the cytoplasm upon glucose deprivation
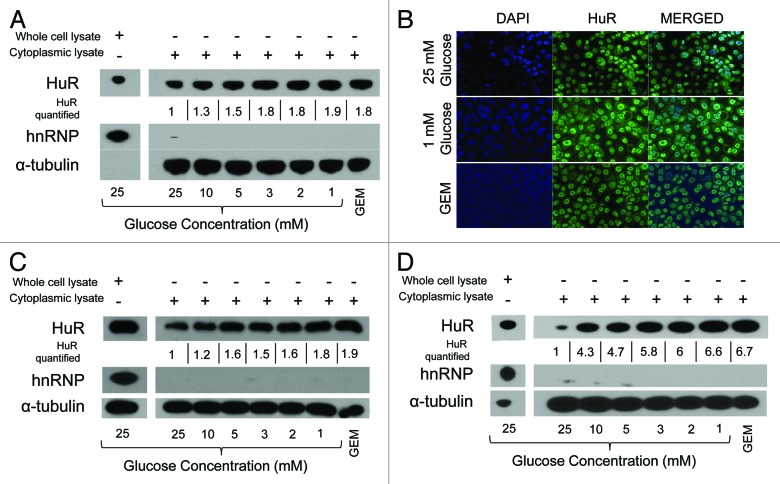
The effect of glucose deprivation on HuR translocation from the nucleus to the cytoplasm (i.e., HuR “activation”) was assessed at 6 h (this time point was chosen based on multiple time course experiments, with a representative immunoblot provided in Fig. S1). Increased cytoplasmic HuR protein expression was observed with glucose concentrations below 10 mM for each of the tested pancreatic cancer cell lines (MiaPaCa2, BxPC3 and PANC1), and was greatest at 3 mM glucose or less (Fig. 2A, C and D). For each cell line, enhanced HuR cytoplasmic expression under glucose deprivation was comparable to HuR activation as a result of gemcitabine treatment (a first-line chemotherapeutic agent against pancreatic cancer and an established inducer of cytoplasmic HuR).30 HuR cytoplasmic expression due to glucose deprivation was also observed by immunofluorescence (Fig. 2B).
Figure 2. HuR protein is activated to the cytoplasm under glucose deprivation. (A) Immunoblots of whole cell (left) and cytoplasmic (right) lysates from MiaPaCa2 cells cultured for 6 h in media with the indicated glucose concentrations. Gemcitabine (GEM) (1 µM) was used as a positive control for cytoplasmic HuR activation. HuR (and GEM) levels are quantified at each glucose concentration and numeric values are displayed normalized to control glucose conditions below the corresponding cytoplasmic HuR band. Immunofluorescence at 12 h (B) and immunoblots at 6 h (C) of BxPC3 cells in normal DMEM (25 mM glucose), 1 mM glucose DMEM and 1 µM gemcitabine (positive control). Nuclei are counterstained in blue (DAPI). Immunoblots of (D) PANC1 cells after incubation in the indicated glucose concentrations for 6 h. Relative HuR levels are quantified, as described for panel (A).
Lactate levels in the media are altered by HuR expression
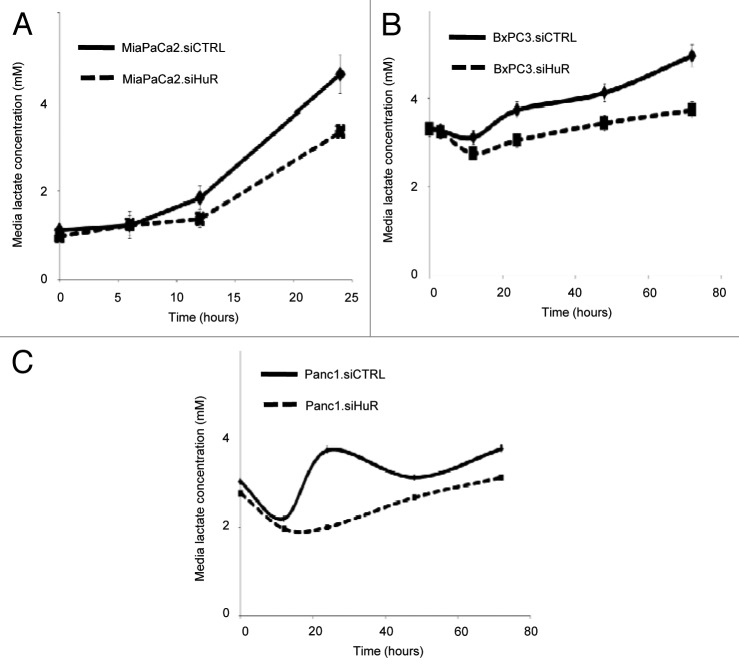
In order to determine whether HuR expression had implications on the metabolic profile of cells, we measured lactate levels in the media in pancreatic cancer cell lines (MiaPaCa2, BxPC3 and PANC1) over time, with a starting glucose concentration of 5 mM. Data was normalized by total cell number as measured by PicoGreen analysis. For each cell line, lactate production was lower in HuR-deficient cells, as compared with HuR-proficient control cells (Figs. 3A–C). The impact of HuR silencing on lactate production was evaluated at 10 mM starting glucose concentrations as well for each of the three pancreatic cancer cell lines, and the results were consistent; HuR expression was associated with increased lactate production (Fig. S2).
Figure 3. HuR expression affects lactate production of pancreatic cancer cells. Lactate levels in the media over time. siCTRL are solid lines; siHuR are dashed lines. The starting glucose concentration for these experiments was 5 mM. (A) MiaPaCa2 cells, (B) BXPC3 cells and (C) PANC1 cells. Lactate levels are normalized to total cell count in each well.
Glucose levels in the media are altered by HuR expression
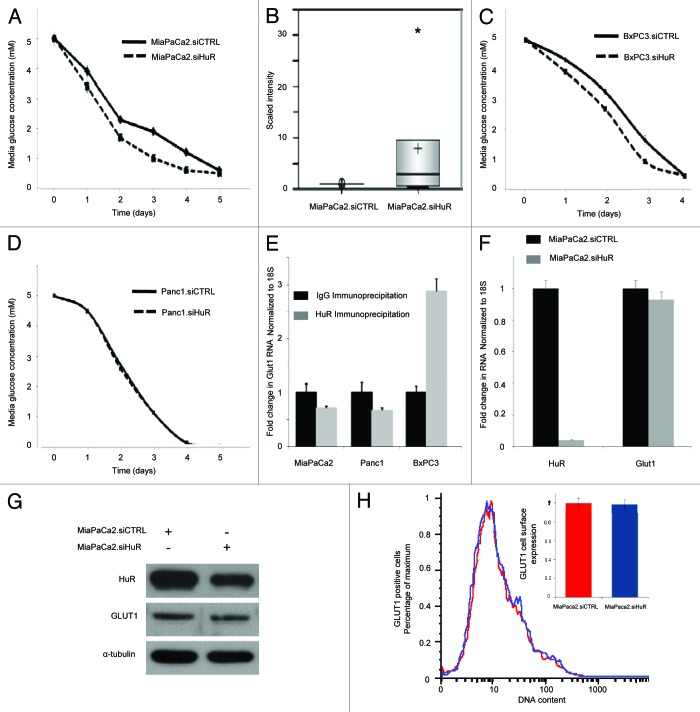
Glucose concentrations were measured in the media over 5 d as an estimate of glucose uptake by pancreatic cancer cells. In a manner similar to lactate studies, data was normalized to cell number as measured by PicoGreen analysis. Surprisingly, the glucose levels diminished more rapidly in MiaPaCa2.siHuR cells, as compared with MiaPaCa2.siCTRL cells. The experiment presented in Figure 4A was performed with a starting glucose concentration of 5 mM. A replicate experiment performed at 10 mM glucose is provided in Fig. S2. Findings were also consistent with six separate experiments performed at starting glucose concentrations ranging between 2.5–20 mM (data not shown); cells with silenced HuR consumed more glucose than their isogenic counterparts. Notably, when these results are considered in the context of survival data demonstrated in Figure 1, our assay likely underestimates the degree to which HuR activity minimizes glucose uptake relative to HuR-deficient cells.
Figure 4. HuR expression affects glucose uptake into pancreatic cancer cells independent of GLUT1. In glucose concentration curves: siCTRL are solid lines; siHuR are dashed lines. (A) Glucose levels in the media (5 mM starting glucose) over time in MiaPaCa2 cell culture. (B) Box plot of intracellular glucose levels detected by GC/MS and LC/MS/MS platforms in MiaPaCa2 cells incubated in 25 mM glucose (*p < 0.1). Glucose levels in the media (5 mM starting glucose) over time in (C) BxPC3 cells and (D) PANC1 cells. (E) Ribonucleotide immunoprecipitation and qPCR for GLUT1 demonstrating that GLUT1 does not bind HuR (gray bars), relative to IgG (black bars) in MiaPaCa2 and PANC1 cell lines. BxPC3 cells demonstrated mild enrichment of GLUT1 mRNA in the HuR sample relative to the IgG sample. The HuR target, dCK, served as a positive control (data not shown, MiaPaCa2: 8 fold increased binding; PANC1: 4 fold increased binding; BxPC3: 3-fold increased binding). (F) qPCR of GLUT1 mRNA in MiaPaCa2.siHuR (gray bars) and MiaPaCa2.siCTRL cells (black bars). (G) Immunoblots for HuR, GLUT1 and α tubulin of whole cell MiaPaCa2 lysates 72 h following transfection with siRNA oligs. (H) Flow cytometric analysis of GLUT1 cell surface expression of MiaPaCa2.siHuR (blue) and MiaPaCa2.siCTRL cells (red, set to a value of 1 in the bar graph).
Metabolomic studies (GC/MS and LC/MS/MS) were performed in MiaPaCa2 cells at 25 mM glucose (MiaPaCa2.siCTRL vs. MiaPaCa2.siHuR cells). The concentrations of many cellular metabolic intermediates were similar between the two groups (see Table S2). However, intracellular glucose levels were noted to be 7-fold greater in MiaPaCa2.siHuR cells, as compared with MiaPaCa2.siCTRL (Fig. 4B), supporting the findings from glucose measurements obtained from the media over time (Fig. 4A). These data confirm that HuR expression reduces glucose uptake into MiaPaCa2 cells. A relative increase in glucose uptake was also observed in BxPC3.siHuR cells, as compared with BxPC3.siCTRL cells (Fig. 4C), although this trend did not extend to the PANC1 cell line (Fig. 4D).
Assessment of the candidate HuR target, GLUT1, in pancreatic cancer cells
GLUT1 is the best characterized glucose transporter in cancer cells and has been implicated as an HuR target in one previous study performed in adipocytes.31 GLUT1 is believed to be an important regulatory target of key oncogenes involved in metabolic reprogramming of cancer cells and responsible for altered glucose uptake.32,33 Therefore, we hypothesized that changes in glucose uptake related to HuR expression may result from HuR binding and post-transcriptional regulation of GLUT1. We performed a ribonucleotide-immunoprecipitation (RNP-IP) experiment under glucose deprivation (1 mM glucose) with HuR antibody (Fig. 4E). We detected no enrichment of GLUT1 mRNA in the HuR sample (as compared with the IgG sample) in two of three cell lines. We were able to detect minimal enrichment in one cell line (BxPC3). In these experiments, the known HuR target, dCK, was used as a positive control.30 In line with these findings, there was no change in GLUT1 mRNA (Fig. 4F) or protein expression (Figs. 4G and H) in MiaPaCa2.siHuR cells, as compared with MiaPaCa2.siCTRL cells. These data demonstrate that HuR’s effects on glucose utilization by pancreatic cancer cells must be independent of GLUT1.
Identification of novel metabolic targets of HuR
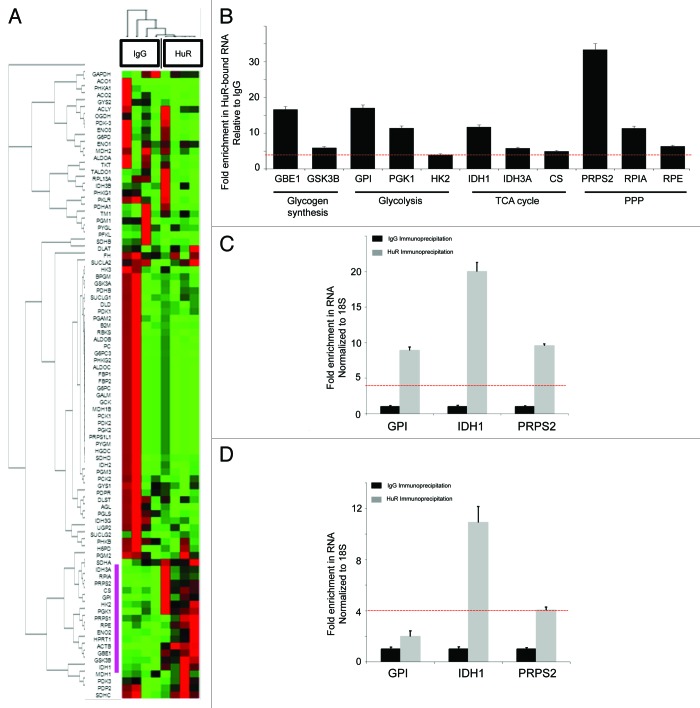
RNP-IP assays with HuR followed by qPCR Array analysis of bound mRNA were performed with MiaPaCa2 cells after 6 h of glucose deprivation (1 mM). RNP-IP with IgG antibody was performed in parallel as a negative control. A heat map and clustergram based on four independent RNP-IP experiments is shown in Figure 5A (detailed results are provided in Table S3). There were 11 metabolic transcripts (~13% of the genes on the glucose metabolism array) which were enriched in the HuR sample by more than 4-fold over IgG (p < 0.05) (Fig. 5B), representing novel HuR-bound mRNA targets. These HuR binders did not cluster into any specific branch of glucose metabolism, but rather, were distributed throughout multiple metabolic pathways, including: glycogen synthesis [glucan (1,4-α-), branching enzyme 1, GBE1; glycogen synthase kinase 3 β, GSK3B]; glycolysis (glucose-6-phosphate isomerase, GPI; phosphoglycerate kinase 1, PGK1; hexokinase 2, HK2); tricarboxylic acid cycle and related pathways (isocitrate dehydrogenase 1, IDH1; isocitrate dehydrogenase 3A, IDH3A; citrate synthase, CS); and the pentose phosphate pathway (phosphoribosyl pyrophosphate synthetase 2; PRPS2, ribose 5-phosphate isomerase A, RPIA; ribulose-5-phosphate-3-epimerase, RPE).
Figure 5. Identification of HuR metabolic target mRNAs through ribonucleotide immunoprecipitation assays. mRNA enriched by HuR immunoprecipitation were compared with an IgG control sample in MiaPaCa2 cells with an 84-gene glucose metabolism qPCR Array. (A) Heatmap/clustergram: red represents increased transcript levels with HuR RNP-IP compared with control, and green represents decreased levels. Genes enriched in the HuR sample at levels 4-fold or greater than the IgG sample are highlighted with a purple bar to the left of the heat map. (B) Eleven HuR targets are presented and grouped by pathway. RNA levels represent fold-change enrichment to HuR, relative to IgG. Data are normalized to GAPDH and the red dashed line (also present in panels C and D) indicate the predetermined cutoff used to define highly enriched HuR targets (greater than 4-fold enrichment in the HuR sample over IgG control). IDH1 was grouped with TCA cycle enzymes for the purposes of presentation, but technically is an isoenzyme of mitochondrial IDH and resides in the cytosol. GPI, IDH1 and PRPS2 were validated as HuR-bound mRNA targets by HuR RNP-IP and q-PCR in (C) BxPC3 and (D) PANC1 cell lines. HuR RNP-IP (gray bars) and IgG RNP-IP (black bars). PCR Arrays were normalized to GAPDH. qPCR assays were normalized to 18S.
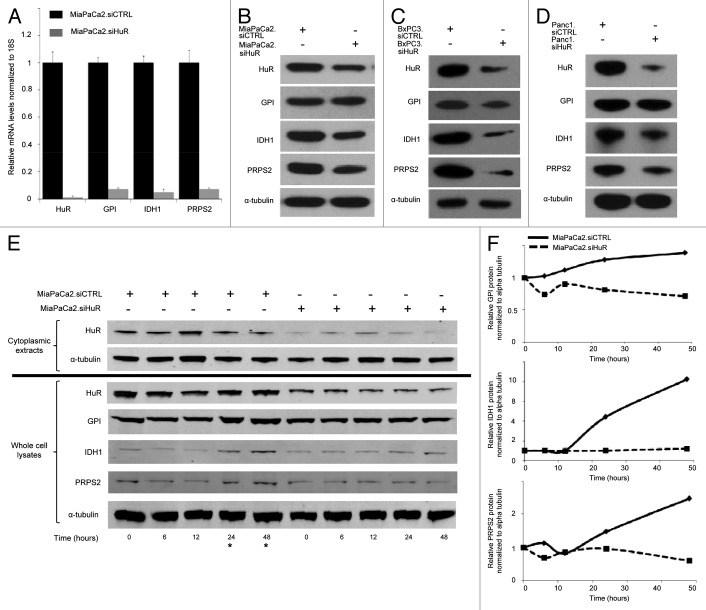
Three HuR metabolic target genes were selected for further validation studies (GPI, IDH1, and PRPS2). First, the transcripts were validated by HuR enrichment in RNP-IP/qPCR experiments in two additional pancreatic cancer cell lines. In BxPC3 cells, all three metabolic genes were enriched after extraction with the HuR antibody, compared with the IgG control (Figs. 5C). IDH1 and PRPS2 were also validated in PANC1 cells, while the fold-change of enrichment in the HuR sample for GPI did not achieve the pre-determined cutoff value used to define HuR binders (Fig. 5D). Second, we silenced HuR in MiaPaCa2 cells (MiaPaCa2.siHuR) and observed a corresponding decrease in mRNA expression in all three transcripts (> 85% reduction) compared with MiaPaCa2.siCTRL cells (Fig. 6A, an additional negative control for this experiment, again using GLUT1, is shown in a separate replicate as Fig. S4). Third, we observed a decrease in protein expression of the three targets with HuR silencing in all three pancreatic cancer cell lines (Fig. 6B–D), providing evidence of protein transcriptional changes directly related to HuR regulation. Levels of IDH1 and PRPS2 appeared most affected by direct silencing alone, while GPI appeared to be impacted the least in response to direct HuR silencing.
Figure 6. HuR silencing alters expression of metabolic targets. (A) Gene target mRNA expression 48 h after transfection (normalized to 18S) in MiaPaCa2.siCTRL (black bars) and MiaPaCa2.siHuR cells (gray bars). Whole cell protein lysates were assayed for metabolic proteins at 72 h in (B) MiaPaCa2, (C) BxPC3 and (D) PANC1 cells. (E) MiaPaCa2 cells were incubated in 1 mM glucose for the indicated time points and metabolic target protein expression was determined. (F) Protein expression of HuR targets over time from panel (E) are quantified and graphed. MiaPaCa2.siCTRL are solid lines; MiaPaCa2.siHuR are dashed lines. * highlights the later time points in panel (E) with increased target expression in MiaPaca.siCTRL cells.
In addition, we tested for HuR-directed changes of target protein expression (GPI, IDH1 and PRPS2) as a function of glucose deprivation (Fig. 6E). This experiment was meant to simulate austere conditions encountered by cancer cells in human tumors. MiaPaCa2.siCTRL and MiaPaC2.siHuR cells were incubated in media with 1 mM glucose for different time intervals. Cytoplasmic HuR peaked by 12 h in the MiaPaCa2.siCTRL cells (Fig. 6E, left). Subsequently, HuR returned to baseline levels, as has been previously described with other stressors.30 Importantly, protein expression of metabolic targets increased above baseline levels after 24–48 h of glucose deprivation (and 12–24 h after HuR translocation to the cytoplasm) in MiaPaCa2.siCTRL cells (left side of Fig. 6E and F). In contrast, changes in target protein expression were not observed in the MiaPaCa2.siHuR cells (Fig. 6E, right), providing evidence that the protein expression findings of HuR target genes in MiaPaCa2.siCTRL cells were in fact due to post-transcriptional changes mediated by HuR.
Discussion
To date, research on cancer cell metabolism has predominantly attributed metabolic reprogramming to dysregulation of certain cancer genes involved in cell proliferation (“classic” oncogenes and tumor suppressor genes), principally through somatic mutations.11,13,17 The current study presents an alternative strategy used by cancer cells to overcome acute metabolic stress (e.g., glucose deprivation). These data suggest that pancreatic cancer cells exploit RNA binding protein biology to affect the expression of metabolic enzymes, resulting in altered metabolism and improved cancer cell survival under glucose deprived conditions. Specifically, we demonstrate that (1) HuR is engaged in the acute stress response and translocates to the cytoplasm upon acute glucose withdrawal, (2) HuR protects cells in low glucose conditions resulting in improved survival and less apoptosis, (3) HuR-proficient cells demonstrated increased lactate production and diminished glucose uptake and (4) HuR binds and regulates multiple core metabolic target transcripts. Importantly, despite differences in the degree to which each cell line is impacted, these core differences were preserved across three genetically and phenotypically diverse pancreatic cancer cell lines. The role of RNA-binding proteins as central metabolic regulators remains an unexplored concept, yet is consistent with a widely accepted idea that HuR is a master-regulator of pro-survival pathways and becomes activated by acute cellular stress.26 In contrast to genetic changes that are acquired in cancer genes and provide cancer cells a survival advantage over a protracted time interval, HuR-directed survival mechanisms are rapid and reversible.19,26,28 These biological features of HuR are well suited for the unpredictable, changing and heterogeneous tumor microenvironment.34
While HuR has been indirectly linked to cancer cell metabolism in a handful of studies,31,35-40 no studies have examined HuR as regulatory hub for cancer cell metabolism.4 One study in non-cancer cells demonstrated that HuR stabilizes phosphoenolpyruvate carboxykinase mRNA (an enzyme that catalyzes the first committed step of gluconeogenesis and was previously thought to be transcriptionally regulated) in the setting of an acute drop in pH.41 A separate study concluded that the mRNA transcript of the glucose receptor, GLUT1, binds to HuR in adipocytes.31 Based on this study, we hypothesized that HuR’s effect on glucose uptake in pancreatic cancer cells (Fig. 4) was mediated through GLUT1. However, we were not able to validate GLUT1 mRNA as a true HuR target in all of our cell lines, nor were we able to demonstrate GLUT1 protein changes with HuR manipulation in isogenic cell lines (Fig. 4E–H). The discrepant results between the present study in pancreatic cancer cells and the prior study in adipocytes raises an intriguing possibility that HuR’s target mRNA substrates are tissue- (and perhaps even cancer-) specific, dependent on the available transcriptome in a given cell. This idea may explain certain inconsistencies observed between different pancreatic cell lines in the present study, such as RIP-qPCR results for GPI (Fig. 4). A recent study in prostate cancer cells demonstrated that total glucose withdrawal resulted in degradation of HuR through ubiquitin-dependent proteolysis within 24 h.35 While the findings from that study could be interpreted as conflicting with a model placing HuR at the center of an acute metabolic stress response, we notably did not observe any HuR degradation in pancreatic cancer cells even out to 72 h (Fig. S3A). Time points beyond 72 h could not be assessed due to extensive cell death under severe hypoglycemic stress (Fig. S3B). We also caution against an experimental model where glucose is removed completely from the system, as that deviates from physiologic conditions in tumors.
An unexpected finding in our work was that HuR expression was actually associated with decreased glucose uptake from the media (Fig. 4). On the surface, the observation contradicts an accepted principle that cancer cells utilize more glucose than normal cells. Baseline increases in glucose uptake by cancer cells has been linked to certain activated oncogenes (e.g., Kras, PI3K, AKT and NOTCH)13,32,42,43 and is thought to fuel increased glycolytic and biosynthetic activity associated with cancer cell proliferation.44 This biologic phenomenon is exploited clinically by PET imaging, which detects increased (F-18) flurodeoxyglucose uptake by tumor cells.45
The present data, which demonstrates decreased glucose uptake as a result of a different oncoprotein, HuR, should be interpreted with the following points in mind. First, comparisons in this study were made between cancer cells with silenced (e.g., MiaPaCa2.siHuR) and endogenous levels of HuR (e.g., MiaPaCa2.siCTRL), and not between cancer cells and normal cells. Therefore, we cannot make any conclusions regarding glucose utilization in cancers with activated HuR relative to normal tissues. Second, despite decreased glucose uptake in HuR proficient cells, our data suggest that HuR expression still could support a glycolytic phenotype, as lactate production was elevated (Fig. 3A–C). Third, the findings are consistent with previous studies showing an association between increased glucose uptake related to oncogene activation (e.g., AKT activation) and increased sensitivity to glucose withdrawal.42 Accelerated glucose consumption was previously noted to put cancer cells at risk when nutrients are scarce, presumably due to rapid depletion of energy substrates. Therefore, it stands to reason that pancreatic cancer cells would have mechanisms to counteract this action under austere conditions. A unifying model puts HuR as a key regulator of cancer cell metabolism in the setting of acute metabolic stress (e.g., glucose deprivation, acidification,41 hypoxia36,40), while “classic” oncogenic or tumor suppressor proteins regulate cancer cell metabolism under more stable conditions with a goal to maximize cell proliferation.
While changes in intracellular glucose levels were the most striking finding with the metabolomics analysis (Fig. 4B), other subtle differences were noted (Table S2). Increased glucose-6 phosphate levels were observed in the MiaPaCa2.siCTRL cells (compared with MiaPaCa2.siHuR cells), even in the setting of less glucose substrate. Stabilization of hexokinase (the first step in glycolysis, Fig. 5B) by HuR likely accounted for this result. Similarly, 6-phosphogluconate (a proximal metabolite in the pentose phosphate pathway and downstream product of glucose 6-phosphate) was increased in MiaPaCa2.siCTRL cells. Notably, three central enzymes in the non-oxidative pentose phosphate pathway (RPE, RPIA and PRPS2) were identified as HuR targets (Fig. 5B). These findings highlight the pentose phosphate pathway as a potential focal point for HuR-directed metabolic reprogramming, and suggest the identification of a candidate, “non-canonical” pathway specific for pancreatic cancer cells that could be targeted.
In summary, this study provides a foundation for a completely new line of investigation in cancer biology focused on post-transcriptional regulation of core metabolic enzymes by RNA binding proteins. We anticipate future studies to show that HuR enhances overall metabolic efficiency under stress (e.g., a cellular “adrenaline” of sorts) through its numerous targets involved in multiple metabolic pathways. Improved metabolic performance due to activated HuR enables pancreatic cancer cells to adapt to harsh metabolic conditions by conserving nutrient availability while still maintaining a glycolytic phenotype. Future studies, such as RNP-IP-sequencing experiments and targeted metabolomic studies will provide additional metabolic pathway insights. Elucidation of the most important metabolic targets of the HuR stress response should uncover novel therapeutic opportunities aimed at blocking pathways that are critical for pancreatic cancer cell survival within its unique metabolic milieu.
Materials and Methods
Pancreatic cancer cell lines, cell culture and transfections
Pancreatic cancer cell lines (MiaPaCa2, PANC1 and BxPC3) were purchased from ATCC and grown at 37°C and 5% CO2 in 75 cm2 flasks. Under control conditions, standard DMEM (25 mM glucose, or 450 mg/dl) was used for MiaPaCa2 and PANC1 cells, and RPMI (11.11 mM glucose, or 200 mg/dl) for BxPC3 cells. Media was supplemented with 10% fetal bovine serum, 1% L-glutamine and 1% penicillin/streptomycin (Invitrogen). Low glucose media was used to simulate glucose deprivation commonly encountered in the tumor microenvironment. The most severe hypoglycemic stress used in these experiments was 1 mM glucose (18 mg/dL) to approximate in vivo metabolic stress as observed in prior studies.46,47 Physiologic glucose levels in human serum are between 4 and 6 mM (80 and 110 mg/dl).
We performed transient silencing of HuR expression using siRNA oligos purchased from Invitrogen (siRNA sequence used against HuR available upon request). Cells were suspended in 10 ml of Opti-mem® (Invitrogen) at a concentration of 3 million cells per 150 cm2 flask. Transfections were performed using Oligofectamine™ (Invitrogen) with a final siRNA concentration of 25 nM. For each experiment, gene silencing was confirmed 24–48 h after transfections by immunoblotting with an HuR antibody (Santa Cruz Biotechnology, sc-5621) or qPCR analysis of HuR mRNA as previously described.29,30 For transient overexpression of HuR, the HuR gene (OriGene Technologies, pCMV6-XL5) was cloned into a pcDNA3.1 plasmid vector (Invitrogen, V790-20) and transfected into cell lines using Lipofectamine™ (Invitrogen, 18324) at a plasmid concentration of 10 µg cDNA per 3 million cells as previously described.30
Apoptosis, cell death, survival and growth assays
Cell lines were plated into 75 cm2 flasks and allowed to adhere. Cells were washed with PBS, glucose-free DMEM twice and then incubated with media containing the indicated glucose concentrations. To estimate cell death, cells were trypsinized and counted after Trypan blue staining (Invitrogen, 15250-061) with a Hausser bright-line hemocytometer (Fisher Scientific). Annexin V® labeling was measure to estimate apoptosis using a flow cytometry staining kit (Invitrogen, V13242) on a BD Biosciences FACS Calibur system (BD Biosciences).
Cell survival was measured using a method similar to drug sensitivity assays, as previously described.30 Briefly, 1,000 cells per well were plated in triplicate. The cells were incubated in media with the indicated glucose concentration for seven days, lysed with de-ionized water and cell viability was detected using Quanti-iT PicoGreen® (Invitrogen, P7581) to stain double-stranded DNA. Results are reported as the percentage of viable cells at each glucose concentration, relative to control glucose conditions.
Soft-agar anchorage independence growth-assays were performed in 60 mm culture dishes (Fisher Scientific). A 5 ml bottom layer of 0.75% agar was prepared in DMEM with the indicated glucose concentrations and allowed to solidify. A top layer was added with 30,000 cells in 3 ml of 0.36% agar dissolved in DMEM containing the indicated glucose concentrations. After three weeks, colonies were examined and counted.
Immunoblot and immunofluorescence
Detection of HuR cytoplasmic protein expression was assessed by immunoblot and immunofluorescence. For immunoblotting, cells were cultured in 75 cm2 flasks, trypsinized, washed with PBS and lysed. RIPA buffer (Santa Cruz Biotechnology, SC-24948) was used to extract whole-cell protein and a digitonin-based lysis buffer was used to prepare cytoplasmic extracts.48 Protein concentrations were quantified by Bradford assay (Bio-Rad Laboratories, 500-0001) and equal amounts were loaded on an SDS-PAGE gel (Invitrogen, NP315BOX). The protein was transferred to a membrane and blocked for 1 h in 5% milk, followed by primary incubation with mouse anti-α tubulin (loading control) and mouse anti-HuR antibodies overnight (Santa Cruz Biotechnology, sc-8035 and sc-5621).30 Immunoblotting was also used to validate metabolic enzymes [e.g., GPI (TA501137), PRPS2 (TA308129), and IDH1(TA500610); OriGene Technologies], as well as GLUT1 (SLC2A1: Santa Cruz Biotechnology, sc-1605).
Immunofluorescence was performed using 4-well chamber slides, and cells were incubated under the indicated glucose concentrations for six hours. The cells were washed with PBS, fixed with 4% paraformaldehyde (Fisher Scientific, AC41678-0030), and permeabilized with Triton X-100 (Sigma-Aldrich, X100-500). Slides were blocked with 5% normal goat serum (Vector labs, 005-000-121) diluted in PBS for 1 h, and incubated with the primary anti-HuR antibody overnight at 4°C. Slides were washed and incubated with secondary goat anti-mouse antibody labeled with Alexa-Fluor® (Invitrogen, A11001) for 2 h. After washing with PBS, DAPI (Invitrogen, D1306) was added to counterstain the nuclei. Protein labeling was visualized with a Nikon confocal microscope (Nikon Corporation).
Glucose and lactate assays
Cells were pipetted into 96-well plates and allowed to adhere for 36 h. Subsequently, the cells were washed twice with glucose-free DMEM and 200 µl of media (containing the indicated glucose concentration) were added to each well. Plates were returned to the incubator and assayed over time for glucose (glucCELL Device, Bellco) and lactate (D-Lactate assay kit, Eton Bioscience Inc.,1200024002) per manufacturer instructions. Concentrations of glucose and lactate were normalized to total cell number in each well (quantified by Quant-iT PicoGreen as detailed above).
Intracellular metabolites (i.e., metabolomics) were measured in MiaPaCa2 cells using GC/MS, and LC/MS/MS platforms (Metabolon Corporation). Prior to assay preparation, 2 million cells per 150 cm2 flask were transfected with siRNA oligos (scrambled or against HuR) as described above. Following transfection, media was replaced with standard DMEM and cells were allowed to grow for 60 h. Cells were trypsinized, collected in standard DMEM and counted. A total of 6 million cells were washed in PBS twice and snap frozen in liquid nitrogen. Metabolite analysis was then performed as previously described.49,50 Metabolite levels were normalized to total cellular protein and data from each experimental group were analyzed as a group of 5 replicates. Welch’s t-test was used to compare metabolite levels between groups, with statistical significance accepted for p < 0.1. The q-value is provided in the Table S2 as an additional statistical measure of confidence, but was not used as a criterion for the present analysis.50
Ribonucleoprotein-immunoprecipitation (RNP-IP) assay, qPCR Array and RT-qPCR
Cells were plated at a concentration of 2 × 106 cells per 150 cm2 flask and allowed to adhere overnight. The flasks were washed with PBS and incubated for six hours in media containing 1 mM glucose, trypsinized and collected. RNP-IP was performed from cytoplasmic lysates with anti-HuR or control rabbit anti-IgG antibodies (MBL International Corporation, 07-468 and 6401-05).30 RNA bound to either HuR or control IgG was extracted, treated with DNase (SABiosciences, PA-001) and converted to a cDNA library using the RT2 First Strand Kit (SABiosciences, 330401). The cDNA library was then probed with a glucose metabolism qPCR Array consisting of 84 metabolic probes and 12 control probes (SABiosciences, PAHS-006Z). A total of 4 replicate RNP-IP qPCR Array experiments were performed and analyzed using the manufacturer recommended software (RT2 Profiler™; SABiosciences; www.sabiosciences.com/pcrarraydataanalysis.php). Candidate HuR binders were identified as transcripts with at least 4-fold expression after HuR RNP-IP extraction, compared with IgG controls.
Selected candidate genes (GLUT1, GPI, IDH1 and PRPS2) were evaluated as HuR mRNA targets by RT-qPCR of extracted RNA after independent RNP-IP experiments on an ABI 7500 Fast Analyzer (Life Technologies Corp.). The selected enzymes were selected with the goal of testing genes from different metabolic pathways. Results were normalized against 18S expression levels.
Flow cytometry analysis of GLUT1
Cells were trypsinized, washed and resuspended in FACS buffer (1% BSA, 0.05% sodium azide in PBS) at a concentration of 200,000 cells per 500 µL, and incubated with a GLUT1 antibody (Santa Cruz Biotechnology, sc-1605) for 1 h at room temperature. Cells were washed with buffer and incubated with a secondary antibody (Alexa Fluor® 488 goat anti-rabbit IgG; Invitrogen, A11008) for 1 h at room temperature in the dark. Samples were analyzed by flow cytometry (BD Bioscience FACS Calibur) and histograms created using the FlowJo software package (Tree Star, Inc.).
Supplementary Material
Acknowledgments and Grant Support
This work was supported in part by resources provided by the AACR-Pancreatic Cancer Action Network (AACR-PanCAN grant # 10-20-25-BROD), The American Cancer Society (ACS grant # RSG-10-119-01-CDD), the Lisa Waller Hayes Foundation, the “Fund-a-Cure for Pancreatic Cancer” project and Friends against Pancreatic Cancer (Dan and Heather Mendelow and Josh Donfeld).
Disclosure of Potential Conflicts of Interest
Dr Karoly is an employee of Metabolon Inc., which assisted in the metabolomics experiments included in this manuscript. Otherwise, these authors have no conflicts of interest to disclose regarding this manuscript.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/25274
References
- 1.Boeck S, Ankerst DP, Heinemann V. The role of adjuvant chemotherapy for patients with resected pancreatic cancer: systematic review of randomized controlled trials and meta-analysis. Oncology. 2007;72:314–21. doi: 10.1159/000113054. [DOI] [PubMed] [Google Scholar]
- 2.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–15. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 3.Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer. Gut. 2011;60:861–8. doi: 10.1136/gut.2010.226092. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland RM. Cell and environment interactions in tumor microregions: the multicell spheroid model. Science. 1988;240:177–84. doi: 10.1126/science.2451290. [DOI] [PubMed] [Google Scholar]
- 5.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–61. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erkan M, Michalski CW, Rieder S, Reiser-Erkan C, Abiatari I, Kolb A, et al. The activated stroma index is a novel and independent prognostic marker in pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2008;6:1155–61. doi: 10.1016/j.cgh.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Mesker WE, Junggeburt JM, Szuhai K, de Heer P, Morreau H, Tanke HJ, et al. The carcinoma-stromal ratio of colon carcinoma is an independent factor for survival compared to lymph node status and tumor stage. Cell Oncol. 2007;29:387–98. doi: 10.1155/2007/175276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parham DM, Robertson AJ, Brown RA. Morphometric analysis of breast carcinoma: association with survival. J Clin Pathol. 1988;41:173–7. doi: 10.1136/jcp.41.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Izuishi K, Kato K, Ogura T, Kinoshita T, Esumi H. Remarkable tolerance of tumor cells to nutrient deprivation: possible new biochemical target for cancer therapy. Cancer Res. 2000;60:6201–7. [PubMed] [Google Scholar]
- 11.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72. doi: 10.1016/j.cell.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–70. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, et al. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–34. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz V, Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene. 2010;29:4369–77. doi: 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 20.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–50. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, et al. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–65. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–54. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter JM, Dixon DA, Brody JR. HuR. Springer, 2012. [Google Scholar]
- 27.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214–29. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B, et al. HuR’s post-transcriptional regulation of Death Receptor 5 in pancreatic cancer cells. Cancer Biol Ther. 2012;13:946–55. doi: 10.4161/cbt.20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–72. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gantt KR, Cherry J, Richardson M, Karschner V, Atasoy U, Pekala PH. The regulation of glucose transporter (GLUT1) expression by the RNA binding protein HuR. J Cell Biochem. 2006;99:565–74. doi: 10.1002/jcb.20950. [DOI] [PubMed] [Google Scholar]
- 32.Landor SK, Mutvei AP, Mamaeva V, Jin S, Busk M, Borra R, et al. Hypo- and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc Natl Acad Sci USA. 2011;108:18814–9. doi: 10.1073/pnas.1104943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–76. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu PC, Chuang HC, Kulp SK, Chen CS. The mRNA-stabilizing factor HuR protein is targeted by β-TrCP protein for degradation in response to glycolysis inhibition. J Biol Chem. 2012;287:43639–50. doi: 10.1074/jbc.M112.393678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–23. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 37.Raspaglio G, De Maria I, Filippetti F, Martinelli E, Zannoni GF, Prislei S, et al. HuR regulates beta-tubulin isotype expression in ovarian cancer. Cancer Res. 2010;70:5891–900. doi: 10.1158/0008-5472.CAN-09-4656. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Fan J, Yang X, Fürer-Galban S, Lopez de Silanes I, von Kobbe C, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol. 2002;22:3425–36. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yaman I, Fernandez J, Sarkar B, Schneider RJ, Snider MD, Nagy LE, et al. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J Biol Chem. 2002;277:41539–46. doi: 10.1074/jbc.M204850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galbán S, Kuwano Y, Pullmann R, Jr., Martindale JL, Kim HH, Lal A, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gummadi L, Taylor L, Curthoys NP. Concurrent binding and modifications of AUF1 and HuR mediate the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in kidney cells. Am J Physiol Renal Physiol. 2012;303:F1545–54. doi: 10.1152/ajprenal.00400.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buzzai M, Bauer DE, Jones RG, Deberardinis RJ, Hatzivassiliou G, Elstrom RL, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–73. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 43.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–77. doi: 10.1016/S1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 44.Costello LC, Franklin RB. ‘Why do tumour cells glycolyse?’: from glycolysis through citrate to lipogenesis. Mol Cell Biochem. 2005;280:1–8. doi: 10.1007/s11010-005-8841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683–93. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 46.Ziebart T, Walenta S, Kunkel M, Reichert TE, Wagner W, Mueller-Klieser W. Metabolic and proteomic differentials in head and neck squamous cell carcinomas and normal gingival tissue. J Cancer Res Clin Oncol. 2011;137:193–9. doi: 10.1007/s00432-010-0875-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tannock IF, Kopelyan I. Influence of glucose concentration on growth and formation of necrosis in spheroids derived from a human bladder cancer cell line. Cancer Res. 1986;46:3105–10. [PubMed] [Google Scholar]
- 48.Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha. EMBO J. 2005;24:1852–62. doi: 10.1038/sj.emboj.7600661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 50.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, et al. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Natl Acad Sci USA. 2011;108:3270–5. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.